Highlights
-
•
All +RNA viruses induce replication organelles to shield viral RNA from innate immune surveillance.
-
•
Recent literature suggests that non-self or aberrant-self membrane structures can be tagged with LC3 or ubiquitin.
-
•
Interferon-induced GTPases then recognize these tags and destroy the membrane structures, thereby exposing PAMPs.
-
•
More research will have to indicate whether this is a general antiviral mechanism affecting +RNA virus infections.
Abbreviations: +RNA, positive-strand RNA; 5′ppp-RNA, 5′ tri-phosphorylated RNA; DENV, Dengue virus; DMV, double membrane vesicle; EAV, equine arteritis virus; eIF, eukaryotic translation initiation factor; EndoU, endonuclease; GBP, guanylate-binding protein; HCV, Hepatitis C virus; HIV, human immunodeficiency virus; IFN, interferon; Inv, invaginated vesicle/spherule; IRF, interferon regulating factor; IRG, immunity-related p47 GTPase; ISG, interferon-stimulated gene; JAK/STAT, Janus kinase-signal transducer and activator of transcription; JEV, Japanese encephalitis virus; LC3, microtubule-associated protein 1A/1B-light chain 3; MAM, mitochondrion-associated membrane; MAVS, mitochondrial antiviral signaling; MDA 5, melanoma differentiation associated factor 5; MERS-CoV, Middle East respiratory coronavirus; MHV, mouse hepatitis virus; MNV, murine norovirus; Mx, myxovirus resistance proteins; MyD88, myeloid differentiation primary response gene 88; nsp, non-structural protein; OAS, 2′,5′-oligoadenylate synthetase; PAMPs, pathogen-associated molecular patterns; PE, phosphatidylethanolamine; PKR, protein kinase R; PLP, papain-like protease; PV, pathogen-containing vacuole; RIG-I, retinoic acid-inducible gene I; RLR, RIG-I like receptor; RNP, ribonucleoprotein; RO, replication organelle; SARS-CoV, severe acute respiratory coronavirus; SG, stress granule; STING, stimulator of interferon genes; T. gondii, Toxoplasma gondii; TAG, targeting by AutophaGy proteins; TIR, Toll/interleukin-1 receptor; TLR, toll-like receptor; TRIF, TIR-domain- containing adapter-inducing interferon-β; VLIGs, very large IFN-inducible GTPases; WNV, West Nile virus
Keywords: +RNA virus, GTPases, Replication organelles, Interferon
Abstract
The potential health risks associated with (re-)emerging positive-strand RNA (+RNA) viruses emphasizes the need for understanding host-pathogen interactions for these viruses. The innate immune system forms the first line of defense against pathogenic organisms like these and is responsible for detecting pathogen-associated molecular patterns (PAMPs). Viral RNA is a potent inducer of antiviral innate immune signaling, provoking an antiviral state by directing expression of interferons (IFNs) and pro-inflammatory cytokines. However, +RNA viruses developed various methods to avoid detection and downstream signaling, including isolation of viral RNA replication in membranous viral replication organelles (ROs). These structures therefore play a central role in infection, and consequently, loss of RO integrity might simultaneously result in impaired viral replication and enhanced antiviral signaling. This review summarizes the first indications that the innate immune system indeed has tools to disrupt viral ROs and other non- or aberrant-self membrane structures, and may do this by marking these membranes with proteins such as microtubule-associated protein 1A/1B-light chain 3 (LC3) and ubiquitin, resulting in the recruitment of IFN-inducible GTPases. Further studies should evaluate whether this process forms a general effector mechanism in +RNA virus infection, thereby creating the opportunity for development of novel antiviral therapies.
1. Introduction
The innate immune system forms the first line of defense against pathogens and its initial function is to recognize pathogen-associated molecular patterns (PAMPs) [1], which ultimately leads to induction of the antiviral state that effectively hampers spread of the infection. The adaptive immune system then kicks in to (in most cases) fully clear the virus and build up memory. Viral RNA is a very potent inducer of innate antiviral signaling [2], [3]. Therefore, detection of viral RNA and the subsequent induction of antiviral effector mechanisms play an important part in the onset of an antiviral state in the context of RNA virus infections. The study of PAMP recognition and signaling by cytosolic and membrane bound sensors has intensified tremendously in the last decade. Several comprehensive reviews on this subject have been published lately [4], [5]. Additionally, investigation of the involvement of intracellular organelle membranes of mitochondria, ER, and peroxisomes and their mutual interactions indicated that these are important signaling platforms in innate immunity [6], [7]. Together, this has resulted in a better understanding of the details of innate immune responses that target RNA viruses.
In this review, we will focus on the interaction of the innate immune system with viruses that have a positive-strand RNA (+RNA) genome. In response to innate immune reactions, virtually all +RNA viruses interfere to delay antiviral innate immune signaling in several ways [1], [8], [9]. A key feature of immune evasion by +RNA viruses, and simultaneously the hallmark of +RNA virus infection, is rearrangement of host membranes into viral replication organelles (ROs). As these structures are thought to shield viral RNA from the host innate immune system and additionally seem to play a fundamental role in viral RNA replication, ROs in this sense seem to have a central and dual function in viral replication [10], [11], [12], [13], [14]. Therefore, disrupting integrity of ROs might simultaneously result in impaired viral replication and enhanced antiviral immune signaling, which would be a beneficial effect from an antiviral immunity point-of-view. However, whether host cells possess effector mechanisms to disrupt +RNA virus ROs has hardly been investigated yet, and only quite recently some studies have shed more light on this, which will be the focus of this review together with the body of literature that surrounds it.
2. Positive-strand RNA viruses: societal impact, taxonomy, and virus-induced replication organelles
Positive-strand RNA viruses have caused multiple outbreaks during the past decade: severe acute respiratory coronavirus (SARS-CoV) infected more than 8000 individuals in 2003 of which almost 800 died [15]. Its close relative Middle East respiratory syndrome coronavirus (MERS-CoV) emerged in Saudi Arabia in 2012. While SARS-CoV infections have not been reported in humans after 2004, MERS-CoV currently still regularly occurs in dromedary camels as well as in humans, and the virus displays a lethality rate of around 35% in the latter [16]. Zika virus, which received a lot of societal attention lately, is an example of a re-emerged +RNA virus with significant impact [17]. These outbreaks, and the lack of tailored antiviral strategies against them, clearly illustrate the potential health risks associated with (re-)emerging +RNA viruses, and emphasize the need for a precise understanding of host-pathogen interactions to facilitate development of novel antiviral therapies or vaccination methods. Over the last decades, many +RNA viruses have been extensively studied for their intriguing biology, including coronaviruses as those mentioned above, picornaviruses such as coxsackievirus and poliovirus, and flaviviruses such as West Nile virus (WNV), Hepatitis C virus (HCV), dengue virus (DENV), Zika virus, and Japanese encephalitis virus (JEV), as well as the family of togaviridae, including rubella virus and chikungunya virus.
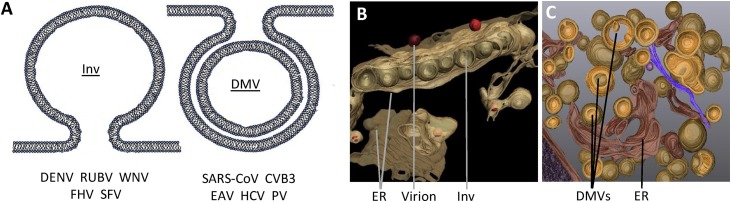
One of the intriguing features that is shared by all +RNA viruses is the rearrangement of host membranes into viral ROs, whereby particular cellular organelles (depending on the virus) are used as membrane donors. Concerning their role in viral RNA replication, it is thought that the structures form a scaffold that improves efficiency of enzymatic reactions and provides a spatiotemporal regulation of the different stages in the virus life cycle. Besides this, it is thought that ROs have an important role in shielding viral RNA products from the RNA sensors of the innate immune system. ROs are therefore believed to have a dual role in +RNA virus infection and innate immune evasion, which will be elaborated on further in this review. Several comprehensive reviews have recently described the current knowledge of these structures [12], [18], [19], [20], therefore we will only briefly summarize this below. Most studies until now focused on the architecture of structures induced by different viruses, and based on electron microscopy and electron tomography, ROs were categorized into two different classes: the invaginated vesicle/spherule (Inv) type, and the double membrane vesicle (DMV) type (Fig. 1 ).
Fig. 1.
