Abstract
Background
The epidemiology of respiratory tract infections (RTIs) in a daycare cohort has not been explored using molecular techniques.
Objectives
(1) Determine the overall incidence of RTIs in a daycare cohort using real-time reverse transcriptase polymerase chain reaction (RT-PCR). (2) Determine the relative incidence and impact of specific respiratory viruses, and characterize and compare clinical features associated with these pathogens.
Study design
In this prospective cohort study conducted from February 2006 to April 2008, nasal swabs were obtained from symptomatic children ages 0–30 months enrolled in fulltime daycare.
RT-PCR was performed to detect respiratory syncytial virus (RSV), human metapneumovirus (MPV), influenza (Flu) viruses A and B, parainfluenza (PIV), adenovirus (AdV), human coronaviruses (CoV) and rhinovirus (RhV). Symptom diaries were completed for each illness.
Results
We followed 119 children (mean age 10 months; range 2–24 months) for 115 child years. The mean annual incidence of RTI per child was 4.2 the first year and 1.2 the second year of the study. At least 1 virus was identified in 67% RTIs. Co-infections were common (27% RTIs), with RhV, CoV, and AdV the most common co-pathogens. PIV was identified in 12% of RTIs with a high incidence of PIV4. The viruses with the greatest impact on our population were RSV, RhV and AdV.
Conclusions
Using molecular techniques, viruses were identified in approximately twice as many RTIs as previously reported in a daycare cohort. Infections with newly identified viruses, such as HMPV and CoV subtypes were less frequent and severe than infections with RSV, AdV and RhV.
Abbreviations: RTIs, respiratory tract infections; RT-PCR, real-time reverse transcriptase polymerase chain reaction; HMPV, human metapneumovirus; RhV, rhinovirus; HCoV, human coronavirus; PIV, parainfluenza viruses; AdV, adenovirus; flu, influenza virus
Keywords: Respiratory infection, Viral, Daycare, Epidemiology, PCR
1. Background
Currently, 61% of United States children ages 0–6 years attend daycare, and 36.1% attend center based care, including 19.6% of children ages 0–2 years.1 Respiratory tract infections (RTIs) are a major cause of illness in daycare attendees.2, 3, 4 RTIs occur more often, and cause more hospitalizations, in children attending daycare centers compared to home care.4, 5, 6, 7
Given the burden of RTIs in daycare centers, effort should be directed towards the pathogens causing the greatest impact. Few longitudinal studies have evaluated the epidemiology of viral RTIs in daycare, and none have used modern molecular diagnostic techniques. A previous epidemiological daycare study spanning 16 years from 1966 to 1982 identified a viral agent in only 31% of illness events, using standard methods.2 Newly identified viruses, such as human metapneumovirus (HMPV), the newly identified human coronaviruses, and difficult to culture viruses such as rhinovirus (RhV) and parainfluenza virus (PIV) type 4 have not been similarly examined in the daycare setting.
2. Objectives
We devised a prospective cohort study of RTIs in young children attending daycare. A sensitive real-time reverse transcription polymerase chain reaction (RT-PCR) panel was used to detect 14 viruses during acute RTIs in our cohort. Our objective was to use this molecular technique to measure the relative impact of these viruses in our cohort.
3. Study design
We recruited children attending two large daycare centers located on Fort Lewis, WA. Children who attended for at least 20 h/week were eligible. Parents provided informed consent, and the Madigan Army Medical Center and Seattle Children's Hospital Institutional Review Boards approved the study. Between 1 February 2006 and 28 April 2008, all eligible children between 0 and 30 months were recruited upon enrollment into daycare. Children who discontinued fulltime daycare during the study period were removed from the study.
