Highlights
-
•
50 event-based Internet biosurveillance systems were identified.
-
•
There is an increase in internet data source diversity in newly identified systems.
-
•
Event-based Internet biosurveillance may supplement traditional systems for a more comprehensive estimate of disease burden.
Keywords: Biosurveillance, Disease surveillance, Public health
Abstract
Background
Internet access and usage has changed how people seek and report health information. Meanwhile,infectious diseases continue to threaten humanity. The analysis of Big Data, or vast digital data, presents an opportunity to improve disease surveillance and epidemic intelligence. Epidemic intelligence contains two components: indicator based and event-based. A relatively new surveillance type has emerged called event-based Internet biosurveillance systems. These systems use information on events impacting health from Internet sources, such as social media or news aggregates. These systems circumvent the limitations of traditional reporting systems by being inexpensive, transparent, and flexible. Yet, innovations and the functionality of these systems can change rapidly.
Aim
To update the current state of knowledge on event-based Internet biosurveillance systems by identifying all systems, including current functionality, with hopes to aid decision makers with whether to incorporate new methods into comprehensive programmes of surveillance.
Methods
A systematic review was performed through PubMed, Scopus, and Google Scholar databases, while also including grey literature and other publication types.
Results
50 event-based Internet systems were identified, including an extraction of 15 attributes for each system, described in 99 articles. Each system uses different innovative technology and data sources to gather data, process, and disseminate data to detect infectious disease outbreaks.
Conclusions
The review emphasises the importance of using both formal and informal sources for timely and accurate infectious disease outbreak surveillance, cataloguing all event-based Internet biosurveillance systems. By doing so, future researchers will be able to use this review as a library for referencing systems, with hopes of learning, building, and expanding Internet-based surveillance systems. Event-based Internet biosurveillance should act as an extension of traditional systems, to be utilised as an additional, supplemental data source to have a more comprehensive estimate of disease burden.
1. Introduction
The large-scale spread of infectious diseases has a significant impact on individuals and society [1]. Data systems are important to producing an efficient approach to prevent, detect, respond to, and manage infectious disease outbreaks of plants, animals, and humans [3], [4]. Due to the nature of epidemics, there is a vital need for timely data collection and processing [5]. Large quantities of infectious disease data are continuously compiled and analysed by various laboratories, health providers, and government agencies at local, national, and international levels with increasing complexity [5]. In practice, infectious disease data collection and analysis is complicated and encompasses a multi-stage process with several stakeholders across many organisational boundaries [5].
The main objective of infectious disease surveillance is to identify changes in incidence, either in the form of an acute outbreak or a change in long-term trends [6]. Epidemic intelligence includes all activities related to prompt identification of potential health hazards and their verification, assessment and investigation to enable public health control recommendations [7]. Epidemic intelligence incorporates two components: an indicator-based component and an event-based component [7]. The indicator-based component refers to structured (or formal) data collected through routine surveillance systems, such as the number or rates of cases based on standard case definitions, and the computation of indicators upon which abnormal disease patterns to investigate are detected [7], [8]. The goal of indicator-based surveillance is to find increased numbers or clusters at a specific time, period, and/or location that may indicate a threat [9]. Statistical methods set against thresholds of increased cases or clusters are essential to determining potential health effects [9]. In some cases, non-specific syndromes are monitored as markers for specific diseases, termed ‘syndromic surveillance’.
