Abstract
Human rhinovirus type C is a recently discovered species that has been associated with respiratory tract infections of unusual severity in some cases. However, the precise type of diseases associated with this new human rhinovirus needs to be investigated. In the present report, we used adapted real-time PCR assays to screen different clinical specimens collected from a 14-month-old boy presenting an acute lower respiratory tract disease complicated by a severe pericarditis. RT-PCR identified picornavirus RNA in the bronchoalveolar lavage (BAL) specimen, pericardial fluid, plasma and stools. This supported the existence of a disseminated viral infection that extended to the pericardial space. 5′UTR and VP1 sequence analysis performed directly from the BAL sample allowed genotyping of the virus as a human rhinovirus C. This observation highlights the need for adapted diagnostic tools and the potential for the new rhinovirus species C to cause complications, including pericarditis.
Keywords: Human rhinovirus C, Lower respiratory tract, Disseminated disease, Pericarditis
1. Introduction
Human rhinoviruses (HRV) and enteroviruses (HEV) are leading causes of infection in children. These viruses share a similar genomic organisation, but harbor significant phenotypic differences.1, 2 Infection is restricted to the respiratory tract for HRV, whereas HEV target primarily the gastrointestinal tract and, less frequently, the respiratory tract. HEV can cause viremia and secondarily seed other sites, notably the central nervous system (the first cause of meningitis) and the heart (causing mainly pericarditis).3 The diversity of HRV and HEV is not restricted to previously known serotypes and new serotypes and even new species have been identified recently (http://www.picornaviridae.com/enterovirus/enterovirus.htm). At least eight studies4, 5, 6, 7, 8, 9, 10, 11, 12, 13 have reported a new HRV species, HRV-C, that is phylogenetically and structurally distinct from the A and B rhinovirus groups.14 Until now, this virus has been identified only in respiratory tract specimens and, in some cases, it was associated with lower respiratory diseases of unusual severity.6, 8, 9, 13 Given its novelty, this virus can only be detected by adapted diagnostic tools. We describe the case of severe respiratory and pericardial disease in an infant infected by the new HRV-C. This suggests that the tropism of this virus in vivo is not strictly restricted to the respiratory tract.
2. Case report
2.1. Case description
In April 2008, a 14-month-old male child was hospitalized with fever and respiratory symptoms. He had been referred to the emergency room for acute rhinitis, cough and sleep disturbance 2 weeks earlier. An acute upper and lower viral respiratory tract infection was diagnosed, but no specific therapy was introduced. Persisting fever and worsening of symptoms, including shortness of breath, led to subsequent admission. The history was remarkable for a severe hypoglycemia (1.4 mmol/l) at birth, pyelo-ureteral dilatation, a small patent ductus arteriosus, and a Sotos syndrome (slight motor developmental delay with a macrocephaly) confirmed at the genetic level. His temperature was 37.3 °C, heart and respiratory rates were 157 bpm and 50 per min, respectively, a moderate pansystolic parasternal murmur as well as grunting were audible, and orthopnea was observed. The white blood cell count was 12.8 g/l (55% neutrophils, 31% lymphocytes and 7% monocytes), C-reactive protein level was elevated at 180 mg/l (normal value <10), and procalcitonin at 2.29 μg/l (normal value <0.04). The chest radiograph revealed a cardiomegaly with a left lung infiltrate and pleural effusions, and intravenous ceftriaxone was administered. Cardiac echography revealed a circonferential 25 mm-thick, fibrinous pericardial effusion, and a tiny patent ductus arteriosus. There was no significant impairment of systolic function. All abnormalities were confirmed by a thoracic CT-scan (Fig. 1 ). A bronchoalveolar lavage (BAL) and surgical drainage of the pericardial effusion were performed. The pericard was fibrinous with a blood-tinged effusion within the pericardial space. His general condition improved progressively and he was discharged after 14 days. The BAL and pericardial specimens as well as blood cultures remained negative for both bacteria and mycobacteria. The pericardial biopsy showed a subacute and chronic inflammation characterized by a moderate infiltrate of mononuclear cells with superficial fibrin deposits overlaying and intermingled in an early granulation tissue. No bacteria, mycobacteria or fungi were observed on specific stains. Extensive serological work-up, including enteroviral serologies by complement fixations, did not point to any acute viral disease. Follow-up at 2 months was characterized by an excellent clinical evolution with resolution of fever and any echographic sign of pericarditis.
Fig. 1.
