Abstract
Vertebrate innate immunity is characterized by an effective immune surveillance apparatus, evolved to sense foreign structures, such as proteins or nucleic acids of invading microbes. RIG-I-like receptors (RLRs) are key sensors of viral RNA species in the host cell cytoplasm. Activation of RLRs in response to viral RNA triggers an antiviral defense program through the production of hundreds of antiviral effector proteins including cytokines, chemokines, and host restriction factors that directly interfere with distinct steps in the virus life cycle. To avoid premature or abnormal antiviral and proinflammatory responses, which could have harmful consequences for the host, the signaling activities of RLRs and their common adaptor molecule, MAVS, are delicately controlled by cell-intrinsic regulatory mechanisms. Furthermore, viruses have evolved multiple strategies to modulate RLR-MAVS signal transduction to escape from immune surveillance. Here, we summarize recent progress in our understanding of the regulation of RLR signaling through host factors and viral antagonistic proteins.
Keywords: Innate immunity, Type-I interferon, RIG-I, MDA5, Viral immune evasion
1. Introduction
The innate immune system is an organism's first line of defense against microbial pathogens and is composed of a defined set of germline-encoded pattern-recognition receptors (PRRs). PRRs sense specific microbial structural components or nucleic acid, commonly termed pathogen-associated molecular patterns (PAMPs), and subsequently initiate an antimicrobial defense program. This immune control program is characterized by PRR-induced production of type-I and -III interferons (IFNs), which, once secreted, alert surrounding cells of the microbial infection by inducing the expression of hundreds of IFN-stimulated genes (ISGs). These ISGs encode antiviral restriction factors, key molecules in innate immune signaling pathways, chemokines, and cytokines, which together lead to the establishment of an ‘antiviral state’ in infected and uninfected cells, and also stimulate adaptive immunity.
PRRs are distinguished by the PAMPs they recognize and the subcellular compartments in which they are found. Members of two distinct PRR families have been described to detect viral RNA: Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) [1]. TLR3 and TLR7/8 are found primarily in the membrane of endosomes, where they are positioned to detect the double-stranded (ds) and single-stranded (ss) RNA of incoming virions, respectively. While it has long been recognized that viral dsRNA also accumulates intracellularly during viral replication, the PRRs that detect these cytosolic RNA replication products were not discovered until relatively recently. In 2004, Yoneyama et al. identified the RNA helicase RIG-I (retinoic acid-inducible gene-I) as a sensor of viral RNA using a cDNA library screen of molecules that potently enhanced type-I IFN production in response to dsRNA transfection [2]. This study also demonstrated that RIG-I functions upstream of the transcription factors essential for IFN-β promoter activation, IFN regulatory factor 3 (IRF3) and NF-κB. In the same year, Andrejeva et al. discovered a second cytosolic receptor of viral RNA, MDA5 (melanoma differentiation-associated protein 5) [3]. Based on the observation that paramyxovirus V proteins effectively block the production of IFN-β in response to intracellular dsRNA, Andrejeva et al. sought to identify the cellular protein targeted by the V protein in hopes of discovering key molecules involved in the TLR-independent IFN induction pathway. Mass spectrometry analysis of a protein that co-immunoprecipitated with the V protein of parainfluenza virus 5 (PIV5), formerly called simian virus 5, identified MDA5 as a cytosolic viral RNA sensor required for antiviral IFN induction. In 2005, the third RLR member was identified, LGP2 (laboratory of genetics and physiology 2), which appears to exert a regulatory role in RLR signaling [4].
RLRs are essential for innate immune detection of RNA virus infection in nearly all cell types, including fibroblasts, epithelial cells, and conventional dendritic cells [5]. Since the discovery of RLRs, immense progress has been made toward understanding the molecular details of how these receptors sense viral infection and coordinate an effective antiviral defense program. Recent findings demonstrated that the signaling activities of RLRs are delicately controlled through a multi-step regulatory program to avoid excessive and uncoordinated cytokine production. Furthermore, viruses have developed a myriad of mechanisms to manipulate and dampen the immune response initiated by RLRs. Here, we detail these recent discoveries of the cell-intrinsic regulatory circuits that lead to balanced RLR signaling, as well as the viral mechanisms to disrupt RLR signal transduction for immune escape.
2. RLR structures and signaling pathway
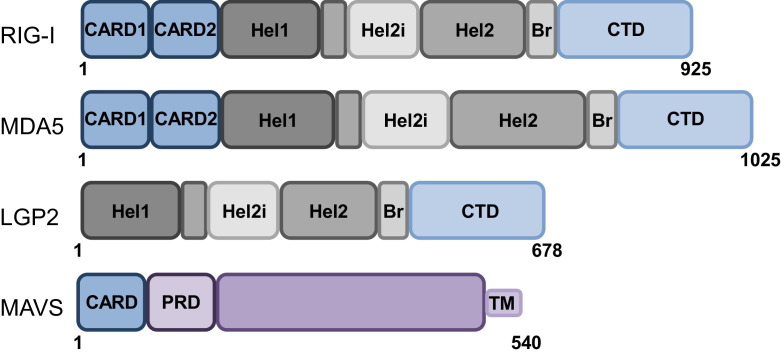
RIG-I and MDA5 belong to the DExD/H-box RNA helicase family and possess two RNA binding modules, a helicase core (Hel) and a C-terminal domain (CTD), which are linked through a bridging/pincer domain (Br) (Fig. 1 ). The RLR helicase core is comprised of two helicase domains, Hel1 and Hel2 with a unique insertion (Hel2i) in Hel2. Together these domains work in concert to surround and make tight interactions with RNA ligands and also have ATPase activity [6], [7]. In addition, RIG-I and MDA5 possess two N-terminal caspase activation and recruitment domains (CARDs) that mediate interactions with downstream signaling partners, thereby inducing IFN production [8]. LGP2 is composed of a helicase domain and a CTD but lacks the N-terminal CARDs and is therefore incapable of direct signaling. Studies attempting to elucidate the role of LGP2 in the RLR signaling pathway have described LGP2 as a negative regulator of RLR signaling in vitro [4], [9], but as a positive RLR regulator in vivo [10], [11]. Thus, further investigation will be needed to define the precise role of LGP2 in RLR signal transduction.
Fig. 1.
Schematic representation of RLR and MAVS domain structures. RIG-I and MDA5, but not LGP2, possess tandem caspase activation and recruitment domains (CARDs), a signaling module allowing for MAVS binding and IFN-α/β induction. In addition, all three RLR members have a helicase core consisting of two helicase domains (Hel1 and Hel2), a helicase insertion domain within Hel2 (Hel2i) with ATPase activity, a bridging domain (Br), and a C-terminal domain (CTD). Both the helicase and the CTD have RNA binding abilities. MAVS is comprised of a single CARD, a proline-rich domain (PRD), and a transmembrane (TM) domain that anchors it to mitochondria, peroxisomes, and MAM.
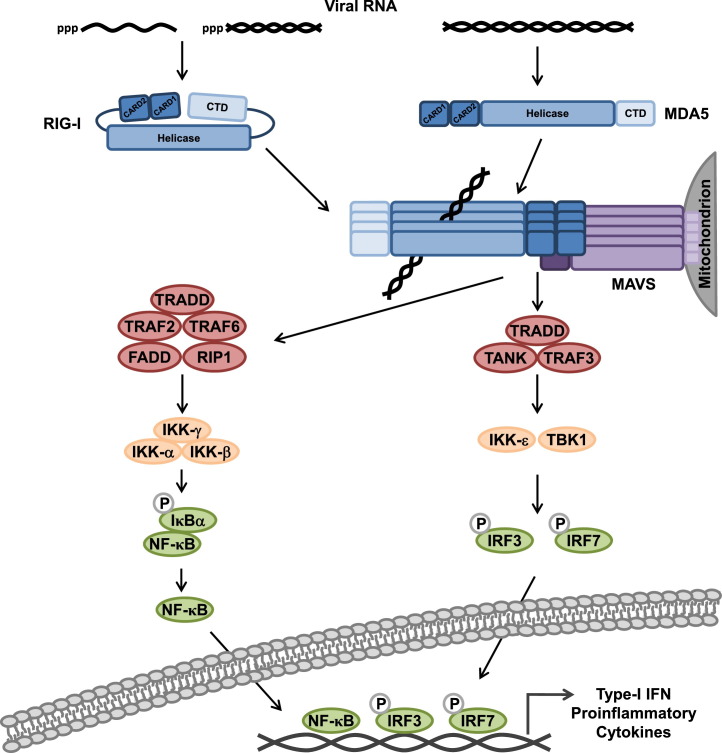
Soon after the identification of RIG-I and MDA5, their common downstream adaptor, the mitochondrial antiviral signaling protein (MAVS) (also known as VISA, IPS-1 or Cardif), was identified by four independent groups [12], [13], [14], [15]. MAVS is comprised of a single CARD at the N-terminus, a central proline-rich domain (PRD), and a C-terminal transmembrane domain (TM) that contains a mitochondrial localization signal. Once activated, RIG-I and MDA5 bind MAVS through homotypic CARD–CARD interactions (Fig. 2 ). Recent studies indicated that MAVS is not only localized to the outer membrane of mitochondria, but also resides in peroxisomal membranes, and mitochondrial-associated membranes (MAM) which connect the endoplasmic reticulum (ER) to mitochondria [16], [17]. There is new evidence suggesting that MAVS signaling from peroxisomes occurs rapidly upon viral detection and leads to the expression of a subset of ISGs in the absence of type-I IFN production. In contrast, mitochondrion-localized MAVS signals later during infection, triggering type-I IFN production and IFN-dependent ISG expression. Thus, the current view is that peroxisomal MAVS is responsible for the establishment of an antiviral state early in infection while mitochondrial MAVS facilitates a sustained IFN-dependent immune response. MAVS at the MAM is proposed to organize an ‘innate immune synapse’ for RLR-MAVS signaling by coordinating MAVS localization in peroxisomal membranes and mitochondria.
Fig. 2.
Schematic model of the signal transduction pathway induced by RLRs, leading to gene expression of type-I IFN and proinflammatory cytokines. The specific details of the model are described in the text.
Activation of MAVS facilitates the assembly of a number of proteins that induce downstream signaling (Fig. 2). This ‘MAVS signalosome’ includes the TNF receptor-associated factor (TRAF) 2, 3, and 6, as well as tumor necrosis factor receptor-associated death domain (TRADD), TRAF family member-associated NF-κB activator (TANK), Fas-associated death domain (FADD), and receptor interacting protein 1 (RIP1) [18]. Additionally, the mitochondria- and ER-associated protein stimulator of interferon genes (STING/MITA), a critical molecule in the sensing pathway of cytoplasmic viral DNA [19], was reported to interact with the MAVS signalosome, promoting RIG-I signal transduction [20], [21]. Recently, it has been shown that MAVS forms large prion-like aggregates, which represent the signaling-active form of MAVS [22]. MAVS signaling ultimately results in the activation of TANK-binding kinase 1 (TBK1) and IκB kinase-ɛ (IKK-ɛ) as well as the IKK-α/β/γ complex, resulting in IRF3/7 and NF-κB activation [1], [8]. IRF3/7 and NF-κB translocate from the cytoplasm to the nucleus, where they – together with the activating transcription factor 2 (ATF2)/c-Jun – induce the transcriptional activation of genes encoding IFN-α/β, proinflammatory cytokines, and many other antiviral proteins, including RIG-I and MDA5 themselves, creating a positive feedback loop that amplifies the RLR response.
