Abstract
Background:
Severe acute respiratory syndrome (SARS) is caused by infection with SARS-associated coronavirus (CoV). Amino acid residues 450–650 of the spike (S) glycoprotein of SARS-CoV (S450-650) contains dominant epitopes for anti-viral antibodies (Abs) in patient sera.
Objectives:
To develop and evaluate an ELISA system for detection of anti-S Abs in patient sera.
Study design:
Express recombinant S450-650 in E. Coli and evaluate the sensitivity and specificity of an ELISA system based on the S450-650 polypeptide.
Results:
The S450-650-based ELISA detected IgG Abs in 41 out of 51 serum samples from 22 hospitalized patients with probable SARS, a result closely correlated with that obtained with a virus-based ELISA (r = 0.75, k = 0.8). Differential anti-S IgG responses were observed amongst SARS patients. Some of them produced anti-S Abs early during their infection, while others failed to make IgG Abs against the S450-650 polypeptide. None of the serum samples from 100 healthy blood donors was positive in the S450-650-based assay.
Conclusion:
The S450-650-based ELISA can detect anti-S IgG Abs with high sensitivity and specificity.
Keywords: SARS-coronavirus, Spike glycoprotein, ELISA, Antibodies
1. Introduction
Severe acute respiratory syndrome (SARS) is caused by SARS-associated coronavirus (SARS-CoV), an enveloped, positive-stranded RNA virus of the Coronaviridae family (Rota et al., 2003, Peiris et al., 2003). The genome of SARS-CoV encodes several structural proteins including the spike (S) glycoprotein, nucleocapsid protein, membrane protein and envelope protein (Marra et al., 2003). Membrane fusion between SARS-CoV and the host cell is mediated by its S glycoprotein (Li et al., 2003, Xiao et al., 2003, Dimitrov, 2003, Wong et al., 2004, Wang et al., 2004, Sui et al., 2004), which consists of 1255 amino acid residues with approximately 25% homology to that of the other human CoVs (Spiga et al., 2003, Ho et al., 2004). The S1 subunit (amino acids 1–680) of the S glycoprotein contains a receptor-binding domain and is apparently the main target for neutralizing Abs in patient sera (Xiao et al., 2003, Sui et al., 2004, Wong et al., 2004, Wang et al., 2004, Babcock et al., 2004). Western blot (WB) and indirect immunofluorescence assays have been developed for detection of Abs against the S protein (Tan et al., 2004, Chang et al., 2004, Wu et al., 2004, Woo et al., 2004, Ho et al., 2004, He et al., 2004). However, so far there is no enzyme-linked immunosorbent assay (ELISA) available for easier and more sensitive detection of anti-S Abs. Our computer-assisted analysis suggested that amino acid residues 450–650 of the S glycoprotein (S450-650) of SARS-CoV is largely solvent accessible and likely to contain dominant B cell epitopes. This coincided with recently published results by Lu et al. (2004) showing that residues 441–700 of the S protein of SARS-CoV contained dominant epitope(s) for anti-S Abs in patient sera, as determined in WB assays. Sequences outside the 441–700 region were relatively poorly recognized by patient sera (Lu et al., 2004). It is also supported by findings of Zhou et al. (2004) that residues 485–625 of the S protein of SARS-CoV elicited neutralizing Abs against the virus. In this study, the S450-650 fragment was expressed in E. Coli and used as Ag in an ELISA system for detection of anti-S Abs.
2. Materials and methods
2.1. Molecular biology reagents
Restriction enzymes and T4 ligase were from Invitrogen (USA). A kit for DNA extraction and purification was from Qiagen (Hilden, Germany). E. coli strains of BL21 (OE3) and DH5α were obtained from Novagen (Germany) and Invitrogen (USA), respectively. Complementary DNA encoding full length S of SARS-CoV was a gift from China CDC.