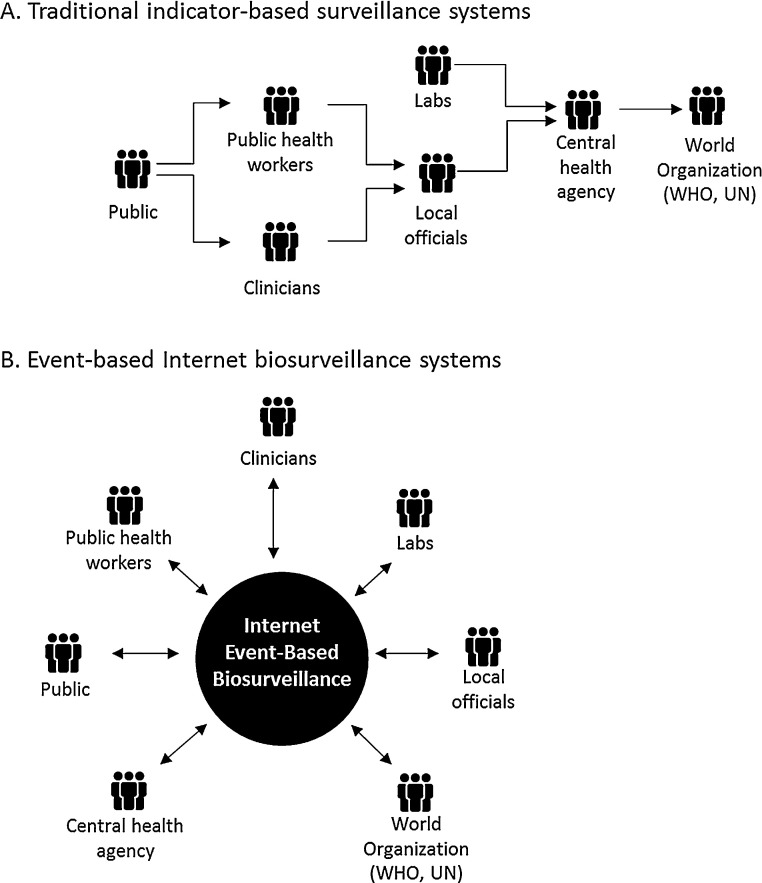
Traditional indicator-based surveillance systems are based on the obligatory reporting of certain diagnosed diseases to a central health agency. Most of these systems depend on data from physician visits and laboratory confirmations, which can be costly and require a formal public health structure (see Fig. 1 ) [10]. Though this data is typically very accurate; data gathering can be slow [10], [11]. Substantial lags, sometimes weeks or months, between an event and its notification are common; a consequence of late or failed reporting and the hierarchical structure of these systems [11].
Fig. 1.
Traditional reporting versus new systems (i.e. Internet-based) and technologies.
The ‘event-based component' of epidemic intelligence refers to unstructured data gathered and collected from sources of intelligence of virtually any nature [12]. Thereby, the detection of public health events is based on the capture of ad-hoc unstructured reports issued by formal or informal sources [12]. Rather than relying on formal official sources, information is received directly from the witnesses of real-time events or indirectly from reports sent through different communication channels, such as social media or established alert systems, and information channels, such as news, public health networks, and nongovernmental organisations [12].
1.1. Big data and digital infectious disease surveillance
The availability of health-related information on the Internet has changed how people seek information about health [11]. The United States alone generates eight million search queries for health-related information daily [11]. The analysis of digital data, such as Google search queries, has been used to monitor communicable and non-communicable diseases, as well as mental health, illegal drug use, health policy impact, and behaviours with potential health implications [11]. In addition, social media is an easily approachable, highly cost-effective and interoperable system, that provides real-time online data with high geographical resolution that can be systematically mined, aggregated and analysed to inform public health agents [13].
Event-based Internet surveillance, also known as digital surveillance, could improve both the sensitivity and timeliness of detection of health events [8]. Event-based Internet biosurveillance systems are systems that use information on events impacting human health or the economy from Internet sources, instantaneously incorporating diverse streams of data [14]. They contain the event-based component described above, but with the Internet as the means for data sources, processes, and dissemination and analysis of health information stored digitally. The data reported is aggregated and visualised in real-time, which enables immediate feedback to the users, the public, and officials [15]. Several surveillance systems use non-structured, event-based, digital data, such as the Global Public Health Intelligence Health Network (GPHIN), which detected severe acute respiratory syndrome (SARS) more than two months before the first publications by the World Health Organization (WHO) [11].
Event-based surveillance systems either can broadly be categorised as news aggregators, automatic systems, or moderated systems [9]. News aggregates collect articles and news from sources, aggregates them, either by location or topic, and filters them. The result is provided as an Rich Site Summary (RSS) feed. Automatic systems further advance systems like news aggregators by adding a series of steps of analysis [9]. Non-moderated systems can search the web and display new articles without time delay in an unbiased manner than moderated systems [16]. Often, the data is not structured or collated, and therefore epidemiologists must spend more time and energy determining their relevance to a specific situation of interest [9]. Moderated systems rely on some component of human input or analysis, either solely, or after data is first processed automatically [9]. Moderated systems offer a chance of screening for relevance by public health practitioners before it is disseminated [9]. Thereby, moderated systems might show less irrelevant news items, fewer false positives than the non-moderated systems, but are subject to moderator bias [16]. Systems without human moderation often focus on data sources that already have been validated and, therefore, do not help aid those interested in the early warning and alert potential for unknown or new outbreaks or diseases [9].
Until now, a comprehensive systematic literature review on event-based Internet biosurveillance surveillance systems, including unevaluated systems, participatory surveillance systems, social media, and mobile applications, has not been completed. A summary of previous studies can be found in Table 1 . Therefore, the purpose of this study is to systematically review and update the current state of knowledge on event-based Internet biosurveillance systems by identifying all of these systems, including current functionality, with hopes to aid decision makers with whether to incorporate new methods into their existing surveillance programmes and provide researchers a catalogue of systems.