Subjects with onset of at least 2 of 5 symptoms including cough, rhinorrhea, wheezing, fever (tympanic or rectal temperature of ≥100.4, or axillary of ≥99.4), and nasal congestion underwent a posterior nasal swab for PCR, generally within 48 h of symptom onset. The study nurse was contacted if any of these symptoms were noted by the child's parent or daycare. A daily symptom diary was completed by the child's parent for 10 days following illness onset. The nurse also conducted phone follow-ups. One of 2 study physicians documented medical visits related to the illnesses using a standardized form.
Respiratory secretions were sampled by inserting nylon flocked swabs (Copan Diagnostics, Corona, CA) into the posterior nasopharynx, rotating 180°, and rinsing in 0.5 mL of lysis buffer.8 Total nucleic acid was extracted from 200 μL of the buffer specimen by adding 200 μL of Tris–EDTA, pH 8.0 buffer and 200 μL of lysis buffer containing dithiothreitol and external control and processed as previously described.9 Specimens were tested for HMPV, respiratory syncytial virus (RSV), PIV1–4, influenza viruses (Flu A and B), RhV, human coronaviruses (HCoV) Group 1 (229E and NL63) and Group 2 (OC43 and HKU1), and adenovirus (AdV) by separate RT-PCR assays.8, 9, 10, 11 Although the coronavirus assay detected all 4 HCoV groups, it did not distinguish among the 4 types. Swabs were repeated at 7–10 day intervals until resolution of symptoms. Enrollment swabs were obtained on all subjects enrolled after 1 May 2006. If the subjects were ill on enrollment, diaries were obtained and swab results were categorized as an illness event.
3.1. Statistical methods
Data was analyzed using STATA 10.1 (College Station, Texas). Demographics were analyzed using descriptive analysis. Frequency, incidence and jackknife 95% confidence interval were calculated overall and individually for age and gender subgroups. Cox regression models were used to determine independent risk factors for illness overall and for virus-specific infections. An indicator term was included in the Cox regression models to adjust for respiratory season and robust standard errors were used to account for multiple illnesses per child. Differences in the presence and duration of symptoms up to 10 days were evaluated using Generalized Estimating Equations with a robust estimator, accounting for multiple illnesses in individual children as described previously.12 The prevalence of each respiratory virus at enrollment of asymptomatic children was compared to the prevalence at each child's first RTI using McNemar's test. A two-sided p value of <0.05 was considered significant. In analyzing viral loads, the maximal viral load for each discrete illness episode was utilized.
4. Results
We followed 119 children from 1 February 2006 to 28 April 2008 for a total of 115.3 child years. On a monthly basis, the percentage of children enrolled averaged 56% (range 35–71%). Children were followed for an average of 12 months (range 11 days to 2.2 years). The mean age at enrollment was 10.4 months (range 1.6–24.9 months), with 56 (47%) females. Racial background included 57 (48%) Caucasians, 38 (32%) African Americans, and 4 (4%) Asians. Siblings were present in 67 (56%) households. Single parents headed 32 (27%) of households. Children spent a mean of 44 h/week (range 20–60) in daycare.
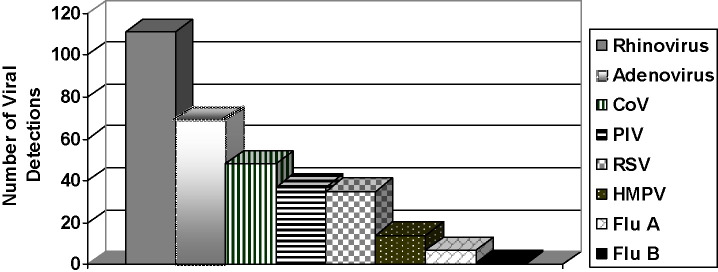
We evaluated 318 RTIs, for a mean annualized incidence of 2.8 per child (95%C.I. 2.3, 3.3); children had RTI symptoms an average of 11% of the time. The incidence of infection was lower after a year in daycare, with a mean annual incidence of 4.2 RTI/child (95%C.I. 3.6, 5.0) in the first year of study participation and 1.2 RTI/child (95%C.I. 0.9, 1.6) after one year (p < 0.001). On univariate hazard analysis, enrollment at under 12 months of age (Table 1 ) and Hispanic descent were the only demographic factors significantly associated with RTI (HR 1.7, p = 0.001 and HR 1.4, p = 0.03, respectively). Male sex was significantly associated with RhV infections (HR 1.8, p = 0.01), and this association was not dependent on age.We detected viruses in 213/318 (67%) RTIs (Fig. 1 ). Table 2 displays clinical characteristics associated with identified pathogens. RhV was detected more than any other virus (111/318, 35%, p < .01). Only 7 Flu A and 1 Flu B infections were identified. HMPV was also detected infrequently (4% of RTIs). The viruses with the overall largest impacts on our cohort were RSV, RhV and AdV. These 3 viruses accounted for 215 (67%) of single viral infections.