2.2. Subjects and blood samples
From March 24, 2003, a major outbreak of SARS had taken place in Beijing, China. We collected sequential blood samples (297 samples in total) from 122 patients (both sexes, 18–51 years of age, average 35.5 years), admitted to the First Affiliated Hospital of Peking University, Beijing, China between 15th April and 5th June, 2003. All patients fulfilled the WHO definition of probable SARS (fever 38 °C or higher, cough, new pulmonary infiltrates on chest radiography in the absence of an alternative diagnosis to explain the clinical presentation). The blood samples were processed within 18 h of collection. All patient sera were tested for anti-SARS-CoV IgG Abs using an ELISA kit produced by Huada Institute (see below). A total of 45 serum samples from 18 patients, randomly selected from the above patient group, were tested using our S450-650-based ELISA kit. In addition, sera from 100 healthy blood donors (both sexes, 22–45 years of age, collected between May and July 2002) were provided by Beijing Red Cross Blood Center.
A small outbreak of SARS took place in April 2004 involving nine patients in Anhui and Beijing, China. Sequential serum samples from four of the confirmed patients (second or third generation cases), admitted to Ditan Hospital between 15th April and 10th June 2004, were also included in this study.
2.3. Construction of expression plasmids
DNA coding for S450-650 of SARS CoV S protein was PCR amplified using high fidelity Taq DNA polymerase (TaKaRa Biotech Co. Ltd., Japan). The sequences of the primers employed in the PCR reaction were S450-650 5: CGC GGA TCC ATG CCC TTT GAG AGA GAC ATA TCT (forward primer, carrying a BamHI restriction site) and S450-650 3: CCC GAA TTC TTA AAT GTC GCA CTC ATA AGA AGT G (reverse primer, carrying an EcoRI site). The PCR product was gel-purified and cloned into expression vector PET-28a (Novagen, Germany). The resultant construct, PET28a-S450/650, encodes for the S450-650 fragment with a histidine (His) tag (6 His) at the N terminus. DNA sequence of the insert was determined and compared with the S gene sequence of SARS-CoV strain Urbani (accession number AY278741) using DNASTAR software.
2.4. Expression and purification of recombinant proteins
PET28a-S450/650 was transformed into E. Coli BL21 cells for expansion. A bacterial colony harboring the plasmid was inoculated into 2YT medium containing kanamycin (25 μg/ml) and cultured, with continuous shaking, at 37 °C to appropriate density. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.1 mM to induce the expression of the recombinant protein (for further 3.5 h). After centrifugation at 4000 × g for 15 min, the pellet was resuspended in PBS and subjected to sonication in an ice bath for 30 min. The inclusion bodies were solubilized with 8 M urea, after centrifugation at 12000 × g at 4 °C for 30 min, the supernatant was applied to an equilibrated Ni column (Novagen, Germany). After on-column refolding procedure, proteins bound to the column were eluted with 800 mM imidazole in Tris–buffered saline (pH 7.9). The eluted fractions were examined by SDS-PAGE and the proteins were either stained with Coomassie blue or transferred to nitro-cellulose membrane for WB assays.
2.5. Western blot assays
The nitro-cellulose membranes, on which protein bands had been transferred, were blocked at room temperature for 2 h with block solution (5% non-fat dry milk) and then incubated for 2 h at room temperature with serum sample. After washing in Tris–buffered saline (TBS, pH 8.0) containing 0.05% Tween 20, the membranes were incubated with HRP-labeled goat anti-human IgG Ab (Zhongshan Biotech Co., China). 3,3′-diaminobenzidine tetrahydrochloride (DAB, Sigma) was used to visualize the reaction.
2.6. ELISAs
ELISA plates (Nunc, Demark) were coated at 4 °C overnight with recombinant protein (2.5 pmol/well) in carbonate buffer (pH 9.6). Immediately before use, the coated plates were incubated with blocking solution (2% BSA in PBS) for 2 h at 37 °C and then washed 4 times with PBS containing 0.05% Tween 20 (PBS-T). Serum samples (1/100 diluted, 100 μl/well) were added in triplicate and incubated for 90 min at 37 °C. After washes with PBS-T, horseradish peroxidase (HRP)-labeled goat anti-human IgG Ab was added and incubated for 1 h at 37 °C. ortho-Phenylenediamine (OPD) substrate was used (100 μl/well) as substrate with 2 M H2SO4 solution as stop buffer. The optical density (OD) was immediately read at 492 nm.