Viral replication organelles. (A) Schematic representation of viral replication organelles (ROs), modified from [18]. Membranous structures that form ROs can be categorized as the invaginated vesicle/spherule (Inv) type (left), and the double membrane vesicle (DMV) type (right). Inv ROs are predominantly observed during infection with alphaviruses, such as Semliki Forest virus (SFV), nodavirideae such as flock house virus (FHV), and flaviviridae such as rubella virus (RUBV), dengue virus (DENV), and West Nile virus (WNV). Alternatively, double membrane vesicle (DMV) type ROs are formed by enteroviruses coxsackievirus B3 (CVB3) and poliovirus (PV), coronaviruses severe acute respiratory coronavirus (SARS-CoV) and equine arteritis virus (EAV), and the flavivirus hepatitis C virus (HCV). (B) 3D reconstruction of Inv ROs upon DENV infection, modified from [21] (C) 3D reconstruction of DMV ROs upon HCV infection modified from [27].
Inv ROs are predominantly observed during infection with alphaviruses, such as Semliki Forest virus and Sindbis virus, nodavirideae such as Flock House virus, bromoviridae such as bromovirus, and flaviviridae such as rubella virus, DENV, and WNV [19], [21]. Although the location and dynamics of the formation of Inv ROs varies per virus, the result is a single membrane invagination of which the content is connected to the cytosol. In addition, replicase proteins and newly synthesized viral RNA were observed in these spherules, strongly suggesting that Inv ROs are sites of viral RNA replication [22], [23], [24]. Furthermore, ribosomes are found in close proximity of some Inv ROs [25], suggesting that viral RNA replication and translation are spatially separated. A similar organization of Inv ROs was found for Flock House virus, rubella virus, DENV, and WNV. Moreover, a recent study visualized virus budding in close association with Inv ROs, suggesting a direct role for these structures in coordinating virus assembly [26].
Alternatively, DMV ROs are known to be formed by enteroviruses such as coxsackievirus B3 and poliovirus, coronaviruses such as SARS-CoV and MERS-CoV, arteriviruses such as equine arteritis virus (EAV), and the flavivirus HCV [19], [27]. Besides DMVs, for most of these viruses other kinds of structures are found during infection, mostly in close proximity to the DMVs themselves, such as single membrane vesicles, multi-membrane vesicles, vesicle packets, tubular structures or zippered ER membrane. As for Inv ROs, replicases and viral RNA of poliovirus, EAV, and coxsackievirus B3 were demonstrated to localize to DMVs, suggesting that these structures might also support viral RNA replication. Likewise, for HCV replicase proteins and viral RNA were predominantly found in DMVs, and the number of DMVs positively correlated to the amount of viral RNA produced, suggesting that DMVs indeed serve as sites of RNA replication. However, for coronaviruses, the number of DMVs is not necessarily proportional to replication capacity [28], [29]. Also, whereas SARS-CoV dsRNA (a replication intermediate) was found inside DMVs, replicase proteins were more abundant in surrounding convoluted membranes, raising questions regarding the spatial organization of SARS-CoV RNA replication. The lack of clear connections between the inside of corona- and arterivirus-induced DMVs and the cytosol (where the RNA products have to go for translation and particle formation) further complicates our current understanding of coronavirus DMV functionality [30], [31]. Together, these data continue to cause debate on the exact function of DMVs and other features of DMV ROs. The notion that expression of combinations of non-structural viral proteins (nsps) for some of these viruses mimics the formation of membrane alterations as observed during infection confirms the viral induction of these structures and opens possibilities for detailed study of this particular feature of the infection [32], [33], [34].
3. Innate immune recognition and responses targeted towards viral RNA replication, transcription, and translation
3.1. Innate immune detection of viral RNA establishes an antiviral state
Rapid production of interferons (IFNs) and pro-inflammatory cytokines is an important consequence of virus detection, as it contributes to an antiviral state in both the infected host cell and the (un)infected surrounding cells. In addition, IFNs play an essential role in coordinating the antiviral adaptive immune response, which has been reviewed elsewhere [35]. Three types of IFNs have been described. Type I IFNs consist of 13 subtypes of IFN-α and a single subtype of IFN-β, IFN-δ, IFN-ε, IFN-κ, IFN-τ and IFN-ω. Type II IFN only contains one subtype of IFN-γ, and type III IFNs comprise of IFN-λ1 through −λ4. Whereas it is known that most cell types produce type I IFNs in response to viral infection, type II IFNs are only produced after antigenic stimulation of an expanding group of certain immune cells, including T-cells, natural killer cells, dendritic cells, and macrophages [36], [37]. In contrast, little is known about type III IFN production in vitro and in vivo, although it is believed that most cell types that produce type I IFNs are capable of producing type III IFNs as well [38]. Most cells are able to respond to IFN-I and −II, whereas IFN-III receptors are mainly found on epithelial cells [39]. The protective role of IFNs during viral infection is for example illustrated by inhibitory effects of IFN-α, IFN-β, and IFN-γ on SARS-CoV replication in vitro and in vivo [40], [41], [42], [43]. Importantly, correct timing and amount of IFN and subsequent pro-inflammatory cytokine expression is essential for an effective antiviral immune response, as delayed and/or elevated induction may stimulate immunopathological outcomes [35].
Below we will focus on the interactions of the innate immune system with the RNA replication stages of +RNA virus infection, including exposure of viral RNA to the cytosol after the unpacking of virus particles, and formation of ROs by the virus. We will also discuss examples of viral evasion surrounding these processes. IFN-I and –III production is triggered after host cells detect viral RNA, primarily using cytosolic RIG-I like Receptors (RLRs) and membrane-bound Toll-like receptors (TLRs). RLRs retinoic acid-inducible gene I (RIG-I) and melanoma differentiation associated factor 5 (MDA5) have been studied extensively (reviewed in [44], [45]). Besides their role in RNA metabolism, RIG-I and MDA5 recognize viral RNA by binding phosphorylated 5′ termini (5′ppp-RNA) in combination with dsRNA or ssRNA motifs. Detection of viral RNA activates RLRs and triggers downstream signaling through the mitochondrial antiviral signaling (MAVS) adaptor, which is localized on the outer mitochondrial membrane [46], [47], [48], [49]. The importance of MAVS is underscored by the observation that silencing of this protein by RNA interference abolishes expression of type I IFNs [46]. Subsequently, MAVS recruits various adaptor molecules such as Stimulator of interferon genes (STING) and TNF receptor-associated factors, resulting in the formation of large signaling complexes [50]. Ultimately, this leads to activation of kinase complexes IKKε/TBK1 and IKKα/IKKβ/IKKγ, resulting in activation of interferon regulating factor 3 (IRF3), 7 (IRF7) and NF-κB. These transcription factors then translocate to the nucleus and initiate the expression of IFNs and pro-inflammatory cytokines.
Besides the role of RLRs, other RNA sensing molecules have been shown to participate in antiviral responses. For example, of the 10 types of TLRs described in humans, endosomal TLRs 3, 7, and 8 are known to have a role in viral RNA detection. A detailed overview on the role of TLRs in antiviral signaling can be found elsewhere [51]. Briefly, TLRs interact with various adaptor molecules after stimulation, and the combination of recruited adaptors influences downstream signaling events. TLR3 eventually recruits common adaptor molecule TIR-domain-containing adapter-inducing interferon-β (TRIF), whereas TLRs 7 and 8 engage myeloid differentiation primary response gene 88 (MyD88). As in RLR signaling, this leads to activation of kinase complexes IKKε/TBK1 and IKKα/IKKβ/IKKγ. Furthermore, Protein kinase R (PKR) and 2′,5′-oligoadenylate synthetase (OAS) have been demonstrated to induce antiviral activities upon binding of dsRNA, as is described in detail elsewhere [45]. Well-established examples of antiviral activity provoked by PKR include inhibition of translation and inflammasome activation, and OAS activates RNase L to initiate degradation of host and viral RNA. In addition, PKR and OAS have been suggested to amplify RLR-mediated antiviral immune signaling [45].
The production and release of IFNs and pro-inflammatory cytokines contributes to an antiviral state in both infected host cells and in (un)infected surrounding cells. Despite the presence of multiple IFN and receptor types [52], the Janus kinase-signal transducer and activator of transcription (JAK/STAT) pathway is utilized by all IFNs to establish expression of interferon-stimulated genes (ISGs). The observation that a deficiency in separate IFN receptors did not affect disease progression during murine SARS-CoV infection, in contrast to a deficiency in common signaling molecule STAT1, illustrates this high degree of redundancy [53]. At the moment, hundreds of ISGs have been identified. However, an exact function has only been clarified for a relatively low number of the corresponding proteins, a description of which can be found elsewhere [54], [55], [56]. Nonetheless, these studies suggest that ISGs exhibit overlapping inhibitory activity towards most components of virus replication, including virus entry, uncoating, translation of viral proteins, RNA replication, and egress [54]. In summary, host cells detect viral RNA by several detection mechanisms to establish an antiviral state via the induction of IFNs and pro-inflammatory cytokines, leading to expression of ISGs.
