1. Background
The non-enveloped Saffold viruses are presently nine picornavirus genotypes assigned to the species Theilovirus, genus Cardiovirus. SAFV-1 was first defined in 2007 having been the putative cause of fever of unknown origin in a child in 1981.1 SAFVs include an approximately 8 kb positive-sense single-stranded RNA genome (Fig. 1 ). Since the first definition, SAFV-2,2 SAFV-3,3 SAFV-4–SAFV-84 and SAFV-95 have been described along with a suggestion that more genotypes remain to be characterised.6 Cell culture is in general known to be an inefficient method of routine virus detection but it can be used for study of SAFV. The SAFVs are predicted to encode a single polyprotein and in silico analysis suggest that a classical picornavirus post-translational proteolytic cleavage pattern ensues (Fig. 1).
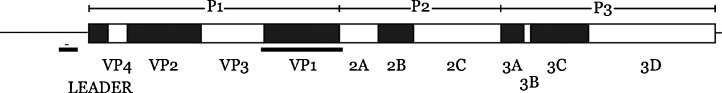
Fig. 1.
Schematic of the SAFV genome drawn to scale, based on GenBank accession No. NC_009448. The site of the RT-rtPCR diagnostic, VP1 Genotyping assays and the in vitro transcribed RNA template region are underlined.
Most SAFV genotypes have been identified from acute gastroenteritis (AGE) cases; only the SAFV-2 genotype was initially discovered in the airways; the nasopharynx of children with a common cold, otitis media and pneumonia.2 Interestingly, the distinctly respiratory human rhinoviruses (HRV) have also been found in the gut7, 8, 9 calling into question any automatic assumption that SAFVs are restricted to causing AGE just because they are found in stools. Other SAFV detections have been made in the nasopharynx of patients with tonsillitis,10, 11 pharyngitis,11 herpangina11 and in cerebrospinal fluid specimens.12, 13, 14 These are often accompanied by fever, cough and rhinorrhoea. Currently, SAFVs are described as orphan viruses lacking a consensus disease association15, 16 despite >90% of the population having serological evidence of past exposure.15 It is therefore important to examine a range of disease states for this ubiquitous virus, to establish a link between detection and illness. The first step is the development of assays that can detect viral variants and their implementation by others.
2. Objectives
We sought to design are reverse transcriptase real-time PCR (RT-rtPCR), for application to respiratory tract specimens, that could concurrently detect all known SAFV genotypes with confirmation using a VP1-based conventional PCR genotyping assay. Our diagnostic workflow may support enhanced epidemiology studies worldwide.
3. Study design
3.1. RT-rtPCR screening assay and conventional genotyping RT-PCR assays
We developed a diagnostic assay targeting a portion of the 5′ untranslated region (5′ UTR) that is predicted to detect all SAFVs. Its design was based on all publicly available 5′ UTR sequences (alignments included known SAFV-1, 2, 3, 5, 6 genotypes and a range of unclassified SAFV sequences prepared in Geneious Pro 5.6. Defined SAFV-4, 7–9 5′ UTRs remain absent from public databases). The newly developed SAFV RT-rtPCR employed 0.4 μM of each oligonucleotide primer (TIB-MOLBIOL, Germany; Table 1 ) and 0.1 μM of each dual-labelled fluorogenic oligoprobe (SAFV_TM) in a 20 μL one-step RT-PCR reaction mix (SensiFAST OneStep Mix, Bioline, Australia) including RNase inhibitor and 5 mM MgCl2. After 2 μL of nucleic acid extract was added and reverse transcribed for 20 min at 45 °C, the mixes were incubated at 94 °C for 2 min then cycled through 55 rounds of 94 °C for 15 s and 60 °C for 60 s using a RotorGene 3000, 6000 or RGQ (QIAGEN, Australia) to acquire fluorescence data at the final (annealing and extension) step of each cycle. Assay sensitivity was determined using a specific in vitro transcribed RNA (ivtRNA; Fig. 1, Table 1) created based on a previously described approach.17 Briefly, ivtRNA was subjected to two treatments with DNase (Turbo, Life Technologies), each followed by column purification (ISOLATE RNA Mini Kit, Bioline). The stock RNA was used to determine the analytical sensitivity of each assay after testing a 10-fold dilution series. The last dilution to yield a positive result (defined below) was taken as the limit of analytical sensitivity. The ivtRNA copy number was calculated using the optical density to approximate the mass of RNA in the stock solution at 260 nm, determining the number of moles by establishing the molecular weight of the ivtRNA and then multiplying by 6.02 × 1023 (Avogadro's number). A positive diagnostic result was defined by the presence of a sigmoidal curve that crossed an arbitrary threshold of 0.05, before 45 cycles, during an experiment in which duplicate non-template controls did not cross the threshold.