For ELISAs using the kit produced by Huada Institute (Beijing, China), the manufacturers’ instruction was followed. Briefly, dilution buffer (100 μl/well) was added to pre-coated wells followed by 10 μl serum and incubated for 30 min at 37 °C. After washes, HRP-labeled detection Ab (1/2000 diluted) was dispensed (100 μl/well) and incubated for 20 min at 37 °C before further washes. Substrate buffer containing ABTS [2,2′-Azino-di-(3-ethylbenzo-thiazoline sulfonate)] was then added and allowed to develop for 10 min. After stop buffer was added and the plates were read at 450 nm.
2.7. Statistical analysis
All experiments describe here have been repeated at least three times. Results obtained using S450-650 recombinant protein-based ELISA and SARS-CoV-specific kit were compared using the CORREL module of Microsoft Excel software. The Cohen kappa test was performed to analyze the agreement between the results of the two ELISA kits using SPSS software.
3. Results
3.1. Expression and purification of the S450-650 fragment of S protein
Complementary DNA encoding S450-650 of SARS-CoV was cloned into expression vector PET-28a for expression in E. Coli BL21. The His-tag-containing recombinant protein was purified to more than 95% homogeneity using a Ni column. SDS-PAGE analysis of the affinity purified product revealed protein band of expected molecular weight (Fig. 1 ). In WB assays, convalescent serum from a SARS patient (PT-LX) and anti-His-tag mAb specifically recognized the recombinant S450-650 (Fig. 1). In the subsequent study, recombinant S450-650 was employed as coating Ag for an ELISA kit with HRP-labeled goat-anti-human IgG Abs as secondary Ab. Checker-board titration experiments were carried out to optimize the concentration of the coating Ag (S450-650) and secondary Ab (data not shown).
Fig. 1.
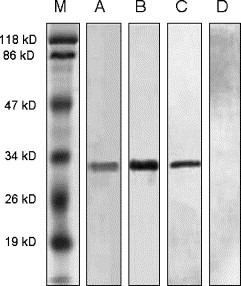
SDS-PAGE and WB analysis of the recombinant S450-650 fragment. Affinity purified recombinant S450-650 was run in 2 identical SDS PAGE 12% gels with molecular weight markers in Lane M. One of the gels was stained with Coomassie blue (Lanes M and A). Protein bands in the other gel were transferred onto cellulose nitrite membranes for WB with convalescent serum from patient PT-LX (Lane B), or anti-His-tag mAb (Lane C), or serum from a healthy subject (Lane D), as the first Ab.
3.2. SARS-CoV-specific IgG Abs in patient sera
A SARS-CoV-specific ELISA kit, developed by Huada Institute, Beijing, China, has been licensed by the Ministry for Public Health of China. It employs the lysate of SARS-CoV- infected Vero-E6 cells as coating Ags and has been widely used in China for SARS-CoV-specific Ab testing with reliable results. By using the Huada kit, we analyzed sequential serum samples (total 297 sera) from 122 hospitalized patients with probable SARS and the results are summarized in Table 1 . Half of the patients seroconverted for IgG Abs against SARS-CoV by Day 14 after the onset of illness. Nearly 90% of them produced virus-specific serum Abs by Days 29–50.
Table 1.
Anti-SARS-CoV IgG Abs in sera of patients with probable SARSa
| Days after onset of illness |
|||||
|---|---|---|---|---|---|
| 2–7 days | 8–14 days | 15–21 days | 22–28 days | 29–50 days | |
| Number of samples tested | 22 | 44 | 68 | 79 | 87 |
| Percent Positive | 14 | 50 | 70 | 84 | 89 |
Tested using the Huada ELISA kit. Cutoff value: OD450 = 0.18.
3.3. Specificity and sensitivity of the S450-650-based ELISA
Sera from three convalescent SARS patients and two healthy individuals were serial diluted and tested in the S450-650-based ELISAs, which detected anti-S IgG Abs in a specific and sensitive manner, with the reactivity end point from 1:400 to 1:800 diluted patient sera (Fig. 2 ). Serum samples from 100 healthy blood donors were subsequently screened and none of them was positive (Fig. 3A), suggesting a high specificity for the S450-650-based system.
Fig. 2.