Table 1.
Previous Systematic Reviews.
| Author | Years Covered | Purpose | Results |
|---|---|---|---|
| Bravata et al. [24] | 1985–2002 | Critically evaluate the potential utility of existing surveillance systems for illnesses and syndromes related to bioterrorism only.34 | Included 115 systems, mostly non-Internet based, that collect surveillance reports, including nine syndromic surveillance systems, 20 systems collecting bioterrorism detector data, 13 systems collecting influenza-related data, and 23 systems collecting laboratory and antimicrobial resistance data.34 Only three systems have had both sensitivity and specificity evaluated.34 |
| Vrbova et al. [18] | 1992–2006 | To find public health surveillance initiatives for emerging zoonoses and what criteria has been used to evaluate these systems.35 | Revealed that out of 221 systems identified, 17 systems were evaluated.35 |
| Velesco et al .[9] | 1990–early 2011 | To review event-based surveillance systems to 2011 and uncover which have been evaluated and which are being utilised by national surveillance systems. [21] | 13 event-based systems were identified, and 10 out of 13 evaluated. [21] |
| Gajewski et al. [14] | 1994–2012 | To systemically assess electronic event-based biosurveillance system evaluations to identify uncertainties about current systems and guide development to exploit web-based information.30 | Identified 11 electronic event-based biosurveillance systems that have been evaluated.30 |
2. Methods
A preliminary literature review was conducted through Google Scholar with general topic terms such as ‘digital disease’ and ‘detection’ and ‘surveillance’ and by author to identify scope, previous work, and to aid with keyword generation. Keyword generation was adapted from previous systematic reviews with the addition of supplementary terms from preliminary search results. A list of keywords can be found in Appendix A (in Supplementary material). A literature search was conducted in July 2015 by the author through the databases PUBMED; SCOPUS; and Google Scholar from May 2011 to July 2015. The search was a combination of all the key terms generated by using Boolean functions of ‘and’ and ‘or.’ Grey literature results relevant to event-based Internet biosurveillance systems; including mobile applications and participatory systems; was perfomed via Google and Google Scholar (not time restricted); which also included patents. Finally; when a system was mentioned in any of the above capacities; a general Google query with the system’s name; or creator of the system; and the first 1000 entries were analysed by relevance in order to further discover details and functionality of systems.
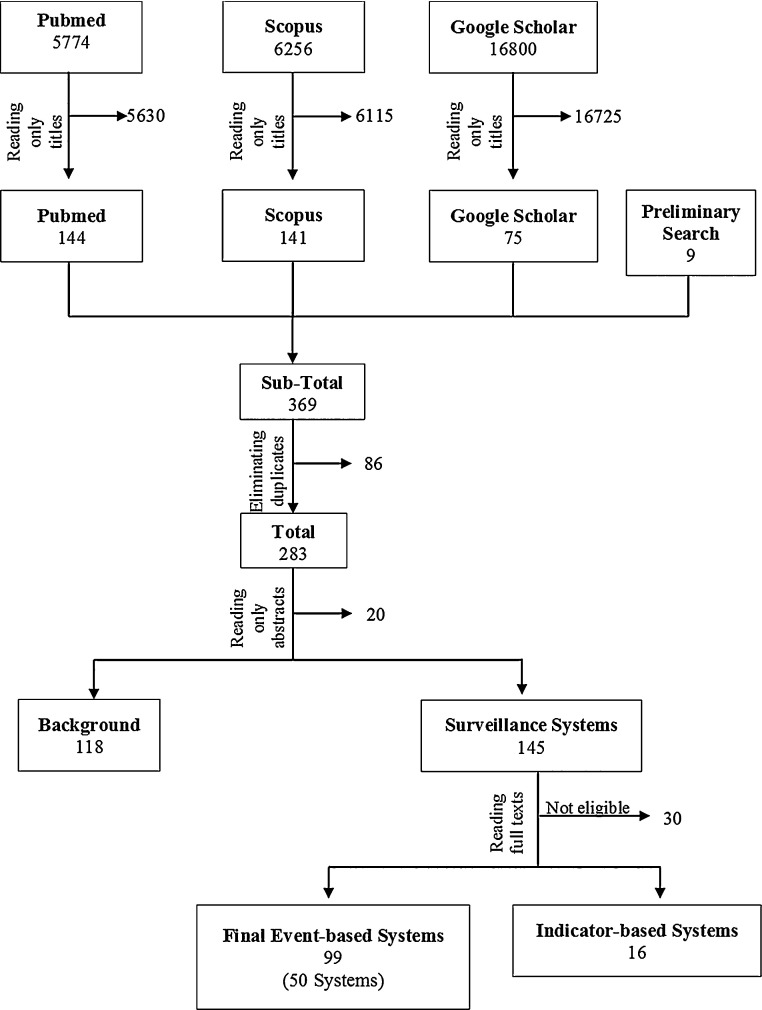
The electronic databases were selected based on their relevance to the subject matter and previous reviews. Reference lists from the selected literature were examined to include articles of relevance. Titles were screened first, then abstracts, based on inclusion and exclusion criteria. Questionable eligibility based on title and abstract would then be read in full and judged for eligibility. Duplicates were eliminated and irrelevant articles were excluded from the review. Process for the article selection and screening can be found in the flowchart, Fig. 2 , which is similar to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram methodology. Extraction criteria were used to collect comparable data on each system.
