Abstract
Clinical microbiology laboratories increasingly rely on molecular diagnostic techniques. The various formats of nucleic acid amplification are the most frequently used molecular tests in the diagnosis of infectious diseases. In many clinical settings, polymerase chain reaction (PCR) is clearly the method of choice due to its exquisite sensitivity and specificity. Today, many conventional PCR methods are being replaced by real-time PCR, which allows more rapid detection and quantification of the PCR product, as well as detection of different strains of the pathogen by melting curve analysis. The ability to measure the quantity of microbe by quantitative PCR has become increasingly important, providing information on the progression and prognosis of disease, and effectiveness of treatment. Other widely used molecular diagnostic techniques are isothermal amplification methods and nucleic acid hybridization techniques. Microarray is a technique which holds promise and has an exceptional sensitivity and the capacity to detect several pathogens simultaneously. However, microarrays are currently too expensive to be adapted for routine diagnostics, and their diagnostic use requires broad-based nucleic acid amplification prior to analysis which is not well established. Several molecular methods can be used for genotyping, which allows the identification of different subtypes of the pathogen; genotyping plays a role in the risk assessment and management of infections. Clinicians need to recognize the enhanced accuracy and speed of the molecular diagnostic techniques for the diagnosis of infections, but also to understand their limitations. Laboratory results should always be interpreted in the context of the clinical presentation of the patient, and appropriate site, quality, and timing of specimen collection are required for reliable test results.
Keywords: genotype, mass spectrometry, microarray analysis, nucleic acid amplification techniques, nucleic acid hybridization
What's new?
-
•
Conventional PCR methods are being replaced by real-time PCR, which allows real-time detection and quantification of the PCR product, as well as detection of different strains of the pathogen
-
•
Quantitative PCR has become an important tool, providing information on the progression and prognosis of disease, and effectiveness of treatment
Introduction
Clinical microbiology laboratories use a wide range of different techniques to support clinical decision-making in diagnosis, treatment and prognosis of infections. Two broad approaches to diagnosis are used. The gold standard is the direct detection of the pathogen itself in clinical samples collected from the cases. A range of methods such as culture, electron microscopy, and pathogen genome detection by polymerase chain reaction (PCR) are used. Key performance issues are summarized in Table 1 . Infection is often diagnosed indirectly by the detection of the patient's immune response to infection. The detection of pathogen specific IgM is a commonly used method for confirming acute infection. Today, clinical microbiology diagnostics increasingly rely on molecular techniques. This brief review will focus on the key features influencing the clinical use of molecular tests. The most widely used techniques are described. The advantages and disadvantages of the techniques are outlined in Table 2 .
Table 1.
Comparison of laboratory methods for direct detection of infections
| Technique | Sensitivity | Turnaround | Availability |
|---|---|---|---|
| Culture | Moderate | > 3 days |
|
| Electron microscopy | Low | <1 day |
|
| Antigen detection | Moderate | < 1 day |
|
| Direct immunofluorescence | Moderate | <1 day |
|
| Polymerase chain reaction | High | <1 day |
|
Table 2.
Advantages and disadvantages of different molecular diagnostic techniques
| Technique | Advantages | Disadvantages |
|---|---|---|
| PCR and its modifications |
|
|
| Isothermal amplification |
|
|
| Fluorescence in situ hybridization |
|
|
| Genotyping |
|
|
| Mass spectrometry |
|
|
| Microarrays |
|
|
PCR: polymerase chain reaction, NASBA: nucleic acid sequence based amplification, MALDI: matrix-assisted laser desorption/ionization.
Nucleic acid amplification techniques
Cycling amplification
Polymerase chain reaction (PCR): The various forms of PCR are the most frequently used molecular diagnostic techniques in the diagnosis of infectious pathogens. In certain clinical situations, such as diagnosis of viral encephalitis or monitoring of cytomegalovirus (CMV) and Epstein–Barr virus (EBV) in organ transplant patients, PCR has transformed laboratory investigations because of its exquisite sensitivity and specificity, and is clearly the method of choice. PCR allows the amplification of millions of identical DNA copies from an originally small amount of pathogen genome in a clinical sample. In principal, the target DNA is first extracted then denatured at high temperature. Specific oligonucleotide primers are annealed to the DNA in a lower temperature, followed by an extension phase during which the DNA polymerase enzyme copies the template strand. This cycle is repeated, usually 30–40 times, resulting in millions of identical DNA copies. For the principles of PCR see Figure 1 .