Table 1.
Age at enrollment as a risk factor for infection in the most commonly identified viruses.
| n | 0–6 months at enrollment | >6–12 months at enrollment |
12–24 months at enrollment |
||
|---|---|---|---|---|---|
| Incidence/child year (95%C.I.) | Incidence/child year (95%C.I.) | Hazard ratioa; p value | Incidence/child year (95%C.I.) | Hazard ratio; p value | |
| All RTIs (318) | 3.5 (2.8, 4.3) | 3.5 (2.7, 4.4) | 1.00; p = 0.99 | 1.8 (1.4, 2.3) | 0.57; p = 0.01 |
| RhV (111) | 1.3 (0.9, 1.8) | 1.3 (1.0, 1.8) | 1.02; p = 0.92 | 0.5 (0.3, 1.0) | 0.43; p = 0.01 |
| AdV (69) | 0.9 (.0.6, 1.6) | 0.8 (4.6, 1.3) | 0.83; p = 0.55 | 2.5 (0.1, 0.5) | 0.30; p = 0.001 |
| CoV (48) | 0.6 (0.4, 1.0) | 0.5 (0.3, 1.1) | 0.90; p = 0.79 | 0.2 (0.1, 0.5) | 0.40; p = 0.04 |
| PIV (37) | 0.3 (0.2, 0.6) | 0.6 (0.3, 1.0) | 1.80; p = 0.15 | 0.2 (0.1, 0.4) | 0.61; p = 0.25 |
| RSV (35) | 0.5 (0.3, 0.8) | 0.3 (0.2, 0.6) | 0.76; p = 0.44 | 0.2 (0.09, 0.3) | 0.46; p = 0.04 |
| MPV (14) | 0.2 (0.05, 0.8) | 0.2 (0.07, 0.4) | 1.02; p = 0.98 | 0.06 (0.02, 0.3) | 0.46; p = 0.33 |
RTIs, respiratory tract infections; RhV, rhinovirus; AdV, adenovirus; CoV, human coronavirus; PIV, parainfluenza viruses; RSV, respiratory syncytial virus; MPV, human metapneumovirus.
Hazard ratios are calculated in comparison to the 0–6 months age group.
Fig. 1.
Viral identification.
Table 2.
Characteristics of 318 respiratory tract infections.