Fig. 2.
Flowchart of search procedure for literature review.
For eligibility, inclusion and exclusion criteria were created. These can be found in Appendix A (in Supplementary material) and were inspired and adapted from Velasco et al. [9], [17]. The publication type included journal articles in addition to conference abstracts and presentations, letters to the editor, commentaries, and patents.
The articles were then categorised for classification. These categories are 1) ‘background’ (which are articles not directly describing an event-based surveillance system, but rather surveillance systems in general) or 2) ‘system’ (articles describing at least one event-based surveillance system). Those placed in ‘system’ were further distinguished by either ‘indicator-based systems’ or ‘event-based systems’. ‘Event-based systems’ was also further subdivided if it was a participatory surveillance system. Participatory surveillance systems enable the public to report directly on diseases through the Internet, such as through crowdsourcing [15]. These systems encourage the regular, voluntary submission of syndromic, health-related information by the general public using technology like computers, tablets, or smartphones [15].
A specified set of evaluation criteria to describe all of the resulting systems uncovered in this review are an adaptation of the work of Vrbova et al., and Velasco et al., with certain fields being added or deleted [9], [17], [18]. The final 15 criteria can be found in Appendix A (in Supplementary material). These criteria served as the basis of the extraction of relevant data on event-based systems and were used to collect comparable data on each system.
3. Results
The systematic review, covering over 28,830 articles, discovered 50 event-based Internet systems, including subsystems, described in 99 articles (see Fig. 2 for the flowchart of these results and methodology). Thirty-two of the 50 systems have been evaluated in the literature in some capacity. Table 2 includes a list of all identified systems.
Table 2.
List of Event-Based Internet Biosurveillence Systems Identified.
| System Name | Category | Founding Country | Year Started | Currently online | Prototype |
|---|---|---|---|---|---|
| Argus | Moderated | USA | 2004 | No | No |
| BioCaster | Automatic | Japan | 2006 | No | No |
| GeniDB | Automatic | Japan | 2012 | No | No |
| GODSN | Automatic | USA | 2006 | No | No |
| MiTAP | Automatic | USA | 2001 | No | Yes |
| EWRS | Moderated | EU | 1998 | Yes | No |
| Proteus-BIO | Automatic | USA | 2000 | No | Yes |
| MedCOLLECTOR | Automatic | Portugal | 2009 | Yes | No |
| nEmesis | Moderated | USA | 2013 | No | Yes |
| EpiSPIDER | Automatic | USA | 2006 | No | No |
| Digital Disease Detection Dashboard | Moderated | USA | 2014 | No | Yes |
| MedISys and PULS | Automatic | EU | 2004 | Yes | No |
| GPHIN | Moderated | Canada | 1997 | Yes | No |
| HealthMap | Automatic | USA | 2006 | Yes | No |
| InSTEDD | Moderated | USA | 2006 | Yes | No |
| ProMED-mail | Moderated | USA | 1994 | Yes | No |
| Foodborne Chicago | Moderated | USA | 2013 | Yes | No |
| GET WELL | Automatic | Sweden | 2010 | Yes | No |
| Google Trends | Automatic | USA | 2009 | Yes | No |
| De Grote GriepMetinga | Moderated | Netherlands Belgium |
2003 | Yes | No |
| Gripeneta | Moderated | Portugal | 2005 | Yes | No |
| Influweba | Moderated | Italy | 2008 | Yes | No |
| FluSurveya | Moderated | UK | 2009 | Yes | No |
| InfluenzaNeta | Moderated | EU | 2009 | Yes | No |
| Influmetera | Moderated | Denmark | 2012 | Yes | No |
| GripeNeta | Moderated | Spain | 2012 | Yes | No |
| Grippeneta | Moderated | France | 2011 | Yes | No |
| Halsorapporta | Moderated | Sweden | 2011 | Yes | No |
| FluSurvey.Iea | Moderated | Ireland | 2013 | Yes | No |
| FluTracking | Moderated | Australia | 2006 | Yes | No |
| Reporta | Moderated | Mexico | 2009 | Yes | No |