Figure 1.
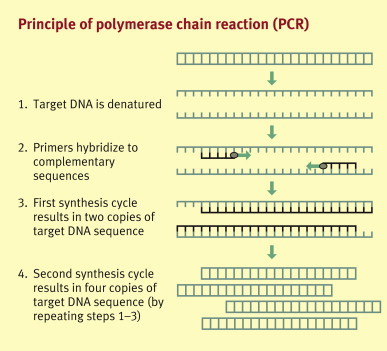
Principle of polymerase chain reaction (PCR)
PCR is increasingly applied in the diagnosis of viruses, bacteriae, parasites, and fungi. Qualitative PCR is widely used for a range of infections including herpesviruses (HSV, VZV, CMV, EBV), enteroviruses, parvoviruses, respiratory viruses, SARS-CoV, and poxviruses, as well as general or specific bacterial screening, such as Neisseria meningitidis, Streptococcus pneumoniae, Bordetella pertussis, and Borrelia spp. Quantitative PCR measures nucleic acid copy numbers of the sample by comparison to a control with a known copy level, and can be applied to, for example, CMV, EBV, HBV, HCV, and HIV. Quantification provides information on the progression and prognosis of disease, and effectiveness of antiviral treatment.
Today, real-time PCR is the favoured methodology. Real-time PCR takes advantage of a fluorescent reporter, which gives signal in direct proportion to the amount of PCR amplicon. The method allows real-time detection and quantification of PCR amplicons following each thermal cycle. A melting curve analysis method differentiates PCR amplicons according to length and different guanine–cytosine contents. This can be used as an alternative to sequencing for detection of the different strains of the pathogen or gene mutations related to antibiotic resistance (e.g. meticillin∗ -resistant Staphylococcus aureus and vancomycin-resistant enterococci). Real-time PCR is rapid with high sample throughput, and usually is more sensitive and specific in comparison to conventional methods.
Both conventional and real-time PCR can be used in a multiplex format, which allows the detection of several pathogens simultaneously. However, in multiplex PCR it is usually more difficult to optimize sensitivity and specificity, and the method may be preferential to certain amplification products. Clinical examples of multiplex PCR are screening of positive blood cultures for bacterial pathogens causing sepsis,1 and screening of respiratory viruses from nasopharyngeal swabs.2
Ligase chain reaction (LCR): LCR is a modification of PCR. In this method, two adjacent probes hybridize to one strand of the target DNA. The small gap between the two adjacent primers is recognized by a highly specific thermostable DNA ligase, and ligated to form a single probe. The ligated products then serve as templates for the amplification process. LCR allows the detection of only single base pair mutations, and it is very specific. The most common application of LCR is in the diagnostics of Chlamydia trachomatis in cervical and urine samples.3
Isothermal amplification
Unlike PCR, isothermal amplification techniques are based on a distinct enzyme that does not require thermocycling (heating and cooling cycles), which can take several hours. Isothermal amplification is rapid; it usually requires less than an hour to perform. However, the technique is currently fairly expensive. We describe here some of the best-known isothermal amplification methods.
Nucleic acid sequence-based amplification (NASBA): In NASBA analysis (Figure 2 ), which is performed in isothermal conditions, the target RNA is converted to double-stranded DNA by using T7 RNA polymerase, RNaseH, and a primer with a T7 promoter. The DNA acts as a template to produce multiple copies of RNA with a polarity opposite that of the target, which can be used for the production of additional DNA templates. NASBA is also suitable for DNA, with slight modifications in the early steps of the process. A recent example of NASBA technology in clinical diagnostics is a real-time multiplex NASBA assay for the detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Legionella spp. in respiratory specimens.4
Figure 2.