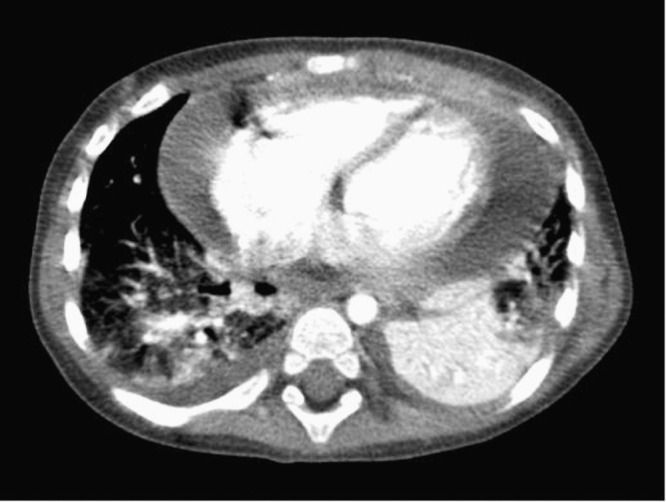
Thoracic CT-scan confirmed a 20 mm-thick, high-density pericardial effusion, an inferior left lobe condensation, a right inferior lobe infiltrate, and a small bilateral pleural effusion.
2.2. Virological investigations
RNA was extracted with the most appropriate method for each type of specimen: HCV Amplicor (Roche, Rotkreuz, Switzerland) for the pericardial fluid and plasma as previously described15; TRIzol (Invitrogen, Basel, Switzerland) for the BAL sample; both TRIzol and QIAamp Viral RNA Mini Kit (Qiagen, Hombrechtikon, Switzerland) for the stool sample; and Absolutely RNA FFPE kit (Qiagen) for the pericardial biopsy. Procedures were performed according to the manufacturers’ instructions. RT (Superscript II, Invitrogen) was performed with random hexamers as previously described.15 Real-time PCR assays targeting HRV and HEV (“Panenterhino real-time PCR”16) were positive for the BAL, the pericardial fluids, and the two plasma specimens. The assay was also weakly positive on stools collected 1 week later (Table 1 ). Each specimen was extracted and analyzed at least twice. Based on cycle values, the viral load was significantly higher in the BAL fluid compared to the plasma and stools that had very low viral loads, and a slight inhibition was observed in the pericardial fluid. HEV-specific PCR15 was negative for all specimens. RT-PCR targeting other respiratory viruses (respiratory syncytial virus A and B, bocavirus, coronavirus NL63, E229, OC43 and HKU1, influenza A, B and C, human metapneumovirus, and parainfluenza virus 1, 2, 3 and 417) remained negative in the BAL specimen. Viral cultures conducted on all specimens, except for the blood and pericardial biopsy, were all negative.
Table 1.
Overview of real-time PCR and viral culture results according to the type of specimen and timing.
| Specimen | Plasma 1 | Plasma 2 | BAL fluid | aPericardial effusion | bPericardial biopsy | Stools |
| Date (2008) | April 15 | April 16 | April 17 | April 17 | April 17 | April 24 |
| Panenterhino RT-PCR | cPositive | cPositive | d,ePositive | cPositive | Negative | cPositive |
| HEV-specific PCR | Negative | Negative | Negative | Negative | Negative | Negative |
| Viral culture | Not done | Not done | Negative | Negative | Not done | Negative |
BAL = Bronchoalveolar lavage.
Pericardial effusion fluid obtained during a surgical procedure. The RT-PCR was performed on two separate aliquots processed independently.
The pericardial biopsy was parafin-embedded.
Cycle values above 40.
cycle values between 22 and 25.
The 5′UTR (forward primer 11 and reverse primer 23) and VP1 (forward primer P1.161 and reverse primer P2.69) regions18 were amplified from the BAL sample and sequenced as previously described.2 The sequences are available at GenBank under accession FJ392316 and FJ392317. The VP1 sequence was aligned with corresponding sequences of available HRV-C strains (GenBank EF582385-7, NAT001, NAT045, EF186077, EF512666-82) using the vector NTI alignX module (Invitrogen). The identity table application was then used to calculate the percentage of homology. This VP1 sequence comparison revealed a nucleotide homology ranging from 62% to 83% with other HRV-C genotypes, EF582386 being the closest (data not shown).