3.2. Antiviral innate immune signaling is spatially organized
As mentioned earlier, MAVS localized on mitochondria is a key player in antiviral signaling. Therefore, studies demonstrating that the MAVS adaptor molecule STING is located on the ER implied that earlier identified contacts between mitochondrial membranes and ER membranes (mitochondrion-associated membranes: MAMs) were possibly involved in antiviral signaling [57]. In support of this theory, expression of a MAM-enriched marker protein followed by cell fractionation revealed that MAVS localizes to MAMs, which was supported by immunofluorescence assays of MAVS and MAM-enriched proteins [58]. Additionally, immunoprecipitation analysis of the MAM fraction of Sendai virus-infected hepatocytes confirmed that MAM-localized MAVS interacts with RIG-I and signaling cofactor TRAF3 [58], indicating that MAMs are involved in the RLR-mediated antiviral immune signaling pathway. Therefore, MAMs function as important signaling platforms that govern expression of type I and III IFNs. Interestingly, MAVS is also expressed on peroxisomal membranes, and interacts with stimulated RLRs during viral infection [59]. However, in contrast to its mitochondrial counterpart, peroxisomal MAVS was found to induce expression of ISGs by an IFN-independent mechanism, and this relatively rapid process seems to provide short-term protection until the IFN-dependent expression of ISGs mediated by other viral RNA-sensing mechanisms is established [59]. Interestingly, recent studies found that direct contacts between peroxisomes and ER membranes were involved in maintaining peroxisomal function [60], [61], [62], raising the question whether these contact sites might also regulate antiviral immunity-related processes. In summary, peroxisomal and mitochondrial membranes and their contact-sites with the ER are important regions for converting viral RNA detection by RLRs to downstream events required for establishing an antiviral state.
Besides MAMs, increasing evidence suggest a role for stress granules (SGs) in antiviral immune signaling. SGs are non-membranous compartments in the cell that contain aggregates of ribonucleoprotein (RNP) and mRNA, and are formed as a physiological response to various stress stimuli that cause temporal stalling of mRNA translation, suggestively preventing accumulation [63]. Consistent with the function of this structure, inhibition of translation through the inactivation of eukaryotic translation initiation factor (eIF)2a by phosphorylation is an important cue for SG formation [63]. Considering that various RNA virus infections result in shutdown of host translation by inactivation of eIF2a by for example PKR, it is now known that formation of SGs occurs to different extends during infection with various RNA viruses, including HCV, DENV, and SFV [63], [64]. Interestingly, multiple lines of evidence have demonstrated that SGs in encephalomyocarditis virus- or influenza virus-infected cells contain multiple RNA sensing molecules that were found to contribute to IFN production in vitro, including RIG-I, MDA5, PKR, OAS, RNaseL, and dsRNA [65], [66]. However, it should be noted that the contribution of SGs to antiviral signaling differs per virus, as SG-localized MDA5, an RLR that greatly contributes to IFN induction upon encephalomyocarditis virus infection, was not found to support IFN expression upon infection with this virus [67]. Nonetheless, these findings suggest an important role for SGs as detection platforms for viral RNA. Taken together, increasing evidence suggests that the antiviral innate immune signaling cascade is spatially organized, and since different structures in the cell seem to be specialized in a particular aspect of this process, it can be hypothesized that interaction between these structures plays a fundamental role in orchestrating the antiviral innate immune response.
4. Positive-strand RNA viruses interfere with antiviral innate immune signaling
4.1. Shielding viral RNA species away from innate immune sensors
Positive-strand RNA viruses developed multiple mechanisms to impair RNA recognition and antiviral signaling (also reviewed in [8]). As mentioned above, the presence of viral RNA in ROs suggests a role for ROs in shielding viral RNA from the innate immune detection machinery. While there is no unequivocal evidence supporting this theory, the finding that isolated viral RNA-containing membrane alterations only become sensitive to nuclease treatment after membrane disruption by nonionic detergents supports this hypothesis [68], [69], [70], [71]. In addition, a positive correlation between cytosolic exposure of viral RNA and IFN induction was recently found by an in vitro comparison of JEV and DENV infections [72]. Another important strategy by which RNA viruses avoid recognition is their modification of viral RNA molecules to mimic eukaryotic mRNA. For example, the formation of a 5′ cap is catalyzed by viral phosphatases and methyltransferases in some viral families such as the coronaviruses. From work on mouse hepatitis virus (MHV) it became clear that coronaviruses need to take care of N-linked as well as 2′O-linked methylation on the CAP structure, in order to avoid recognition by innate immune sensors [73]. Mimicry of eukaryotic RNA in this way was shown to be an important immune evasion strategy by +RNA viruses such as SARS-CoV and JEV [74], [75]. A recent report indicated that coronaviruses also evade dsRNA mediated recognition by PKR and OAS by using their endonuclease (EndoU), which cleaves free viral (and cellular) RNA that is somehow exposed to the cytosol, in order to prevent antiviral signaling [76].
4.2. Active interference with innate immune signaling by viral proteins
In addition to avoidance of recognition, +RNA viruses actively interfere with antiviral signaling components to impair expression of IFNs and pro- inflammatory cytokines. Very often, the viral proteases that are expressed by +RNA viruses, and which mostly have a primary function in cleavage of viral replicase polyproteins, seem to have a prominent role in this. For example, HCV NS3/4A and DENV NS4A cleave MAVS [47], [77], [78], [79], [80], [81], [82], and the DENV protease complex and HCV NS4B inhibit adaptor molecule STING by cleaving it [83], [84], [85], [86], [87]. Additionally, human CoV NL63 and SARS-CoV nsp3 possess a papain-like protease (PLP) domain that prevents dimerization of STING and its complex formation with MAVS and IKKε, and almost all nidovirus-encoded PLPs have been shown to display deubiquitinating activity [88]. Several have been suggested to facilitate deubiquitination of innate immune factors such as STING, RIG-I, TBK1 and IRF3 in order to halt innate immune signaling [8]. Moreover, in collaboration with others, our lab demonstrated that EAV and MERS-CoV virus mutants that lack the DUB activity of their PLPs while retaining polyprotein cleavage functions suppress IFN production less efficiently during infection ([89], [90] and our unpublished data). Furthermore, viral proteins such as DENV NS2B/3 and the SARS-CoV membrane glycoprotein interfere with complex formation of IKKε/TBK1 and IKKα/IKKβ/IKKγ or downstream transcription factors, thereby effectively inhibiting both RLR and TLR signaling [91], [92]. Interestingly, accessory proteins encoded by open reading frames of CoVs have been shown to be involved in inhibition of IRF3, IRF7, and NF-κB by undefined mechanisms [93], [94]. In addition, several methods are employed by +RNA viruses to interfere with JAK/STAT signaling. For instance, JEV NS5 and WNV NS4 B inhibit phosphorylation and activation of JAK1, and suppressors of JAK1 are induced by viruses such as WNV, JEV, and chikungunya virus [95], [96], [97], [98], [99]. However, STAT1 and STAT2 are most heavily targeted. JEV NS5, WNV and DENV NS4B, and JEV NS4A have been shown to inhibit STAT activation. In addition, DENV NS5 promotes proteasomal degradation of STAT2. Interestingly, SARS-CoV inhibits STAT1 signaling in three different ways. Firstly, SARS-CoV nsp1 binds STAT1 to inhibit its phosphorylation [100]. Secondly, the accessory protein encoded by SARS-CoV open reading frame 6 prevents nuclear transport of STAT1 [93], [101]. Thirdly, SARS-CoV PLP induces expression of E3 ubiquitin ligase E2–25k, which promotes proteasomal degradation of extracellular signal-regulated kinase 1, a protein responsible for activation of STAT1 [102]. To our knowledge, no studies have been published (yet) about specific interference of +RNA viruses with IFN-II production.