| Flu Near You | Moderated | USA | 2011 | Yes | No |
| Salud Boricua | Moderated | Puerto Rico | 2012 | Yes | No |
| Dengue na Web | Moderated | Brazil | 2011 | Yes | No |
| Crowdbreaks | Moderated | USA | 2012 | Yes | Yes |
| Doctor Me App | Moderated | Thailand | 2011 | Yes | No |
| GermTrax | Automatic | USA | 2012 | Yes | No |
| SickCity | Automatic | USA | 2009 | No | No |
| FluDetector | Automatic | UK | 2009 | No | No |
| Infovigil | Moderated | Canada | 2009 | No | Yes |
| M-Eco | Automatic | EU | 2010 | No | Yes |
| NowTrending (previously MappyHealth) | Automatic | USA | 2012 | Yes | No |
| FluTrackers.com | Moderated | USA | 2006 | Yes | No |
| GoViral | Moderated | USA | 2013 | Yes | Yes |
| BlueDot (previously BioDiaspora) | Moderated | Canada | 2008 | Yes | No |
| Mo-Buzz | Moderated | Singapore | 2013 | Yes | No |
| Sickweather | Automatic | USA | 2011 | Yes | No |
| FluCaster | Automatic | USA | 2013 | Yes | No |
| GOARN | Moderated | USA | 2000 | Yes | No |
| HealthTweets.org | Automatic | USA | 2013 | Yes | No |
Collectively can be called InfluenzaNet.
The data extractions of individual systems and analyses that are more detailed can be found in Appendix B (in Supplementary material), which should serve as a database for other researchers in this field for future studies.
3.1. System category
The systematic review found no systems that can only be classified as news aggregators, although most utilise them, such as HealthMap. Of the 50 systems, nine (38%) automatic systems and 31 (62%) moderated systems.
3.2. Coordinating organisations
The systematic review revealed four types of coordinating organisations: 1) university based or in collaboration with a university (n = 24, 48%) 2) government agency (n = 15, 30%) 3) NGO or non-profit based (n = 3, 6%) and 4) private corporation based (n = 8, 16%).
3.3. Currently online
Of all the systems, 13 (26%) were not currently online, meaning likely no longer in existence, and 37 (74%) are fully functioning and online. Google Trends is no longer publishing their system’s findings, but rather going to provide the data to a select number of systems that utilise their data. It is unclear if this will affect systems that utilise Google Trends’ estimates.
3.4. Purpose
Each of the systems has a different purpose and aim, but three overarching themes can be illuminated: 1) to improve and enhance early detection (n = 45, 90%) 2) to improve communication or collaboration between actors, users, and parties involved (n = 3, 6%) 3) to supplement other existing systems (n = 2, 4%). EpiSPIDER supplements the Program for Monitoring Emerging Diseases (ProMED-mail) and GeniDB supplements BioCaster. Many systems are a blend of these three themes.
3.5. Stage
Eight (16%) of the systems can be classified as prototypes (experimental or beta), whereas 42 (84%) can be classified as fully developed.
3.6. Geographic scope
The geographic scope of the system varies. A system could operate and cover one particular area of a nation, the entire nation, continent, or the globe. Further, coverage may be restricted or confined to a particular region or state (n = 28, 56%). Of the total systems, 22 (44%) monitor internationally. The following countries had systems based in the country: United States (n = 23, 46%), European Union (n = 16, 32%), Canada (n = 3, 6%), Japan (n = 2, 4%). 6 (12%) are based elsewhere, such as Australia, Brazil, Singapore, Mexico, and Thailand.
3.7. Languages
Thirty four (68%) systems collect or disseminate their data only in one language, with the majority being English (n = 21), whereas 16 (32%) are multilingual–with the highest being Medical Information System (MedISys) with 43 languages.
3.8. Disease type
Of the 20 (40%) that focus only on one disease type, 17 of which monitor influenza-like-illness. Thirty (60%) focus on multiple infectious diseases and types, with the highest being HealthMap with over 170 disease types.