3. Discussion
This paper reports the case of a 14-month-old boy presenting a complicated pericarditis following an acute respiratory illness. Based on 5′UTR and VP1 sequences, our investigations revealed that the lower respiratory tract was infected with a rhinovirus belonging to the HRV-C species. Surprisingly, although at a low level, RNA was found in the pericardial fluid, the blood and the gastrointestinal tract. Although we cannot exclude that this RNA could be a spill over from the main respiratory site, dissemination via blood viremia is a hypothesis to consider in this case. The infection was evidenced by nucleic acid detection and, as in previous reports; we failed to isolate this new HRV-C (Table 1, Table 2 ). However, the presence of RNA in multiple different sites, confirmed in duplicated experiments, corroborates the presence of a disseminated infection. According to the virological findings and medical history, the virus possibly reached the pericardial space via a blood viremia following a respiratory tract infection, and our results suggest that the HRV-C infection contributed to the acute pericardial disease.
Table 2.
Summary of available studies describing the clinical features associated with HRV-C.
| Period analyzed | 1999–2001 | 2001–2004 | 2003–2006 | 2003 | 2003 | 2004 | 2004–2005 | 2004–2005 |
| Number of HRV-C cases | 9 | 5 | 30 | 6 | 17 | 8 | 21 | 9 |
| Age range | Infants | Adults | <5 years | Adults | All ages | All ages | Children | Children |
| Type of patient | Community | Community | Hospitalized | Hospitalized | Hospitalized and community | Community | Hospitalized | Hospitalized and community |
| Clinical syndromes | Asthma | Asthma | Upper and lower respiratory tract infections | Unspecified respiratory tract infections | Upper and lower respiratory tract infections | Influenza-like illness | Upper and lower respiratory tract infections | Upper respiratory tract infections, respiratory infections with wheezing or distress |
| Specimen analyzed | Nasal secretions | Nasal lavage | Nasopharyngeal | Throat swab | Nasopharyngeal | Narines nasopharyngeal swabs | Nasopharyngeal | Nasopharyngeal wash |
| Diagnosis method | RT-PCR | DNA microarray | MassTag PCR | RT-PCR | RT-PCR | PCR and MassTag PCR | RT-PCR | MassTag-PCR |
| Site | Wisconsin, USA | San Francisco, USA | Germany | California, USA | Australia | New York, USA | Hong Kong SAR | Colorado, USA |
| References | 10 | 26 | 13 | 6 | 11 | 8 | 9 | 27 |
Most HRV-C infections are self-limited, but a review of available studies (Table 2) suggests that a substantial number of cases appear to present lower respiratory tract complications, as observed for other rhinoviruses.19, 20, 21 Our observation highlights that HRV-C might be associated not only with lower respiratory tract complications, but also with pericarditis. As HRV is the most frequent viral respiratory infection experienced during the first year of life,22 and since HRV-C are widely circulating around the world,23 its role as a causative agent of complicated diseases requires careful study. Although frequent in HEV infections, viremia is an unusual occurrence for HRV. At least two studies24, 25 have documented rhinovirus viremia in young children or infants but, to the best of our knowledge, HRV has not been described yet in pericardial effusion. Whether HRV-C genotypes could harbor specific phenotypic abilities similar to some entero- or coxsackie viruses is suggested also by the presence of viral RNA in stools. All these observations are consistent with an “entero-like” tropism, but we cannot rule out unusual host susceptibility. In addition, since the viral load was low in plasma and stools, we cannot exclude a RNA spill over from the respiratory tract. This possibility seems however unlikely given the number of positive sites and samples.
Beyond the present case our observation highlights that diagnostic tools need to be adapted to the detection of HRV-C not only in lower respiratory specimens, but also in other biological fluids. The fact that we used a PCR assay with the ability to detect all HRV and HEV in the presence of a negative HEV-specific assay allowed to suspect HRV-C. The 5′UTR and VP1 sequencing confirmed this hypothesis. It is therefore of importance to adapt diagnostic tools to further assess the potential ability of new picornaviruses to exhibit an unusual tropism.
Conflict of interest
None declared.
Acknowledgments
We thank Rosemary Sudan for editorial assistance, Sandra Van Belle and Lara Turin for technical help, and Alessandro Diana, Aline Prina-Rosso and Armand Bottani for their contribution to this report. This study was supported by the Swiss National Science Foundation (Nos. 3200B0-101670 to L.K.), the Department of Medicine of the University Hospitals of Geneva, the University of Geneva Dean's programme for the promotion of women in science (C.T).
