Graphical abstract
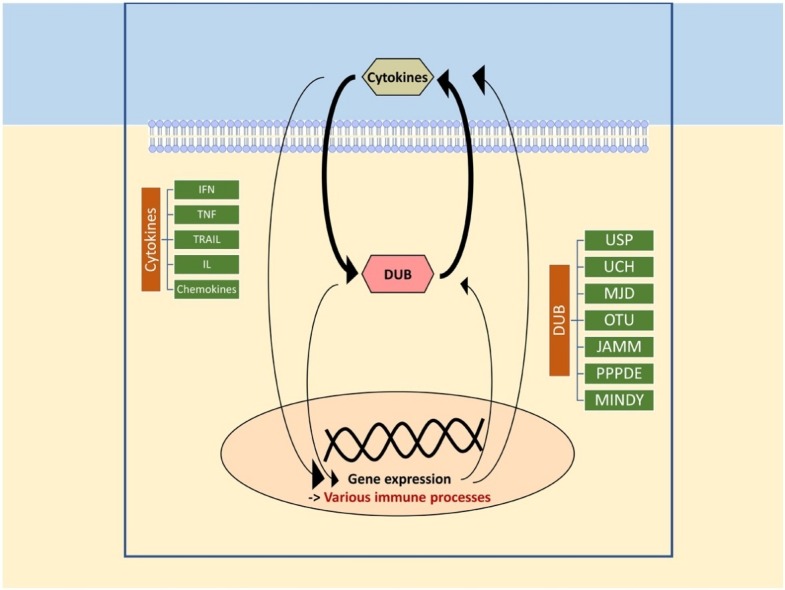
Keywords: Cytokine-inducible, DUB, IFN, Interleukin, TNF, Ubiquitination
Abstract
Deubiquitinating enzymes (DUBs) are cysteine protease proteins that reverse the ubiquitination by removing ubiquitins from the target protein. With over 100 DUBs identified and categorized into at least 7 families, many DUBs interact with one or more cytokines, influencing cellular processes, such as antiviral responses, inflammatory responses, apoptosis, etc. While some DUBs influence cytokine pathway or production, some DUBs are cytokine-inducible. In this article, we summarize a list of DUBs, their interaction with cytokines, target proteins and mechanisms of action.
1. Introduction
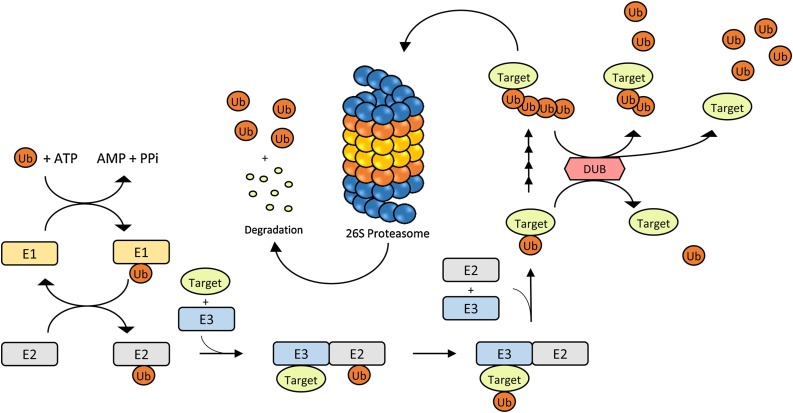
Ubiquitination and deubiquitination are post-translational modifications for numerous proteins, which in turn affect many physiological processes. Ubiquitination is defined as an attachment of one or more ubiquitin (Ub) molecules onto the target protein through the function of a series of proteins: E1, E2 and E3 (Fig. 1 ) [1]. Seven lysine residues have been identified (K6, K11, K27, K29, K33, K48 and K63) on the ubiquitin molecule [2]. Also, additional Ub molecules can be attached onto one of the seven lysine residues or the N-terminal methionine to form polyubiquitin chains (polyUb) [2,3]. Perhaps, Ub is most well-known as the crucial marker of the Ubiquitin-Proteasome System (UPS), in which ubiquitinated proteins enter proteasomal degradation via 26S proteasome (Fig. 1) [3]. However, Ub also affects many other aspects of tagged proteins, such as localization, protein interaction, function, etc. [3]. On the other hand, deubiquitination refers to the process that reverses ubiquitination via deubiquitinating enzymes (DUBs) (Fig. 1). Little less than 100 human DUBs have been identified so far [4]. DUBs were categorized into five different families in the past [5], but at least seven different families are identified as of now, which include the ubiquitin-specific protease (USP), ubiquitin carboxyl-terminal hydrolase (UCH), Machado-Josephin disease protein (MJD), ovarian tumor (OTU), JAB1/MPN/Mov34 (JAMM), permutated papain fold peptidases of dsRNA viruses and eukaryotes (PPPDE) and MIU-containing novel DUB family (MINDY) [6,7].
Fig. 1.
Mechanism of action of ubiquitin proteasome system and deubiquitinating enzymes. Ub attaches to the target protein by going through a series of reaction with E1 (ubiquitin activating), E2 (ubiquitin conjugating) and E3 (ubiquitin ligating) enzymes. A target protein could be ubiquitinated once or multiple times on lysine residues. 26S proteasome identifies target proteins with polyUb chain and degrades them into amino acid segments and reusable Ub. Ubiquitinated proteins could also be deubiquitinated by DUBs, resulting in a different fate.
Cytokines are groups of small proteins that play a role in cell signaling and immune system by binding to their respective receptors. Since DUBs regulate diverse physiological processes, it was to be expected that DUBs and cytokines affect one another. As anticipated, the more studies were performed regarding the functions of DUBs, the more interaction between DUBs and cytokines were revealed. Recently, several reviews dealing with how DUBs affect pathway of a specific cytokine were published [[8], [9], [10]], but none has yet introduced as a whole the interaction between DUBs and cytokines. In this review, we wish to provide a brief overview of the DUBs discovered to regulate cytokine signaling pathways and cytokine-inducible DUBs. We will discuss the DUBs that influence the pathways of interferons (IFN), tumor necrosis factors (TNF), TNF-related apoptosis-inducing ligand (TRAIL), interleukins (IL) and chemokines.