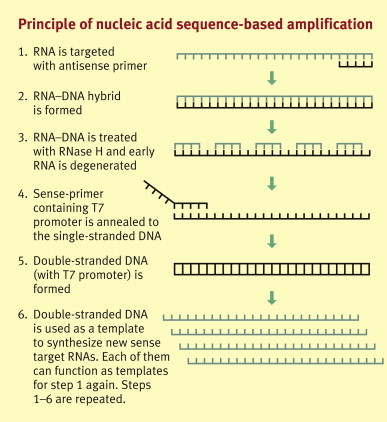
Principle of nucleic acid sequence-based amplification
Transcription-mediated amplification (TMA): The principle of TMA is similar to NASBA, except that it uses the RNaseH activity of reverse transcriptase, whereas NASBA uses a separate enzyme for that. Due to the introduction of West Nile virus (WNV) in to the US since 1999, screening of primary WNV infection in blood donors was initiated. Currently, TMA is referred to as the most sensitive nucleic acid test commercially available for WNV infection,5 and is used on a large scale for blood donor screening in the US. Another clinical example of a large-scale and worldwide use of TMA technology is in the detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine and urogenital swab specimens.6
Strand displacement amplification (SDA): SDA is another isothermal amplification method. The first set of primers containing a restriction site is annealed to DNA template. Second primers are then annealed adjacent to the first ones and start amplification, after which restriction enzyme HincII is introduced in order to nick the synthesized DNA. An exonuclease-deficient form of the Klenow fragment of Escherichia coli DNA polymerase I starts the amplification again, displacing the newly synthesized strands. Similar to other isothermal amplification techniques, the amplification process of SDA is very efficient. SDA is used, for example, in the diagnosis of N. gonorrhoeae infection from female endocervical swabs and male first-void urine samples.
Nucleic acid hybridization techniques
Fluorescence in situ hybridization (FISH)
The FISH technique allows the detection of microbial nucleic acid directly from the sample (or cultured sample) without prior nucleic acid amplification. Briefly, the technique consists of specimen fixation on a microscope slide, hybridizing the prepared sample with a specific fluorescent-labelled probe, and visual detection of the hybridization with a fluorescent microscope. In medical microbiology, the technique is used, e.g. in determination of antibiotic resistance and detection of Helicobacter pylori from gastric biopsy specimens.7 Identifying bacteria from CSF,8 or Staphylococcus aureus 9 from blood cultures.
Line probe assay
The line probe assay (LiPA) is another nucleic acid hybridization test, in which specific oligonucleotide probes are attached at known locations on a nitrocellulose strip as parallel lines and hybridized with biotin-labelled PCR products. One of the widely used LiPA applications is rapid detection of rifampicin resistance in Mycobacterium tuberculosis.10 The LiPA technique is also applied in the detection of antiviral drug-resistant mutations of HBV.11
Microarrays
Microarrays consist of a two-dimensional matrix of biomolecules which are printed or synthesized on a glass, silicon, plastic or nylon membrane. Microarrays can detect both nucleic acids and antibodies, which attach to the immobilized biomolecule, which can be, e.g., an oligonucleotide or a protein. Positive reaction can be detected with highly advanced scanners by use of target labelling with fluorescent probes or antibodies. A schematic illustration on one of the microarray modifications (by Baek et al., 200812) is shown in Figure 3 . The variety of microarray modifications is vast, varying from immobilization of DNA probes in three-dimensional gel drops to electrochemical detection of hybridization without preceding PCR amplification. It is foreseen that both DNA and serological microarrays will play an important role in the future technologies applied in clinical microbiology laboratories. Several applications have already been proposed and introduced, including detection of rifampicin- and isoniazid-resistant M. tuberculosis,13 detection of chloroquine-resistant Plasmodium falciparum,14 and herpesvirus detection from CSF.15
Figure 3.
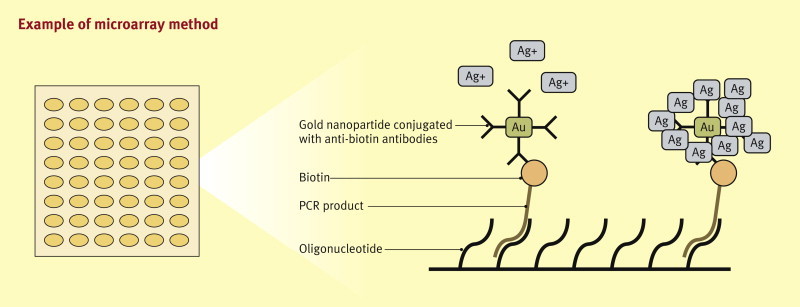
This example is a modification of a microarray for the detection of human papilloma virus. In this example, biotin-labeled PCR products are hybridized with immobilized oligonucleotides. The chip is incubated in an anti-biotin antibody-conjugated gold nanoparticle solution. The silver enhancement by the gold nanoparticles bound to the biotin precipitates silver metal particles at the chip surfaces. This blocks light irradiated from above, which can be measured, allowing detection and quantitative analysis of the target DNA.