| Characteristic | Single | Coinf | None | RhV | Adv | CoV | PIV | RSV | Flu A | HMPV |
|---|---|---|---|---|---|---|---|---|---|---|
| Total (%) RTIs | 126 (40) | 87 (27) | 105 (33) | 111 (35) | 69 (22) | 48 (15) | 37 (12) | 35 (11) | 7 (2) | 14 (4) |
| Mean age (months) | 13.2 | 11.5 | 12.3 | 12.4 | 10.4 | 11.8 | 13.1 | 12.4 | 17.9 | 12.0 |
| n (%) male | 65 (52) | 50 (57) | 56 (53) | 69 (62) | 40 (58) | 29 (60) | 14 (38) | 17 (49) | 2 (29) | 8 (57) |
| Single infections only | ||||||||||
| Number (%) RTIs as single virus identified | 126 (100) | N/A | N/A | 50 (16) | 17 (5) | 14 (4) | 14 (4) | 20 (6) | 5 (2) | 5 (2) |
| Mean (range) of peak viral load (log copies/mL) | N/A | 5.0 (2.9, 7.9) | 6.5 (4.5, 9.2) | 6.2 (3.5, 8.5) | 7.9 (5.4, 9.4) | 7.1 (6.1, 8.1) | 6.9 (5.5, 9.4) | |||
| Mean duration (days) | 8.2 | 8.7 | 7.4a | 8.2 | 9.1 | 7.9 | 8.4 | 8.6 | 7.8 | 6.2 |
| n (%) fever | 67 (53) | 39 (46) | 44 (42) | 25 (50) | 8 (47) | 4 (29) | 6 (43) | 16 (80)b | 5 (100)c | 3 (60) |
| n (%) wheezing | 43 (34) | 34 (39) | 34 (32) | 17 (34) | 6 (35) | 3 (21) | 4 (29) | 10 (50)d | 0 (0) | 3 (60) |
| Mean days DC missed | 1.1 | 1.2 | 1.1 | 0.7b | 1.1 | 1.7 | 0.9 | 1.4 | 3 | 1 |
| Mean days work missed | 0.9 | 1.2 | 1.1 | 0.7d | 0.7 | 0.9 | 0.9 | 1.4 | 2.8 | 1 |
| n (%) HCP visits | 62 (49) | 44 (51) | 41 (39) | 18 (36) | 8 (47) | 5 (36) | 9 (64) | 16 (80)a | 2 (40) | 3 (60) |
| Co-infections only | ||||||||||
| Number (%) of RTIs with virus as co-infection | 61 (19) | 52 (16) | 34 (11) | 23 (7) | 15 (5) | 2 (0.6) | 9 (0.3) | |||
| Mean (range) of peak viral load, log copies/mL | N/A | 4.6 (2.5, 7.6) | 6.7 (2.8, 10.1) | 6.2 (2.5, 9.8) | 7.3 (3.7, 9.0) | 6.1 (3.6, 8.6) | 6.3 (4.0, 8.4) | |||
| Mean duration (days) | 8.6c | 8.6 | 8.8c | 8.3 | 9.1c | 9 | 9.3e | |||
| n (%) fever | 29 (48) | 22 (44) | 14 (41) | 10 (43) | 8 (53) | 1 (50) | 4 (44) | |||
| n (%) wheezing | 22 (36) | 19 (37) | 14 (41) | 8 (35) | 10 (67)f | 1 (50) | 4 (44) | |||
| Mean days DC missed | 1.2 | 1.2 | 1.2 | 1.3 | 1.8 | 0.5 | 1.8 | |||
| Mean days work missed | 1.2 | 1.1 | 1.1 | 1.3 | 1.9 | 0.5 | 1.1 | |||
| n (%) HCP visits | 30 (49) | 23 (44) | 16 (47) | 16 (70)b | 10 (67)d | 0 (0) | 5 (56) | |||
RhV, rhinovirus; AdV, adenovirus; CoV, human coronavirus; PIV, parainfluenza viruses; RSV, respiratory syncytial virus; flu A, influenza virus A; MPV, human metapneumovirus; RTIs, respiratory tract infections.
p = 0.001.
p = 0.002.
p = 0.02.
p = 0.01.
p = 0.007.
p = 0.009.