Table 1.
Oligonucleotides used to detect and genotype SAFV.
| Oligonucleotide name | Oligonucleotide sequence | PCR product (bp) | Reference | |
|---|---|---|---|---|
| SAFV RT-rtPCR | ||||
| SAFV_S | TCGAAACAGCTGTAGCGACC | 199 | This study | Primers |
| SAFV_AS | CTTCAGGACATTCTTGGCTTCTC | |||
| SAFV_TM | FAM-ACAGCAGTGGATCTTATCCACGGGGC-BBQ | This study | Probe | |
| In vitro transcription RNA control primers | ||||
| SAFV_T7_S | AAAATAATACGACTCACTATAGGGCCGGAAACGGTGAAGA | 397 | This study | Primers |
| SAFV_T7_AS | TATCCGTGTTTGCACGCCAT | |||
| VP1 Genotyping primers | ||||
| SAFV_VP1_S1 | ACWCTTGGTTTCDGGHGG | 1043 | Blinkova et al.4 | Primers |
| SAFV_VP1_AS1 | TCGCCCATRCASACRAGRA | |||
| SAFV_VP1_S2 | GACTTYACYCTBAGAATGCC | 955 | Blinkova et al.4 | primers |
| SAFV_VP1_AS2 | ACTGTTCTAYCRTGAACTTTGTA | |||
T7 RNA polymerase promoter region-underlined; S/AS – sense/antisense primers; S1/AS1 – round 1 sense/antisense primer set; S2/AS2 – round 2 sense/antisense, nested primer set.
In order to confirm the SAFV genotype of a positive patient specimen, nucleic acids from each were added to a conventional nested RT-PCR targeting VP1 (underlined, Fig. 1).4 Apart from the RT step (as above) both rounds of PCR cycled mixes through four rounds of 95 °C for 60 s, 55 °C for 60 s and 68 °C for 90 s followed by 35 rounds of 95 °C for 30 s, 53 °C for 30 s, 68 °C for 90 s.4 A final extension incubation at 68 °C occurred for 10 min. The first round PCR contained 3 mM MgCl2, the second contained 5 mM.