Genotyping
Genotyping refers to the identification of variations of different strains or subtypes of the pathogen at nucleic acid level. Genotyping of PCR amplicons can be performed by direct sequencing, by reverse hybridization to genotype-specific probes, by restriction fragment length polymorphism, or by detection of single nucleotide polymorphisms with mass spectrometry. Direct sequencing is usually considered as the gold standard as it provides accurate and extensive data. Genotyping plays an important role in the management of HCV infection, as response to treatment considerably varies between different genotypes; patients with HCV genotype 2 or 3 infection respond better to treatment than those with genotype 1 infection. Both direct sequencing and probe hybridization approaches are widely used for HCV genotyping.16 An example of genotyping currently moving into routine practice is human papilloma virus (HPV) typing. Eight oncogenic HPV types (16, 18, 31, 33, 35, 45, 52, and 58) are responsible for 80% of cervical cancers and cervical intraepithelial neoplasia worldwide; distinct patterns are seen in different regions. Genotyping for the oncogenic HPV types enables risk assessment, personalized clinical management and early intervention in women with mildly abnormal Pap results. Genotyping is also widely used for molecular epidemiology and in identifying antimicrobial resistance and high-risk strains.
Mass spectrometry
Mass spectrometry allows the measurement of the molecular mass of a sample, which can be utilized in various clinical diagnostic settings. In this technique, the sample is first ionized, the ions are separated according to their mass-to-charge ratios, and finally the separated ions are detected according to the ion current. Ionization of the sample can be done in various ways, of which matrix-assisted laser desorption/ionization (MALDI) is widely used in the genotyping of single nucleotide polymorphisms. MALDI mass spectrometry is an important tool in typing of bacteria and viruses, which is useful, e.g. in detecting hypervirulent or antimicrobial drug resistant strains. The technique has been introduced, e.g. for genotyping of HCV and monitoring of quasispecies (a cloud of variant RNA viruses that arise from mutations over time within a viral isolate) in chronic HCV infection.17 It is also used in the detection of hypervirulent N. meningitidis strains18; only five of the 13 serogroups are frequently associated with invasive disease.19 Recently, a method based on broad-range RT-PCR followed by electrospray ionization mass spectrometry has been applied for monitoring of global spread of novel influenza virus genotypes.20
Key factors affecting test performance and reliability
As with all laboratory testing, microbiological laboratory results are never definitive, and the clinical significance of the test result should always be placed in the context of the patient's clinical presentation. Molecular diagnostic techniques are no exception to this rule. Indeed the exquisite sensitivity of PCR adds new issues of clinical interpretation of laboratory results. Many infectious agents cause clinical and asymptomatic infection; for example, norovirus, a cause of gastroenteritis, has a high rate of asymptomatic infections. Systematic application of the PCR to both respiratory and enteric illness has revealed that mixed infections are common (5–10%), which means attributing causality to a single agent is difficult.
Sensitivity, specificity, and predictive values
The performance of a laboratory method is defined by specificity, sensitivity, and most importantly by predictive values. The sensitivity of the assay refers to its ability to correctly identify affected individuals, whereas specificity refers to its ability to correctly identify non-affected individuals. False-positive and/or false-negative results in molecular diagnostic techniques can occur either due to suboptimal assay performance, sample contamination, wrong specimen material, or wrong timing of the sample collection. In addition, a positive result may be without clinical significance, sometimes due to the low detection threshold of these techniques.
The positive and negative predictive values of the assay refer to the performance of the test at a population level, i.e. the probability that an individual testing positive is truly affected and that an individual testing negative is truly non-affected. The predictive values of the test are not only dependent on the sensitivity and specificity of the assay, but also on the prevalence of the disease in the population. For example, the performance of any given HIV antibody test is different when used in high-risk groups as compared to general population; the positive predictive value will fall as the prevalence of the disease falls.