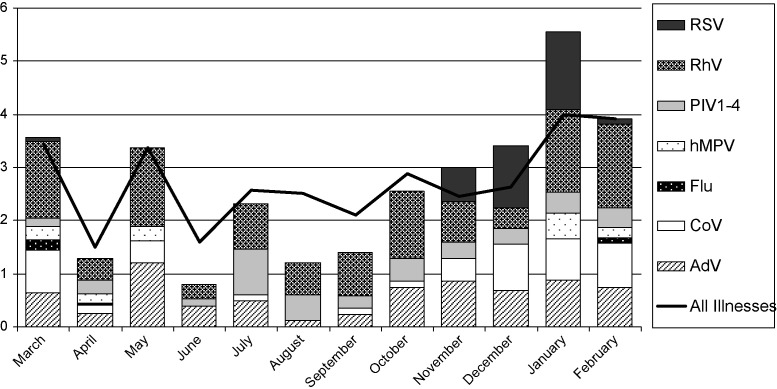
Seasonal distribution of detected viruses is displayed in Fig. 2 . Viruses were most often detected in the winter months. Influenza (winter), RSV (fall/winter) and HMPV (winter/spring) had clear seasonal patterns with some overlap. HCoV and ADV were detected less frequently in the summer months. The other viruses did not have a seasonal pattern of detection.
Fig. 2.
Seasonal pattern of individual viral detection consolidated over 2 years.
Single viral infections due to RSV were more likely to include fever (p = .002) than single viral illnesses without RSV, and the rate of fever (80%) in RTIs with RSV as the only identified virus was higher than the rate of fever in RTIs with all other viruses except influenza A. Wheezing was also more likely to be noted with single RSV infections (p = .01). Of all the study viruses, RSV was the only single infection significantly associated with a visit to the health care provider (p = .001). AdV had the longest mean duration of symptoms of any single viral infection (9.1 days). Even though the relative severity of RhV infection was not significant, RhV caused the largest burden of disease compared to the other viruses. RhV was the virus detected in the highest total number of both febrile and wheezing episodes, caused the highest total number of infections, and the highest total number of visits to a health care provider. Unlike RSV, RhV was detectable year round.
Co-infections with multiple respiratory viruses were common, occurring in 87/318 (27%) RTIs, with RhV, CoV and AdV being the most common co-pathogens. Co-infections with RhV, CoV, RSV and HMPV each resulted in significantly longer duration of illness than RTI's with other pathogens (Table 2). Severity of illness was not worse with co-infections. However, co-infections with PIV or RSV both resulted in a significantly higher number of visits to health care providers than PIV- or RSV-negative illnesses, respectively.
Of 37 parainfluenza virus infections, 8 (22%) were due to PIV4, 25 (67.6%) were due to PIV3, 3 (0.1%) to PIV1, and 1 (0.03%) to PIV2. When compared to the other PIV subtypes, PIV4 infections had the following characteristics: mean duration of symptoms lasted 8.8 days versus 8.2 days; percentage with fever was 50% versus 41%; the mean number of days missed from daycare was 1.5 versus 1.1; and the percent of infections requiring a visit to a health care provider was 88% versus 62%. Only one of the PIV4 infections was associated with croup.
Forty-five children were asymptomatic on enrollment, with viruses identified in 19 (42%). The most commonly isolated viruses were RhV (n = 11; 24%), AdV (n = 5; 11%) and CoV (n = 3; 7%). Neither RSV nor HMPV was identified. Twenty-five children with asymptomatic enrollment swabs had subsequent illness swabs available for comparison at a median of 24 days after asymptomatic enrollment (Table 3 ), resulting in a significantly higher isolation of any virus, RSV, and PIV. Isolation of RhV from illness swabs also approached significance in comparison to the previous asymptomatic swabs.
Table 3.