4.3. Dysregulation in the spatial organization of the antiviral innate immune system
Interference of +RNA viruses with antiviral signaling also has consequences for the spatial organization of the antiviral innate immune system. For instance, since SGs suggestively have an antiviral role, interfering with SG formation might constitute an opportunity for viruses to hamper the innate immune response. Indeed, various proteins of poliovirus, coxsackievirus, encephalomyocarditis virus, DENV, WNV, MERS-CoV, chikungunya virus, and JEV have been shown to prevent the formation of SGs and thereby suppress IFN production [63], [66], [103], [104], [105], [106], [107], [108], [109]. Thus, dysregulation of SGs seems to be a common strategy for +RNA viruses in order to dampen the innate antiviral immune response. In addition, the finding that MAMs are involved in the RLR-mediated antiviral immune signaling pathway shedded new light on the immunosuppressive role of viral proteases. By performing colocalization analysis with a MAM-enriched marker protein and cell fractionation experiments in both uninfected and HCV-infected cells, it was found that NS3/4A also localizes to MAMs [58]. Intriguingly, despite the clear function for peroxisomal MAVS in antiviral signaling, cleaved MAVS was found solely in MAM-enriched cell fractions. In addition, recent findings suggest that MAMs are also physically disrupted during DENV infection [110]. Thus, these data indicate that MAMs are critical locations for antiviral signaling and have an important role in expression of type I and III IFNs. Moreover, increasing evidence suggests that at least some +RNA viruses in fact occupy or hijack MAM-membranes during infection, as MAMs of HCV-infected cells were found to contain proteins involved in virus assembly and fully assembled virions [111]. It remains to be investigated whether MAMs also serve as platforms for viral assembly of other +RNA viruses. In addition, MAM disruption as a consequence of DENV infection was followed by the formation of convoluted membranes at the same location, which supported DENV replication [110]. Moreover, promoting the formation of convoluted membranes further repressed the IFN response, underscoring the importance of MAM disruption in both the replication and immune evasion of +RNA viruses [110]. Taken together, these studies underscore the importance of MAMs as key antiviral signaling platforms, and disruption of MAMs constitutes an effective immune evasion strategy for +RNA viruses. In conclusion, +RNA viruses developed divergent methods to delay antiviral innate immune signaling at multiple levels. In addition, interference of +RNA viruses with antiviral signaling leads to spatial disorganization of the antiviral innate immune system, and future studies will hopefully elucidate the consequences of this phenomenon in the context of other cellular compartments and viruses.
5. Specific responses towards viral replication organelles
Assuming that ROs impair antiviral signaling by shielding viral RNA from the host, it is tempting to hypothesize the existence of host cell mechanisms aimed to disrupt ROs, thereby exposing viral RNA and promoting antiviral signaling. The fact that +RNA viruses display such elaborate activities to inhibit viral RNA recognition and subsequent signaling, as detailed in the former paragraphs, supports the view that disruption of RO integrity is a situation these viruses anticipate dealing with. However, evidence suggesting targeting of ROs by the innate immune system is still scarce, although it was reported that HCV ROs are attacked by the ISG Viperin [94]. Additionally, 25-hydroxycholesterol, a product produced by the ISG cholesterol 25-hydroxylase also modifies HCV replication organelles [112], [113]. Recently, our lab published a study suggesting that IFN-β signaling also negatively influences arterivirus ROs, since significantly less were formed and remaining structures showed drastically different morphology after IFN-β treatment. The results suggested that the treatment interrupted biogenesis of these membrane structures rather than breaking down structures that were already made [114]. Interestingly, neither Viperin nor 25-hydroxycholesterol was involved in the observed effects, and the mechanistic details therefore have to be further investigated.
5.1. The innate immune system disrupts pathogen-containing membrane rearrangements to expose PAMPs
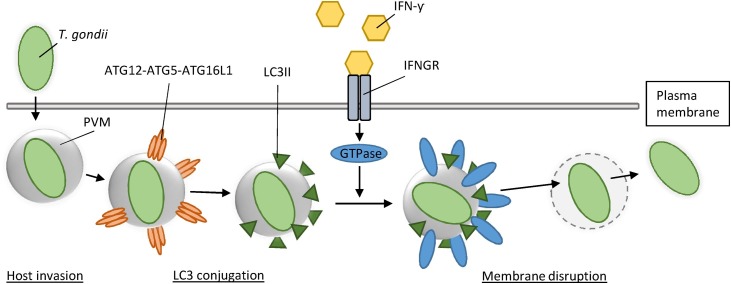
Other recent data suggest that there may indeed be specific mechanisms by which the innate immune system tags and disrupts so-called non- or aberrant-self membrane structures in the cell [115], which could include viral ROs. Most data originate from studies focusing on bacteria, fungi, or parasites that reside in rearranged membranes, known as pathogen-containing vacuoles (PVs), which prevent detection of cytosolic innate immune sensors in a similar way as viral ROs are thought to do. Multiple studies found that enzymes capable of disrupting PVs are GTPases, such as effector immunity-related p47 GTPases (IRGs) and guanylate-binding proteins (GBPs), which are part of a family which we will refer to as IFN-inducible GTPases [116]. Expression of IRGs and GBPs is induced upon IFN-γ stimulation, and leads to their accumulation on PVs. Subsequently, activation of these GTPases due to the exchange of GDP for GTP results in membrane disruption as a consequence of their dynamin-like activity [117]. To prevent aspecific disruption, host cell membranes contain a set of proteins that inhibit GTPase activity, known as ‘’guard proteins’’ [118]. Well-studied pathogens in the context of this process are the protozoan parasite Toxoplasma gondii (T. gondii), the bacterium Chlamydia trachomatis and the microsporidian Encephalitozoon cuniculi [119], [120], [121], [122], [123], [124], [125]. Although it remains unclear for most of these pathogens how IFN-inducible GTPases are recruited towards PVs, recent studies on T. gondii infection demonstrated that host cells label these membrane structures, for example with various forms of microtubule-associated protein 1A/1B-light chain 3 (LC3) to initiate this process [122], [123], [124], [125] (Fig. 2 ). LC3 is a well-known factor in the autophagy pathway, and several forms of LC3 exist in the human proteome, which seem to have overlapping as well as distinct functions. The human genome encodes homologs LC3A, LC3B, and LC3C and LC3-like homologs GABARAP, GABARAPL1, GABARAPL2, and GABARAPL3. We will refer to all of these as LC3 unless stated otherwise. LC3 was initially discovered as an essential protein for autophagosome formation, which requires a covalent interaction between cytosolic LC3 (LC3-I) and the phospholipid phosphatidylethanolamine (PE), after which LC3 is referred to as LC3-II or lipidated LC3 [126]. However, recent studies have now revealed that labeling of (foreign) membrane compartments by LC3 conjugation can also have various autophagy-unrelated consequences, as has been reviewed elsewhere [127], [128]. Interestingly, multiple studies demonstrated that labeling of LC3 on PVs recruits IFN-inducible GTPases upon IFN-γ expression during T. gondii infection, resulting in exposure of T. gondii to the host cell cytosol, thereby triggering anti-bacterial immune responses [122], [123], [125]. Considering the function of LC3 in autophagosome formation, Jayoung Choi and co-workers [124] termed this process Targeting by AutophaGy proteins (TAG). In addition, a similar role was recently found for ubiquitin, the versatile regulator of numerous important cellular processes, and also a well-known autophagy-related protein [129]. A study on T. gondii and Chlamydia trachomatis infection demonstrated that PVs are also recognized and labeled by the ubiquitination pathway upon IFN-γ expression, resulting in recruitment and activation of IFN-inducible GTPases [130]. Importantly, it was shown that virulent strains of T. gondii and Chlamydia trachomatis interfere with the deposition of IFN-inducible GTPases on PVs, underscoring the importance of this mechanism in clearance of associated infections [119], [131], [132]. Collectively, these studies demonstrate the existence of various innate immune responses that result in tagging and subsequent disruption of non- or aberrant-self membrane structures in the cell, thereby exposing PAMPs and promoting immune signaling. Importantly, the findings suggest that IFN-mediated membrane disruption might be a common principle in clearance of pathogens that use rearranged membranes to support replication and avoid innate immune detection.
Fig. 2.
LC3 recruits IFN-γ inducible GTPases to disrupt the parasitophorus vacuole of Toxoplasma gondii. Upon invasion of the host cell, Toxoplasma gondii (T. gondii) resides in a host-derived membranous structure, termed the parasitophorus vacuole membrane (PVM), to evade immune detection. Recent studies demonstrate that LC3 is conjugated to the PVM by the LC3 conjugation system, leading to recruitment of IFN-γ inducible GTPases to the PVM upon IFN-γ stimulation. As a consequence, activation of IFN-γ inducible GTPases leads to membrane disruption of the PVM, and exposure of T. gondii to the innate immune system.