3.2. Specimen details and statistics
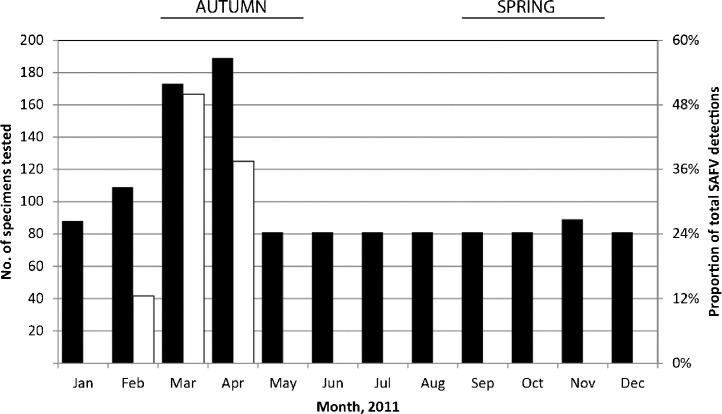
Specimens (n = 1, 215) for respiratory virus testing originated from a convenience population of inpatients and outpatients (53.0% male) aged two days to 95 years (mean 22.6 years, median 4.0, inter quartile range 44.5) presenting to Queensland hospitals with symptoms of ARI. Specimens (81–189 per month; Fig. 2 ) were mostly nasopharyngeal aspirates (48.1% of specimens) and nasopharyngeal swabs (31.8%) and most specimens were tested from autumn (36.5% of all specimens; Fig. 2). Other samples included nasal swabs (n = 66; 5.4%), throat swabs (n = 43; 3.5%) and bronchoalveolar lavage (n = 17; 1.4%). Nucleic acids were extracted as previously described.18
Fig. 2.
Timeline of all the tested specimens (left y-axis; filled) and the proportion of total SAFV-positive samples (right y-axis; unfilled) occurring from specimens represnting all seasons of 2011.
Association between virus detection and sex or between single versus multiple viruses per specimen was considered using 2 × 2 contingency tables with Fisher's exact test (http://statpages.org/ctab2x2.html). Results are presented as relative risk (RR) with 95% confidence intervals (CIs). Significance level was set at p = 0.05.
4. Results
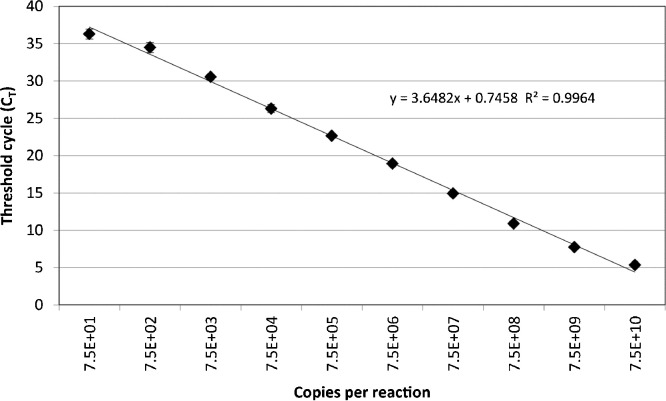
The specificity of the SAFV RT-rtPCR was evaluated using clinical specimens previously tested, some of which were and found to be positive for each of the commonly considered respiratory viruses (rhinovirus [n = 419 positives], HPIV-1 [n = 4], HPIV-2 [n = 9],HPIV-3 [n = 51], human respiratory syncytial virus [n = 167], adenoviruses [n = 74], influenza A[n = 52] and B [n = 25] viruses, enteroviruses [n = 113] and meta pneumo virus [n = 31]) and it did not cross-react with any of these viruses. The assay's analytical sensitivity was between 40 and 100 RNA copies per 20 μL reaction and the assay was linear across a 10 log10 dynamic range (Fig. 3 ). The RT-rtPCR identified 8 distinct cases of SAFV. Most cases had fever reported on presentation; one case (QPID11-0005) had coinciding sore throat, headache, neck stiffness, diarrhoea and vomiting. Five cases were able to be genotyped (four SAFV-2s and a SAFV-3; GenBank Accession numbers JQ820263–JQ820267) and three were not. The latter may have been related to mismatches we observed between the VP1 primers and some SAFV genotypes or lower viral loads (all typed specimens had C T values ≤33 while the three untypeable specimens had C T values >33). No SAFV detections were made from winter or spring (May–December; Fig. 2). Most (n = 6; 75%) of the SAFV-positive specimens were from children two years of age or younger; an age group that comprised 39.2% of the entire specimen population. Specimens from this age group were 3.9 times as likely as those from older patients to be positive for SAFV but the risk failed to attain statistical significance (1.2% vs 0.4%, RR 3.6, 95% CI 0.66–25.4, p = 0.152).