2. IFN signaling pathways
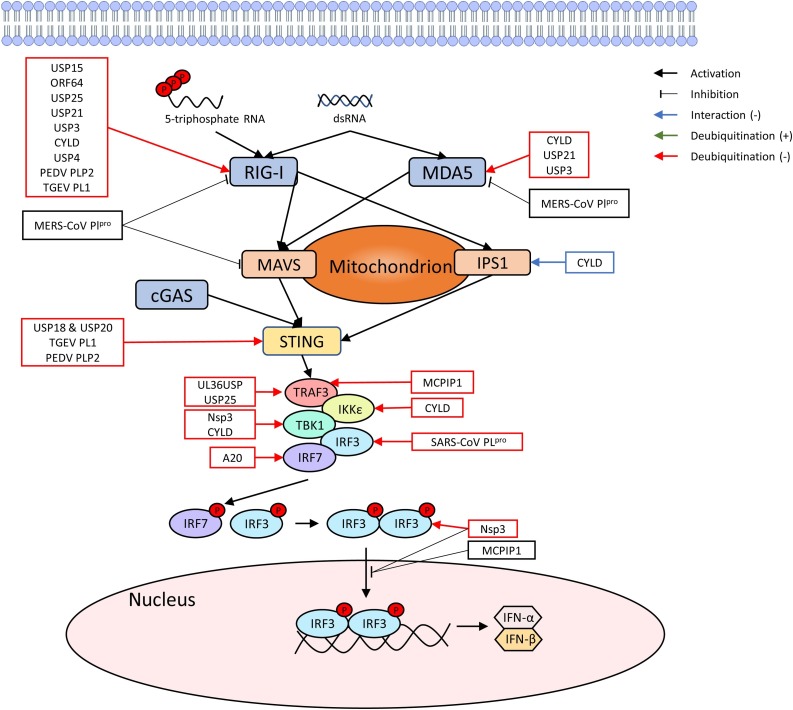
IFN-α and IFN-β cytokines belong to type I IFN family that are essential for antiviral responses, cancer, inflammation, etc. [11]. When a cell recognizes a viral infection through detecting IFN-stimulating signaling molecules or foreign double stranded DNA in the cytosol, retinoic acid-inducible gene-I (RIG-I) is activated, triggering the cascade of the second messenger system to activate and translate IFN-α and IFN-β signaling pathways (Fig. 2 ) [12]. DUBs interact with some of the key molecules in the IFN signaling pathway, which include, but are not limited to, RIG-I, stimulator of interferon genes (STING), tumor necrosis factor receptor-associated factors (TRAFs), interferon regulatory factor 1 (IRFs) and IκB kinases (IKKs) ( Fig. 3 ). Because type I IFN from the host antagonizes viral infection, viruses are in need of downregulating or inhibiting the production of IFN. Consistent with this, many discovered virally encoded DUBs antagonize the production of IFN, which are summarized in Table 1 . We will discuss the DUBs in the order of IFN signaling pathway shown in Fig. 1.
Fig. 2.
Viral genome induced IFN production pathway via RIG-I and interacting DUBs. Upon sensing viral dsRNA, RIG-I and MDA5 activate an IFN production cascade. The name and the effects of DUBs identified in IFN related studies are mapped to show the mechanism of their action.
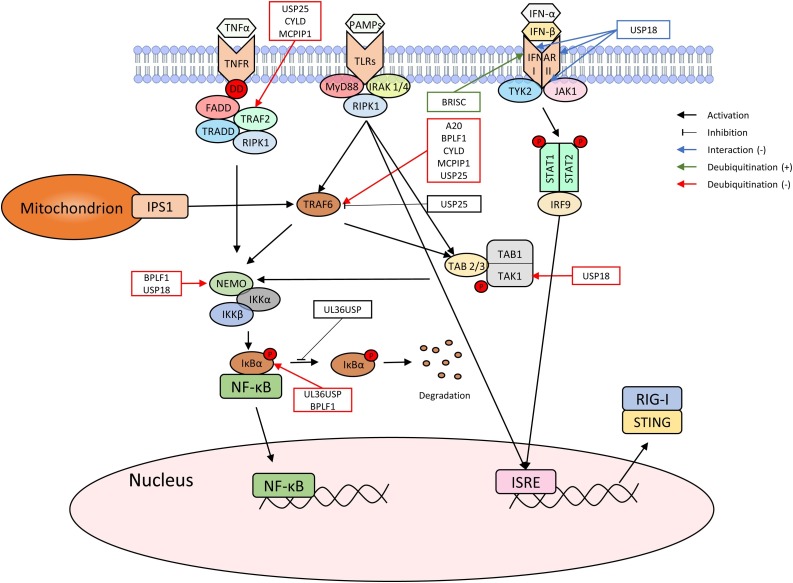
Fig. 3.
TLRs, IFNARI and IFNARII induced IFN production pathway. PAMPs, IFN-α and IFN-β stimulate TLR4 and IFNAR I & II receptors respectively to induce IFN production as well as NF-κB activation. DUBs that play a role in these pathways are indicated in the figure to show the mechanism of their action.
Table 1.
Interferon-, TNF- and TRAIL-inducing DUBs and their mechanisms.