3. RLR ligands
The question of how RLRs distinguish viral RNA from host RNA has been an area of intense study. Using synthetic RNAs, such as the dsRNA analogue polyinosine-polycytidylic acid [poly(I:C)] and in vitro-transcribed RNAs, basic features of RLR agonists have been identified. Interestingly, despite their homologous domain architecture, RIG-I and MDA5 have been shown to recognize distinct RNA species. It is now well established that the key signature of RNAs recognized by RIG-I is a 5′triphosphate (5′ppp) moiety combined with adjacent stretches of blunt-end base pairing of ∼20 nucleotides, as found in the genomes of many RNA viruses (reviewed in further detail in [23], [24]). Furthermore, there is increasing evidence suggesting that specific sequence motifs in the RNA ligand, such as poly-U/UC, are needed for optimal RIG-I activation [25], [26]. Structural studies have further defined the molecular determinants of RNA recognition by RIG-I. A basic groove in the CTD of RIG-I has been shown to bind the negative charge of the 5′ppp group [27], [28], while the adjacent short dsRNA stretches stabilize the association of the CTD with the RNA [6], [7]. In addition to RIG-I's function as a sensor of RNA viruses, there is new evidence that RIG-I also detects various DNA viruses by sensing small viral RNA species produced during replication [29], [30], [31]. Specifically, RIG-I was shown to bind the Epstein–Barr virus (EBV)-encoded RNAs (EBERs), small 5′ppp-containing untranslated RNAs synthesized by RNA polymerase III, as well as adenovirus-associated RNA (VA). Furthermore, it was reported that RIG-I is also implicated in the detection of herpes simplex virus-1 (HSV-1); however, the physiological ligand(s) for RIG-I in the context of HSV-1 infection remains elusive.
Compared with those of RIG-I, the molecular features of viral PAMPs that trigger MDA5 activation are less well understood. MDA5 does not require a 5′ppp moiety for RNA recognition, and instead, is activated upon binding to longer dsRNAs, as well as web-like RNA aggregates as found, for instance, in cells infected with the picornavirus encephalomyocarditis virus (EMCV) [32], [33]. More recent studies have described cooperative binding and multimerization of MDA5 on viral RNA as a mechanism by which MDA5 discriminates between dsRNA species of different lengths [34], [35], [36]. This activity of MDA5 to bind to the RNA helix along its length, forming large filaments, is dependent on the ATPase function of its helicase domain. Structural studies showed that not only the helicase but also the CTD of MDA5 coordinates the formation of MDA5 filaments and subsequent interaction with MAVS [37]. In addition to this length-dependent mechanism of RNA recognition, it has been proposed that MDA5 can discriminate host and viral RNA based on the 2′-O-methylation status of their 5′cap structures [38].
Initial infection studies performed in mice deficient in RIG-I (Ddx58 −/−) or MDA5 (Ifih1 −/−) indicated that these two sensors recognize different subsets of viruses. Viruses found to be solely detected by RIG-I were influenza A virus (IAV), vesicular stomatitis virus (VSV), arenaviruses, and multiple paramyxoviruses, including Sendai virus (SeV), New Castle Disease virus (NDV), and measles virus (MV). In contrast, MDA5 was shown to be activated primarily in response to picornaviruses as well as RNA intermediates produced during vaccinia virus infection [39], [40]. Furthermore, detailed studies in cells deficient in either RIG-I or MDA5 revealed partially redundant roles of these sensors in the detection of reoviruses and flaviviruses such as dengue virus (DenV) and West Nile virus (WNV) [41]. While these investigations greatly expanded our knowledge about the roles of RIG-I and MDA5 in antiviral immunity to distinct viral pathogens, the fact that many viruses encode potent RLR antagonists, which could obscure the contribution of RIG-I or MDA5 to virus recognition, was often not considered. For example, initial infection studies suggested that RIG-I, but not MDA5, is the primary sensor for the detection of RNA species during paramyxovirus infection; however, it has been demonstrated that the V proteins of many paramyxoviruses specifically inhibit MDA5 activation. Indeed, detailed in vivo studies showed that SeV infection of Ifih −/− mice was more highly pathogenic and elicited much lower levels of pro-inflammatory cytokines compared to infection of wild-type mice [42]. Additional studies strengthened that indeed MDA5's contribution to sensing of paramyxovirus infection was not apparent in previous infection studies due to the antagonistic function of the V protein; when RIG-I knockdown cells were infected with a recombinant MV lacking the V protein, high IFN-β induction was observed, which was triggered by MDA5 [43]. In support of this, Runge et al. recently identified both distinct and common RNA species bound by RIG-I and MDA5 in MV-infected cells by using a novel protein-RNA cross-linking approach followed by next generation sequencing [44]. Both RLRs were able to recognize defective interfering (DI) copy-back RNA products and showed preference for AU-rich regions from the L gene. RIG-I also bound to both negative-stranded genomic RNA and positive-stranded mRNA transcripts, likely dependent on the presence of a 5′ppp. MDA5 on the other hand, was only found associated with positive-stranded mRNA transcripts as well as replication intermediates [44]. A similar RNA pull-down and deep sequencing approach had previously defined shorter 5′ppp-containing RNA segments of the IAV genome as well as DI genomes of both IAV and SeV as physiological ligands for RIG-I in infected cells [45]. More recently, it has been shown that the nucleoprotein-encapsidated viral genomic RNA of incoming virions can also trigger RIG-I activation [46].
Furthermore, recent studies have implied that RIG-I and MDA5 may respond to viral infection in a temporal manner. WNV-infected Ddx58 −/− mice were able to produce IFN, but in a delayed manner compared to wild-type mice. Indeed, MDA5 was able to compensate for the lack of RIG-I, but was not activated as quickly in response to WNV [47]. This effect has been shown to be due to the recognition of two distinct RNA species that are present in WNV infection: RIG-I responded to early RNA replication intermediates in a 5′ppp-dependent manner, whereas MDA5 responded to viral RNA species that accumulated as the infection progressed [48]. It is thus tempting to speculate that in the context of many viral infections, during which a variety of viral RNA species are likely produced, RIG-I and MDA5 act in concert to sense viral PAMPs in a temporally distinct manner. Further studies will be required to identify these physiological PAMPs and the individual and cooperative contributions of RIG-I and MDA5 to IFN induction in response to viral infection.
4. Regulation of RLR-MAVS signal transduction by host proteins
An effective antiviral immune response is characterized by high levels of cytokine production and the broad activation of antiviral effector genes that control many important cellular processes including apoptosis and protein synthesis; as such, dysfunctions in this process can be severely detrimental to the host [1]. Thus, immune responses to infection require a fine balance between viral clearance and host preservation. Insufficient IFN-induced cell death may prevent complete clearance of the virus, leading to persistent infection, while an overactive inflammatory response may cause tissue damage and reduced overall fitness of the host. Over the past few years, many host regulatory proteins have been identified that act either at the level of RLRs or at other important checkpoints in the RLR pathway, ultimately maintaining a balanced – yet effective – antiviral immune response [41]. Here, we focus on recent findings concerning the molecular mechanisms that control the antiviral signaling activities of RIG-I, MDA5 and MAVS.
4.1. Regulation of RIG-I
A recent series of structural and biochemical studies has demonstrated a multi-step model of RIG-I activation that is subject to several intricate regulatory mechanisms. Control of RIG-I activation is primarily mediated by conformational changes and host enzyme-induced post-translational modifications (PTMs) (Fig. 3 ). During the initial characterization of RIG-I as a viral sensor, Yoneyama et al. observed that the overexpressed CARDs of RIG-I were constitutively active and robustly induced signaling without viral RNA stimulation [2]; in contrast, full-length RIG-I had a low basal signaling activity in the absence of viral PAMPs, providing the first hint that RIG-I is maintained in an inactive, signaling-repressed state in uninfected cells. Subsequent studies showed that the deletion of the CTD of RIG-I leads to its constitutive activation, while overexpressed CTD alone had a dominant-negative effect on RIG-I-induced signal transduction [49]. This suggested a model in which the CTD functions as a regulatory/repressor domain (RD) by keeping RIG-I in an inactive conformation in which the CARDs are masked and thus unable to signal. The model for RIG-I autorepression was further refined by crystal structure data that demonstrated distinct conformational states for RIG-I in the absence of PAMPs and upon activation through viral RNA binding [6]. These findings indicated that in the repressed state, interactions between the helicase core – in particular Hel2i – and the CARD2 of RIG-I inhibit MAVS binding and downstream signaling.
Fig. 3.
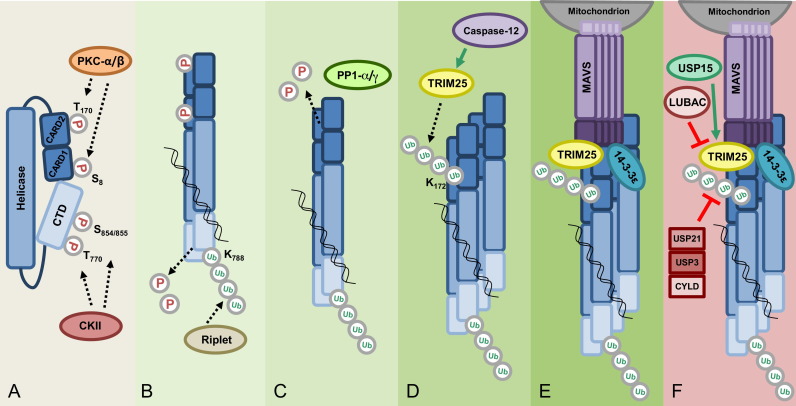
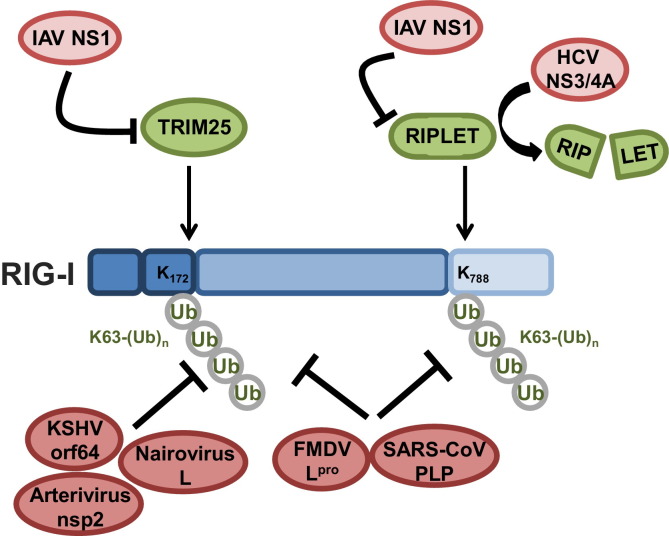
Model of RIG-I activation and regulation. (A) In uninfected cells, RIG-I is kept inactive via two mechanisms: a closed conformation mediated by the helicase domain and phosphorylation at specific Thr and Ser residues in the CTD and CARDs mediated by casein kinase II (CKII) and protein kinase C (PKC)-α/β, respectively. (B) Upon viral infection, RIG-I binds to its RNA ligand, which facilitates RIG-I conformational changes and multimerization. The E3 ligase Riplet mediates the Lys63-linked polyubiquitination of Lys788 in the CTD, which contributes to the release of RIG-I from its autorepressed state. Dephosphorylation of the CTD is also thought to be required for RIG-I activation although the phosphatase responsible has not been identified. (C) Release of RIG-I from the autorepressed conformation renders the CARDs accessible to the phosphatases PP1-α/γ, which dephosphorylate Ser8 and Thr170 in the CARDs. (D) Dephosphorylation of the CARDs of RIG-I allows for interaction with the E3 ligase TRIM25, possibly by inducing a conformational rearrangement within the tandem CARD. TRIM25 mediates Lys63-linked polyubiquitination of Lys172 in RIG-I CARD2. (E) Ubiquitination of RIG-I facilitates further multimerization and the recruitment of the mitochondrial targeting chaperone 14-3-3ɛ, inducing the translocation of RIG-I to mitochondria-associated membranes (MAMs) where RIG-I interacts with MAVS. (F) Negative regulators of RIG-I signaling include the deubiquitinating enzymes (DUBs) CYLD, USP3, and USP21, which remove Lys63-linked ubiquitin chains from RIG-I. LUBAC, comprised of the E3 ligases HOIL-1L and HOIP, downregulates RIG-I signaling by targeting TRIM25 for proteasomal degradation. The DUB USP15 counteracts the activity of LUBAC by removing Lys48-linked ubiquitin chains from TRIM25, thus stabilizing TRIM25 during viral infection.