Fig. 3.
Dynamic range and sensitivity (y-intercept) of the SAFV RT-rtPCR when the threshold cycle (CT) is plotted against log10 of the initial ivtRNA copy number. Each ivtRNA dilution was amplified on three distinct occasions each time using a fresh dilution of ivtRNA stock and each dilution was tested in triplicate. Error bars represent +/− one standard deviation.
Specimens from males were 1.5 times as likely as those from females to be positive for SAFV (0.9% vs 0.6%, RR 1.5, 95% CI 0.3–7.7, p = 0.73). Over half of SAFV detections (n = 5; 62.5%) were in the presence of another virus (0.8% vs 0.7%, RR 1.159, 95% CI 0.244–6.087, p = 1.00). Similar or greater numbers of SAFV detections were made than of influenza C virus (n = 1), HPIV-1 (n = 9), HPIV-2 (n = 4), coronavirus 229E/HKU1 (n = 0), NL63 (n = 3), or HPIV-4 (n = 9) detections were apparent.
5. Discussion
We present a useful SAFVRT-rtPCR that identified the co-circulation of two genotypes in the respiratory tracts of young children during 2011 in and around Brisbane, Australia. This assay will be an important tool in augmenting the culture, serological, conventional or SYBR green RT-rt PCR methods that have underpinned most SAFV detections to date. The assay's performance on SAFV genotypes lacking 5′ UTR sequence on public databases or with undefined 5′ UTR sequences that may represent these types (SAFV-4, 7–9), remains to be evaluated. Other SAFV RT-rtPCRs designed for broad reactivity only identified a single SAFV type circulating in the CSF of children13 or contain some mismatches3 which our assay sought to minimise. It may be that certain SAFV genotypes impart a tissue-specific pathogenicity, but much more research will be required to investigate this theory. Commonly, use of PCR documents one or no SAFV genotypes in the airways,3, 10, 11, 19 our assay concurrently identified both SAFV-2 and SAFV-3 in the airways of children during 2011. We believe this assay is likely to detect all SAFVs with characterised 5′ UTRs and will permit better understanding of the role of SAFVs in human disease.
Funding
This study was funded by Queensland Children's Foundation Project Grant 50028.
Competing interests
None.
Ethical approval
Queensland Children's Health Services human research ethics committee (#HREC/10/QRCH/95) and University of Queensland Medical Research Ethics Committee (#2010001381).
Acknowledgments
We thank Olfert Landt, Tib-Molbiol for assay design assistance and Pathology Queensland Central for the provision of clinical specimens.
References
- 1.Jones M.S., Lukashov V.V., Ganac R.D., Schnurr D.P. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J Clin Microbiol. 2007;45:2144–2150. doi: 10.1128/JCM.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abed Y., Boivin G. New Saffold cardioviruses in 3 children, Canada. Emerg Infect Dis. 2008;14:834–836. doi: 10.3201/eid1405.071675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drexler J.F., Luna L.K., Stocker A., Almeida P.S., Ribeiro T.C., Petersen N. Circulation of 3 lineages of a novel Saffold cardiovirus in humans. Emerg Infect Dis. 2008;14:1398–1405. doi: 10.3201/eid1409.080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blinkova O., Kapoor A., Victoria J., Jones M., Wolfe N., Naeem A. Cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J Virol. 2009;83:4631–4641. doi: 10.1128/JVI.02085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowles N.J. 2012. Saffold Virus.http://www.picornaviridae.com/cardiovirus/theilovirus/safv.htm Ref Type: Electronic Citation. [Google Scholar]