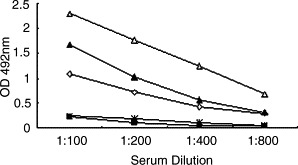
Sensitivity of the S450-650-based ELISA. ELISA plates were coated with recombinant S450-650. Serum samples from 3 convalescent SARS patients (▴, ♢, ▵) and 2 healthy individuals (■, □) were serial diluted and dispensed, in triplicates, into the wells. HRP-labeled goat anti-human IgG was used as secondary Ab with OPD as substrate. The results are expressed as absorbance reading at 492 nm wavelength.
Fig. 3.
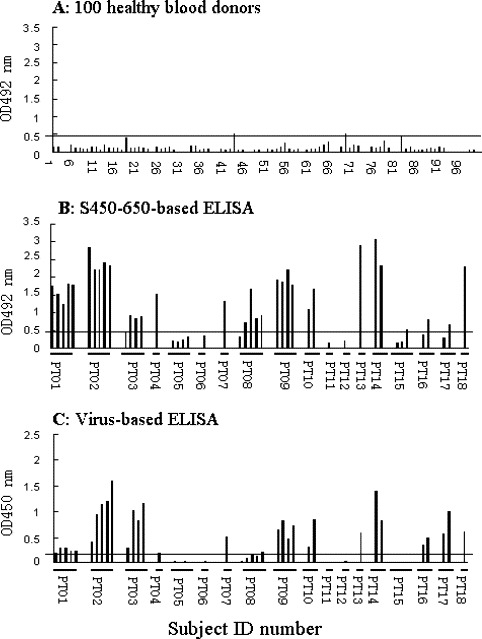
Comparison of S450-650-based and virus-based ELISA results. The S450-650-based ELISA kit was used to screen serum samples from (A) 100 healthy blood donors and (B) 18 patients (PT01 to PT18, collected between 10 and 42 days from the onset of illness) with probable SARS. The serum samples were 100 folds diluted, results are expressed as absorbance reading at 492 nm wavelength. Cutoff value was 0.46. ELISA results obtained with the Huada kit are shown in (C) for comparison. In this case, serum samples were 10 folds diluted, results are expressed as absorbance reading at wavelength of 450 nm. Cutoff value was 0.139. Each bar represents one serum sample, samples from each patient were group together in the order of collection time (days) after onset of illness.
When sequential serum samples (45 total, 1/100 diluted) from 18 patients (randomly selected from the above-mentioned 122 patient set) with probable SARS were analyzed using the S450-650-baesd assays, 33 were positive (Fig. 3B). The Huada kit detected anti-viral IgG Abs in 32 out of the 45 samples (Fig. 3C), 29 of which were positive in both assays. When the S450-650-based and virus-based ELISA results were plotted against each other, a linear correlation (first degree regression, r = 0.75) was observed (Fig. 4 ), the Cohen kappa test also confirmed an agreement between them (κ = 0.8).
Fig. 4.
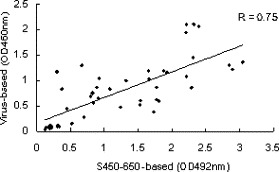
Correlation between S450-650-based and SARS-CoV-based ELISA results. The ELISA results shown in Fig. 3B and 3C were plotted against each other. First-degree regression (r = 0.75) shows a linear correlation between the results of the two ELISA tests.
3.4. Profiles of IgG responses against the S protein in patients
Time courses of IgG responses against the S protein and the whole virus in 7 out of the 18 patients are compared in Fig. 5 A and B. Sera from patients PT01, PT02 and PT08 contained high titer anti-S but low titer anti-virus IgG Abs. In contrast, patient PT03 was a high responder against the whole virus but low responder against S450-650. Patient PT03 seroconverted 36 days after the onset of symptoms, suggesting a possible case of SARS-CoV infection being acquired in the hospital ward. Patient PT05 remained negative in both tests 2 months after the onset of illness and was eventually excluded as a SARS patient.
Fig. 5.