3.9. Accessibility
Access levels vary from system to system, partly because of the scope of the system and the intended audience. Systems are freely available to the public (n = 31, 62%), paid subscriptions (n = 1, 2%), free subscriptions (n = 5, 10%), and restricted access (n = 12, 24%), either to the coordinating entity or denied to the outsiders of the intended jurisdiction such as the European Union within a closed network. MedISys offers multiple access levels, freely available and restricted.
While it is important to offer freely accessible information, some sensitive information (personal data or other confidential data) is often filtered in specific ways among public health officials with specific restricted access. GPHIN has restricted access for organisations with an established public health mandate, with access varying according to factors like the organisation’s size and number of users [9].
3.10. Data collection and process
Data collection and acquisition is different for each system. However, general categories exist: 1) systems that collect information directly from RSS feeds or mailing lists (n = 13), 2) those that mine social media (n = 13), 3) those that mine search engine queries (n = 3), 4) those that collect data from both formal and informal sources (n = 4), and 5) those that are crowdsourced (or participatory), where data is collected or submitted by users (n = 19). Many systems are combinations of the above, however most systems incorporate the use of RSS feeds and news aggregators. Data processing is either automated or moderated, as divided in the system category section.
For example, MedISys monitors at least 50,000 articles per day, mostly through RSS feeds. These types of systems all use text-mining technology to extract relevant data and most have sophisticated algorithms for processing, filtering, and classifying relevant disease information [9]. The text extraction process utilises document heuristics, an experience-based method for machine learning that is applied to the information to enable an intelligent, and more accurate, decision about its relevance [9]. Monitoring is improved over time, as the heuristics learn as their output is confirmed against a set threshold for the epidemiological attributes of extracted health events [9]. Prior to returning the extracted information, the system aggregates the extracted events into outbreaks, across several documents and sources [9].
Some systems, such as HealthMap, relieve noise by integrating data from an assortment of online sources that have been moderated already (see Appendix B (in Supplementary material)) [9]. HealthMap’s informal source feeds vary, such as using ProMed-Mail and official-validated outbreak RSS feeds and alerts. These sources feed into a classification engine, such as a parser, which utilises the information to generate disease and location output codes [9]. Systems like HealthMap then filter the articles into a category and then store them in a database [9].
3.11. Dissemination of data
Systems may disseminate data by four identified trends: 1) via a display onto a map interface or as a time series graph (n = 32, 64%) 2) through a secured or restricted portal (n = 8, 16%) 3) to a website or newsgroup (n = 8, 16%) and 4) as a dashboard, where the dashboard is an interactive multi-tooled system, sometimes with plug-and-play features added (n = 2, 4%). HealthMap is an example of data results being illustrated through a map interface.
4. Discussion
The review emphasises the importance of utilising both formal and informal sources for timely and accurate infectious disease outbreak surveillance, cataloguing all event-based Internet biosurveillance systems. The review covers 50 systems, ten of those may be classified as subsystems with similar characteristics, or part of a larger umbrella organisation, with a detailed analysis of 15 attributes of each system.
North America and Europe are leading in event-based Internet biosurveillance systems, whereas Africa, Asia, Australia, and South America possess little or no such systems to monitor their epidemic threats. There lies an opportunity for these systems in varying income countries to aid with emerging diseases in vulnerable countries or encompass people who may not have access to healthcare systems.
There is a rise in the private sector as the creator or coordinator for the system. Perhaps there is profit making potential in this field that has yet to be seen, or perhaps technology can excel more quickly when left with reduced bureaucracy. Coincidentally, a large amount of systems are not currently online or ceased to function. Various reasons can be postulated, such as funding issues, lack of manpower, little utilisation or lack of results.
Event-based Internet biosurveillance is a recognised effective approach for infectious disease detection. Inevitably, the number of online data sources will increase, and as it does, event-based Internet biosurveillance will increasingly become more important in infectious disease surveillance. These systems are attractive from a logistic, economic, and epidemiologic viewpoint [19]. They are intuitive, flexible, function close to real-time, and many are freely available [19]. Once they are established, they are relatively inexpensive to run and sustain [19].
Many of the systems allow citizens to report public health events via social media platforms or electronic communication channels independently of governments [14]. Therefore, these systems do not rely on the formal healthcare system or hierarchal organisation structures to provide, analyse, or disseminate data, or to advise the international community of emerging infectious disease concerns. Governments are no longer in sole control of their public health information, making it substantially harder to hide or delay outbreak or event reports [14], [20]. However, the same aspects of event-based Internet biosurveillance systems that make them an important new surveillance tool also may makes them a less reliable tool [14]. Since public health professionals do not verify some sources of data, these systems are prone to noise and false alarms [14], [21]. Further, there is difficulty differentiating signal from noise. Sometimes, they lack specificity and results from these systems differ from official sources [14], [21].