5.2. Indications for disruption of +RNA virus ROs by the innate immune system
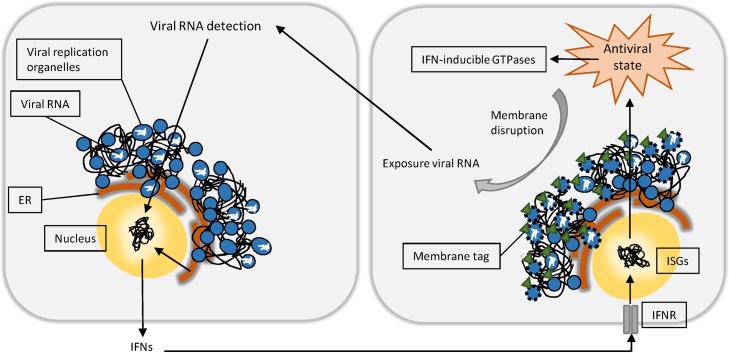
Like PVs, ROs induced by +RNA viruses are membranous compartments that constitute an environment for efficient replication, simultaneously avoiding immune detection by the host. Therefore, similar mechanisms might be aimed towards viral ROs. The observation that ROs induced by various +RNA viruses consist of a double membrane initially suggested a role for the autophagy machinery in the formation of these structures. However, only nonlipidated LC3, and not an intact autophagy pathway, was found to be essential for viral RO formation and viral replication upon infection with EAV and MHV [133], [134], thereby contradicting this hypothesis. Interestingly, lipidated LC3 was found to support IFN-γ mediated protection against murine norovirus (MNV) infection by autophagy-unrelated means [135]. Furthermore, the induction of viral membranous structures by expression of MNV nsps led to a high colocalization between ATG16L1, a protein involved in LC3 conjugation, and the MNV polymerase. Since ATG16L1 determines the site for LC3 conjugation [136], and the MNV polymerase is associated with ROs, this further supports the possibility that LC3 conjugation occurs on ROs. Other supporting in vitro experiments show that conditional KO of ATG4B inhibits replication of MNV and T. gondii more effectively compared to wildtype cells [123], [135]. In contrast to the multiple autophagins involved in pre-processing of LC3 for conjugation, ATG4B is the only human homolog of yeast ATG4 that efficiently deconjugates LC3 from membranes [137], and therefore plays a major role in negatively regulating the fraction of conjugated LC3. Several groups have reported enhanced conjugation of LC3 to membranes during knockdown of ATG4B in various cell types [138], [139], [140], [141], including those used by aforementioned studies [123], [135]. Thus, it is likely that increased lipidated LC3 was present on ROs and PVMs during experiments in ATG4B-deficient cells, thereby explaining the enhanced inhibition of replication. Interestingly, findings in other studies are contradictory in the sense that only LC3-I was observed on ROs of MNV, EAV, and MHV-infected cells [133], [134], [135]. On this note, it is worth mentioning that LC3-I does not support IFN-γ mediated inhibition of viral replication, as IFN-γ mediated inhibition of MNV-infected cells does not occur in the case of ATG7 deficiency, one of the proteins responsible for LC3 conjugation [135]. In conclusion, although there is no unequivocal evidence regarding the presence of conjugated LC3 on +RNA virus induced ROs, the studies described above provoke the hypothesis that membrane disruption of viral ROs mediated by IFN-γ inducible GTPases may be part of the innate immune response against +RNA viruses (Figure 3 ).
Fig. 3.
A possible role for IFN-inducible GTPases in the targeting of viral replication organelles. Upon infection, +RNA viruses hamper IFN and ISG induction at multiple levels to decelerate innate immune detection. Viral replication organelles (ROs) are believed to have a dual role in +RNA virus infection and innate immune evasion, as they facilitate efficient RNA replication while shielding viral RNA from the host. Based on current knowledge, we hypothesize the existence of host cell IFN-inducible GTPases aimed to disrupt ROs, thereby exposing viral RNA and promoting antiviral signaling. IFNR: interferon receptor.
5.3. A general role for IFN-inducible GTPases in the antiviral innate immune response?
Interestingly, current knowledge of IFN-inducible GTPases suggests that membrane disruption of ROs by IFN-γ inducible GTPases might be part of a common antiviral mechanism that utilizes IFN-inducible GTPases to combat viral infection. Aside from IRGs and GBPs, other families of dynamin-like IFN-inducible GTPases exist, such as myxovirus resistance proteins (Mx) and the very large IFN-inducible GTPases (VLIGs) [116]. These enzymes are also known to have antiviral properties against a variety of viruses (reviewed in [116]). For example, the protective role of Mx proteins has been demonstrated for orthomyxoviruses such as influenza, lentiviruses such as human immunodeficiency virus (HIV) 1, and, interestingly, several +RNA viruses belonging to the picornaviridae and togaviridae [116]. Moreover, based on evolutionary studies, it was recently suggested that Mx proteins mediate protection against even a wider variety of viruses [142]. In contrast to IFN-γ inducible IRGs and GBPs, expression of Mx proteins is induced by IFNs type I and III, implying that all IFN types are capable of inducing IFN-inducible GTPases [116]. Therefore, all cell types known to respond to IFNs could theoretically induce a variety of IFN-inducible GTPases in response to viral infection. In the case of +RNA viruses, it is intriguing that multiple studies demonstrated how MxB suppresses replication of HIV-1 by targeting the viral capsid protein [143], [144], [145], given the parallels between retroviral capsids and +RNA virus ROs [146]. In conclusion, these findings again suggest a general function for IFN-inducible GTPase effectors in the targeting of viral intracellular membrane structures that function to shield away viral components away from the cytosol.
6. Concluding remarks
Rearrangement of host membranes into ROs is considered a hallmark of +RNA virus infection. ROs are believed to have at least a dual role in +RNA virus infection and innate immune evasion, as they facilitate efficient RNA replication while shielding viral RNA from the host antiviral response machinery. As viral RNA is a very potent inducer of antiviral signaling, we focused on how both the biochemical and spatial organization allows the innate immune system to convert detection of viral RNA to an antiviral state. In addition, we described part of the divergent methods by which +RNA viruses impair antiviral signaling surrounding their RNA replication, transcription, and translation in order to establish infection. At last, based on recent studies that demonstrated how IFN-γ inducible GTPases are capable of disrupting PVs, we discussed the possibility of a general function of IFN-inducible GTPases in the targeting of viral ROs. In summary, upon infection, +RNA viruses hamper IFN and ISG induction at multiple levels to decelerate antiviral innate immune signaling. In this process, the formation of ROs enables rapid viral RNA replication while masking it from the host. However, in case of sufficient activation of IFN-inducible GTPases by IFN-I and –III originating from various cell types, or IFN-II produced by specialized immune cells, disruption of ROs by IFN-inducible GTPases may result in enhanced exposure of viral RNA, thereby amplifying the innate immune response and ensuring efficient clearance of the virus. However, considering the differences in morphology and origin of ROs, the variety of infection dynamics within the category of +RNA viruses, and a lack of knowledge regarding the exact mechanism(s) of disruption of non/aberrant-self membrane structures by IFN-inducible GTPases and possibly other factors, extensive further research is required to elucidate whether this process plays a general role in +RNA virus infection, and might open possibilities for development of novel antiviral therapies.
Conflict of interest
None
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Biographies

Enzo Scutigliani (1993) obtained a BSc in Biomedical Sciences with honor at the University of Amsterdam (The Netherlands) in 2014, and is currently in the final process of becoming a MSc with a specialization in cell biology and advanced microscopy. Over the last years, he developed a particular interest for microbiology, and his ambition is to develop novel treatment or vaccination methods by understanding pathogens at the molecular level. To achieve this goal, he is currently specializing in combining microbiological research and advanced microscopy.

A Molecular Sciences study at Wageningen University and Research Center (The Netherlands) motivated Marjolein Kikkert (1969) to pursue a Ph.D. degree, which she achieved in 1999 at the Department of Virology of this University. Her thesis was entitled “Role of the envelope glycoproteins in the infection of tomato spotted wilt virus”. She then switched to mammalian Virology, became a post-doc at the National Institute of Health and the Environment in Bilthoven, the Netherlands with prof. Emmanuel Wiertz, and subsequently at Leiden University Medical Center (LUMC), working on immune evasion strategies of human cytomegalovirus. After this she started working with Prof. Eric Snijder at the LUMC as a post-doc, and developed into an Assistant Professor in the Department of Medical Microbiology. Her research currently focuses on the replication organelles and related virus-host interactions of nidoviruses and other +RNA viruses, and the innate immune evasion mechanisms which these viruses employ. The knowledge gained from her research is being used for development of novel antiviral vaccines and −drugs.