In addition to conformational auto-repression of RIG-I, multi-site Ser/Thr phosphorylation has been shown to play an important role in preventing aberrant RIG-I signaling in uninfected cells [50]. RIG-I is constitutively phosphorylated at the CARDs and the CTD in uninfected cells, and rapidly dephosphorylated upon viral infection, indicating that a balance of phosphorylation and dephosphorylation regulates RIG-I signal transducing activity. Phosphorylation of the residues Thr770 and Ser854/855 in the RIG-I CTD by casein kinase II (CKII) was shown to be required for maintaining the autorepressed conformation [51]. Phosphorylation of Ser8 and Thr170 in the RIG-I CARDs induced by protein kinase C-α (PKC-α) or PKC-β inhibits CARD-dependent downstream signaling by preventing the interaction of RIG-I with MAVS [52], [53], [54]. In contrast, dephosphorylation of both Ser8 and Thr170 allows efficient MAVS binding, possibly caused by a distinct conformation of the tandem CARD triggered by dephosphorylation of these residues. Extensive research has been performed to determine the molecular details of how RIG-I is released from its autorepressed state in response to viral infection. The current view is that RIG-I activation requires (i) Binding of 5′ppp-containing dsRNA to the CTD and helicase domain, and (ii) a series of dephosphorylation and Lys63-linked ubiquitination events both in the CTD and the CARDs. As described above, the RIG-I CTD and helicase domain bind to the 5′ppp moiety and duplex RNA, respectively [6], [7], [55], [56], [57]. Thermodynamic studies further supported the model that the CARDs contribute to dsRNA binding specificity by controlling RNA access to the helicase domain [58]. Upon infection, viral RNA binding is believed to induce conformational changes that release the CARDs, rendering them accessible to downstream interaction partners.
Studies addressing the role of inhibitory phosphorylation marks in RIG-I demonstrated that binding of viral RNA is insufficient for RIG-I activation, but that dephosphorylation of the RIG-I CARDs (at Ser8 and Thr170) and CTD (at Thr770, Ser854/855) is required for RIG-I signaling [51], [52], [53], [54]. These studies thus indicated that an unknown phosphatase(s) is likely recruited to RIG-I in response to viral RNA binding, leading to RIG-I activation. Wies et al. performed a phosphatome RNAi screen that targeted 257 human phosphatases, including Ser/Thr phosphatases, Tyr phosphatases and dual-specificity phosphatases as well as regulatory subunits of phosphatases. This screen identified two isoforms of the phosphoprotein phosphatase 1 (PP1), PP1-α and PP1-γ, as essential regulators of RIG-I (and also MDA5) signaling [59]. The PP1 subfamily is a group of Ser/Thr phosphatases that in mammals consists of the isoforms PP1-α, PP1-β, and PP1-γ, and that are involved in many cellular processes, including cell cycle regulation, metabolism, protein synthesis, and muscle function [60]. However, a role of PP1 in innate immune sensing or the type-I IFN response had not been shown. This study demonstrated that PP1-α/γ are required for the RIG-I- and MDA5-mediated type-I IFN response, by dephosphorylating specific CARD residues (Ser8 and Thr170 in RIG-I; Ser88 in MDA5), thus allowing MAVS binding and downstream signaling. Gene silencing of PP1-α/γ reduced RLR-meditated IFN and ISG production and, consistent with this, enhanced the replication of VSV and DenV. The activity of PP1-α/γ toward RIG-I and MDA5 was specific, as PP1 had no effect on TLR3-induced signaling. These studies further indicated that PP1-α and PP1-γ, which arose by gene duplication and share more than 90% identity at the amino acid level, have redundant functions in phosphorylating RLRs. Studies to address the mechanistic details of how PP1-α/γ are activated in response to viral infection, and the molecular determinants of specificity toward RLRs are currently under investigation. Furthermore, although the dephosphorylation of the sites in the RIG-I CTD is also thought to be required for RIG-I activation [51], the responsible phosphatase(s) is still unknown.
In addition to phosphorylation/dephosphorylation, RIG-I's signal transducing activity is tightly regulated by Lys63-linked ubiquitin chains which, in contrast to Lys48-linked ubiquitination, do not target proteins for proteasomal degradation but rather modulate their activity (Fig. 3). Over the past few years, a growing number of substrates for Lys63-linked ubiquitination have been described that are involved in diverse processes,including DNA repair, protein transport, translation and innate immune signaling [61]. Using mass spectrometry analysis, Gack et al. identified that the RIG-I CARDs, purified from human cells, undergo covalent Lys63-linked ubiquitination at six lysine residues (Lys 99, 169, 172, 181, 190 and 193). Mutational analysis identified that Lys63-linked polyubiquitin attached to Lys172 in RIG-I is necessary for RIG-I's ability to bind MAVS and to induce type-I IFN [62]. Mass spectrometry analysis of RIG-I CARD-interacting proteins further identified tripartite motif protein 25 (TRIM25) as being responsible for catalyzing the covalent Lys63-linked ubiquitination of the RIG-I CARDs. TRIM25 belongs to the TRIM protein family of ubiquitin E3 ligases with more than 80 members in humans, many of which play important roles in antiviral immune responses by either acting as restriction factors or modulators of innate signaling pathways [63], [64]. All TRIM proteins are comprised of a conserved domain structure containing an N-terminal RING domain with ubiquitin E3 ligase activity, one or two B boxes, and a central coiled-coiled domain (CCD) for homo- or hetero-oligomerization. In addition, the majority of TRIM proteins harbor a PRY-SPRY domain at the C-terminus, which often dictates substrate specificity by mediating binding of the TRIM proteins to their substrates. Mapping studies of TRIM25 showed that the C-terminal SPRY domain of TRIM25 binds to CARD1 of RIG-I, allowing TRIM25 to ubiquitinate Lys172 in CARD2 [65].
More recently, the ubiquitin E3 ligase, Riplet (also known as RNF135 or REUL), has been shown to facilitate Lys63-linked ubiquitination of the RIG-I CTD [66], [67]. Multiple lysines in the CTD were shown to be ubiquitinated by Riplet; however, Lys63-linked ubiquitination at the specific residue Lys788 was critical for RIG-I activation [68]. Mechanistically, Riplet-induced Lys63-linked ubiquitination does not affect the RNA binding activity of RIG-I, but allows for subsequent ubiquitination of the CARDs by TRIM25, potentially by facilitating an overall conformational change in RIG-I that exposes the CARDs. The importance of covalent Lys63-linked ubiquitination for RIG-I activation was supported by the identification of a splice variant of RIG-I that lacks amino acids 36–80 within CARD1 and thereby loses TRIM25 interaction, CARD ubiquitination, and antiviral signaling activity. This RIG-I splice variant is specifically upregulated upon virus infection or IFN-β stimulation and acts as a dominant-negative feedback inhibitor by hetero-oligomerizing with RIG-I, thereby blocking its interaction with MAVS [65]. Furthermore, several cellular proteins have been shown to modulate the TRIM25- and Riplet-induced Lys63-linked ubiquitination of RIG-I. Three deubiquitinating enzymes (DUBs) have been identified that remove Lys63-linked ubiquitin chains from RIG-I, thus dampening RIG-I activity. Tumor suppressor protein cylindromatosis (CYLD) reduced the baseline Lys63-linked ubiquitination of RIG-I in uninfected cells [69]. Viral infection and antiviral cytokine production reduces CYLD protein levels, thereby relieving its repressive activity on RIG-I. The effect of CYLD was not specific to RIG-I, as CYLD also deubiquitinated and inhibited the signaling activities of TBK1 and IKKɛ. Another DUB for RIG-I, USP3, was identified in a screen for DUBs that negatively regulated RIG-I-mediated IFN-β induction [70]. This study further showed that USP3 acts not only as a negative regulator of RIG-I, but also MDA5. In contrast to CYLD, USP3 was shown to act on RIG-I and MDA5 specifically after viral infection, as the interaction of USP3 with both sensors was observed only in response to viral infection or viral RNA stimulation. A similar screen of DUBs identified USP21 as an inhibitor of RIG-I signaling by removing its Lys63-linked ubiquitination [71]. Usp21 knockout mice showed enhanced antiviral responses to SeV and VSV, but unlike USP3, viral infection was not necessary to facilitate the interaction between RIG-I and USP21. In contrast to these negative regulators of RIG-I Lys63-linked ubiquitination, Caspase-12 has been identified as a factor that facilitates the covalent Lys63-linked ubiquitination of the RIG-I CARDs by promoting binding of TRIM25 to RIG-I [72]. In addition to covalent Lys63-linked polyubiquitination, the RIG-I CARDs have been shown to bind short, unanchored Lys63-linked polyubiquitin chains in vitro [73]. TRIM25, together with the E2 ubiquitin-conjugating enzyme Ubc5 or Ubc13, were responsible for the synthesis of free polyubiquitin chains, which upon binding to the CARDs facilitated RIG-I multimerization and engagement with MAVS [74]. The role of non-covalent Lys63-ubiquitination of RIG-I in the context of an authentic infection, as well as the functional contribution of covalent versus non-covalent Lys63-linked ubiquitination in RIG-I activation have been a subject of debate. Recently, a crystal structure of the tetrameric 2CARD of human RIG-I revealed that both covalent and non-covalent Lys63-linked ubiquitin chains act synergistically in promoting RIG-I 2CARD oligomer assembly and signaling ability [75]. Furthermore, it has been unclear how RIG-I, upon its ubiquitination, engages MAVS to induce antiviral signal transduction. Ubiquitinated, activated RIG-I as well as TRIM25 were recently reported to form a complex with the mitochondrial targeting chaperone protein 14-3-3ɛ. This ‘translocon’ complex triggered the redistribution of RIG-I from the cytosol to the membranes where MAVS is found [76].
Moreover, several proteins have been identified that regulate the stability of RIG-I or its upstream regulators. The ubiquitin E3 ligase RNF125 (ring-finger protein 125) induces Lys48-linked ubiquitination and subsequent proteasomal degradation of RIG-I (and also MDA5 and MAVS) [77]. Since RNF125 expression is IFN-inducible, its repression of RLR signaling may serve as a negative-feedback loop to prevent excessive cytokine production. The ubiquitin-editing protein, A20, also negatively regulates RIG-I signaling, specifically through its C-terminal ubiquitinating domain; however, the precise mechanism by which it does so is not known [78]. On the other hand, ubiquitin-specific protease 4 (USP4) removes Lys48-linked ubiquitin chains from RIG-I, thus stabilizing the sensor [79]. TRIM25's protein levels are also tightly regulated by a fine balance of Lys48-linked ubiquitination and deubiquitination. The linear ubiquitin assembly complex (LUBAC), composed of the two E3 ligases HOIL-1L and HOIP, inhibits the TRIM25-RIG-I axis by inducing Lys48-linked ubiquitination of TRIM25 at its C-terminal SPRY domain, thereby targeting TRIM25 for degradation [80]. In addition, this study proposed a second mechanism of how LUBAC inhibits TRIM25-RIG-I signaling: the interference of LUBAC with TRIM25 binding to RIG-I. Recently, USP15 has been identified as a counterplayer of LUBAC in TRIM25 regulation. USP15 bound to TRIM25 specifically late during infection, removing Lys48-linked ubiquitin chains from TRIM25 and thus stabilizing it [81]. Virus replication assays further indicated that USP15 is important for a sustained IFN-β-mediated antiviral response against VSV, NDV and SeV.