| Cytokine | DUB | Effects (cell line/organism) | Mechanism | References |
|---|---|---|---|---|
| Interferon | USP15 | - (HEK293 T) | deubiquitinates K63-polyUb of RIG-I | [16] |
| ORF64 | - (HEK293 T) | deubiquitinates RIG-I (TRIM25 dependent) | [18] | |
| - (C57BL/6 mice) | inhibits STING-mediated IFN production | [19] | ||
| USP25 | - (HEK293 T) | deubiquitinates RIG-I, TRAF2 and TRAF6 | [20] | |
| - (BMDC, MEF) | deubiquitinates K48-Ub of TRAF3 | [21,22] | ||
| - (MEF) | stabilizes, but not deubiquitinates K48-Ub of TRAF6 | [21] | ||
| USP21 | - (HEK293 T) | deubiquitinates K63-polyUb of RIG-I | [24] | |
| USP3 | - (HEK293 T) | deubiquitinates K63-polyUb of RIG-I and MDA5 | [25] | |
| CYLD | - (MC) | deubiquitinates K63-Ub of RIG-I and MDA5 | [26] | |
| - (293 EBNA) | deubiquitinates RIG-I, TBK1 and IKKε | [27] | ||
| negatively regulates IPS-1 | ||||
| - (HEK293) | deubiquitinates TRAF2 | [29] | ||
| - (HEK293 T) | deubiquitinates TRAF6 | [30] | ||
| - (U2OS/NOD2) | deubiquitinates RIPK2 | [31] | ||
| PEDV PLP2 | - (HEK293 T) | deubiquitinates RIG-I and STING | [35] | |
| TGEV PL1 | - (HEK293 T) | deubiquitinates RIG-I and STING | [36] | |
| MERS-CoV PLpro | - (HEK293 T) | targets RIG-I, MDA5 and MAVS (DUB activity dependent) | [33,34] | |
| SARS-CoV PLpro | - (HEK293 T) | deubiquitinates IRF3 | [38] | |
| USP20 | - (MEF) | deubiquitinates K33- or K48-Ub of STING (USP18 dependent) | [41] | |
| USP18 (UBP43) | - (MEF) | recruits USP20 to form a complex with USP20 and STING (DUB activity independent) | [41] | |
| - (293 T) | deubiquitinates K63-Ub of TAK1 and NEMO | [42] | ||
| - (Th17) | deubiquitinates K63-Ub of TAK1 | [43] | ||
| - (293 T) | interacts with IFNAR2 to inhibit JAK's tyrosine kinase activity (DUB activity independent) | [45] | ||
| - (U5A) | interferes with IFNAR2 to reduce its recruitment of IFNAR1 | [46] | ||
| UL36USP | - (HEK293 T) | deubiquitinates TRAF3 | [48] | |
| - (HFF) | deubiquitinates and decreases degradation of IκBα | [49] | ||
| USP25 | - (HEK293 T) | decreases IRF3 phosphorylation | [21] | |
| - (BMDM) | deubiquitinates K48-Ub of TRAF3 | [22] | ||
| Nsp3 | - (HEK293 T) | deubiquitnates IRF3 to inhibit its nuclear translocation | [51] | |
| - (HEK293 T, MEF) | deubiquitinates K63-polyUb of TBK1 | [52] | ||
| MCPIP1 | - (HEK293) | deubiquitinates TRAF2, TRAF3 and TRAF6 | [53] | |
| - (HEK293 T, HeLa) | interacts with IRF3 and inhibits its nuclear translocation | [54] | ||
| A20 | - (HEK293 T) | deubiquitinates K63-Ub of TRAF6 | [30] | |
| - (Raji) | deubiquitinates IRF7 | [55] | ||
| BPLF1 | - (293 T) | deubiquitinates K63-Ub of TRAF6 and NEMO | [58] | |
| deubiquitinates K48-Ub of IκBα | [58] | |||
| BRISC | + (2fTGH, MEF) | forms a complex with SHMT and deubiquinates K63-Ub of IFNAR1 | [59] | |
| TNF | USP4 | - (microglia of Sprague-Dawley rats) | deubiquitinates TRAF6 | [60] |
| - (HEK293 T) | deubiquitinates TRAF2 and TRAF6 | [61] | ||
| - (HEK293 T) | deubiquitinates TAK1 | [62] | ||
| - (A549) | inhibits degradation of IκBα | [61] | ||
| A20 | - (C57BL/6 J) | inhibits ubiquitination of K48- and K63-Ub of RIPK1 | [66] | |
| + (IEC) | forms a dimer to bind to Ripoptosome, which hinders deubiquitination of RIPK1 | [67] | ||
| Cezanne | - (HEK293, HEK293 T) | deubiquitinates RIPK1 | [68,69] | |
| - (HUVEC) | deubiquitinates TRAF6 | [70] | ||
| USP48 (USP31) | - (beas2B) | deubiquitinates TRAF2 of JNK pathway | [71] | |
| OTULIN | - (U2OS) | deubiquitinates RIPK1 | [83] | |
| TRAIL | BAP1 | + (H226) | knockout decreases DR4 and DR5 expression | [72] |
| + (H2818) | knockout decreases DR4 expression | [72] | ||
| USP35iso1 | - (HEK293 FIpIn) | delays caspase-8 process in TRAIL-induced apoptosis (DUB activity dependent) | [73] | |
| USP35iso2 | + (U2OS FIpIn, HeLa) | Induces ER stress, which activates apoptosis through DR5 | [73] | |
| USP14 and/or UCHL5 | - (A549, HCT116, H460) | b-AP15 elevates DR5 levels | [77] | |
| MCPIP1 | - (MDA-MB-231) | deubiquitinates DR5 and enhances autophagic/lysosomal degradation of DR5 | [79] |
The first group of DUBs are those that deubiquitinate RIG-I to inhibit the production of IFN. RIG-I is a cytosolic protein that plays a significant role in IFN signaling by detecting viral DNA and RNA [13,14], which is then ubiquitinated by tripartite motif (TRIM) to activate the signaling cascade to synthesize IFN [15]. ORF64, USP25, USP21, USP15, USP3, Cylindromatosis (CYLD), porcine epidemic diarrhea virus papain-like protease 2 (PEDV PLP2) and transmissible gastroenteritis virus papain-like protease1 (TGEV PL1) are the DUBs found to deubiquitinate RIG-I. We will discuss them one by one.
2.1. USP15
In a study using HEK293 T cells and Sendai virus (SeV), knockdown of the gene transcribing USP15 resulted in upregulation of type I IFN, while overexpression of USP15 decreased type I interferon as USP15 showed dose-dependent inhibition of IFN-β [16]. Further experiment supported the idea that USP15 deubiquitinates K63-polyUb from RIG-I [16]. However, when the effects of USP15 with a mutated catalytic site and wild type USP15 were compared, the results were surprisingly similar, indicating that USP15’s catalytic activity is not necessary for it to inhibit IFN synthesis [16].
2.2. ORF64
ORF64 is a DUB activity containing tegument protein, found within Kaposi’s sarcoma-associated herpesvirus (KSHV) and murine gamma herpesvirus 68 (MHV68) [17,18]. The expression of KSHV ORF64 in HEK293 T cells led to suppression of both RIG-I-induced and SeV infection-induced IFN-β promoter activation [18]. On the contrary, KSHV ORF64-C29 G mutant, with defective deubiquitinating activity, resulted in lesser to no suppression, confirming the influence of ORF64 on the IFN synthesis [18]. Also, the overexpression of TRIM25, but not the mutant TRIM25, reversed the ORF64’s effect on IFN production and reverted the ubiquitination of RIG-I, further confirming the result [18]. It is also noteworthy that ORF64 was not capable of suppressing MAVS-induced activation of IFN-β production [18]. When MHV68 infected bone marrow derived dendritic cells (BMDC) were induced by MCMV and HSV-1 for type I IFN induction, no IFN was detected, but TNF-α, IL-6 and IL-1β were expressed upon high dose stimulation [19]. This provided evidence that MHV68 induced innate immunity of the host to a lesser extent [19]. On the other hand, ORF64 mutant MHV68 stimulated innate immune response [19]. By utilizing DUB activity of ORF64, MHV68 blocked viral DNA induced, STING-mediated IFN production [19].