References
- 1.Mackay I.M. Human rhinoviruses: the cold wars resume. J Clin Virol. 2008;42:297–320. doi: 10.1016/j.jcv.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tapparel C., Junier T., Gerlach D., Cordey S., Van Belle S., Perrin L. New complete genome sequences of human rhinoviruses shed light on their phylogeny and genomic features. BMC Genomics. 2007;8:224. doi: 10.1186/1471-2164-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Racaniello V.R. Picornaviridae: the viruses and their replication. In: Knipe D., Howley P.M., Griffin D.E., editors. 5th edition. vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 795–898. (Fields virology). [Google Scholar]
- 4.Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerna G., Percivalle E., Sarasini A., Capanini G., Piralla A., Rovida F. Human respiratory coronavirus HKU1 versus other coronavirus infections in Italian hospitalised patients. J Clin Virol. 2007;38:244–250. doi: 10.1016/j.jcv.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiang D., Yagi S., Kantardjieff K.A., Kim E.J., Louie J.K., Schnurr D.P. Molecular characterization of a variant rhinovirus from an outbreak associated with uncommonly high mortality. J Clin Virol. 2007;38:227–237. doi: 10.1016/j.jcv.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Kistler A.L., Webster D.R., Rouskin S., Magrini V., Credle J.J., Schnurr D.J. Genome-wide diversity and selective pressure in the human rhinovirus. Virol J. 2007;4:40. doi: 10.1186/1743-422X-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau S.K., Yip C.C., Tsoi H.W., Lee R.A., So L.Y., Lau Y.L. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee W.M., Kiesner C., Pappas T., Lee I., Grindle K., Jartti I. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE. 2007;2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McErlean P., Shackelton L.A., Lambert S.B., Nissen M.D., Sloots T.P., Mackay I.M. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McErlean P., Shackelton L.A., Andrews E., Webster D.R., Lambert S.B., NIssen M.D. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C) PLoS ONE. 2008;3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renwick N., Schweiger B., Kapoor V., Liu Z., Villari A., Bullmann R. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J Infect Dis. 2007;196:1754–1760. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordey S., Gerlach D., Junier T., Zdobnov E.M., Kaiser L., Tapparel C. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA. 2008;14:1568–1578. doi: 10.1261/rna.1031408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deffernez C., Wunderli W., Thomas Y., Yerly S., Perrin L., Kaiser L. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J Clin Microbiol. 2004;42:3212–3218. doi: 10.1128/JCM.42.7.3212-3218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tapparel C, Cordey S, Van Belle S, Turin L, Lee W-M, Regamey N, et al. New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J Clin Microbiol; 2009; (April 1) [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 17.Garbino J., Soccal P.M., Aubert J.D., Rochat T., Meylan P., Thomas Y. Respiratory viruses in bronchoalveolar lavage: a hospital-based cohort study in adults. Thorax. 2009;(January 27) doi: 10.1136/thx.2008.105155. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Tapparel C., Junier T., Gerlach D., Van Belle S., Turin L., Cordey S. New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis. 2009;15:719–726. doi: 10.3201/eid1505.081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malmstrom K., Pitkaranta A., Carpen O., Pelkonen A., Malmberg L.P., Turpeinen M. Human rhinovirus in bronchial epithelium of infants with recurrent respiratory symptoms. J Allergy Clin Immunol. 2006;118:591–596. doi: 10.1016/j.jaci.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 20.Papadopoulos N.G. Do rhinoviruses cause pneumonia in children? Paediatr Respir Rev. 2004;5(Suppl. A):S191–S195. doi: 10.1016/s1526-0542(04)90036-x. [DOI] [PubMed] [Google Scholar]
- 21.Papadopoulos N.G., Sanderson G., Hunter J., Johnston S.L. Rhinoviruses replicate effectively at lower airway temperatures. J Med Virol. 1999;58:100–104. doi: 10.1002/(sici)1096-9071(199905)58:1<100::aid-jmv16>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 22.Regamey N., Kaiser L., Roiha H.L., Deffernez C., Kuehni C.E., Latzin P. Rhino- and coronavirus infections and lower respiratory tract symptoms in infants: a population-based cohort study. Swiss Med Wkly. 2006;136:13S. [Google Scholar]
- 23.Briese T., Renwick N., Venter M., Jarman R.G., Ghosh D., Göndken S. Global distribution of novel rhinovirus genotype. Emerg Infect Dis. 2008;14:944–947. doi: 10.3201/eid1406.080271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urquhart G.E., Stott E.J. Rhinoviraemia. BMJ. 1970;4:28–30. doi: 10.1136/bmj.4.5726.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xatzipsalti M., Kyrana S., Tsolia M., Psarras S., Bossios A., Laza-Stanca V. Rhinovirus viremia in children with respiratory infections. Am J Respir Crit Care Med. 2005;172:1037–1040. doi: 10.1164/rccm.200502-315OC. [DOI] [PubMed] [Google Scholar]
- 26.Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominguez S.R., Briese T., Palacios G., Hui J., Villari J., Kapoor V. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J Clin Virol. 2008;43:219–222. doi: 10.1016/j.jcv.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