4.2. Regulation of MDA5
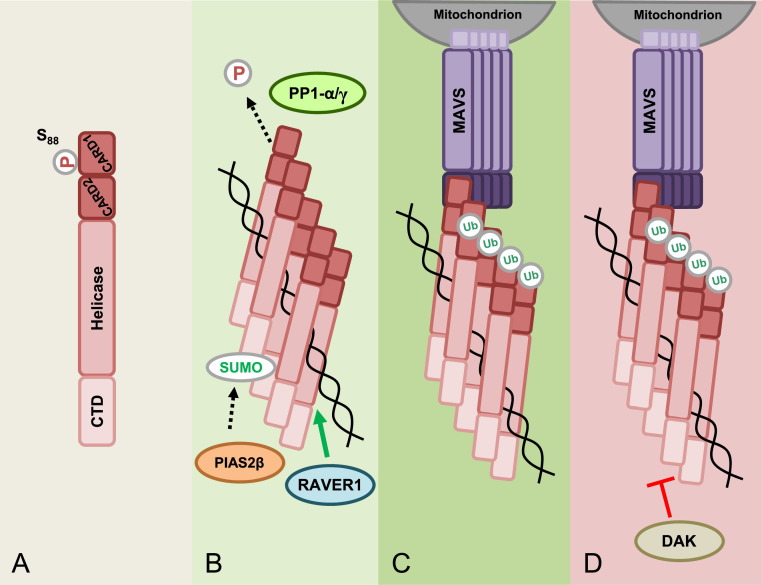
Compared with those of RIG-I, the mechanisms of MDA5 activation are not as well characterized, but what is known suggests that some of the regulatory steps leading to MDA5 activation may be quite different from those of RIG-I. Unlike RIG-I, MDA5 is not kept inactive by an autorepressed conformation [82], suggesting that PTMs may play an even more important role in controlling MDA5 signaling activity (Fig. 4 ). One commonality of RIG-I and MDA5 activation is the removal of repressive phosphorylation marks in the CARDs [59]. Mass spectrometry analysis of purified human MDA5 2CARD revealed phosphorylations at Ser88 and Ser104, located in the CARD1 and short CARD–CARD linker region of MDA5, respectively. Mutational analysis indicated that phosphorylation at Ser88, but not Ser104, modulates the CARD-dependent signaling activity of MDA5: phospho-mimetic Ser88 mutants of MDA5 were unable to signal downstream, while a S88A mutant of MDA5 had optimal signaling and IFN-β induction activity. Through the use of a phospho-Ser88-MDA5 specific antibody, this study also showed that endogenous MDA5 was robustly phosphorylated at Ser88 in uninfected cells; however, MDA5 stimulation by EMCV infection or poly(I:C) transfection induced rapid dephosphorylation of MDA5. As described above, the phosphatases PP1-α/γ have been identified as being responsible for not only dephosphorylating RIG-I, but also MDA5 at Ser88, leading to RLR activation. Sequence analysis identified two potential PP1-binding sites in MDA5, one located in the CARDs and one in the helicase domain. Mutation of each of these PP1-binding motifs in MDA5 abolished PP1-α/γ interaction, keeping MDA5 in the phosphorylated, signaling-repressed state [59]. In infected cells, specific recruitment of PP1-α/γ to endogenous MDA5 induced dephosphorylation of Ser88, promoting MDA5 binding to MAVS and type-I IFN induction. These results demonstrated that phosphorylation/dephosphorylation of specific Ser/Thr residues in the N-terminal CARDs regulates MDA5 and RIG-I signaling abilities, and also identified the phosphatases PP1-α/γ as common activators of both sensors.
Fig. 4.
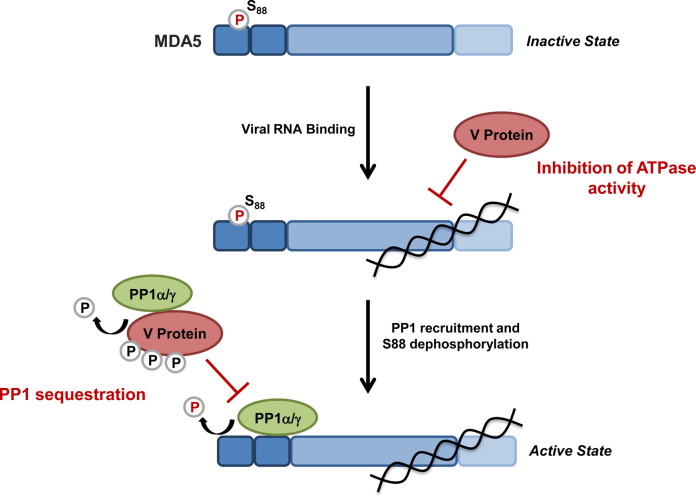
Model of MDA5 activation and regulation. (A) In uninfected cells, MDA5 is kept inactive by phosphorylation at residue Ser88 in CARD1. (B) Upon viral infection, MDA5 binds to dsRNA which allows for the recruitment of PP1α/γ to the CARDs, which then induce Ser88 dephosphorylation. In addition, RNA binding facilitates MDA5 multimerization. The ribonucleoprotein binding protein RAVER1 is thought to enhance the binding of MDA5 to its RNA ligand, thereby enhancing MDA5 signaling. The CTD also undergoes SUMOylation mediated by the E3 ligase PIAS2β, which has been shown to enhance MDA5 signaling. (C) A study indicated that Lys63-linked ubiquitin chains associated with the MDA5 CARDs facilitate MDA5 multimerization and activation; however, other studies showed that Lys63-linked ubiquitin is not required for MDA5 activation. Activated MDA5 is then able to interact with its downstream signaling partner, MAVS. (D) DAK is a negative regulator of MDA5 signaling; however, the precise mechanism of MDA5 inhibition by DAK is unknown.
While it is now well-established that Lys63-linked ubiquitination is essential for RIG-I signaling, the requirement for Lys63-linked ubiquitin chains in MDA5 activation has been a subject of debate. No covalent Lys63-linked ubiquitination of MDA5 CARDs purified from human cells has been detected [62]. Furthermore, while a study suggested that unanchored Lys63-ubiquitin chains promote MDA5 oligomerization and filament formation in vitro [74]; others have not observed a requirement for unanchored Lys63-linked ubiquitin chains in MDA5 activation [83].
Only a few upstream regulators of MDA5 have been described to date (Fig. 4). A yeast two-hybrid screen identified dihydroacetone kinase (DAK) as an interaction partner of MDA5, inhibiting its downstream signaling. Notably, the inhibitory role of DAK on MDA5 signaling was not associated with its kinase activity, but proposed to be attributed to its ability to prevent the binding of MDA5 to dsRNA or downstream signaling proteins [84]. The E3 ligase PIAS2β was shown to enhance MDA5 signaling by facilitating the conjugation of SUMO to the MDA5 CTD [85]; however the molecular mechanism by which SUMOylation activates MDA5 has not been addressed. Another positive regulator of MDA5 is ribonucleoprotein PTB-binding 1 (RAVER1), which was shown to interact with MDA5 and to increase the binding of MDA5 to poly(I:C), thereby enhancing MDA5 signaling [86].
4.3. Regulation of MAVS
Numerous cellular factors have been shown to modulate MAVS signal transduction through diverse mechanisms, including steric hinderance of MAVS binding to RLRs or other components of the MAVS signalosome (Table 1 ). The mitochondrial NOD-like receptor X 1 (NLRX1) was initially identified as a mitochondrial outer membrane protein that acts as a negative regulator of RIG-I signaling by competing with RIG-I for MAVS binding [87]. Other studies, however, have suggested an alternative mechanism of NLRX1 action and reported that NLRX1 is localized to the mitochondrial matrix [88], [89]. Furthermore, in vivo studies using different strains of Nlrx1 −/− mice have provided inconsistent results on the inhibitory role of NLRX1 in type-I IFN signaling [90], [91], [92]. Thus, the role of NLRX1 in MAVS regulation warrants further investigation. Furthermore, Nlrx1 −/− mice showed reduced autophagy in response to viral infection. Mass spectrometry analysis of NLRX1-interacting proteins identified the mitochondrial Tu translation elongation factor (TUFM) [93]. TUFM was shown to act as a molecular bridge between the type-I IFN signaling pathway and autophagy by facilitating the formation of a complex comprised of TUFM, MAVS, RIG-I and the autophagy-associated proteins Atg5-Atg12 and Atg16L1. In this complex, TUFM was found to inhibit type-I IFN signaling through MAVS, leading to enhanced VSV replication. Mitofusin 2 (Mfn2), a mitochondrial outer-membrane protein, inhibits MAVS signaling through an interaction with the TM domain of MAVS [94]. The complement receptor gC1qR was also reported to have a dampening effect on RLR signaling through MAVS [95]. Upon viral infection, gC1qR is relocalized to mitochondria where it interacts with MAVS, suppressing its interaction with RLRs. Another cellular protein that has been reported to antagonize antiviral signaling by inhibiting the interaction between RIG-I and MAVS is the autophagy-regulatory protein conjugate Atg5-Atg12, which binds to the CARDs of both RIG-I and MAVS, thereby preventing their interaction and downstream signaling [96]. The proteins SEC14L1 and NLRC5 have been shown to interact with the CARDs of RIG-I and/or MDA5, competing with RLR binding to MAVS [97], [98]. Furthermore, the ubiquitin-regulatory-X-domain-containing protein 1 (UBXN1) was found to be relocalized to mitochondria upon viral infection, where it competes with the signalosome proteins TRAF3 and TRAF6 for binding to MAVS [99].
Table 1.
Cellular regulators of MAVS signaling activity. Cellular regulators of MAVS, grouped by their mechanism of action, are detailed.
| Mechanism | Host regulator | |
|---|---|---|
| RLR-MAVS binding inhibition | NLRX1 | Moore et al., 2008 [87] |
| gC1qR | Xu et al., 2009 [95] | |
| SEC14L1 | Li et al., 2013 [97] | |
| NLRC5 | Cui et al., 2010 [98] | |
| Atg5-Atg12 | Jounai et al., 2007 [96] | |
| MAVS1a | Lad et al., 2008 [100] | |
| MAVS-TRAF2/6 binding inhibition | Mini-MAVS | Brubaker et al., 2014 [101] |
| MAVS degradation | PCBP2, AIP4 | You et al., 2009 [102] |
| Ndfip, Smurf1 | Wang et al., 2012 [103] | |
| RNF5 | Zhong et al., 2010 [104] | |
Moreover, variants of MAVS have been described to act as suppressors of MAVS downstream signaling. During the initial identification of MAVS, smaller versions of the protein were detected and assumed to be splice variants or degradation products of MAVS. A splice variant of MAVS, known as MAVS1a, was identified which strongly bound to RIG-I, thereby inhibiting its interaction with MAVS [100]. A recent study reported that the MAVS mRNA transcript is bicistronic, and that one of the smaller MAVS variants, known as ‘mini-MAVS,’ is produced by alternative translation [101]. Mini-MAVS has been shown to be involved in downregulating the type-I IFN response and promoting cell death. It has been proposed that mini-MAVS competes with full-length MAVS for binding its downstream signaling partners TRAF2 and TRAF6; however, the exact mechanism by which mini-MAVS inhibits MAVS signaling is unclear.
MAVS signal transduction is further regulated by proteins that modulate the stability of MAVS. At least three ubiquitin E3 ligases have been reported to induce Lys48-linked ubiquitination of MAVS, promoting its proteasomal degradation. The HECT E3 ligase AIP4 is recruited to MAVS by the poly(rC) binding protein 2 (PCBP2), which relocalizes to mitochondria upon viral infection [102]. Upon recruitment, AIP4 induces Lys48-linked ubiquitination at Lys371 and Lys420 in MAVS, triggering its proteasomal degradation. A similar mechanism has been described for the Nedd4 family interacting protein-1 (Ndfip1), in which Ndfip1 promotes Lys48-linked ubiquitination and degradation of MAVS by recruiting the ubiquitin E3 ligase Smurf1 (SMAD ubiquitination regulatory factor 1) [103]. The E3 ubiquitin ligase RING-finger protein 5 (RNF5) has also been reported to interact with MAVS upon viral infection, ubiquitinating and targeting MAVS for proteasomal degradation [104].