2.3. USP25
In a study performed by Zhong et el, USP25, too, was found to deubiquitinate RIG-I and reduce SeV-induced IFN-β production in HEK293 T cell line [20]. Knockdown of USP25 gene also led to augmentation of ISRE promoter upon SeV induction [20]. Mutating the catalytic residue of USP25 was sufficient to block USP25’s effect on IFN-β induction, supporting that IFN-β suppression via USP25 is DUB activity dependent [20]. USP25 targeted not only RIG-I for deubiquitination, but also extended to TRAF2 [20], TRAF3 [21,22] and TRAF6 [20] and to affect IFN signaling. USP25 also deubiquitinated TRAF5 and TRAF6 to regulate in IL-17 signaling [23]. However, some studies have given different results regarding USP25’s ability to deubiquitinate TRAF6. Lin et al. stimulated Usp25 knockout BMDC with SeV or HSV-1 infection and added wild type (WT) USP25 or mutant USP25, but K48-Ub of TRAF6 did not differ from one another [21]. However, Zhong et al.’s study using HEK293 T cells supported deubiquitination of TRAF6 by USP25 [20]. This variance in the result may be caused by the difference of the cell line used for the studies. Furthermore, USP25 showed its ability to suppress phosphorylation of interferon regulatory factor 3 (IRF3) and p65, also contributing to inhibition of IFN promoter activation [20].
2.4. USP21
Fan et al., knowing that USP21 inhibits RIG-I-induced IFN-β production, searched for its mechanism [24]. They unveiled that USP21 inhibited ISRE reporter activity induced by SeV and RIG-I-CARD, but not by TANK-binding kinase 1 (TBK1) in mouse embryonic fibroblasts (MEF) cells [24]. USP21 deubiquitinated RIG-I in HEK293 T cells [24]. Also, they found that USP21’s function regarding antiviral response is compatible in MEF and HEK293 T cell lines by introducing each cell line’s USP21 to the other cell line and observing the effect [24]. USP21’s specificity to RIG-I was also confirmed in HeLa cells through coimmunoprecipitation (co-IP) of USP21 with RIG-I, using rabbit polyclonal antibodies against USP21 [24]. USP21 also deubiquitinated MDA5 to inhibit antiviral response [24].
2.5. USP3
USP3 is also a DUB that deubiquitinates K63-polyUb chain of both RIG-I and MDA5 and suppresses IFN-β activation [25]. USP3’s effect was found viable in 293 T, THP-1, human peripheral blood mononuclear cells (PBMCs) and RAW264.7 cells, supporting that USP3’s activity is viable in both human and murine cells [25]. USP3 did not inhibit MAVS, STING, TBK1, IRF3 and TIRF, as demonstrated by ISRE-luc activity induction test [25]. Also co-IP demonstrated the interaction between USP3 and stimulated RIG-I or MDA5, but not the unstimulated ones, supporting that ligand stimulation is required for USP3 to interact with RIG-I or MDA5 [25]. More specifically, Poly(I:C) (LMW) stimulation leads USP3 to have a strong interaction with RIG-I, but a weak one with MDA5, while Poly(I:C) (HMW) stimulation leads USP3 to have a strong interaction with MDA5, but a weak one with RIG-I [25].
2.6. CYLD
CYLD is another DUB that removes K63-Ub chain from RIG-I to decrease the IFN production [26,27], but TBK1 and IKKε were also identified as the target of the deubiquitination of CYLD in 293 EBNA cells [27], resulting in the same effect. CYLD also interacted with IPS-1 to negatively regulate it, but did not deubiquitinate it [27]. Schmid et al. found that in brain and peripheral blood of C57BL/6, the mRNA level of IFN- γ gene decreased with the knockdown of CYLD, while the serum concentration of IFN-γ increased [28]. A study conducted using human kidney mesangial cells (MC) showed slightly different results: silencing CYLD in MC cells and stimulating them with poly IC increased the toll-like receptor 3 (TLR3)-induced activation of RIG-I and MDA5 [26]; however, the level of mRNA of RIG-I and MDA5 actually decreased [26]. The authors speculated this difference to be caused by the change in cell line used [26], but further study is necessary to determine the cause. CYLD also decreased IFN promotor activation by deubiquitinating TRAF2 and TRAF6 in HEK293 T cells, respectively [29,30].
CYLD in U2OS/NOD2 cells were found to deubiquitinate K63-Ub of RIPK proteins, especially RIPK2, to suppress NOD2-induced NF-κB activation [31]. When CYLD was suppressed, ubiquitinated receptor interacting protein kinase 1 (RIPK1), also called RIP1, and RIPK2 proteins accumulated within cells [31].
2.7. PLPs
PLPs, first discovered in Coronavirus in 2005 [32], are multifunctional proteins with DUB activity that are synthesized by many families of viruses that regulate IFN signaling pathway by interacting with RIG-I [[33], [34], [35], [36]]. Recently, the mechanism by which PEDV PLP2 suppresses IFN production in the host cell was identified. In HEK293 T cells, PEDV PLP2 was found to deubiquitinate RIG-I and STING, thereby affecting its downstream pathway, resulting in suppression of IFN production [35]. TGEV PL1 also was revealed to bind and deubiquitinate both RIG-I and STING in HEK293 T cells [36]. Studies on Middle East respiratory syndrome coronavirus encoded papain-like protease (MERS-CoV PLpro) showed that it also has a DUB function [37]. This was supported by a study by Bailey-Elkin et al., in which they obtained the crystal structure of PLpro-Ub complex and showed that WT MERS-CoV PLpro, but not the DUB mutant PLpro, suppresses IFN-β promotor activity [33]. The targets of MERS-CoV PLpro were identified as RIG-I, MDA5 and MAVS [33,34]. MERS-CoV PLpro and severe acute respiratory syndrome coronavirus (SARS)-CoV PLpro inhibited the proinflammatory signaling in HEK293 T cells upon MDA5 stimulation, which included decreased expression of CCL5 and IFN-β and decreased level of CXCL10 mRNA [34]. Another study on SARS-CoV PLpro found that IRF3 is ubiquitinated and that deubiquitinating activity of SARS-CoV PLpro was required for it to affect IRF3 [38]. SARS-CoV PLpro did not affect IRF3 in other means, such as dimerization or nuclear translocation [38]. DUB domain mutated PLP2 of Equine Arteritis virus (EAV) also increased in expression of IFN-β and IL-8 in equine long fibroblasts (ELF) [39]. PLP domain was also found in nsp3 protein, which will be discussed in a later section.