References
- 1.Beachboard D.C., Horner S.M. Innate immune evasion strategies of DNA and RNA viruses. Curr. Opin. Microbiol. 2016;32:113–119. doi: 10.1016/j.mib.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 3.Gantier M.P., Williams B.R.G. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007;18:363–371. doi: 10.1016/j.cytogfr.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brubaker S.W., Bonham K.S., Zanoni I., Kagan J.C. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Errett J.S., Gale M. Emerging complexity and new roles for the RIG-I-like receptors in innate antiviral immunity. Virol. Sin. 2015;30:163–173. doi: 10.1007/s12250-015-3604-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Vliet A.R., Verfaillie T., Agostinis P. New functions of mitochondria associated membranes in cellular signaling. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843:2253–2262. doi: 10.1016/j.bbamcr.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Giacomello M., Pellegrini L. The coming of age of the mitochondria–ER contact: a matter of thickness. Cell Death Differ. 2016;23:1417–1427. doi: 10.1038/cdd.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma D.Y., Suthar M.S. Mechanisms of innate immune evasion in re-emerging RNA viruses. Curr. Opin. Virol. 2015;12:26–37. doi: 10.1016/j.coviro.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kai Chan Y., Gack M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Publ. Gr. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Den Boon J.A., Diaz A., Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe. 2010;8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuman B.W., Angelini M.M., Buchmeier M.J. Does form meet function in the coronavirus replicative organelle? Trends Microbiol. 2014;22:642–647. doi: 10.1016/j.tim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Hoeven B., Oudshoorn D., Koster A.J., Snijder E.J., Kikkert M., Bárcena M. Biogenesis and architecture of arterivirus replication organelles. Virus Res. 2016;220:70–90. doi: 10.1016/j.virusres.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groneberg D.A., Zhang L., Welte T., Zabel P., Chung K.F. Severe acute respiratory syndrome: global initiatives for disease diagnosis. QJM. 2003;96:845–852. doi: 10.1093/qjmed/hcg146. http://www.ncbi.nlm.nih.gov/pubmed/14566040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aleanizy F.S., Mohmed N., Alqahtani F.Y., El Hadi Mohamed R.A. Outbreak of Middle East respiratory syndrome coronavirus in Saudi Arabia: a retrospective study. BMC Infect. Dis. 2017;17:23. doi: 10.1186/s12879-016-2137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kay M., Allen T., Frank V., Santhana S., Dye C. Zika: the origin and spread of a mosquito-borne virus. World Heal. Organ. 2016:1–18. doi: 10.2471/BLT.16.171082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shulla A., Randall G. (+) RNA virus replication compartments: a safe home for (most) viral replication. Curr. Opin. Microbiol. 2016;32:82–88. doi: 10.1016/j.mib.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul D., Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. May. 2013;12:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.V’kovski P., Al-Mulla H., Thiel V., Neuman B.W. New insights on the role of paired membrane structures in coronavirus replication. Virus Res. 2015;202:33–40. doi: 10.1016/j.virusres.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K.E., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimley P.M., Berezesky I.K., Friedman R.M. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J. Virol. 1968;2:1326–1338. doi: 10.1128/jvi.2.11.1326-1338.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kujala P., Ikaheimonen A., Ehsani N., Vihinen H., Auvinen P., Kaariainen L. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001;75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. (07-PLBI-RA-0823 [pii]\r10.1371/journal.pbio.0050220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Froshauer S., Kartenbeck J., Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988;107:2075–2086. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Junjhon J., Pennington J.G., Edwards T.J., Perera R., Lanman J., Kuhn R.J. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 2014;88:4687–4697. doi: 10.1128/JVI.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero-Brey I., Merz A., Chiramel A., Lee J.Y., Chlanda P., Haselman U., Santarella-Mellwig R., Habermann A., Hoppe S., Kallis S., Walther P., Antony C., Krijnse-Locker J., Bartenschlager R. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beachboard D.C., Anderson-Daniels J.M., Denison M.R. Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications. J. Virol. 2015;89:2080–2089. doi: 10.1128/JVI.02776-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Mulla H.M.N., Turrell L., Smith N.M., Payne L., Baliji S., Züst R., Thiel V., Baker S.C., Siddell S.G., Neuman B.W. Competitive fitness in coronaviruses is not correlated with size or number of double-membrane vesicles under reduced-temperature growth conditions. MBio. 2014;5 doi: 10.1128/mBio.01107-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knoops K., Kikkert M., Van Den Worm S.H.E., Zevenhoven-Dobbe J.C., Van Der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:1957–1974. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knoops K., Barcena M., Limpens R.W. a. L., Koster a. J., Mommaas a. M., Snijder E.J. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egger D., Wölk B., Gosert R., Bianchi L., Blum H.E., Moradpour D., Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snijder E.J., Van Tol H., Roos N., Pedersen K.W. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 2001;82:985–994. doi: 10.1099/vir.0.17499-0. [DOI] [PubMed] [Google Scholar]
- 34.Angelini M., Akhlaghpour M. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 Induce double-membrane vesicles. MBio. 2013;4:1–10. doi: 10.1128/mBio.00524-13. Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mcnab F., Mayer-barber K., Sher A., Wack A., Garra A.O. Type I interferons in infectious disease. Nat. Publ. Gr. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darwich L., Coma G., Peña R., Bellido R., Blanco E.J.J., Este J.A., Borras F.E., Clotet B., Ruiz L., Rosell A., Andreo F., Parkhouse R.M.E., Bofill M. Secretion of interferon-(by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology. 2009;126:386–393. doi: 10.1111/j.1365-2567.2008.02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoenborn J.R., Wilson C.B. Regulation of interferon-γ during innate and adaptive immune responses. Adv. Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- 38.Iversen M.B., Paludan S.R. Mechanisms of type III interferon expression. J. Interferon Cytokine Res. 2010;30:573–578. doi: 10.1089/jir.2010.0063. [DOI] [PubMed] [Google Scholar]
- 39.Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-??) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ströher U., DiCaro A., Li Y., Strong J.E., Aoki F., Plummer F., Jones S.M., Feldmann H. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon- alpha. J. Infect. Dis. 2004;189:1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sainz B., Mossel E.C., Peters C.J., Garry R.F. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329:11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K.S., Sidwell R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-CoV inhibitors for inhibition of SARS-CoV replication in BALB/c mice, Antivir. Chem. Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 43.Kumaki Y., Ennis J., Rahbar R., Turner J.D., Wandersee M.K., Smith A.J., Bailey K.W., Vest Z.G., Madsen J.R., Li J.K.K., Barnard D.L. Single-dose intranasal administration with mDEF201 (adenovirus vectored mouse interferon-alpha) confers protection from mortality in a lethal SARS-CoV BALB/c mouse model. Antiviral Res. 2011;89:75–82. doi: 10.1016/j.antiviral.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loo Y.M., Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sparrer K.M.J., Gack M.U. Intracellular detection of viral nucleic acids. Curr. Opin. Microbiol. 2015;26:1–9. doi: 10.1016/j.mib.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 47.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 48.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-??B and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B. VISA is an adapter protein required for virus-triggered IFN-?.?. signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 50.West A.P., Shadel G.S., Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lester S.N., Li K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014;426:1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Q., Yoo D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res. 2016 doi: 10.1016/j.virusres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frieman M.B., Chen J., Morrison T.E., Whitmore A., Funkhouser W., Ward J.M., Lamirande E.W., Roberts A., Heise M., Subbarao K., Baric R.S. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 2010;6:e1000849. doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadler A.J., Williams B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borden E.C., Williams B.R. Interferon-stimulated genes and their protein products: what and how? J. Interferon Cytokine Res. 2011;31:1–4. doi: 10.1089/jir.2010.0129. [DOI] [PubMed] [Google Scholar]