Social media data, and other online data, are often closed to the public or agencies, but companies may pay large amounts of money to acquire user information [22]. To further facilitate meaningful data mining in social networking, more open-source data or increased data sharing is required [22]. In addition, regulations are needed in social media as a data source, and all online data sources, to ensure good governance of the data and that individuals’ privacy is not violated.
More synergy among systems is needed. Systems often operate in silos and may also compete with one another, especially those that are run by a private corporation. Now that the systems exist, ways to complement one another should be researched, such as integrating indicator-based systems to these new digital systems to enhance accuracy and surveillance, thereby creating a hybrid disease surveillance system.
The systematic review has several limitations. The scope of the study excluded bioterrorism and animal related systems, so potential relevant systems may have been neglected or not included. Data on some systems was not publicly available, due to restricted access, thus complete information or key factors may be missing from the attribute list. Further, there is a possibility of systems existing outside of academic literature or search engine realms, such as those developed by military or closed environments. Additionally, there is inconsistent application of the term ‘surveillance’ here and throughout the literature. Some system information provided suggests that the system is a monitoring system rather than a surveillance system, and many articles did not contain enough information to correctly distinguish between the two, reflecting a lack of overall surveillance theory in public health. Further, as this review was limited to articles in English, the results may be biased towards including systems from English-speaking areas.
Ongoing evaluation, validation and verification of event-based Internet biosurveillance systems with epidemiological and clinical data by users, developers, and agencies will greatly increase the robustness of these systems for infectious disease detection and monitoring [23]. Yet, the willingness to integrate these systems into public health surveillance programmes is rooted in the effectiveness studies and evaluations, but such effectiveness evaluation studies can only be proved through integration—a circular dilemma [9]. Big Data has the chance to revolutionise infectious disease outbreak detection and management. If we, as a society, are going to allow this to occur, we must collaborate as academics, health professionals, and civil society, to achieve it.
Conflicts of interest
None.
Author contribution
All authors have made substantial contributions to all of the following: (1) the conception and design of the study, or acquisition of data, or analysis and interpretation of data, (2) drafting the article or revising it critically for important intellectual content, (3) final approval of the version to be submitted.
Summary points
What was known before this study?
-
•
Innovative event-based Internet biosurveillance systems are being utilised, assessed, and popularized with varying sources of data.
-
•
Previous reviews focused primarily on systems that have been evaluated in the literature and did not include new surveillance systems such as participatory systems.
What did this study add to our body of knowledge?
-
•
The current review contributes with an extensive overview of all event-based Internet biosurveillance systems, including those that use social media, participatory surveillance, mobile applications, and those which have not been evaluated yet.
-
•
Results yielded the largest catalogue of systems, with 15 attributes described for each system, and each using different technology and data sources to gather data, process, and disseminate data to detect infectious disease outbreaks.
-
•
The review points out several areas in need of more research such as ongoing evaluation, validation, and verification of systems and meaningful data mining and data sharing to facilitate synergy.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijmedinf.2017.01.019.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.McNeill W. Anchor Press; New York: 1976. Plagues and Peoples: A Natural History of Infectious Diseases. [Google Scholar]
- 3.Damianos L., Ponte J., Wohlever S., Reeder F., Day D., Wilson G., Hirschman L. AAAI/IAAI; 2002. Mitap, Text and Audio Processing for Bio-security: A Case Study. [Google Scholar]
- 4.Centers for Disease Control and Prevention Updated guidelines for evaluating public health surveillance systems. MMWR. Morb. Mortal. Wkly. Rep. 2001;50:1–35. [Google Scholar]
- 5.Zeng D., Chen H., Lynch C., Millicent D., Gotham I. Infectious disease informatics and outbreak detection. In: Chen H., Fuller S., Friedman C., Hersch W., editors. Medical Informatics, Knowledge Management and Data Mining in Medicine. Springer Science + Business Media, Inc; New York, NY: 2005. pp. 359–395. [Google Scholar]