- 6.Blinkova O., Rosario K., Li L., Kapoor A., Slikas B., Bernardin F. Frequent detection of highly diverse variants of cardiovirus, cosavirus, bocavirus, and circovirus in sewage samples collected in the United States. J Clin Microbiol. 2009;47:3507–3513. doi: 10.1128/JCM.01062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stott E.J., Eadie M.B., Grist N.R. Rhinovirus infections of children in hospital: isolation of three possibly new rhinovirus serotypes. Am J Epidemiol. 1969;90:45–52. doi: 10.1093/oxfordjournals.aje.a121048. [DOI] [PubMed] [Google Scholar]
- 8.Lau S.K., Yip C.C., Lung D.C., Lee P., Que T.L., Lau Y.L. Detection of human rhinovirus C in fecal samples of children with gastroenteritis. J Clin Virol. 2012 doi: 10.1016/j.jcv.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvala H., McIntyre C.L., McLeish N.J., Kondracka J., Palmer J., Molyneaux P. High detection frequency and viral loads of human rhinovirus species A to C in fecal samples; diagnostic and clinical implications. J Med Virol. 2012;84:536–542. doi: 10.1002/jmv.23203. [DOI] [PubMed] [Google Scholar]
- 10.Tsukagoshi H., Masuda Y., Mizutani T., Mizuta K., Saitoh M., Morita Y. Sequencing and phylogenetic analyses of Saffold cardiovirus (SAFV) genotype 3 isolates from children with upper respiratory infection in Gunma, Japan. Jpn J Infect Dis. 2010;63:378–380. [PubMed] [Google Scholar]
- 11.Itagaki T., Abiko C., Ikeda T., Aoki Y., Seto J., Mizuta K. Sequence and phylogenetic analyses of Saffold cardiovirus from children with exudative tonsillitis in Yamagata, Japan. Scand J Infect Dis. 2010;42:950–952. doi: 10.3109/00365548.2010.496791. [DOI] [PubMed] [Google Scholar]
- 12.Himeda T., Hosomi T., Asif N., Shimizu H., Okuwa T., Muraki Y. The preparation of an infectious full-length cDNA clone of Saffold virus. Virol J. 2011;8:110. doi: 10.1186/1743-422X-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen A.C., Bottiger B., Banner J., Hoffmann T., Nielsen L.P. Serious invasive Saffold virus infections in children, 2009. Emerg Infect Dis. 2012;18:7–12. doi: 10.3201/eid1801.110725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drexler J.F., Baumgarte S., Eschbach-Bludau M., Simon A., Kemen C., Bode U. Human cardioviruses, meningitis, and sudden infant death syndrome in children. Emerg Infect Dis. 2011;17:2313–2315. doi: 10.3201/eid1712.111037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoll J., Erkens H.S., Lanke K., Verduyn L.F., Melchers W.J., Schoondermark-van de V.E. Saffold virus, a human Theiler's-like cardiovirus, is ubiquitous and causes infection early in life. PLoS Pathog. 2009;5:e1000416. doi: 10.1371/journal.ppat.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Z.Q., Cheng W.X., Qi H.M., Cui S.X., Jin Y., Duan Z.J. New Saffold cardiovirus in children, China. Emerg Infect Dis. 2009;15:993–994. doi: 10.3201/eid1506.090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith G., Smith I., Harrower B., Warrilow D., Bletchly C. A simple method for preparing synthetic controls for conventional and real-time PCR for the identification of endemic and exotic disease agents. J Virol Methods. 2006;135:229–234. doi: 10.1016/j.jviromet.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackay I.M., Bialasiewicz S., Jacob K.C., McQueen E., Arden K.E., Nissen M.D. Genetic diversity of human metapneumovirus over 4 consecutive years in Australia. J Infect Dis. 2006;193 doi: 10.1086/504260. [DOI] [PubMed] [Google Scholar]
- 19.Chiu C.Y., Greninger A.L., Kanada K., Kwok T., Fischer K.F., Runckel C. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc Natl Acad Sci USA. 2008;105:14124–14129. doi: 10.1073/pnas.0805968105. [DOI] [PMC free article] [PubMed] [Google Scholar]