Contamination
Cross-contamination of the specimen is a source of error and can occur during sampling, due to specimen-to-specimen contamination during sample processing, or due to contamination of laboratory reagents. For PCR the main issue is avoiding contamination from the products of PCR which have a high concentration after amplification. This was a major issue when PCR was first introduced, and has led to laboratory work areas for different steps of PCR being physically separated to avoid contamination. In optimal conditions, the same personnel will never work with both amplified products and clean reagents. Aseptic and meticulous working techniques are required to avoid specimen-to-specimen contamination. The introduction of real-time PCR has reduced the potential for contamination by reducing handling. Regular quality control of the assays is vital, including negative and internal positive controls, as well as continual trend analyses, which must be reflected in the epidemiology of the pathogen in question.
Specimen selection
Molecular analyses can be performed on a wide variety of specimens, including serum, whole blood, cerebrospinal fluid (CSF), urine, genital swabs, respiratory swabs, stool, and different tissue biopsies. The clinical interpretation of the result must be adjusted to the site of pathology and type of specimen. Detecting a potential pathogen (e.g. rotavirus) in a faecal sample from a case of meningitis may not be significant. Quantity of pathogen present is also important; for example, detection of CMV nucleic acid in blood is a sign of ongoing infection, but may be without clinical significance as healthy individuals may also undergo reactivations; therefore, quantitative CMV PCR is the method of choice for use in immunocompromised patients to guide treatment.
Specimen collection
Appropriate collection, handling, and delivery of the specimen to the laboratory are vital to avoid false-negative results. Quality and timing of sample collection are of high importance. A fine example of this is PCR testing of C. trachomatis in urine. Too early post-treatment testing may result in clinically false-positive results due to detectable traces of nucleic acid by PCR without presence of infectious pathogen, since shedding can be detected for much longer by PCR than culture. Wrong timing may also lead to false-negative results, such as PCR testing of respiratory and stool samples collected late in the disease process, when the viral nucleic acid has already cleared. In this case, serological investigation may help in the diagnosis.
Practice points.
-
•
Appropriate site, quality, and timing of specimen collection are vital for reliable molecular diagnostic analyses
-
•
To fully exploit the potential of new molecular tests, clinicians need to recognize their enhanced opportunities for diagnosis, but also their constraints.
-
•
Laboratory results should always be interpreted in the context of the clinical presentation of the patient
Acknowledgements
The authors would like to thank Dr Katrin Leitmeyer for commenting on the manuscript. Dr Kurkela is affiliated as a Fellow in the European Public Health Microbiology Training Programme (EUPHEM), which is developed and endorsed by the Collaborative Laboratory Response Network of the European Network for Diagnostics of ‘Imported’ Viral Diseases (ENIVD-CLRN) and the European Centre for Disease Prevention and Control (ECDC).
Footnotes
In line with the British National Formulary, the spelling ‘meticillin’ has been used throughout these issues, rather than ‘methicillin’.
References
- 1.Wellinghausen N., Wirths B., Essig A., Wassill L. Evaluation of the hyplex bloodscreen multiplex PCR-enzyme-linked immunosorbent assay system for direct identification of gram-positive cocci and gram-negative bacilli from positive blood cultures. J Clin Microbiol. 2004;42:3147–3152. doi: 10.1128/JCM.42.7.3147-3152.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SR, Ki CS, Lee NY. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system-based multiplex PCR assay. J Virol Methods 2009; 156: 111–16. [DOI] [PMC free article] [PubMed]
- 3.Marrazzo J.M., Johnson R.E., Green T.A. Impact of patient characteristics on performance of nucleic acid amplification tests and DNA probe for detection of Chlamydia trachomatis in women with genital infections. J Clin Microbiol. 2005;43:577–584. doi: 10.1128/JCM.43.2.577-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loens K., Beck T., Ursi D. Development of real-time multiplex nucleic acid sequence-based amplification for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Legionella spp. in respiratory specimens. J Clin Microbiol. 2008;46:185–191. doi: 10.1128/JCM.00447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziermann R., Sánchez-Guerrero S.A. PROCLEIX® West Nile virus assay based on transcription-mediated amplification. Expert Rev Mol Diagn. 2008;8:239–245. doi: 10.1586/14737159.8.3.239. [DOI] [PubMed] [Google Scholar]