- 57.Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. (0900850106 [pii]\r10.1073/pnas.0900850106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horner S.M., Liu H.M., Park H.S., Briley J., Gale M. Mitochondrial-associated endoplasmic reticulum membranes {(MAM)} form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. {U.S.A.}. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dixit E., Boulant S., Zhang Y., Lee A.S.Y., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M., Nibert M.L., Superti-Furga G., Kagan J.C. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costello J.L., Castro I.G., Hacker C., Schrader T.A., Metz J., Zeuschner D., Azadi A.S., Godinho L.F., Costina V., Findeisen P., Manner A., Islinger M., Schrader M. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol. 2017;216:1–12. doi: 10.1083/jcb.201607055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strzyz P. Organelle dynamics: connections, connections, connections. Nat Rev. Mol. Cell Biol. 2017;18:139. doi: 10.1038/nrm.2017.14. [DOI] [PubMed] [Google Scholar]
- 62.Hua R., Coyaud D.É., Freeman S., Di Pietro E., Wang Y., Vissa A., Yip C.M., Fairn G.D., Braverman N., Brumell J.H., Trimble W.S., Raught B., Kim P.K. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis., Cheng. J. Cell Biol. 2017;216:1–11. doi: 10.1083/jcb.201608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Onomoto K., Yoneyama M., Fung G., Kato H., Fujita T. Antiviral innate immunity and stress granule responses. Trends Immunol. 2014;35:420–428. doi: 10.1016/j.it.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J.-P., Mazur J., Bankhead P., Hiet M.-S., Kallis S., Alvisi G., Samuel C.E., Lohmann V., Kaderali L., Rohr K., Frese M., Stoecklin G., Bartenschlager R. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Onomoto K., Jogi M., Yoo J.S., Narita R., Morimoto S., Takemura A., Sambhara S., Kawaguchi A., Osari S., Nagata K., Matsumiya T., Namiki H., Yoneyama M., Fujita T. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng C.S., Jogi M., Yoo J.-S., Onomoto K., Koike S., Iwasaki T., Yoneyama M., Kato H., Fujita T. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J. Virol. 2013;87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langereis M.A., Feng Q., van Kuppeveld F.J. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J. Virol. 2013;87:6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uchil P.D., Satchidanandam V. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology. 2003;307:358–371. doi: 10.1016/s0042-6822(02)00130-7. (S0042682202001307 [pii]) [DOI] [PubMed] [Google Scholar]
- 69.Uchil P.D., Satchidanandam V. Architecture of the flaviviral replication complex: protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 70.Van Hemert M.J., Van Den Worm S.H.E., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Hemert M.J., De Wilde A.H., Gorbalenya A.E., Snijder E.J. The in vitro RNA synthesizing activity of the isolated arterivirus replication/transcription complex is dependent on a host factor. J. Biol. Chem. 2008;283:16525–16536. doi: 10.1074/jbc.M708136200. [DOI] [PubMed] [Google Scholar]
- 72.Uchida L., Espada-Murao L.A., Takamatsu Y., Okamoto K., Hayasaka D., Yu F., Nabeshima T., Buerano C.C., Morita K. The dengue virus conceals double-stranded RNA in the intracellular membrane to escape from an interferon response. Sci. Rep. 2014;4:7395. doi: 10.1038/srep07395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Shi P.-Y., Diamond M.S. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Menachery V.D., Yount B.L., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of SARS-CoV mutant lacking 2′O methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li S.-H., Dong H., Li X.-F., Xie X., Zhao H., Deng Y.-Q., Wang X.-Y., Ye Q., Zhu S.-Y., Wang H.-J., Zhang B., Leng Q.-B., Zuest R., Qin E.-D., Qin C.-F., Shi P.-Y. Rational design of a flavivirus vaccine by abolishing viral RNA 2′-O methylation. J. Virol. 2013;87:5812–5819. doi: 10.1128/JVI.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., Züst R., Hwang M., V’kovski P., Stalder H., Marti S., Habjan M., Cervantes-Barragan L., Elliot R., Karl N., Gaughan C., van Kuppeveld F.J.M., Silverman R.H., Keller M., Ludewig B., Bergmann C.C., Ziebuhr J., Weiss S.R., Kalinke U., Thiel V. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li X.-D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foy E., Li K., Wang C., Sumpter R., Ikeda M., Lemon S.M., Gale M. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 79.Foy E., Li K., Sumpter R., Loo Y.-M., Johnson C.L., Wang C., Fish P.M., Yoneyama M., Fujita T., Lemon S.M., Gale M. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loo Y.-M., Owen D.M., Li K., Erickson A.K., Johnson C.L., Fish P.M., Carney D.S., Wang T., Ishida H., Yoneyama M., Fujita T., Saito T., Lee W.M., Hagedorn C.H., Lau D.T.-Y., Weinman S.A., Lemon S.M., Gale M. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller S., Kastner S., Krijnse-Locker J., Bühler S., Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2007;282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 82.He Z., Zhu X., Wen W., Yuan J., Hu Y., Chen J., An S., Dong X., Lin C., Yu J., Wu J., Yang Y., Cai J., Li J., Li M. Dengue virus subverts host innate immunity by targeting adaptor protein MAVS. J. Virol. 2016;90:7219–7230. doi: 10.1128/JVI.00221-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rodriguez-Madoz J.R., Belicha-Villanueva A., Bernal-Rubio D., Ashour J., Ayllon J., Fernandez-Sesma A. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J. Virol. 2010;84:9760–9774. doi: 10.1128/JVI.01051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu C.Y., Chang T.H., Liang J.J., Chiang R.L., Lee Y.L., Liao C.L., Lin Y.L. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aguirre S., Maestre A.M., Pagni S., Patel J.R., Savage T., Gutman D., Maringer K., Bernal-Rubio D., Shabman R.S., Simon V., Rodriguez-Madoz J.R., Mulder L.C.F., Barber G.N., Fernandez-Sesma A. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ding Q., Cao X., Lu J., Huang B., Liu Y.J., Kato N., Shu H.B., Zhong J. Hepatitis C virus NS4 B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol. 2013;59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 87.Nitta S., Sakamoto N., Nakagawa M., Kakinuma S., Mishima K., Kusano-Kitazume A., Kiyohashi K., Murakawa M., Nishimura-Sakurai Y., Azuma S., Tasaka-Fujita M., Asahina Y., Yoneyama M., Fujita T., Watanabe M. Hepatitis C virus NS4 B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology. 2013;57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 88.Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Kasteren P.B., a Bailey-Elkin B., James T.W., Ninaber D.K., Beugeling C., Khajehpour M., Snijder E.J., Mark B.L., Kikkert M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bailey-Elkin B.A., Knaap R.C.M., Johnson G.G., Dalebout T.J., Ninaber D.K., Van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the middle east respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Angleró-Rodríguez Y.I., Pantoja P., Sariol C.A. Dengue virus subverts the interferon induction pathway via NS2B/3 protease-I(B kinase (interaction. Clin. Vaccine Immunol. 2014;21:29–38. doi: 10.1128/CVI.00500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fang X., Gao J., Zheng H., Li B., Kong L., Zhang Y., Wang W., Zeng Y., Ye L. The membrane protein of SARS-CoV suppresses NF-kappaB activation. J. Med. Virol. 2007;79:1431–1439. doi: 10.1002/jmv.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kopecky-Bromberg S.a., Martínez-Sobrido L., Frieman M., Baric R.a., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y., Zhao Z., Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mansfield K.L., Johnson N., Cosby S.L., Solomon T., Fooks A.R. Transcriptional upregulation of SOCS 1 and suppressors of cytokine signaling 3 mRNA in the absence of suppressors of cytokine signaling 2 mRNA after infection with West Nile virus or tick-borne encephalitis virus. Vector Borne Zoonotic Dis. 2010;10:649–653. doi: 10.1089/vbz.2009.0259. [DOI] [PubMed] [Google Scholar]
- 96.Patil D.R., Hundekar S.L., Arankalle V.A. Expression profile of immune response genes during acute myopathy induced by chikungunya virus in a mouse model. Microbes Infect. 2012;14:457–469. doi: 10.1016/j.micinf.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 97.Kundu K., Dutta K., Nazmi A., Basu A. Japanese encephalitis virus infection modulates the expression of suppressors of cytokine signaling (SOCS) in macrophages: implications for the hosts’ innate immune response. Cell. Immunol. 2013;285:100–110. doi: 10.1016/j.cellimm.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 98.Guo J.-T., Hayashi J., Seeger C. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 2005;79:1343–1350. doi: 10.1128/JVI.79.3.1343-1350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin R.J., Chang B.L., Yu H.P., Liao C.L., Lin Y.L. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li S.W., Lai C.C., Ping J.F., Tsai F.J., Wan L., Lin Y.J., Kung S.H., Lin C.W. Severe acute respiratory syndrome coronavirus papain-like protease suppressed alpha interferon-induced responses through downregulation of extracellular signal-regulated kinase 1-mediated signalling pathways. J. Gen. Virol. 2011;92:1127–1140. doi: 10.1099/vir.0.028936-0. [DOI] [PubMed] [Google Scholar]