2.8. USP18 & USP20
STING is a transmembrane protein found in mitochondria and endoplasmic reticulum that regulates IFN-promotor activation at the downstream of RIG-I [40]. Zhang et al. studied the effect of USP18 (also known as UBP43) on STING and revealed that USP18 interacts with STING to affect IFN-promotor activity [41]. However, when USP18-/- MEF cells with either WT USP18 or DUB activity-mutated USP18 were induced with HSV-1, HCMV or cytosolic DNA, Ifnb, Ifna4, Tnf, IL-6 or Cxcl1 genes increased in expression, indicating that the deubiquitinating activity of USP18 is not responsible for this phenomenon [41]. Subsequently, they searched for DUBs that interact with USP18 and found that knockdown of USP20 inhibited USP18-induced deubiquitination of STING and knockdown of USP18 inhibited USP20-induced deubiquitination of STING [41]. Immunoprecipitation revealed that STING, USP18 and USP20 are arranged as USP20-USP18-STING, but both USP20 and USP18 were associated with the N-terminus of STING [41]. USP20 deubiquitinated K33- or K48-linked ubiquitin of STING [41]. These results together supported that although USP18 does not deubiquitinate STING itself, USP18 recruits USP20 to deubiquitinate STING to suppress IFN synthesis [41].
Another way that USP18 inhibited NF-κB activation is by deubiquitinating K63-Ub of TAK1 and NEMO [42]. USP18 strongly interacted with TAK1-TAB1 and DUB activity dependently deubiquitinated K63-Ub of TAK1 in 293 T cells [42] and in Th17 cells [43]. USP18 also decreased K63-Ub of NEMO [42].
In a study by Malakhova et al., USP18 inhibited IFN-induced gene activation by affecting JAK-STAT signaling pathway in 293 T cells [44]. Their study showed that USP18 does not interact with IFNAR1, but with Box1-Box2 region of IFNAR2 to disrupt its interaction with JAK to inhibit JAK’s tyrosine kinase activity in a DUB activity independent manner [44]. Consistent with this, USP18 knockout murine cells displayed hyperactivity towards type I IFN signaling, resulting in the increase of the level of phosphorylation of STAT1 and STAT2 [45]. USP18’s interaction with IFNAR2 also interfered with IFNAR2’s ability to recruit IFNAR1, hindering IFN I signaling [46].
2.9. UL36USP
Herpes simplex virus 1 (HSV-1) invades a host and escapes its IFN-mediated innate immunity by encoding a large tegument protein, UL36, which has a motif with DUB activity, named UL36 ubiquitin-specific protease (UL36USP) [47]. When HEK293 T cells were transfected with markers of co-IP and UL36USP or the C40A (a DUB motif mutant) and then infected with SeV, the result showed reduction of ubiquitination of TRAF3 in cells with WT UL36USP, while C40A has no reduction of UL36USP’s ubiquitination [48]. The result supported that UL36USP deubiquitinates TRAF3 molecules to inhibit IFN-promotor activation [48].
In a different study regarding the function of UL36USP, it was found that UL36USP inhibits cGAS and STING dependent IFN-β production [49]. NF-κB activation from overexpressing STING, TBK1, IKKα and IKKβ was also inhibited, but not from overexpressing p65 [49]. In this study, human foreskin fibroblast (HFF) cells were infected with either HSV-1 or HSV-1 C40A mutant and stimulated with IFN stimulatory DNA [49]. As a result, the level of endogenous IκBα in HFF cells with mutant HSV-1 significantly decreased compared to HFF cells with WT HSV-1, supporting that UL36USP decreases the degradation of IκBα in a DUB activity-dependent manner [49]. Additionally, co-IP study in HEK293 T cells showed decrease in ubiquitination of IκBα in those transfected with WT UL36USP, but not in those transfected with C40A mutant [49]. Taken together, UL36USP deubiquitinates IκBα to inhibit its degradation, suppressing NF-κB activity [49].
2.10. USP25
A study by Lin et al. revealed that USP25 is required for both DNA and RNA virus-induced signaling [21]. Supporting this claim, silencing USP25 in MEFs or mouse lymphatic fibroblasts (MLF) led to inhibition of expression of Ifna4, Tnf, and IL-6 upon triggering them with SeV, Vesicular stomatitis virus (VSV) or poly(I:C) [21]. Also, the level of IFN-α and IL-6 was reduced in MLFs, BMDCs or FLT3LpDC cells with USP25 knockdown [21]. USP25’s DUB activity was also found to be necessary for virus-induced signaling, as USP25 knockdown MEFs with WT USP25 reconstitution allowed expression of Ifnb, Ifna4 and IL-6 upon SeV or HSV-1 induction, while those with DUB activity mutant USP25 did not [21]. Overexpressing USP25 in HEK293 T cells resulted in reduction of IRF3 phosphorylation when stimulated with SeV, leading to inhibition of NF-κB activity [20]. ISRE reporter activity was also inhibited by USP25 in a dose-dependent manner [20]. Taking it one step further, Lin et al. uncovered that USP25 stabilizes TRAF3 in a DUB activity dependent manner by deubiquitinating K48-Ub of TRAF3 in bone marrow-derived macrophages (BMDM) cells, inhibiting TLR4 signaling-induced innate immune responses [21].
2.11. Nsp3
The nonstructural protein 3 (nsp3) is a viral protein with deubiquitinating activity in its PLP2 domain, which was found in SCoV and in mouse hepatitis virus A59 (MHV-A59) [50,51]. Infecting MEF cells with MHV-A59 did not result in detectable IFN-β induction, while infecting them with SeV (the control) did result in IFN-β responses [51]. When cells were given variants of nsp3, the IFN-β induction only took place in cells that lacked WT PLP2 domain [51]. When the PLP2 domain was present, IFN-β induction was suppressed upon viral infection [51]. Moreover, polyUb of IRF3, which is necessary for IFN-β induction, was deubiquitinated in the presence of PLP2, which was further confirmed by co-IP indicated formation of a complex of PLP2 and IRF3 [51]. This deubiquitination inhibited nuclear translocation of IRF3 [51]. In HEK293 T cells and MEF cells, K63-polyUb was also deubiquitinated by the PLP2 domain of nsp3, inactivating TBK1-IRF3 complex in the cytoplasm [52].