- 6.Green M.S., Kaufman Z. Surveillance for early detection and monitoring of infectious disease outbreaks associated with bioterrorism. Isr. Med. Assoc. J. 2002;4(July (7)):503–506. [PubMed] [Google Scholar]
- 7.Paquet C., Coulombier D., Kaiser R., Ciotti M. Epidemic intelligence: a new framework for strengthening disease surveillance in Europe. Euro Surveill. 2006;11(12):212–214. [PubMed] [Google Scholar]
- 8.World Health Organization . World Health Organization Western Pacific Regional Office; 2008. A Guide to Establishing Event-Based Surveillance Manila. [Google Scholar]
- 9.Velasco E., Agheneza T., Denecke K., Kirchner G., Eckmanns T. Social media and internet-based data in global systems for public health surveillance: a systematic review. Milbank Q. 2014;92(1):7–33. doi: 10.1111/1468-0009.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nsoesie E. 2014. Digital Disease Detection: A Look at Participatory Surveillance of Dengue. The Disease Daily Research & Policy.http://www.healthmap.org/site/diseasedaily/article/digital-disease-detection-look-participatory-surveillance-dengue-51314 (2014 May 13 [cited 2015 July 20]. Available from:) [Google Scholar]
- 11.Milinovich G.J., Williams G.M., Clements A.C.A., Hu W. Internet-based surveillance systems for monitoring emerging infectious diseases. Lancet Infect. Dis. 2014;14(2):160–168. doi: 10.1016/S1473-3099(13)70244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kass-Hout TA. Riff A Social Network and Collaborative Platform for Public Health D⋯ [Internet]. [cited 2015 July 22]. Available from: http://www.slideshare.net/kasshout/riff-a-social-network-and-collaborative-platform-for-public-health-disease-surveillance
- 13.Guy S., Ratzki-Leewing A., Bahati R., Gwadry-Sridhar F. Social media: a systematic review to understand the evidence and application in infodemiology. Lect. Notes Inst. Comput. Sci. Soc. Inf. Telecommun. Eng. 2012;91:1–8. doi: 10.1007/978-3-642-29262-0_1. [DOI] [Google Scholar]
- 14.Gajewski K.N., Peterson A.E., Chitale R.A., Pavlin J.A., Russell K.L., Chretien J.-A. A review of evaluations of electronic event-based biosurveillance systems. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0111222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wójcik O.P., Brownstein J.S., Chunara R., Johansson M.A. Public health for the people: participatory infectious disease surveillance in the digital age. Emerg. Themes Epidemiol. 2014;11(1):7. doi: 10.1186/1742-7622-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linge J.P., Steinberger R., Weber T.P., Yangarber R., van der Goot E., Al Khudhairy D.H. Internet surveillance systems for early alerting of health threats. Euro Surveill. 2009;14(April (13)):19162. [PubMed] [Google Scholar]
- 17.T. Agheneza, A systematic literature review on even-based public health surveillance systems [master's thesis]. Hamburg: Hamburg University of Applied Sciences; 2014 [cited 2015 June 10]. Available from: www.edoc.sub.uni-hamburg.de/haw/volltexte/2012/1614/pdf/lsab12_27.pdf
- 18.Vrbova L., Stephen C., Kasman N., Boehnke R., Doyle‐Waters M., Chablitt‐Clark A. Systematic review of surveillance systems for emerging zoonoses. Transbound. Emerg. Dis. 2010;57(3):154–161. doi: 10.1111/j.1865-1682.2010.01100.x. [DOI] [PubMed] [Google Scholar]
- 19.Milinovich G.J., Avril S.M.R., Clements A.C.A., Brownstein J.S., Tong S., Hu W. Using internet search queries for infectious disease surveillance: screening diseases for suitability. BMC Infect. Dis. 2014;14(1) doi: 10.1186/s12879-014-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai F., Tseng E., Chan C., Tamashiro H., Motamed S., Rougemont A.C. Is the reporting timeliness gap for avian flu and H1N1 outbreaks in global health surveillance systems associated with country transparency. Glob. Health. 2013;9:14. doi: 10.1186/1744-8603-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller M., Blench M., Tolentino H., Freifeld C.C., Mandl K.D., Mawudeku A. Use of unstructured event-based reports for global infectious disease surveillance. Emerg. Infect. Dis. 2009;15(May (5)):689–695. doi: 10.3201/eid1505.081114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szomszor M., Kostkova P., editors. Springer-Verlag; Heidelberg, Dordrecht, London, New York: 2011. Electronic Healthcare. LNICST 69(9) [Google Scholar]
- 23.Nelson N.P., Yang L., Reilly A.R., Hardin J.E., Hartley D.M. Event-based internet biosurveillance: relation to epidemiological observation. Emerg. Themes Epidemiol. 2012:9. doi: 10.1186/1742-7622-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bravata D.M., McDonald K.M., Smith W.M., Rydzak C., Szeto H., Buckeridge D.L. Systematic review: surveillance systems for early detection of bioterrorism-related diseases. Ann. Intern. Med. 2004;140(11):910–922. doi: 10.7326/0003-4819-140-11-200406010-00013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.