Mitochondrial and cytoskeletal dynamics also play a role in coordinating MAVS-mediated antiviral signaling. Recent studies showed that RLR signaling promotes mitochondrial elongation and fusion, which in turn facilitates the interaction of MAVS with STING and downstream signaling [105]. Mitochondrial fragmentation had the opposite effect, reducing MAVS-STING interaction and inhibiting RLR signaling. The inner-mitochondrial membrane protein cytochrome c oxidase complex subunit 5B (COX5B) has been shown to play a role in the regulation of RLR signaling [106]. ATP production by mitochondria produces reactive oxygen species (ROS) as a by-product, which is a stimulator of MAVS signaling. COX5B, a component of the mitochondrial electron transport chain, negatively regulates MAVS signaling by downregulating ROS production and, in concerted action with Atg5, inhibiting MAVS aggregation. Furthermore, the maintenance of mitochondrial membrane potential has been shown to be critical for RLR signaling through MAVS [107]. A loss of mitochondrial membrane potential may inhibit rearrangements of the MAVS complex that are necessary for signaling. MAVS activation during viral infection has also been linked to virus-induced cytoskeletal perturbations. The focal adhesion kinase (FAK), which is normally localized to cytoskeleton-linked focal adhesions, redistributed to the mitochondrial membrane upon viral infection, where it interacted with MAVS [108]. Cell deficient in FAK (Ptk2-/-) were more susceptible to infection by RNA viruses and had reduced NF-κB and IRF3 activation. In infected Ptk2 wild-type mouse embryonic fibroblasts, MAVS was observed to relocalize into cytoplasmic aggregates. In contrast, Ptk2-/- cells showed abnormal MAVS localization and morphology, both prior to and after viral infection. Although the precise mechanism of MAVS regulation by FAK is unclear, FAK is thought to facilitate proper MAVS localization and mitochondrial morphology to promote antiviral signaling.
5. Regulation of RLR signaling activities by viral proteins
Successful viral pathogens have evolved many different mechanisms for evading the type-I IFN response to prevent their rapid clearance. In the IFN-α/β induction pathway, many different host targets of viral antagonistic strategies have been identified. Not surprisingly, many RNA and DNA viruses dedicate a significant portion of their genome to blocking the RLR-MAVS signal transduction pathway. Perhaps the most direct way to interfere with RLR-induced IFN induction is through preventing recognition of viral PAMPs by RLRs (reviewed elsewhere [23], [109]). In addition, there is increasing evidence that many viruses have developed means of interfering with distinct steps in RLR-MAVS signaling. Here, we focus on summarizing four major viral strategies of RLR immune escape: (i) manipulation of the multi-site PTM program of RLRs, (ii) inhibition of the ATPase activity of RLRs, (iii) sequestration of RLRs, and (iv) virus-induced cleavage of RLRs and MAVS.
5.1. Manipulation of the posttranslational modification (PTM) program of RLRs
Recent studies uncovered that multiple viruses evade the type-I IFN response by manipulating PTMs critical for RLR activation. Several viruses, both RNA and DNA viruses, have been reported to interfere specifically with the covalent Lys63-linked ubiquitination of RIG-I, emphasizing the important role of this modification for RIG-I activation. At least two distinct viral strategies have been identified for manipulating the Lys63-linked ubiquitination of RIG-I: targeting the responsible E3 ligases, TRIM25 and Riplet, or directly removing Lys63-ubiquitin chains through virus-encoded DUBs. Furthermore, recent studies identified that paramyxoviruses have evolved means of manipulating the phosphorylation levels of RLRs, thus keeping RLRs in the phosphorylated, signaling-repressed states.
5.1.1. Viral inhibition of Lys63-linked ubiquitination of RIG-I
The non-structural protein 1 (NS1) of influenza A virus (IAV) is known to block IFN induction through multiple mechanisms and host targets [110]. NS1 has been reported to bind dsRNA via its N-terminal RNA-binding domain (RBD), thus potentially sequestering viral RNA from cellular sensors. Furthermore, through its C-terminal effector domain (ED), NS1 interacts with several host proteins to dampen antiviral responses, including CPSF30 which is targeted by NS1 to block host gene expression. Recently, NS1 has been shown to inhibit the initiation of IFN induction by blocking the TRIM25-mediated Lys63-linked ubiquitination of the RIG-I CARDs (Fig. 5 ) [111]. NS1 directly binds to the CCD of TRIM25 involving residues E96 and E97 in the ED. The E96A/E97A mutation abrogated the ability of NS1 to block RIG-I ubiquitination by TRIM25 and inhibition of IFN production. Mechanistically, the interaction of NS1 with the CCD of TRIM25 prevented CCD-dependent oligomerization of TRIM25 which appears to be critical for its enzymatic activity. These results identified a novel mechanism for inhibition of RLRs, through viral antagonism of the cellular E3 ubiquitin ligase TRIM25 [111]. As IAV infects and can adapt to many different host species, the species-specificity of the TRIM25-NS1 interaction was investigated [112]. Human TRIM25 efficiently interacted with the NS1 proteins of human, mouse-adapted, and avian strains (including the NS1 protein of the highly pathogenic H5N1 strain). Interestingly, while NS1 of avian IAV bound preferentially to avian (chicken) TRIM25, none of the NS1 proteins tested bound to mouse TRIM25. Further study showed that NS1 is able to effectively antagonize RIG-I in mice. Mechanically, in contrast to inhibition of TRIM25 in human cells, in mice, NS1 targets the E3 ligase Riplet, thereby blocking RIG-I ubiquitination at the CTD and thus RIG-I activation [112]. Furthermore, this study indicated that some human strains of IAV have evolved to bind to and target both TRIM25 and Riplet in human cells, strengthening that TRIM25 and Riplet are critical activators of RIG-I.
Fig. 5.
Viral inhibition of RIG-I through modulation of its Lys63-linked ubiquitination. Shown is a schematic of viral proteins that modulate the Lys63-linked ubiquitination levels of RIG-I to suppress IFN gene expression. The NS1 proteins of influenza A viruses (IAV) interact with and inhibit the ubiquitin E3 ligases TRIM25 and Riplet. This abolishes the Lys63-linked ubiquitination of RIG-I at Lys172 in the CARDs (mediated by TRIM25) and at Lys788 in the CTD (mediated by Riplet). The ability of IAV NS1 proteins to bind and target TRIM25 and Riplet is species-specific. The NS1 proteins of human, avian, and mouse-adapted IAV strains are all capable of binding human TRIM25. The NS1 proteins of some human IAV strains have also evolved to target human Riplet. The protease NS3/4A of hepatitis C virus (HCV) cleaves Riplet to suppress Lys63-linked ubiquitination of RIG-I at the CTD. Furthermore, Kaposi's sarcoma-associated herpesvirus (KSHV), nairoviruses, arteriviruses, foot-and-mouth disease virus (FMDV), and SARS coronavirus (SARS-CoV) encode deubiquitinating enzymes (DUBs) that remove Lys63-linked ubiquitin moieties from the RIG-I CARDs and CTD, thereby suppressing RIG-I antiviral signaling. More details on viral inhibition of RIG-I ubiquitination are described in the text.
Flaviviruses, such as Hepatitis C virus (HCV), are positive-stranded RNA viruses whose genome is directly translated into one large poly-protein. This poly-protein is subsequently cleaved by cellular and viral proteases to produce the individual proteins required for viral replication. The NS3/4A protease of HCV has been identified as an IFN antagonist that targets MAVS for cleavage, thereby inhibiting its downstream signaling (further detail below). Recent studies have indicated that NS3/4A also targets factors upstream of MAVS. Oshiumi et al. observed that expression of NS3/4A, either during viral infection or upon its overexpression, was able to suppress RIG-I-mediated signaling even in the presence of a cleavage-resistant mutant of MAVS [68]. The authors further observed that the protein levels of the E3 ligase Riplet were markedly decreased in HCV-infected hepatocytes. Consistent with this, the Lys63-linked ubiquitination of RIG-I was also decreased. Indeed, bioinformatics analysis revealed that Riplet harbors an NS3/4A consensus cleavage site in the RING domain. NS3/4A did not affect the protein levels of a Riplet mutant with a mutated NS3/4A consensus site; however, functional studies were not possible with this Riplet mutant as it was no longer catalytically active [68]. Thus, further studies are needed to determine the physiological role of Riplet cleavage for RIG-I escape by HCV.
Many herpesviruses encode viral DUBs that target cellular proteins for removal of ubiquitin moieties. Kaposi's sarcoma-associated herpesvirus (KSHV), a member of the γ-herpesvirus family, encodes the viral DUB, orf64 [113]. Orf64 is a component of the viral tegument that is important for virion assembly and lytic replication. Recently, a third function has been unveiled for orf64: the dampening of RIG-I antiviral signaling [114]. Orf64, but not its catalytically inactive mutant, was able to inhibit the production of type I-IFN induced by RIG-I. Further analysis revealed that it did so by decreasing the level of covalently-linked Lys63-linked ubiquitination of the RIG-I CARDs. This reduction was overcome by overexpression of TRIM25, indicating that orf64 specifically removes the TRIM25-induced ubiquitination essential for RIG-I activation [114].
Coronaviruses (CoV) are large, enveloped RNA viruses that cause a variety of respiratory illnesses. During infection, the genome is translated into two poly-proteins which are then processed by proteases into individual proteins. Structural and functional analyses revealed that the papain-like protease (PLP) of severe acute respiratory syndrome coronavirus (SARS-CoV) has DUB activity that catalyzes the removal of both ubiquitin and ISG15 from substrates [115], [116]. PLP's DUB activity was shown to play a major role in IFN antagonism [117], [118]. Cell-based deubiquitination assays identified RIG-I, TBK1, IRF3, and STING as targets of deubiquitination by SARS-CoV PLP, leading to downregulation of the type-I IFN response [119]. Another example of a virus-encoded DUB is found in foot-and-mouth disease virus (FMDV), a picornavirus that is responsible for a debilitating disease of cloven-hoofed animals which can have serious economic implications. The leader proteinase (Lpro) of FMDV is a PLP known to be involved in shut-off of host translation [120]. In addition, FMDV Lpro was reported to have multiple roles in antagonism of IFN induction. Independent of its protease function, Lpro leads to decreases in IRF3, IRF7, and p65/RelA protein levels [121]. Bioinformatics studies predicted USP-type DUB activity in Lpro and showed that, like SARS-CoV PLP, Lpro is indeed a functional DUB, and acted on several proteins involved in the IFN induction pathway – RIG-I, TBK1, TRAF3, and TRAF6. The deubiquitination of each of these proteins prevented their activation and downstream signaling leading to production of IFN [122].
Another class of viral DUBs is encoded by two unrelated RNA viruses: the arteriviruses and the nairoviruses. The arterivirus family contains positive-stranded RNA viruses that cause severe infections in livestock, including porcine respiratory and reproductive syndrome virus (PRRSV). Nairoviruses have tripartite negative-stranded RNA genomes and include Crimean-Congo hemorrhagic fever virus (CCHFV), a tick-borne virus that causes sporadic hemorrhagic fever throughout the Middle East, Africa, Asia, and Southeastern Europe. Unlike viral PLPs which have a USP-like DUB structure, bioinformatics approaches revealed the existence of ovarian tumor (OTU)-type DUB domains in the L protein of CCHFV, as well as the nsp2 proteins of PRRSV and the related equine arteritis virus (EAV). Overexpression of these viral OTU proteins (vOTUs) showed dual specificity for deconjugation of polyubiquitin and ISG15 [123], [124]. Protein deubiquitination and deISGylation by vOTUs led to the inhibition of IFN-β and TNFα induction. More recently, studies have revealed that RIG-I is a substrate for deubiquitination by the vOTUs from arteriviruses and nairoviruses. Expression of wild-type vOTUs, but not catalytically-dead mutants, decreased the Lys63-linked ubiquitination levels of RIG-I and thus its downstream signal transducing activity [125].