- 103.White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 104.Garaigorta U., Heim M.H., Boyd B., Wieland S., Chisari F.V. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J. Virol. 2012;86:11043–11056. doi: 10.1128/JVI.07101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Katoh H., Okamoto T., Fukuhara T., Kambara H., Morita E., Mori Y., Kamitani W., Matsuura Y. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with Caprin-1 and facilitates viral propagation. J. Virol. 2013;87:489–502. doi: 10.1128/JVI.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Emara M.M., Brinton M.A. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J.-P., Mazur J., Bankhead P., Hiet M.-S., Kallis S., Alvisi G., Samuel C.E., Lohmann V., Kaderali L., Rohr K., Frese M., Stoecklin G., Bartenschlager R. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scholte F.E.M., Tas A., Albulescu I.C., Žusinaite E., Merits A., Snijder E.J., van Hemert M.J. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J. Virol. 2015;89:4457–4469. doi: 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rabouw H.H., Langereis M.A., Knaap R.C.M., Dalebout T.J., Canton J., Sola I., Enjuanes L., Bredenbeek P.J., Kikkert M., de Groot R.J., van Kuppeveld F.J.M. Middle east respiratory Coronavirus accessory protein 4a inhibits PKR-Mediated antiviral stress responses. PLoS Pathog. 2016;12:1–26. doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chatel-Chaix M., Romero-Brey S., Bender C.J., Neufeldt W., Fischl P., Scaturro N., Schieber Y., Schwab B., Fischer A., Bartenschlager R. Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe. 2016:1–15. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neufeldt C.J., Joyce M.A., Van Buuren N., Levin A., Kirkegaard K., Gale M., Tyrrell D.L.J., Wozniak R.W. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Anggakusuma I., Romero-Brey C., Berger C.C., Colpitts T., Boldanova M., Engelmann D., Todt P.M., Perin P., Behrendt F.W.R., Vondran S., Xu C., Goffinet L.M., Schang M.H., Heim R., Bartenschlager T., Pietschmann Steinmann E. Interferon-inducible cholesterol-25-hydroxylase restricts hepatitis C virus replication through blockage of membranous web formation. Hepatology. 2015;62:702–714. doi: 10.1002/hep.27913. [DOI] [PubMed] [Google Scholar]
- 113.Helbig K.J., Eyre N.S., Yip E., Narayana S., Li K., Fiches G., Mccartney E.M., Jangra R.K., Lemon S.M., Beard M.R. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54:1506–1517. doi: 10.1002/hep.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oudshoorn D., Van der Hoeven B., Limpens R.W.A.L., Beugeling C., Snijder E.J., Bárcena M., Kikkert M. Antiviral innate immune response interferes with the formation of replication-associated membrane structures induced by a +RNA virus. MBio. 2016;7:1–11. doi: 10.1128/mBio.01991-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coers J. Self and non-self discrimination of intracellular membranes by the innate immune system. PLoS Pathog. 2013;9:10–13. doi: 10.1371/journal.ppat.1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pilla-Moffett D., Barber M.F., Taylor G.A., Coers J. Interferon-inducible GTPases in host resistance, inflammation and disease. J. Mol. Biol. 2016;428:3495–3513. doi: 10.1016/j.jmb.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim B.H., Shenoy A.R., Kumar P., Bradfield C.J., MacMicking J.D. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Haldar A.K., Saka H.A., Piro A.S., Da Dunn J., Henry S.C., Taylor G.A., Frickel E.M., Valdivia R.H., Coers J. IRG and GBP host resistance factors target aberrant, non-self vacuoles characterized by the missing of self IRGM proteins. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Coers J., Bernstein-Hanley I., Grotsky D., Parvanova I., Howard J.C., a Taylor G., Dietrich W.F., Starnbach M.N. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J. Immunol. 2008;180:6237–6245. doi: 10.4049/jimmunol.180.9.6237. (180/9/6237 [pii]) [DOI] [PubMed] [Google Scholar]
- 120.da Fonseca Ferreira-da-Silva M., Springer-Frauenhoff H.M., Bohne W., Howard J.C. Identification of the microsporidian encephalitozoon cuniculi as a new target of the IFN-γ inducible IRG resistance system. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Howard J.C., Hunn J.P., Steinfeldt T. The IRG protein-based resistance mechanism in mice and its relation to virulence in Toxoplasma gondii. Curr. Opin. Microbiol. 2011;14:414–421. doi: 10.1016/j.mib.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 122.Zhao Z., Fux B., Goodwin M., Dunay I.R., Strong D., Miller B.C., Cadwell K., Delgado M.A., Ponpuak M., Green K.G., Schmidt R.E., Mizushima N., Deretic V., Sibley L.D., Virgin H.W. Autophagosome-independent essential function for the autophagy protein atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Choi J., Park S., Biering S.B., Selleck E., Liu C.Y., Zhang X., Fujita N., Saitoh T., Akira S., Yoshimori T., Sibley L.D., Hwang S., Virgin H.W. The parasitophorous vacuole membrane of toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014;40:924–935. doi: 10.1016/j.immuni.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Choi J., Biering S.B., Hwang S. Quo vadis?. Interferon-inducible GTPases go to their target membranes via the LC3-conjugation system of autophagy. Small GTPases. 2016;0:1–9. doi: 10.1080/21541248.2016.1213090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park S., Choi J., Biering S.B., Dominici E., Williams L.E., Hwang S. Targeting by AutophaGy proteins (TAG): Targeting of IFNG-inducible GTPases to membranes by the LC3 conjugation system of autophagy. Autophagy. 2016;12:1153–1167. doi: 10.1080/15548627.2016.1178447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. (4451 [pii]) [DOI] [PubMed] [Google Scholar]
- 127.Pimentel-Muiños F.X., Boada-Romero E. Selective autophagy against membranous compartments: canonical and unconventional purposes and mechanisms. Autophagy. 2014;10:397–407. doi: 10.4161/auto.27244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bestebroer J., V’kovski P., Mauthe M., Reggiori F. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic. 2013;14:1029–1041. doi: 10.1111/tra.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Welchman R.L., Gordon C., Mayer R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 130.Haldar A.K., Foltz C., Finethy R., Piro A.S., Feeley E.M., Pilla-Moffett D.M., Komatsu M., Frickel E.-M., Coers J. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc. Natl. Acad. Sci. 2015;112:E5628–E5637. doi: 10.1073/pnas.1515966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhao Y., Ferguson D.J.P., Wilson D., Howard J.C., Sibley L.D., Yap G.S. Virulent toxoplasma gondii evade immunity-related GTPase (IRG)-Mediated parasite vacuole disruption within primed macrophages. J. Immunol. 2010;182:3775–3781. doi: 10.4049/jimmunol.0804190.Virulent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Khaminets A., Hunn J.P., Könen-Waisman S., Zhao Y.O., Preukschat D., Coers J., Boyle J.P., Ong Y.C., Boothroyd J.C., Reichmann G., Howard J.C. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell. Microbiol. 2010;12:939–961. doi: 10.1111/j.1462-5822.2010.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monastyrska I., Ulasli M., Rottier P.J.M., Guan J.L., Reggiori F., De Haan C.A.M. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy. 2013;9:164–174. doi: 10.4161/auto.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., De Haan C.A.M., Molinari M. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hwang S., Maloney N.S., Bruinsma M.W., Goel G., Duan E., Zhang L., Shrestha B., Diamond M.S., Dani A., Sosnovtsev S.V., Green K.Y., Lopez-Otin C., Xavier R.J., Thackray L.B., Virgin H.W. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257\rE07-12-1257[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131\r117/13/2805. (pii) [DOI] [PubMed] [Google Scholar]
- 138.Tanida I., Ueno T., Kominami E. Human light chain 3/MAP1LC3 B Is cleaved at its carboxyl-terminal Met 121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J. Biol. Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 139.Tanida I., Sou Y.S., Ezaki J., Minematsu-Ikeguchi N., Ueno T., Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 140.Martinez-Lopez N., Athonvarangkul D., Mishall P., Sahu S., Singh R. Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 2013;4:2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang Z., Wilkie-Grantham R.P., Yanagi T., Shu C.W., Matsuzawa S.I., Reed J.C. ATG4B (Autophagin-1) phosphorylation modulates autophagy. J. Biol. Chem. 2015;290:26549–26561. doi: 10.1074/jbc.M115.658088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mitchell P.S., Young J.M., Emerman M., Malik H.S. Evolutionary analyses suggest a function of MxB immunity proteins beyond lentivirus restriction. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kane M., Yadav S.S., Bitzegeio J., Kutluay S.B., Zang T., Wilson S.J., Schoggins J.W., Rice C.M., Yamashita M., Hatziioannou T., Bieniasz P.D. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Goujon C., Moncorgé O., Bauby H., Doyle T., Ward C.C., Schaller T., Hué S., Barclay W.S., Schulz R., Malim M.H. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu Z., Pan Q., Ding S., Qian J., Xu F., Zhou J., Cen S., Guo F., Liang C. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 146.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]