Case crossover analysis of asymptomatic viral detection compared to the first subsequent illness in the same subject.
| Virus | Virus-specific prevalence at asymptomatic enrollment, n = 25 n (%) |
Virus-specific prevalence at first study illness, n = 25 n (%) |
p valuea |
|---|---|---|---|
| RhV | 5 (20) | 11 (44) | 0.06 |
| AdV | 5 (20) | 6 (24) | 0.71 |
| CoV | 1 (4) | 3 (12) | 0.32 |
| PIV (types 1–4) | 1 (4) | 5 (20) | 0.046 |
| RSV | 0 (0) | 4 (16) | 0.046 |
| Flu A | 1 (4) | 0 (0) | 0.32 |
| HMPV | 0 (0) | 0 (0) | n.s. |
| Any positive | 11 (44) | 19 (76) | 0.02 |
RhV, rhinovirus; AdV, adenovirus; CoV, human coronavirus; PIV, parainfluenza viruses; RSV, respiratory syncytial virus; flu A, influenza virus A; HMPV, human metapneumovirus.
p value is for matched comparisons of prevalence using McNemar's test.
Viral load information for illnesses is presented in Table 2. We compared viral loads present in single versus co-infections for individual viruses. We did not detect significant differences, with the exception of CoV infections. After accounting for multiple CoV illnesses experienced by individual children, we found that samples from illnesses with only CoV detected had significantly higher viral loads than samples from illnesses with CoV present as part of a co-infection (increase of 0.9 log copies/mL, p < 0.01, GEE).
5. Discussion
Our study contributes new information to the epidemiology of RTIs in daycare by evaluating the importance of HMPV, PIV4 and the newly identified HCoV subtypes, and is the first prospective daycare study to apply molecular techniques. We documented a similar incidence of RTIs as in a daycare study conducted by Denny et al. over 25 years ago.2 Using molecular techniques, we were able to identify pathogens nearly twice as often. Denny et al. reported that RSV, PIV3, AdV and RhV were found most frequently in their cohort, although the relative impact and incidence of each of these viruses is not described. As in our study, influenza viruses were isolated infrequently.
Another recent study using PCR techniques noted that HMPV and HCoV infections were detected in RTIs more frequently in daycare attendees than home care children, suggesting that these viruses may be a significant cause of disease in the daycare population.13 By contrast, HMPV and HCoV were not significant pathogens in our study as measured by clinical and social impact factors. The majority of disease burden in our cohort was due to infection with RhV, AdV and RSV.
In our cohort, PIV4 caused a greater proportion of PIV infections than expected. The epidemiology of PIV4 is poorly defined, as it is difficult to detect by standard techniques. Lindquist et al investigated the impact of PIV4 as a cause of respiratory illness in children using culture.14 In their study, PIV4 accounted for only 3.5% of PIV isolates, compared to 22% in our study. A larger sample size is needed to further characterize disease manifestations associated with PIV4.
The prominent role of RhV in our study correlates with recent data in other settings. With the use of PCR, RhV is increasingly identified as a more significant disease burden than previously believed in healthy young children.13, 15, 16, 17, 18 As in our study, Van der Zalm et al's prospective study of respiratory tract pathogens during the first year of life identified RhV to cause the greatest impact, though not the most severe individual illness.18 Our findings also add to growing evidence that RhV is an important contributor to severe respiratory disease in healthy young children. Several recent publications have suggested that RhV is a significant trigger of asthma and cause of hospitalization, comparable to RSV.19, 20, 21
A potential limitation of our study is the attribution of acute disease to asymptomatic shedding of a virus. We detected a virus as often in asymptomptic enrollment swabs as in illness swabs, most notably RhV, HCoV and AdV. RhV was detected in 20% of well infants by Van Benten et al.,15 and in 22% of asymptomatic individuals enrolled in vaccine trials.22 The results of our asymptomatic swabs support the likelihood that RSV, PIV and HMPV are the cause of illness when identified, but more data is needed on the use of PCR for the identification of HCoV, AdV and RhV as the definitive cause of a particular illness.