5.1.2. Viral manipulation of the phosphorylation state of MDA5
The IFN-suppressive ability of paramyxoviruses is well known and depends on their non-structural V, W, and C proteins, which are encoded by the P/V/C gene [126]. In particular, the V proteins of 13 different paramyxoviruses have been shown to inhibit specifically MDA5; however, the molecular mechanisms of this inhibition have just begun to be investigated. It has been proposed that one mechanism of MDA5 antagonism involves binding of the V protein to the helicase domain of MDA5, inhibiting the hydrolysis of ATP [3], [127], [128], [129], [130]. Recently, it has been discovered that the V proteins of measles virus (MV) and Nipah virus (NiV), but not of PIV5, efficiently block the PP1α/γ-mediated dephosphorylation of MDA5, preventing its CARD-dependent signaling activity ([164]) (Fig. 6 ). Intriguingly, upon MV infection, but not EMCV or DenV infection, MDA5 remained in the Ser88-phosphorylated, inactive state. This phosphorylation-modulatory effect of MV was due to its V protein, and this activity was shared also by the V protein of NiV. Biochemical studies revealed that the V proteins of MV and NiV efficiently bound to PP1-α/γ, but not to PP1-β which is not involved in MDA5 dephosphorylation. Sequence analysis identified a canonical PP1-binding motif in the very C-terminal ‘tail’ region of the MV V protein (288RIWY291). Deletion of the ‘tail’ region containing the PP1-binding site abrogated the interaction of the MV V protein with PP1-α/γ as well as antagonism of the dephosphorylation-mediated activation of MDA5. Generation of a recombinant MV of the Khartoum-Sudan strain, a clinical isolate of MV, harboring a truncated V protein that is unable to bind PP1α/γ demonstrated that PP1 antagonism by the V protein is an important mechansim of type-I IFN inhibition. Unlike the parental virus, the PP1-binding deficient mutant MV showed a strong replication defect in lung epithelial cells, which correlated with enhanced IRF3 activation and stimulation of IFN-β and ISG levels. This study thus identified a novel mechanism of action for the V protein of MV and NiV that is directed against PP1-α/γ and required for efficient inhibition of MDA5 ([164]). Furthermore, this finding combined with previous studies suggests a model in which different paramyxoviruses using their V proteins may employ different strategies to antagonize the sensor MDA5: PIV5 by targeting the ATPase activity of the helicase domain (discussed below), and MV and NiV through targeting PP1 and thereby MDA5 CARD-dependent signaling. This phenomenon of targeting a common host factor of the IFN system by using multiple different mechanisms has been described previously for STAT1/2 inhibition by paramyxoviral V proteins: while some V proteins induce STAT1/2 degradation, others sequester these transcription factors or prevent their nuclear translocation and thus activation [131]. In addition to these mechansims of IFN antgonism through their V proteins, some paramyxoviruses have also developed strategies to inhibit innate immunity in a V-independent manner. Interestingly, in dendritic cells, measles virus targets DC-SIGN signaling to inhibit PP1α/γ, thereby blocking the activation of both MDA5 and RIG-I (Mesman et al., in press).
Fig. 6.
Suppression of MDA5 activity through paramyxoviral modulation of MDA5 phosphorylation and inhibition of its ATPase activity. The V proteins of measles virus (MV) and Nipah virus (NiV) sequester the phosphatases PP1-α/γ from MDA5, preventing the dephosphorylation of Ser88 in the N-terminal CARDs of MDA5. V protein-induced inhibition of MDA5 S88 dephosphorylation prevents MDA5-MAVS binding and downstream signaling. Furthermore, upon binding to PP1-α/γ, the V proteins serve as substrates for PP1-mediated dephosphorylation. The V proteins of several paramyxoviruses, including Parainfluenza virus type 5 (PIV5), bind directly to the helicase domain of MDA5, inducing conformational changes in MDA5, ultimately suppressing the ATP hydrolysis activity of MDA5 and thereby its filament formation on viral dsRNA.
5.2. Inhibition of the ATP hydrolysis activity of RLRs
As mentioned above, the V proteins of several paramyxoviruses have been shown to interact with the helicase of MDA5, inhibiting its ATPase activity [3], [127], [128], [129], [130]. Recently, the co-crystal structure of porcine MDA5 and the V protein of PIV5 showed that the inhibition of the ATPase activity is due to mutual structural unfolding (Fig. 6) [132]. Additionally, the VP35 protein of Ebola virus has been reported to hinder specifically RIG-I's ATPase activity by preventing its interaction with PKR activator (PACT), a protein previously shown to activate RIG-I's ATPase activity and downstream signaling [133], [134].
The newly emerged Middle East Respiratory Syndrome Coronavirus (MERS-CoV) has been identified as the causative agent of a severe respiratory disease with a high mortality rate. Much research has been focused on understanding the pathogenicity of MERS-CoV in an attempt to limit its transmission and disease. These studies showed that while MERS-CoV replication is impaired by IFN treatment in vitro and in vivo, it fails to induce high levels of IFN during infection, indicating that this virus has effective mechanisms of IFN antagonism [135], [136], [137], [138]. The 4a, 4b, and M proteins of MERS-CoV were identified as potent antagonists of the type-I IFN response. The 4a protein specifically blocked IFN induction mediated by MDA5, but not RIG-I, an activity thought to be due to the predicted dsRNA binding ability of 4a [139]. Subsequent studies revealed that the MERS-CoV 4a protein interacts with PACT, thereby preventing optimal ATPase and signaling activities of not only MDA5 but also RIG-I. [140].
5.3. Sequestration of RLRs
Several viruses have developed means of interfering with the CARD–CARD interaction of RLRs and MAVS by sequestering RIG-I and MDA5 from the MAVS signalosome (Table 2 ). SARS-CoV, using its M protein, blocks the RLR-induced IFN-β gene expression by interacting with RIG-I or key proteins in the RLR pathway, such as TANK, TBK1, IKK-ɛ, and TRAF3 [141]. The M protein is localized to membranes associated with the Golgi complex; thus it is hypothesized that binding of the M protein to RIG-I results in sequestration of this sensor away from MAVS at mitochondria to Golgi-associated membranes instead. Two additional SARS-CoV proteins, ORF3b and ORF6, have been shown to block the interaction between RIG-I and MAVS. Both proteins localize to mitochondria where they likely bind to either RIG-I or MAVS to inhibit IFN induction [142], [143].
Table 2.
Viral inhibition of RIG-I and MDA5. Different viral antagonistic strategies are detailed, including information on the virus and the specific protein responsible for RLR antagonism.
| RIG-I | |||
| Inhibition of ATPase activity | Ebola | VP35 | Luthra et al., 2013 [134] |
| MERS-CoV | 4a | Siu et al., 2014 [140] | |
| Sequestration | SARS-CoV | M Protein | Siu et al., 2009 [141] |
| SARS-CoV | ORF3b | Kopecky-Bromberg et al., 2007 [143] | |
| SARS-CoV | ORF6 | Freundt et al., 2009 [142] | |
| NW Arenavirus | Z protein | Fan et al., 2010 [146] | |
| hMPV | G protein | Bao et al., 2008 [148]; Bao et al., 2013 [147] | |
| RSV | N Protein | Lifland et al., 2012 [149] | |
| RSV | NS1/NS2 | Goswami et al., 2013 [151] | |
| Cleavage | Poliovirus | 3Cpro | Barral et al., 2009 [159] |
| Rhinovirus | |||
| Echovirus | |||
| EMCV | |||
| MDA5 | |||
| Inhibition of ATPase activity | Paramyxovirus | V protein | Motz et al., 2013 [132]; Rodriguez and Horvath 2013 [130] |
| MERS-CoV | 4a | Siu et al., 2014 [140] | |
| Sequestration | RSV | N Protein | Lifland et al., 2012 [149] |
| Cleavage | Poliovirus | Barral et al., 2007 [158] | |
| EV71 | 2Apro | Feng et al., 2014 [163] | |
Arenaviruses are classified as New World viruses and Old World viruses, many of which are capable of causing hemorrhagic fever syndromes. The Z proteins of Arenaviruses have been implicated in many virus-host interactions including binding to eIF4E and PML [144], [145]. In regards to their IFN-antagonistic activities, the Z proteins of New World arenaviruses (e.g. Guanarito, Junin, Machupo, and Sabia), but not those of Old World arenaviruses (LCMV or Lassa virus), have been shown to inhibit RIG-I [146]. Mechanistically, binding of the Z protein to RIG-I prevents the CARD–CARD interaction between RIG-I and MAVS, thereby preventing further downstream signaling.
Human metapneumovirus (hMPV) is a leading cause of respiratory infections, particularly in infants, the elderly, and immunocompromised individuals. Recombinant hMPV lacking the glycoprotein (G) attachment factor (hMPV-ΔG) is attenuated in vivo. Recently this attenuation has been attributed to the loss of this protein's ability to escape innate immunity. The hMPV-ΔG mutant virus induced higher levels of IFN compared to the parental virus. This IFN-suppressive effect of the G protein was due to its interaction with the RIG-I CARD signaling module, preventing the redistribution of RIG-I from the cytosol to MAVS on mitochondria [147], [148].
Like hMPV, respiratory syncytial virus (RSV) is a significant cause of debilitating respiratory infections, particularly in infants. Early after RSV infection, large inclusion bodies (IB) are seen, both in in vitro cell cultures and in cells isolated from infected humans and animals. The formation of IBs appears to be dependent on the N and P proteins, as transfection of these proteins is sufficient to induce IB formation. At 6 h post-infection, Lifland et al. observed co-localization of both RIG-I and MDA5 to IBs, with MAVS also accumulating at later time points. This re-localization was dependent upon the N protein, as immunoprecipitation studies showed an interaction between N and MDA5. Formation of these IBs, induced by N and P proteins, significantly decreased the production of IFN-β mRNA in response to NDV infection, indicating a critical role in innate immune evasion [149].
Additionally, the NS1 and NS2 proteins of RSV are known to be antagonists of IFN induction, leading to inhibition or degradation of several key signaling molecules in the IFN induction cascade [150]. Recent work has identified a large multi-protein complex comprised of mitochondria, RSV NS1 and NS2, and several proteasome-specific host proteins. This complex has been dubbed the NS degradasome (NSD), and depends on MAVS for its mitochondrial association. When assembled, the NSD leads to the degradation of many key signaling molecules in the IFN pathway, including RIG-I, TRAF3, IKK-ɛ, IRF3, IRF7, and STAT2. This degradation was shown to be due to the presence of both proteasomal and non-proteasomal proteases in the NSD; however, more detailed insights into the selective degradative activity of the NSD for these molecules are needed [151].
5.4. Virus-induced cleavage of RLRs and MAVS
Many viruses encode proteases that play important roles in their replication cycle. In addition, some of these viral proteases antagonize RLR-mediated host immunity through cleavage of MDA5 and RIG-I or their adaptor molecule MAVS. The NS3/4A proteases of HCV and the related flavivirus, GB virus B (GBV-B), directly cleave MAVS. This cleavage occurs at Cys508 located in the TM domain of MAVS, thereby dislocating it from mitochondria, and suppressing MAVS-induced downstream signaling [152], [153]. Hepatitis A virus (HAV), of the Picornaviridae family, also encodes a protease which cleaves MAVS to downregulate IFN production [154], [155]; however, unlike HCV, HAV does not utilize its mature protease, 3Cpro, but instead a stable, catalytically-active precursor form of this protease (called 3ABC) which is localized at the mitochondrion [156].
Poliovirus, another picornavirus, specifically induces cleavage of MDA5. Unexpectedly, this activity is not due to either of its viral proteases as recombinant viruses expressing catalytically-dead proteases did not have abrogated MDA5-cleaving abilities. Instead, MDA5 cleavage during poliovirus infection was dependent on both the proteasome and cellular caspases. An independent study showed that mouse MDA5 was cleaved during apoptosis [157]. It is therefore conceivable that poliovirus infection induces apoptosis, leading to cleavage of MDA5, which results in decreased levels of IFN production [158].