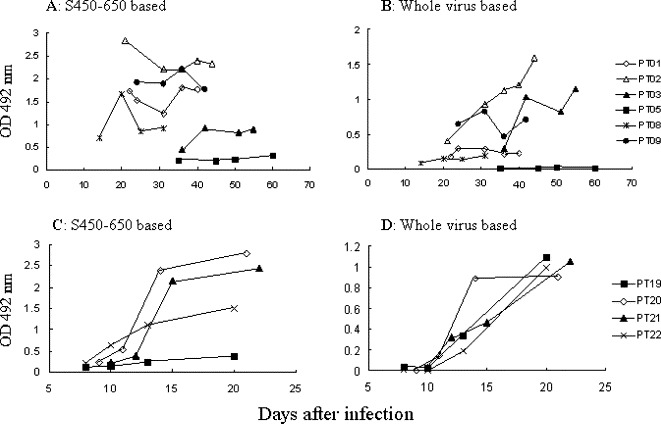
Time course of Ab responses against S450-650 and SARS-CoV in patients. Sequential serum samples from seven patients were tested using the (A and C) S450-650-based and (B and D) Huada ELISA kits. Serum samples were diluted 100 folds in (A) and (C), 10 folds in (B) and (D), and the results are expressed as absorbance reading at 492 or 450 nm, respectively. Cutoff values were (A and C) 0.46 or (B and D) 0.18.
Both the S450-650-based and the virus-based ELISA kits were employed to screen sera from an additional 4 SARS patients infected in April 2004. Three of them were high responders against the S protein, while the other one (PT19) responded poorly to S450-650 but strongly to the whole virus (Fig. 5C and D). In patient PT22, anti-S serum Abs appeared earlier than that specific for other viral proteins.
4. Discussion
Specific and sensitive detection methods for anti-viral Abs in patient sera are of great value in diagnosis as well as research. Virus-based assays have the advantage of being able to detect Abs specific for all structural components of the virus. On the other hand, recombinant protein-based tests would allow analysis of anti-viral humoral immunity in much detail.
The S protein of SARS-CoV is heavily glycosylated and contains many disulfide bonds (Spiga et al., 2003, Krokhin et al., 2003, Tripet et al., 2004, Ying et al., 2004). It is difficult, however, to obtain S glycoprotein expressed in eukaryotic systems, which would otherwise be ideal for detection of anti-S Abs in vitro. Full-length S protein expressed in prokaryotic systems is often insoluble and therefore unsuitable as coating Ag in ELISA systems. Our on-column refolding procedure allowed correct formation of some of the disulfide bonds in the S450-650 fragment, producing soluble recombinant protein at a reasonably high concentration (data not shown). Results reported herein indicate that the S450-650-based ELISA can detect anti-S Abs in patient sera with high sensitivity and specificity.
It has been suggested that the N-glycans of the S glycoprotein could have a significant effect on its antigenicity, as the presence of N-glycans contributes to the correct folding and biological function of glycoproteins. However, recently published results indicate that bacterial expressed fragment of the S protein (non-glycosylated) could maintain its antigenicity (Zhou et al., 2004, Lu et al., 2004). In addition, neutralizing Abs were successfully raised in mice after immunization using prokaryotically expressed S485-625 of SARS-CoV (Zhou et al., 2004).
In this study we wound that most SARS patients developed strong IgG responses against the S glycoprotein of SARS-CoV. Some of them had anti-S Abs in significantly higher titer than that against other structural proteins of the virus (e.g. PT01 and PT08). In this situation, our S protein-based ELISA would be more sensitive than the virus-based assays in detecting antiviral Abs. There were also cases where the S450-650 was poorly recognized by Abs in patient sera (e.g. PT22 in Fig. 4C and D). It should be emphasized, however, that the S450-650 polypeptide covers less than 1/5 of the S protein sequence. A negative result such as this does not rule out possible presence of anti-S Abs against epitopes outside the 450–650 region of the S protein. Moreover, Xu et al. (2004) have reported relatively high frequency of variations in S gene of SARS-CoV. Natural variability in the S protein might also affect the validity of the S450-650-based ELISA, although antigenic variation in S450-650 has not yet been fully characterized.
Acknowledgements
The first two authors contributed equally to this work. This study was supported by grants from the National Key Basic Research Program (2001CB510007), National High Technology Research & Development Program of China (2003AA208412A). Patient's consent was obtained before blood collection.
Contributor Information
Zhendong Zhao, Email: zhaoz@bjmu.edu.cn.