2.12. MCPIP1
Monocyte chemotactic protein-inducing protein 1 (MCPIP1) is a protein, common to human and mouse, with DUB activity toward TRAF2, TRAF3 and TRAF6, thereby inhibiting JNK and NF-κB signaling [53]. A more recent study has uncovered through co-IP in SeV infected HEK293 T and HeLa cells that MCPIP1 interacts with IRF3 and through confocal microscopy that transfection with MCPIP1 inhibited nuclear translocation of IRF3 [54]. Additionally, the presence of MCPIP1 inhibited TRAF3 and TBK1 activated IFN-β expression [54]. Co-IP also revealed possibility of MCPIP1 to interact with IPS-1 and IKKε as well [54].
2.13. A20
A20 (also known as TNFAIP3) inhibited LPS-induced NF-κB activity in MEF cells in a study by Boone et al. [30]. Upon further testing in HEK293 T, they found that WT A20, but not DUB activity domain mutant A20 removed K63-Ub from TRAF6 [30].
In Raji cells, the N-terminal DUB domain of A20 interacted with and deubiquitinated IRF7 [55]. However, in vitro study showed no interaction between A20 and IRF7, which is likely due to requirement of other intracellular proteins [55]. IRF7 has been known to be activated by Epstein-Barr virus (EBV)’s oncoprotein called latent membrane protein 1 (LMP1) [56].
A20 has been known for a long time as a negative regulator of NF-κB pathway mediated by RIG-I. A20 does interact with RIG-I, and suppresses RIG-I-mediated NF-κB pathway [57], but whether A20 deubiquitinates RIG-I or not still requires confirmation.
2.14. BPLF1
Similar to ORF64 in MHV68 [17], BPLF1 is an EBV encoded large tegument protein with DUB activity that opposes TLR signaling in the host [58]. Gent et al.’s research revealed that BPLF1 deubiquitinates TRAF6 and NEMO in 293 T cells [58]. Immunoprecipitation in 293 T cells revealed that K63-Ub of TRAF6 and NEMO was reduced when WT BPLF1 was expressed, while mutant BPLF1 did not [58]. K48-Ub of IκBα was also identified as a target of BPLF1’s DUB activity [58].
2.15. BRISC
The BRCC36 isopeptidase complex (BRISC) is a nuclear DUB complex, composed of Abraxas, BRCC36, BRCC45 and MERIT40, capable of deubiquitinating K63-Ub [59]. In a study by Zheng et al., serine hydroxymethyltransferase (SHMT) formed a complex with BRISC to form BRISC-SHMT complex [59]. SHMT allowed interaction of BRISC with IFNAR1 to deubiquitinate K63-Ub of IFNAR1, reducing IFNAR1’s internalization and degradation by lysosome [59]. Taken together, BRISC is the first DUB complex we discussed that works to actually increase responses to IFN.
3. TNF signaling pathways
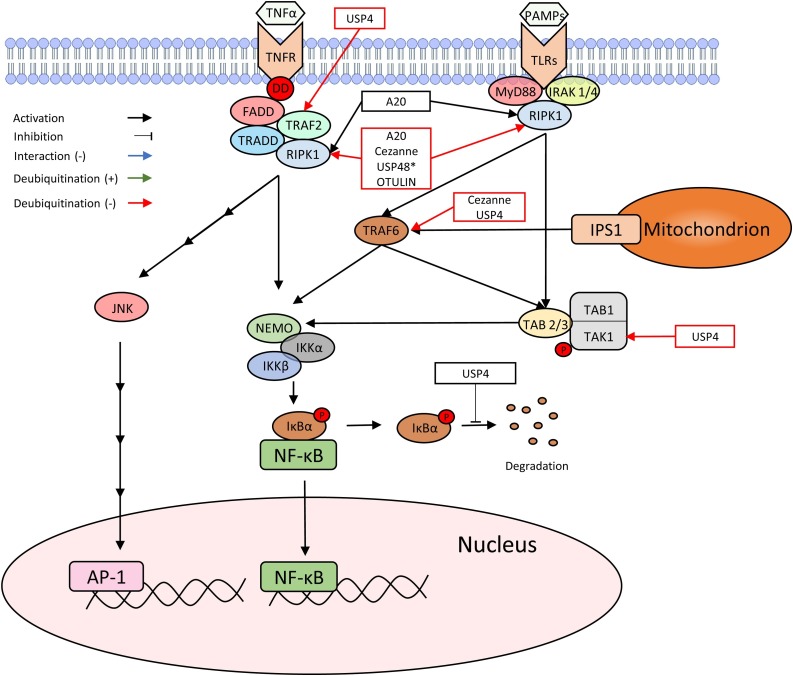
TNF is a cytokine that plays a significant role in inflammation and regulation of immune cells. Since TNF shares some of its pathway with IFN, studies that focused on TNF rather than IFN are included in this section for the purpose of this review (Fig. 4 ) (Table 1).
Fig. 4.
TNFR and TLRs induced TNF signaling pathway. TNF-α and PAMPs stimulate their respective receptors, TNFR and TLRs, and activate NF-κB. DUBs that influence these pathways are marked in the figure to indicate how and where they affect.
3.1. USP4
USP4 was identified by studies to negatively regulate both TNF-α- and IL-1β-induced NF-κB activation [[60], [61], [62]]. Jiang et al. observed that introducing small interfering USP4 (siUSP4) to decrease USP4 level in microglia from the spinal cord of Sprague-Dawley rats led to an increase in p-p65 and TRAF6 expression as well as secretion of TNF-α and IL-1β, all of which decreased upon introduction of HA-USP4 plasmid [60]. Xiao et al.’s study added on to this by demonstrating USP4’s interaction and deubiquitination of TRAF2 and TRAF6, but not TRAF3, both in vivo, in HEK293 T cells, and in vitro [61]. As a result, USP4 negatively influenced TNF-α-induced-NF-κB activation-mediated cytokine induction, including IL-6 and IL-8 in A549 and H1229 cells [61]. USP4 also deubiquitinates TAK1 in HEK293 T cells [62]. USP4 also protected IκBα from degradation, [61], a necessary step for TNF-α-induced NF-κB activation [63]. This was further supported by knockout of USP4 aiding IκBα degradation [61].
3.2. OtuLi
L. infantum otubain (OtuLi) has been shown to induce inflammatory responses in peritoneal macrophages from C57BL/6, shown by production of TNF-α and IL-6 as well as lipid droplet synthesis [64]. Also Otuli demonstrated strong DUB activity on K48-Ub and weak activity on K63-Ub in vitro at pH 7.5 [64].
3.3. USP25
We mentioned in a previous section that USP25 deubiquitinates TRAF3 and interacts with TRAF6, increasing expression of Tnf in HEK293 T cells [21,22], while in MEF cells, USP25 negatively affects TNF-α-induced NF-κB activation [20].