Several other picornavirus-encoded proteases have been shown to cleave multiple members of the RLR signaling cascade. Specifically, the 3Cpro protease of poliovirus, rhinovirus, echovirus, and EMCV has been shown to lead to RIG-I degradation both in vitro and in infected cells [159]. This mechanism is in stark contrast to MDA5 cleavage during poliovirus infection, which is mediated by viral targeting of cellular proteases. Additional studies by the same group identified MAVS as a potential target of 2Apro and 3Cpro -mediated cleavage during rhinovirus infection [160]. More recently, a series of studies has identified both MAVS and MDA5 as targets of proteolytic cleavage during Enterovirus 71 (EV71) infection. First, Wang et al. showed that MAVS at mitochondria was cleaved during EV71 infection. This cleavage was confirmed to be a direct effect of the 2Apro protease, as a catalytically-inactive mutant was unable to cleave MAVS in vitro [161]. Two groups then showed that MDA5 was both important for EV71 detection during infection, but also a target for degradation [162], [163]. The degradation of MDA5 in EV71-infected cells was originally proposed to be due to caspase-mediated cleavage as in poliovirus infection; however, a more recent study reported that MDA5 is also directly cleaved by the virally-encoded 2Apro. The same study also showed that RIG-I, MDA5, and MAVS are all cleaved during EV71 infection by the viral proteases 3Cpro (RIG-I) and 2Apro (MDA5 and MAVS) [163].
6. Conclusion
It is well-established that the intricate interplay between viruses and the host's surveillance machinery dictates the outcome of viral infection and disease. Over the past several years, significant progress has been made toward our understanding of the regulatory mechanisms of RLR-MAVS signaling by host and viral factors. Despite extensive research in this field, many questions remain: Why are so many safeguard mechanisms needed for RLR control? And, are there disorders where RLR activation is dysregulated? In this regard, it is tempting to speculate that the ability of RIG-I and MDA5 to distinguish self and non-self RNAs could be less stringent than anticipated, and that self-RNA present in the cytosol may also be detected under certain conditions. Furthermore, new insights into the regulatory mechanisms of RLR activation and viral antagonistic mechanisms will stimulate drug development for infectious diseases and inflammatory or autoimmune disorders. Indeed there is recent evidence that polymorphisms of RIG-I and MDA5 are associated with autoimmune diseases, such as systemic lupus erythematosus and type-I diabetes; however, whether dysregulated RLR activities are associated with diseases remains to be answered. We eagerly await future discoveries of polymorphisms in regulatory molecules of RLR signaling or their aberrant expressions, and their roles in susceptibility to viral infections and disease development.
Conflict of interest
The authors have nothing to declare.
Acknowledgments
The research on RLR-mediated immunity in the Gack laboratory is supported by grants from the U.S. National Institutes of Health (AI087846 and AI097699), the Giovanni Armenise-Harvard Foundation, and the Alexander and Margaret Stewart Trust Foundation.
Biographies

Jessica Chiang is a Ph.D. candidate in the Virology Graduate Program at Harvard Medical School, Boston. She completed her B.S. in Molecular, Cell, and Developmental Biology at the University of California, Los Angeles. Her research interests include virus entry mechanisms and the strategies employed by RNA and DNA viruses to evade innate immunity. Her previous work focused on the development and characterization of entry inhibitors of HIV-1. Her current research, conducted in the Gack laboratory, is focused on innate immune sensing mechanisms of DNA viruses as well as the strategies used by DNA viruses to escape the type-I interferon-mediated innate immune response.

Meredith Davis graduated with a B.S. in Biochemistry from the University of Missouri-Columbia and is currently a Ph.D. candidate in Virology at Harvard Medical School. Her broad interest in virology includes host–virus interactions and the mechanisms by which viruses usurp or evade critical cellular pathways for their efficient replication. During her undergraduate studies, her research focused on the mechanisms of how viruses utilize the DNA damage response for their own benefit. Since joining the Gack laboratory in 2011, her research has focused on deciphering the mechanisms of how paramyxovirus family members evade antiviral signaling initiated by RIG-I-like receptors. During her thesis research, she identified a novel mechanism utilized by measles and Nipah virus to antagonize innate immune activation by MDA5 through targeting the phosphatases PP1α and PP1γ.

Michaela Gack is an Assistant Professor in the Department of Microbiology and Immunobiology at Harvard Medical School, Boston. Dr. Gack received her Ph.D. degree in Molecular Virology from the collaborative graduate training program between Harvard Medical School and the Friedrich Alexander University Erlangen-Nuremberg, Germany. She completed her postdoctoral training at the University of Southern California. Dr. Gack's research seeks a better understanding of the molecular mechanisms underlying innate immune sensing of RNA viruses and the subsequent type-I interferon response to limit virus replication. Using a combination of proteomics and biochemical, molecular, and cell biological approaches, her laboratory is identifying and characterizing the regulatory mechanisms of RIG-I-like receptor signaling through host factors. Her research interests also include mechanisms of viral evasion as well as the role of TRIM proteins in antiviral defenses. For her academic achievements in the fields of innate immunity and virology, Dr. Gack received several awards including the 2009 GE & Science Prize for Young Life Scientists, the 2013 Junior Investigator Award from the European Society for Virology, and the Ann Palmenberg Junior Investigator Award 2013 from the American Society for Virology. Furthermore, Dr. Gack was awarded with the 2014 Merck Irving S. Sigal Memorial Award from the American Society for Microbiology. Since 2012, Dr. Gack has been an Associate Editor for the journal PLoS Pathogens.
References
- 1.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva J., Childs K.S., Young D.F., Carlos T.S., Stock N., Goodbourn S. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B.G., Schoenemeyer A. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 5.Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Kowalinski E., Lunardi T., McCarthy A., Louber J., Brunel J., Grigorov B. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 7.Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belgnaoui S., Paz S., Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Bruns A.M., Horvath C.M. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol. 2012;47:194–206. doi: 10.3109/10409238.2011.630974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkataraman T., Valdes M., Elsby R., Kakuta S., Caceres G., Saijo S. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 11.Satoh T., Kato H., Kumagai Y., Yoneyama M., Sato S., Matsushita K. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seth R., Sun L., Ea C.-K., Chen Z. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 14.Xu L.-G., Wang Y.-Y., Han K.-J., Li L.-Y., Zhai Z., Shu H.-B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 16.Dixit E., Boulant S., Zhang Y., Lee A., Odendall C., Shum B. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci USA. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West A., Shadel G., Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Hou F., Sun L., Zheng H., Skaug B., Jiang Q.X., Chen Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goubau D., Deddouche S., Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlee M. Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology. 2013;218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito T., Owen D.M., Jiang F., Marcotrigiano J., Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uzri D., Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui S., Eisenacher K., Kirchhofer A., Brzozka K., Lammens A., Lammens K. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 28.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 29.Chiu Y.H., Macmillan J.B., Chen Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K.A., Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minamitani T., Iwakiri D., Takada K. Adenovirus virus-associated RNAs induce type I interferon expression through a RIG-I-mediated pathway. J Virol. 2011;85:4035–4040. doi: 10.1128/JVI.02160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichlmair A., Schulz O., Tan C.-P., Rehwinkel J., Kato H., Takeuchi O. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peisley A., Lin C., Wu B., Orme-Johnson M., Liu M., Walz T. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Q., Hato S.V., Langereis M.A., Zoll J., Virgen-Slane R., Peisley A. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peisley A., Jo M.H., Lin C., Wu B., Orme-Johnson M., Walz T. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci USA. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berke I., Yu X., Modis Y., Egelman E. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci USA. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gitlin L., Barchet W., Gilfillan S., Cella M., Beutler B., Flavell R. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 41.Loo Y.-M., Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gitlin L., Benoit L., Song C., Cella M., Gilfillan S., Holtzman M. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikegame S., Takeda M., Ohno S., Nakatsu Y., Nakanishi Y., Yanagi Y. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J Virol. 2010;84:372–379. doi: 10.1128/JVI.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Runge S., Sparrer K.M.J., Lässig C., Hembach K., Baum A., García-Sastre A. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014:10. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baum A., Sachidanandam R., Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA. 2010:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber M., Gawanbacht A., Habjan M., Rang A., Borner C., Schmidt A.M. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe. 2013;13:336–346. doi: 10.1016/j.chom.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fredericksen B., Keller B., Fornek J., Katze M., Gale M. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Errett J., Suthar M., McMillan A., Diamond M., Gale M. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile Virus infection. J Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito T., Hirai R., Loo Y.M., Owen D., Johnson C.L., Sinha S.C. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gack M.U. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J Virol. 2014;88:5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Z., Ren H., Liu Y., Teeling J.L., Gu J. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J Virol. 2010:85. doi: 10.1128/JVI.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gack M., Nistal-Villán E., Inn K.-S., García-Sastre A., Jung J. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol. 2010;84:3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maharaj N.P., Wies E., Stoll A., Gack M.U. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J Virol. 2012;86:1358–1371. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nistal-Villan E., Gack M.U., Martinez-Delgado G., Maharaj N.P., Inn K.S., Yang H. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem. 2010;285:20252–20261. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohlway A., Luo D., Rawling D.C., Ding S.C., Pyle A.M. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013;14:772–779. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vela A., Fedorova O., Ding S.C., Pyle A.M. The thermodynamic basis for viral RNA detection by the RIG-I innate immune sensor. J Biol Chem. 2012;287:42564–42573. doi: 10.1074/jbc.M112.385146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wies E., Wang M., Maharaj N., Chen K., Zhou S., Finberg R. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cohen P.T. Protein phosphatase 1 – targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 61.Weissman A.M. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 62.Gack M., Shin Y., Joo C.-H., Urano T., Liang C., Sun L. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 63.Rajsbaum R., Garcia-Sastre A., Versteeg G.A. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Versteeg G.A., Rajsbaum R., Sanchez-Aparicio M.T., Maestre A.M., Valdiviezo J., Shi M. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gack M., Kirchhofer A., Shin Y., Inn K.-S., Liang C., Cui S. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci USA. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oshiumi H., Matsumoto M., Hatakeyama S., Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 67.Oshiumi H., Miyashita M., Inoue N., Okabe M., Matsumoto M., Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 68.Oshiumi H., Miyashita M., Matsumoto M., Seya T. A distinct role of Riplet-mediated K63-Linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friedman C.S., O’Donnell M.A., Legarda-Addison D., Ng A., Cardenas W.B., Yount J.S. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cui J., Song Y., Li Y., Zhu Q., Tan P., Qin Y. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 2014;24:400–416. doi: 10.1038/cr.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fan Y., Mao R., Yu Y., Liu S., Shi Z., Cheng J. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med. 2014;211:313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang P., Arjona A., Zhang Y., Sultana H., Dai J., Yang L. Caspase-12 controls West Nile virus infection via the viral RNA receptor RIG-I. Nat Immunol. 2010;11:912–919. doi: 10.1038/ni.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang X., Kinch L., Brautigam C., Chen X., Du F., Grishin N. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peisley A., Wu B., Xu H., Chen Z.J., Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu H., Loo Y.-M., Horner S., Zornetzer G., Katze M., Gale M. The mitochondrial targeting chaperone 14-3-3ɛ regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arimoto K., Takahashi H., Hishiki T., Konishi H., Fujita T., Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin R., Yang L., Nakhaei P., Sun Q., Sharif-Askari E., Julkunen I. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 79.Wang L., Zhao W., Zhang M., Wang P., Zhao K., Zhao X. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J Virol. 2013;87:4507–4515. doi: 10.1128/JVI.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inn K.S., Gack M.U., Tokunaga F., Shi M., Wong L.Y., Iwai K. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pauli E.K., Chan Y.K., Davis M.E., Gableske S., Wang M.K., Feister K.F. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal. 2014;7:ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berke I., Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 84.Diao F., Li S., Tian Y., Zhang M., Xu L.G., Zhang Y. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu J., Xiong Y., Xu Y., Cheng G., Tang H. MDA5 is SUMOylated by PIAS2β in the upregulation of type I interferon signaling. Mol Immunol. 2011;48:415–422. doi: 10.1016/j.molimm.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen H., Li Y., Zhang J., Ran Y., Wei J., Yang Y. RAVER1 is a coactivator of MDA5-mediated cellular antiviral response. J Mol Cell Biol. 2013;5:111–119. doi: 10.1093/jmcb/mjt006. [DOI] [PubMed] [Google Scholar]