Xiao-Ming Gao, Email: xmgao@bjmu.edu.cn.
References
- Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78(9):4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.S., Lu Y.T., Ho S.T., Wu C.C., Wei T.Y., Chen C.J., Hsu Y.T., Chu P.C., Chen C.H., Chu J.M., Jan Y.L., Hung C.C., Fan C.C., Yang Y.C. Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;314(4):931–936. doi: 10.1016/j.bbrc.2003.12.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115(6):652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Chong K.H., Chng H.H., Leung B., Ling A.E., Wei T., Chan S.W., Ooi E.E., Kwang J. Development of a Western blot assay for detection of antibodies against coronavirus causing severe acute respiratory syndrome. Clin. Diagn. Lab. Immunol. 2004;11(2):417–422. doi: 10.1128/CDLI.11.2.417-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.Y., Wu S.L., Cheng S.E., Wei Y.C., Huang S.P., Hsiang C.Y. Antigenicity and receptor-binding ability of recombinant SARS coronavirus spike protein. Biochem. Biophys. Res. Commun. 2004;313(4):938–947. doi: 10.1016/j.bbrc.2003.11.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin O., Li Y., Andonov A., Feldmann H., Flick R., Jones S., Stroeher U., Bastien N., Dasuri K.V., Cheng K., Simonsen J.N., Perreault H., Wilkins J., Ens W., Plummer F., Standing K.G. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell Proteomics. 2003;2(5):346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Manopo I., Leung B.P., Chng H.H., Ling A.E., Chee L.L., Ooi E.E., Chan S.W., Kwang J. Immunological characterization of the spike protein of SARS-coronavirus. J. Clin. Microbiol. 2004;42(4):1570–1576. doi: 10.1128/JCM.42.4.1570-1576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M., Jones S., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y., SARS study group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Spiga O., Bernini A., Ciutti A., Chiellini S., Menciassi N., Finetti F., Causarono V., Anselmi F., Prischi F., Niccolai N. Molecular modeling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem. Biophys. Res. Commun. 2003;310(1):78–83. doi: 10.1016/j.bbrc.2003.08.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 2004;101(8):2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11(2):362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004 doi: 10.1074/jbc.M400759200. March 2 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen J., Zheng A., Nie Y., Shi X., Wang W., Wang G., Luo M., Liu H., Tan L., Song X., Wang Z., Yin X., Qu X., Wang X., Qing T., Ding M., Deng H. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 2004;315(2):439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Chan K.H., Wong B.H., Che X.Y., Tam V.K., Tam S.C., Cheng V.C., Hung I.F., Wong S.S., Zheng B.J., Guan Y., Yuen K.Y. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363(9412):841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.S., Hsieh Y.C., Su I.J., Lin T.H., Lin T.H., Chiu S.C., Hsu Y.F., Lin J.H., Wang M.C., Chen J.Y., Hsiao P.W., Chang G.D., Wang A.H., Ting H.W., Chou C.M., Huang C.J. Early detection of antibodies against various structural proteins of the SARS-associated coronavirus in SARS patients. J. Biomed. Sci. 2004;11(1):117–126. doi: 10.1007/BF02256554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312(4):1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Zhang Z., Wang F.S. SARS-associated coronavirus quasispecies in individual patients. N. Eng. J. Med. 2004;350(13):1366–1367. doi: 10.1056/NEJMc032421. [DOI] [PubMed] [Google Scholar]
- Ying W., Hao Y., Zhang Y., Peng W., Qin E., Cai Y., Wei K., Wang J., Chang G., Sun W., Dai S., Li X., Zhu Y., Li J., Wu S., Guo L., Dai J., Wang J., Wan P., Chen T., Du C., Li D., Wan J., Kuai X., Li W., Shi R., Wei H., Qian X., Zhu Q., He F. Proteomic analysis on structural proteins of severe acute respiratory syndrome coronavirus. Proteomics. 2004;4(2):492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Wang H., Luo D., Rowe T., Wang Z., Hogan R.J., Qiu S., Bunzel R.J., Huang G., Mishra V., Voss T.J., Kimberly R., Luo M. An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies. J. Virol. 2004;78(13):7217–7226. doi: 10.1128/JVI.78.13.7217-7226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