3.4. A20
A20 also affects tumor necrosis factor receptor 1 (TNFR1) signaling pathway. In BMDMs and BMDC, A20 worked together with TAX1BP1 to interact with Ubc13, an E2 enzyme, resulting in the inhibition of E3 ligase activities of TRAF6, TRAF2 and cIAP1 [65]. Futhermore, A20 and TAX1BP1 participated in degrading Ubc13 upon IL-1 and TNF-α stimulation in MEF cells [65].
A20’s ZF4 motif was found to recruit A20 dimers to bind with RIPK1 in the TNFR signaling complexes and inhibit ubiquitination of K48-Ub and K63-Ub chains of RIPK1, hindering TNF signaling [66].
In intestinal epithelial cells (IEC), A20 dimer interacted with the Ripoptosome (also known as complex IIa), which allowed ubiquitins on RIPK1 to sustain, increasing caspase-8 activation to promote TNF-induced apoptosis [67]. However, this effect was not DUB activity dependent [67].
3.5. Cezanne
Cezanne is a DUB that belongs to the A20 subgroup of OTU family. Similar to A20, Cezanne also was shown to suppress NF-κB signaling by deubiquitinating K63-polyUb from RIPK1 [68,69] and TRAF6 [70]. Also, Cezanne was recruited to the activated TNFR prior to deubiquitinating RIPK1, which was dependent on the ubiquitin-associated (UBA) domain of Cezanne [68,69]. Consistnent with the findings, inhibiting Cezanne production via siRNA resulted in an increased production of IL-8 upon TNF-α stimulation [68]. Cezanne’s DUB activity was required for inhibiting phosphorylation and degradation of IκBα [68].
3.6. USP48
USP48 (also known as USP31) interacted with and deubiquitinated TRAF2 in beas2B cells [71]. A noteworthy fact is that TRAF2 in JNK pathway was targeted by USP48, but not TRAF2 in NF-κB signaling [71].
4. TRAIL signaling pathways
TRAIL is a cytokine that binds to death receptors (DR) and induces apoptosis, especially in tumor cells. Its specificity for tumor cells have made TRAIL and its receptor as the targets for anti-cancer therapeutics. TRAIL inducing DUBs are also summarized in Table 1.
4.1. BAP1
In a study of malignant mesothelioma, a loss of function mutation of BRCA associated protein 1 (BAP1) resulted in increased sensitivity fo TRAIL induction [72]. When testing for domains that play a role in TRAIL sensitivity in H226 MM cells, only ASXL1/2 binding site-mutated BAP1 and DUB domain-mutated BAP1 resulted reduction in rTRAIL sensitivity, indicating that ASXL1/2 binding sites play a role in TRAIL sensitivity [72]. This was in congruence with the fact that BAP1 binds to ASXL to form the Polycomb repressive deubiquitinase complex (PR-DUB) that deubiquitinates histone H2A [73]. Also, flow cytometry analysis confirmed that the mutation of C91A, or the deubiquitinating domain, of DUB resulted in decreased expression of DR4 and DR5 in H226 cells [72]. Only DR4 expression increased in H2818 cells upon BAP1 knockout [72].
4.2. USP35
On the same line with BAP1, USP35 knockout also increased TRAIL sensitivity [74]. A study by Leznicki et al. introduced three isoforms of USP3 in HEK293 cells: USP35iso1, USP35iso2 and USP35iso3, although the focus was on the first two [74]. USP35iso2 was revealed as an integral membrane protein on endoplasmic reticulum, while USP35iso1 was identified as a cytosolic protein [74]. Moreover, the proteins that they interacted with also varied [74]. USP35iso2 led to ER stress, BAP31 cleavage and activation of caspase-8 and caspase-3, resulting in apoptosis [74]. USP35iso2 upregulated C/EBP homologous protein (CHOP) and DR5 in U2OSFIpIn and HeLa FIpIn cells [74]. On the other hand, overexpressing USP35iso1 exerted an opposite effect of delaying caspase-8 processing in TRAIL-induced apoptosis [74]. This effect was dependent on DUB activity [74].
4.3. USP14 and/or UCHL5
b-AP15 is a small therapeutic molecule that inhibits USP14 and UCHl5 [75]. b-AP15 has been identified as an agent that increases TRAIL receptors on many types of cancer cells, increasing their likelihood to enter apoptosis via NK cells [76]. Introduction of b-AP15 in A549, HCT116 and Calu-1 cells increased the level of DR5, but not the other death-inducing signaling complex (DISC) components [77]. The increase in the level of DR5 protein was due to reduction in the degradation of DR5, leading to an increase in TRAIL-induced apoptosis [77]. In a different study, caspase-denependent apoptosis was increased in mantle cell lymphoma (MCL) cells when exposed to b-AP15, which was confirmed with addition of pan-caspase inhibitor zVAD-fmk, which resulted in an inhibition of apoptosis [78]. This study indirectly demonstrated that USP14 and/or UCHL5 partakes in decreasing DR5 expression.
4.4. MCPIP1
MCPIP1 is another DUB that decreases DR5 [79]. Exposing MDA-MB-231 cells to doxycycline (DOX) led to induction of MCPIP1, which then led to a decrease in DR5 [79]. Similarly, when A549 human lung cancer cells were exposed to MCP1, MCPIP1 level increased in a dose-dependent manner, also resulting in a decrease in DR5 [79]. Likewise, DR5 level increased when MCPIP was knocked down via short hairpin RNA (siRNA) [79]. MCPIP1 successfully achieved this by deubiquitinating DR5, thereby stimulating lysosomal degradation of DR5 [79]. Also, the increase in DR5 level following MCPIP1 knockdown catalized the formation of DISC during the DR5-induced apoptosis [79].
5. IL family pathways
5.1. USP25
We will now discuss DUBs that induce interleukins, which are listed in Table 2 . USP25 has been shown to deubiquitinate TRAF3 and TLR4 and to interact with TRAF6, increasing expression of IL-6 in MEF cells [21,22].
Table 2.