Co-infections were common, occurring in 27% of RTIs. Similar rates were found in other PCR based studies.13, 19, 23 Evidence about the severity of co-infections compared to single viral infections is conflicting. In our cohort, co-infections were not more severe than single infections. Drews et al. concluded that dual viral RTIS had higher hospitalization rates than single infections.24 However, in their study, 58% of the patients with dual RTIs had chronic lung disease. Aberle et al. noted that in hospitalized children, RSV co-infections correlated with increased severity, but not non-RSV co-infections.23 The severity of co-infection may depend on the pathogens involved. We found that co-infections with CoV, for example, had lower viral loads of CoV than single infections with CoV, suggesting that the some co-infections might represent detection of shedding of CoV rather than a true co-infection. The most severe infection in our cohort was a co-infection, occurring in a child with a triple infection including RSV.
Using molecular techniques, we determined that RhV, RSV and AdV had the greatest impact on young children attending daycare. Further investigation is needed on asymptomatic shedding to better correlate illness with the presence of virus.
Conflicts of interest
JE has received research support from Sanofi Pasteur. EM has received research support from Vioguard, LLC. None of the other authors of this manuscript have any conflicts of interest to report.
Acknowledgements
This work was supported by Grant Number SYN104-05D from the MedImmune, Inc. Investigator Initiated Research Program (PI: Englund).
References
- 1.The Federal Interagency Forum on Child and Family Statistics . 2007. America's children: key national indicators of well-being. Available at: http://childstats.gov/americaschildren/tables.asp, accessed November 1, 2008. Accessibility verified October 22, 2009. [Google Scholar]
- 2.Denny F.M., Collier A.M., Henderson F.W. Acute respiratory infections in daycare. Rev Infect Dis. 1986;8:527–532. doi: 10.1093/clinids/8.4.524. [DOI] [PubMed] [Google Scholar]
- 3.Pacini D.L., Collier A.M., Henderson F.W. Adenovirus infections and respiratory illness in children in group day care. J Infect Dis. 1987;156:920–927. doi: 10.1093/infdis/156.6.920. [DOI] [PubMed] [Google Scholar]
- 4.Dales R.E., Cakmak S., Brand K., Judek S. Respiratory illness in children attending daycare. Pediatr Pulmonol. 2004;38:64–69. doi: 10.1002/ppul.20034. [DOI] [PubMed] [Google Scholar]
- 5.National Institute of Child Health and Human Development Early Child Care Research Network Child care and common communicable illnesses. Arch Pediatr Adolesc Med. 2001;155:481–488. doi: 10.1001/archpedi.155.4.481. [DOI] [PubMed] [Google Scholar]
- 6.Lu N., Shi S.L., Baker S.L., Glover S.H., Sanders J.M. Child day care risks of common infectious diseases revisted. Child: Care, Health Dev. 2004;30:361–368. doi: 10.1111/j.1365-2214.2004.00411.x. [DOI] [PubMed] [Google Scholar]
- 7.Kamper-Jorgensen M., Wohlfart J., Simonsen J., Gronbaek M., Benn C.S. Population-based study of the impact of childcare attendance on hospitalizations for acute respiratory infections. Pediatrics. 2006;118:1439–1446. doi: 10.1542/peds.2006-0373. [DOI] [PubMed] [Google Scholar]
- 8.Kuypers J., Wright N., Morrow R. Evaluation of quantitative and type-specific real-time RT-PCR assays for detection of respiratory syncytial virus in respiratory specimens from children. J Clin Virol. 2004;31:123–129. doi: 10.1016/j.jcv.2004.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuypers J., Wright N., Corey L., Morrow R. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J Clin Virol. 2005;33:299–305. doi: 10.1016/j.jcv.2004.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuypers J., Wright N., Ferrenberg J., Huang M., Cent A., Corey L. Comparison of real-time polymerase chain reaction with fluorescent antibody assay for detection of respiratory virus infections in children. J Clin Microbiol. 2006;44:2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuypers J., Martin E., Heugel J., Wright N., Morrow R., Englund J. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. 2007;119:e70–e76. doi: 10.1542/peds.2006-1406. [DOI] [PubMed] [Google Scholar]