- 6.Boyadzhyan B., Yashina T., Yatabe J.H., Patnaik M., Hill C.S. Comparison of the APTIMA CT and GC assays with the APTIMA Combo 2 assay, the Abbott LCx assay, and direct fluorescent-antibody and culture assays for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J Clin Microbiol. 2004;42:3089–3093. doi: 10.1128/JCM.42.7.3089-3093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Can F., Yilmaz Z., Demirbilek M. Diagnosis of Helicobacter pylori infection and determination of clarithromycin resistance by fluorescence in situ hybridization from formalinfixed, paraffin-embedded gastric biopsy specimens. Can J Microbiol. 2005;51:569–573. doi: 10.1139/w05-035. [DOI] [PubMed] [Google Scholar]
- 8.Poppert S., Essig A., Stoehr B. Rapid diagnosis of bacterial meningitis by real-time PCR and fluorescence in situ hybridization. J Clin Microbiol. 2005;43:3390–3397. doi: 10.1128/JCM.43.7.3390-3397.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmann H., Stender H., Schäfer A., Autenrieth I.B., Kempf V.A. Rapid identification of Staphylococcus aureus in blood cultures by a combination of fluorescence in situ hybridization using peptide nucleic acid probes and flow cytometry. J Clin Microbiol. 2005;43:4855–4857. doi: 10.1128/JCM.43.9.4855-4857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan M., Kalantri S., Flores L., Pai M. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2005;5:62. doi: 10.1186/1471-2334-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degertekin B., Hussain M., Tan J., Oberhelman K., Lon A.S. Sensitivity and accuracy of an updated line probe assay (HBV DR v.3) in detecting mutations associated with hepatitis B antiviral resistance. J Hepatol. 2009;50:42–48. doi: 10.1016/j.jhep.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Baek T.J., Park P.Y., Han K.N., Kwon H.T., Seong G.H. Development of a photodiode array biochip using a bipolar semiconductor and its application to detection of human papilloma virus. Anal Bioanal Chem. 2008;390:1373–1378. doi: 10.1007/s00216-007-1814-x. [DOI] [PubMed] [Google Scholar]
- 13.Gryadunov D., Mikhailovich V., Lapa S. Evaluation of hybridisation on oligonucleotide microarrays for analysis of drug-resistant Mycobacterium tuberculosis. Clin Microbiol Infect. 2005;11:531–539. doi: 10.1111/j.1469-0691.2005.01183.x. [DOI] [PubMed] [Google Scholar]
- 14.Crameri A., Marfurt J., Mugittu K. Rapid microarray-based method for monitoring of all currently known single-nucleotide polymorphisms associated with parasite resistance to antimalaria drugs. J Clin Microbiol. 2007;45:3685–3691. doi: 10.1128/JCM.01178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jääskeläinen A.J., Piiparinen H., Lappalainen M., Vaheri A. Improved multiplex-PCR and microarray for herpesvirus detection from CSF. J Clin Virol. 2008;42:172–175. doi: 10.1016/j.jcv.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Weck K. Molecular methods of hepatitis C genotyping. Expert Rev Mol Diagn. 2005;5:507–520. doi: 10.1586/14737159.5.4.507. [DOI] [PubMed] [Google Scholar]
- 17.Yea C., Bukh J., Ayers M., Roberts E., Krajden M., Tellier R. Monitoring of hepatitis C virus quasispecies in chronic infection by matrix-assisted laser desorption ionization-time of flight mass spectrometry mutation detection. J Clin Microbiol. 2005;43:2810–2815. doi: 10.1128/JCM.02512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowe C.A., Diggle M.A., Clarke S.C. A single nucleotide polymorphism identification assay for the genotypic characterisation of Neisseria meningitidis using MALDI-TOF mass spectrometry. Br J Biomed Sci. 2004;61:8–10. doi: 10.1080/09674845.2004.11732638. [DOI] [PubMed] [Google Scholar]
- 19.Urwin R., Russell J.E., Thompson E.A., Holmes E.C., Feavers I.M., Maiden M.C. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect Immun. 2004;72:5955–5962. doi: 10.1128/IAI.72.10.5955-5962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampath R., Russell K.L., Massire C. Global surveillance of emerging Influenza virus genotypes by mass spectrometry. PLoS ONE. 2007;2:e489. doi: 10.1371/journal.pone.0000489. [DOI] [PMC free article] [PubMed] [Google Scholar]