- 87.Moore C., Bergstralh D., Duncan J., Lei Y., Morrison T., Zimmermann A. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 88.Arnoult D., Soares F., Tattoli I., Castanier C., Philpott D., Girardin S. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci. 2009;122:3161–3168. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tattoli I., Carneiro L., Jéhanno M., Magalhaes J., Shu Y., Philpott D. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soares F., Tattoli I., Wortzman M.E., Arnoult D., Philpott D.J., Girardin S.E. NLRX1 does not inhibit MAVS-dependent antiviral signalling. Innate Immun. 2013;19:438–448. doi: 10.1177/1753425912467383. [DOI] [PubMed] [Google Scholar]
- 91.Allen I., Moore C., Schneider M., Lei Y., Davis B., Scull M. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rebsamen M., Vazquez J., Tardivel A., Guarda G., Curran J., Tschopp J. NLRX1/NOD5 deficiency does not affect MAVS signalling. Cell Death Differ. 2011;18:1387. doi: 10.1038/cdd.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lei Y., Wen H., Yu Y., Taxman D.J., Zhang L., Widman D.G. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–946. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yasukawa K., Oshiumi H., Takeda M., Ishihara N., Yanagi Y., Seya T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 95.Xu L., Xiao N., Liu F., Ren H., Gu J. Inhibition of RIG-I and MDA5-dependent antiviral response by gC1qR at mitochondria. Proc Natl Acad Sci USA. 2009;106:1530–1535. doi: 10.1073/pnas.0811029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li M.T., Di W., Xu H., Yang Y.K., Chen H.W., Zhang F.X. Negative regulation of RIG-I-mediated innate antiviral signaling by SEC14L1. J Virol. 2013;87:10037–10046. doi: 10.1128/JVI.01073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cui J., Zhu L., Xia X., Wang H.Y., Legras X., Hong J. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang P., Yang L., Cheng G., Yang G., Xu Z., You F. UBXN1 interferes with Rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell Rep. 2013;3:1057–1070. doi: 10.1016/j.celrep.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lad S.P., Yang G., Scott D.A., Chao T.H., Correia Jda S., de la Torre J.C. Identification of MAVS splicing variants that interfere with RIGI/MAVS pathway signaling. Mol Immunol. 2008;45:2277–2287. doi: 10.1016/j.molimm.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 101.Brubaker S.W., Gauthier A.E., Mills E.W., Ingolia N.T., Kagan J.C. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell. 2014;156:800–811. doi: 10.1016/j.cell.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.You F., Sun H., Zhou X., Sun W., Liang S., Zhai Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 103.Wang Y., Tong X., Ye X. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J Immunol. 2012;189:5304–5313. doi: 10.4049/jimmunol.1201445. [DOI] [PubMed] [Google Scholar]
- 104.Zhong B., Zhang Y., Tan B., Liu T.T., Wang Y.Y., Shu H.B. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol. 2010;184:6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 105.Castanier C., Garcin D., Vazquez A., Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao Y., Sun X., Nie X., Sun L., Tang T.S., Chen D. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 2012;8:e1003086. doi: 10.1371/journal.ppat.1003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koshiba T., Yasukawa K., Yanagi Y., Kawabata S. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci Signal. 2011;4:ra7. doi: 10.1126/scisignal.2001147. [DOI] [PubMed] [Google Scholar]
- 108.Bozym R., Delorme-Axford E., Harris K., Morosky S., Ikizler M., Dermody T. Focal adhesion kinase is a component of antiviral RIG-I-like receptor signaling. Cell Host Microbe. 2012;11:153–166. doi: 10.1016/j.chom.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bowie A., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.García-Sastre A. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 2011;162:12–18. doi: 10.1016/j.virusres.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gack M., Albrecht R., Urano T., Inn K.-S., Huang I.C., Carnero E. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rajsbaum R., Albrecht R., Wang M., Maharaj N., Versteeg G., Nistal-Villán E. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012:8. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.González C., Wang L., Damania B. Kaposi's sarcoma-associated herpesvirus encodes a viral deubiquitinase. J Virol. 2009;83:10224–10233. doi: 10.1128/JVI.00589-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Inn K.-S., Lee S.-H., Rathbun J., Wong L.-Y., Toth Z., Machida K. Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol. 2011;85:10899–10904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A., Baker S. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ratia K., Saikatendu K., Santarsiero B., Barretto N., Baker S., Stevens R. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Devaraj S., Wang N., Chen Z., Chen Z., Tseng M., Barretto N. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Clementz M., Chen Z., Banach B., Wang Y., Sun L., Ratia K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012:7. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Los Santos T., de Avila Botton S., Weiblen R., Grubman M. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J Virol. 2006;80:1906–1914. doi: 10.1128/JVI.80.4.1906-1914.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hato S., Sorgeloos F., Ricour C., Zoll J., Melchers W., Michiels T. Differential IFN-alpha/beta production suppressing capacities of the leader proteins of mengovirus and foot-and-mouth disease virus. Cell Microbiol. 2010;12:310–317. doi: 10.1111/j.1462-5822.2009.01395.x. [DOI] [PubMed] [Google Scholar]
- 122.Wang D., Fang L., Li P., Sun L., Fan J., Zhang Q. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol. 2011;85:3758–3766. doi: 10.1128/JVI.02589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Frias-Staheli N., Giannakopoulos N., Kikkert M., Taylor S., Bridgen A., Paragas J. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Capodagli G., McKercher M., Baker E., Masters E., Brunzelle J., Pegan S. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J Virol. 2011;85:3621–3630. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Kasteren P., Beugeling C., Ninaber D., Frias-Staheli N., van Boheemen S., García-Sastre A. Arterivirus and nairovirus ovarian tumor domain-containing Deubiquitinases target activated RIG-I to control innate immune signaling. J Virol. 2012;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goodbourn S.R. The regulation of type I interferon production by paramyxoviruses. J Interferon Cytokine Res. 2009 doi: 10.1089/jir.2009.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Childs K., Stock N., Ross C., Andrejeva J., Hilton L., Skinner M. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 128.Parisien J.-P., Bamming D., Komuro A., Ramachandran A., Rodriguez J., Barber G. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J Virol. 2009;83:7252–7260. doi: 10.1128/JVI.00153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Childs K., Andrejeva J., Randall R., Goodbourn S. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J Virol. 2009;83:1465–1473. doi: 10.1128/JVI.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rodriguez K., Horvath C. Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: a single arginine distinguishes MDA5 from RIG-I. J Virol. 2013;87:2974–2978. doi: 10.1128/JVI.02843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Horvath C. Silencing STATs: lessons from paramyxovirus interferon evasion. Cytokine Growth Factor Rev. 2004;15:117127. doi: 10.1016/j.cytogfr.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 132.Motz C., Schuhmann K., Kirchhofer A., Moldt M., Witte G., Conzelmann K.-K. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science (New York, NY) 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- 133.Kok K.-H., Lui P.-Y., Ng M-HJ, Siu K.-L., Au S., Jin D.-Y. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe. 2011;9:299–309. doi: 10.1016/j.chom.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 134.Luthra P., Ramanan P., Mire C., Weisend C., Tsuda Y., Yen B. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe. 2013;14:74–84. doi: 10.1016/j.chom.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A., Matrosovich M. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013;87:5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chan R., Chan M., Agnihothram S., Chan L., Kuok D., Fong J. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87:6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Falzarano D., de Wit E., Rasmussen A., Feldmann F., Okumura A., Scott D. Treatment with interferon-(2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou J., Chu H., Li C., Wong B., Cheng Z.-S., Poon V. Active replication of middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G., Suliman T. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Siu K.-L., Yeung M., Kok K.-H., Yuen K.-S., Kew C., Lui P.-Y. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol. 2014;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 141.Siu K.-L., Kok K.-H., Ng M.-H.J., Poon V., Yuen K.-Y., Zheng B.-J. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Freundt E., Yu L., Park E., Lenardo M., Xu X.-N. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kopecky-Bromberg S., Martínez-Sobrido L., Frieman M., Baric R., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fehling S., Lennartz F., Strecker T. Multifunctional nature of the arenavirus RING finger protein Z. Viruses. 2012;4:2973–3011. doi: 10.3390/v4112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang J., Danzy S., Kumar N., Ly H., Liang Y. Biological roles and functional mechanisms of arenavirus Z protein in viral replication. J Virol. 2012;86:9794–9801. doi: 10.1128/JVI.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fan L., Briese T., Lipkin W. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J Virol. 2010;84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bao X., Kolli D., Ren J., Liu T., Garofalo R., Casola A. Human metapneumovirus glycoprotein G disrupts mitochondrial signaling in airway epithelial cells. PLoS One. 2013:8. doi: 10.1371/journal.pone.0062568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bao X., Liu T., Shan Y., Li K., Garofalo R., Casola A. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog. 2008:4. doi: 10.1371/journal.ppat.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lifland A.W., Jung J., Alonas E., Zurla C., Crowe J.E., Santangelo P.J. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J Virol. 2012:86. doi: 10.1128/JVI.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Barik S. Respiratory syncytial virus mechanisms to interfere with type 1 interferons. Curr Top Microbiol Immunol. 2013;372:173–191. doi: 10.1007/978-3-642-38919-1_9. [DOI] [PubMed] [Google Scholar]
- 151.Goswami R., Majumdar T., Dhar J., Chattopadhyay S., Bandyopadhyay S., Verbovetskaya V. Viral degradasome hijacks mitochondria to suppress innate immunity. Cell Res. 2013;23:1025–1042. doi: 10.1038/cr.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li X.-D., Sun L., Seth R., Pineda G., Chen Z. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen Z., Benureau Y., Rijnbrand R., Yi J., Wang T., Warter L. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J Virol. 2007;81:964–976. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brack K., Berk I., Magulski T., Lederer J., Dotzauer A., Vallbracht A. Hepatitis A virus inhibits cellular antiviral defense mechanisms induced by double-stranded RNA. J Virol. 2002;76:11920–11930. doi: 10.1128/JVI.76.23.11920-11930.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fensterl V., Grotheer D., Berk I., Schlemminger S., Vallbracht A., Dotzauer A. Hepatitis A virus suppresses RIG-I-mediated IRF-3 activation to block induction of beta interferon. J Virol. 2005;79:10968–10977. doi: 10.1128/JVI.79.17.10968-10977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang Y., Liang Y., Qu L., Chen Z., Yi M., Li K. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci USA. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kovacsovics M., Martinon F., Micheau O., Bodmer J., Hofmann K., Tschopp J. Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr Biol: CB. 2002;12:838–843. doi: 10.1016/s0960-9822(02)00842-4. [DOI] [PubMed] [Google Scholar]
- 158.Barral P., Morrison J., Drahos J., Gupta P., Sarkar D., Fisher P. MDA-5 is cleaved in poliovirus-infected cells. J Virol. 2007;81:3677–3684. doi: 10.1128/JVI.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Barral P., Sarkar D., Fisher P., Racaniello V. RIG-I is cleaved during picornavirus infection. Virology. 2009;391:171–176. doi: 10.1016/j.virol.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Drahos J., Racaniello V. Cleavage of IPS-1 in cells infected with human rhinovirus. J Virol. 2009;83:11581–11587. doi: 10.1128/JVI.01490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang B., Xi X., Lei X., Zhang X., Cui S., Wang J. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog. 2013:9. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kuo R.-L., Kao L.-T., Lin S.-J., Wang R., Shih S.-R. MDA5 plays a crucial role in enterovirus 71 RNA-mediated IRF3 activation. PLoS One. 2013:8. doi: 10.1371/journal.pone.0063431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Feng Q., Langereis M., Lork M., Nguyen M., Hato S., Lanke K. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol. 2014;88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Davis M.E., Wang M.K., Rennick L.J., Full F., Mesman A.W., Gringhuis S.I. Antagonism of the Phosphatase PP1 by the Measles Virus V Protein Is Required for Innate Immune Escape of MDA5. Cell Host & Microbe. 2014 doi: 10.1015/j.chom.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]