- 12.Martin E.T., Fairchok M., Kuyper J. Frequent and prolonged shedding of bocavirus in young children attending daycare. J Infect Dis. 2010;201:1625–1632. doi: 10.1086/652405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert S.B., Allen K.M., Druce J.D., Birch C.J., MacKay I.M., Carlin J.B. Community epidemiology of human metapneumovirus, human coranoavirus NL63, and other respiratory viruses in healthy preschool-aged children using parent collected specimens. Pediatrics. 2007;120:e929–e937. doi: 10.1542/peds.2006-3703. [DOI] [PubMed] [Google Scholar]
- 14.Lindquist S.W., Darnule A., Istas A., Demmler G.J. Parainfluenza virus type 4 infection in pediatric patients. Pediatr Inf Dis J. 1997;16:34–38. doi: 10.1097/00006454-199701000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Van Benten I., Koopman L., Niesters B., Hop W., Van Middelkoop B., De Waal L. Predominance of rhinovirus in the nose of symptomatic and asymptomatic infants. Pediatr Allergy Immunol. 2003;14:363–370. doi: 10.1034/j.1399-3038.2003.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legg J.P., Warner J.A., Johnston S.L., Warner J.O. Frequency of detection of picornaviruses and seven other respiratory pathogens in infants. Pediatr Infect Dis J. 2005;24:611–616. doi: 10.1097/01.inf.0000168747.94999.aa. [DOI] [PubMed] [Google Scholar]
- 17.Kusel M.M., deKlerk N.H., Holt P.G., Kebadze T., Johnston S.L., Sly P.D. Role of rhinovirus in acute upper and lower respiratory tract illness in the first year of life: a cohort study. Pediatr Infect Dis J. 2006;25:680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- 18.Van der Zalm M.M., Uiterwaal C.S.P.M., Wilbrink B., de Jong B.M., Verhiej T.J.M., Kimpen J.L.L. Respiratory pathogens in respiratory tract illnesses during the first year of life: a birth cohort study. Pediatr Infect Dis J. 2009;28:472–476. doi: 10.1097/inf.0b013e318195e26e. [DOI] [PubMed] [Google Scholar]
- 19.Calvo C., Garcia-Garcia M.L., Blanco C., Pozo F., Flecha I.C., Perez-Brena P. Role of rhinovirus in hospitalized infants with respiratory tract infections in Spain. Pediatr Infect Dis J. 2007;26:904–908. doi: 10.1097/INF.0b013e31812e52e6. [DOI] [PubMed] [Google Scholar]
- 20.Chung J.Y., Han Th., Kim Sw., Kim C.K., Hwang E.S. Detection of viruses identified recently in children with acute wheezing. J Med Virol. 2007;79:1238–1243. doi: 10.1002/jmv.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sears M.R., Johnston N.W. Understanding the September asthma epidemic. J Allergy Clin Immunol. 2007;120:526–529. doi: 10.1016/j.jaci.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright P.F., Deatly A.M., Karron R.A., Belshe R.B., Shi J.R., Gruber W.C. Comparison of results of detection of rhinovirus by PCR and viral culture in human nasal wash specimens from subjects with and without clinical symptoms of respiratory illness. J Clin Microbiol. 2007;45:2126–2129. doi: 10.1128/JCM.02553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aberle J.H., Aberle J.S., Pracher E., Hutter H.P., Kundi M., Popow-Kraup T. Single versus dual respiratory virus infections in hospitalized infants: impact on clinical course of disease and interferon-gamma response. Pediatr Infect Dis J. 2005;24:605–610. doi: 10.1097/01.inf.0000168741.59747.2d. [DOI] [PubMed] [Google Scholar]
- 24.Drews A.L., Atmar R.L., Glezen W.P., Baxter B.D., Pedra P.A., Greenberg S.B. Dual respiratory virus infections. Clin Infect Dis. 1997;25:1421–1429. doi: 10.1086/516137. [DOI] [PMC free article] [PubMed] [Google Scholar]