Interleukin- and chemokine-inducing DUBs and their effects.
| Cytokine | DUB | Effects (cell line/organism) | References |
|---|---|---|---|
| Interleukins | |||
| IL-1β | ORF64 | increases (C57BL/6) | [19] |
| USP4 | - (HEK293 T) | [60,61,62] | |
| decreases (microglia of Sprague-Dawley rats) | [60] | ||
| Otulin | increases (C57BL/6) | [82] | |
| MCPIP1 | decreases (C57BL/6) | [53] | |
| IL-2 | USP18 | decreases (T cells of C57BL/6) | [43] |
| Otulin | increases (C57BL/6) | [82] | |
| IL-4 | Otulin | increases (C57BL/6) | [82] |
| IL-5 | Otulin | decreases (C57BL/6) | [82] |
| IL-6 | USP25 | increases (MEF) | [21,22] |
| Trabid | increases (C57BL/6 × 129/Sv mixed) | [84] | |
| USP18 | increases (MEF) | [41] | |
| USP25 | increases (MLF,BMDC,FLT3LpDC) | [21] | |
| USP4 | decrease (A549,H1229) | [61] | |
| OtuLi | increases (C57BL/6) | [64] | |
| Otulin | increases (C57BL/6) | [82] | |
| MCPIP1 | decreases (C57BL/6) | [53] | |
| USP8 | - (C57BL/6) | [81] | |
| IL-7Rα | USP8 | decreases (T cells of C57BL/6) | [81] |
| IL-8 | BPLF1 | decreases (293) | [58] |
| EAV-PLP2 | decreases (ELF) | [39] | |
| USP4 | decrease (A549,H1229) | [61] | |
| Cezanne | decrease (HUVEC) | [68] | |
| IL-10 | Otulin | increases (C57BL/6) | [82] |
| IL-12 | Trabid | increases (BMDC) | [84] |
| IL-12p70 | Otulin | increases (C57BL/6) | [82] |
| USP8 | - (C57BL/6) | [81] | |
| IL-13 | Otulin | increases (C57BL/6) | [82] |
| IL-17 | USP25 | - (HEK293 T) | [23] |
| IL-23 | Trabid | increases (BMDC) | [84] |
| Chemokine | |||
| CCL5 | MERS-CoV PLpro | decreases (HEK293 T) | [34] |
| SARS-CoV PLpro | |||
| CYLD | decreases (MC) | [26] | |
| increases (CD8+ T cells of C57BL/6) | [28] | ||
| USP21 | decreases (HEK293 T) | [24] | |
| Ccr7 | USP8 | (thymocytes of C57BL/6) | [81] |
| CXCR3R | CYLD | increases (CD8+ T cells of C57BL/6) | [28] |
| CXCL10 | MERS-CoV PLpro | decreases (HEK293 T) | [34] |
| SARS-CoV PLpro | |||
| CYLD | decreases (MC) | [26] | |
| increases (CD8+ T cells of C57BL/6) | [28] | ||
| ORF64 | increases (C57BL/6 lung homogenate) | [19] | |
Increase = inc. in production, + = induce positive effect.
5.2. ORF64
On top of decreasing production of IFN [17,18], ORF64 in MHV68 also induced the production of IL-1β [19]. IL-1β production was dependent on NLRP3 and ASC, rather than AIM2 [19].
5.3. USP4
USP4 negatively regulates IL-1β-induced NF-κB activation [[60], [61], [62]].
5.4. ESI
Eeyarestatin I (ESI), a small molecule that inhibits deubiquitination, has been found responsible for blocking IL-1β release [80]. Lopex-Castejon et al., who reported this finding speculated UCH37 or USP14 was responsible for this phenomenon, but futher study showed that they do not regulate IL-1β secretion individually or cooperatively [80]. This result left possibility of an uncharacterized DUBs or an additional DUB(s) partaking in the process.
5.5. USP18
USP18 knockout murine splenocytes and naïve T cells produced more IL-2 compared to the WT splenocytes [43]. Under Th17 polarizing condition, IL-2 production was significantly higher in the USP18 knockout naïve CD4+ T cells compared to the WT naïve CD4+ T cells [43]. Also USP18 knockout naïve CD4+ T cells underwent hyperproliferation under Th17 polarizing condition, which was reversed by adding IL-2 neutralizing antibody [43]. Taken together, USP18 downregulates IL-2 synthesis and TCR-induced T cell proliferation [43]. Additional mechanism of action has been discussed in the previous seciton.
5.6. USP8
Dufner et al. found that USP8 was essential in T cell maturation and homeostasis, although it was not required for negative selection [81]. Inhibiting USP8 leads to decrease in IL-7ra mRNA as well as Ccr7 [81]. Also, IL-6, IL-12p70, IFN-γ and TNF levels were increased in the blood of Usp8f/fCd4-Cre mice than USP8f/f [81].
5.7. OTULIN
Deleting Otulin gene in mouse immune cells, T, B, natural killer cells (NK), dendritic cells (DC) and macrophage cells, resulted in production of cytokines specifically responsible for acute systemic inflammation, such as TNF, IL-1β, IL-6, MCP-1, MIP-1α and G-CSF [82]. Cytokines responsible for adaptive immunity were not affected by the deletion [82]. It is noteworthy that this study suggests that although many cytokines partake in the inflammatory response in murine cells without Otulin, the primarily responsible cytokine is TNF [82].
Unlike in immune cells, deleting Otulin in myeloid cells resulted in activation of cytokines responsible for acute and chronic inflammation as well as autoimmunity, showing an increase in the level of 16 out of 25 cytokines tested [82]. Deficiency of Otulin in macrophages resulted in NF-κB activation without an induction, which was due to the inability to manage polyUb chains synthesized by linear ubiquitin chain assembly complex (LUBAC) [82]. Another study found that OTULIN overexpression inhibited TNF-α-induced nuclear translocation of p65 in HEK293 T cells [83]. Interestingly, both WT and DUB domain mutant OTULIN disabled LUBAC-induced NF-κB activation, indicating that OTULIN-mediated Met1-polyUb is not the only factor influencing NF-κB activation [83]. OTULIN has also been identified to deubiquitinate Met1-polyUb of RIPK1 and inhibited the binding of NEMO and RIPK1, blocking the TNF-α-induced NF-κB response [83].
5.8. BPLF1
EBV BPLF1 suppresses IFN production, but IL-8 production by cells upon MALP-2 ligand stimulation was also abrogated upon BPLF1 expression [58]. EBV BPLF1 suppressed production of proinflammatory cytokine IL-8 in 293-TLR2/CD14 cells [58].
5.9. Trabid
Trabid is a DUB from OTU family, translated from the gene Zranb1 [84]. Upon Zranb1 knockout in mice, IL-6, Tnf, IL-12a, IL-12b and IL-23a displayed a decrease in expression [84]. Trabid’s deubiquitinating activity was necessary for recruiting c-Rel and p50 to IL-12 promoter by influencing histone modifications [84]. Knockout of Trabid rendered BMDC incapable of producing IL-12 and IL-23, leading to a decrease in the number of differentiation of CD4 + T cells to TH1 and TH17 cells, which was reversible with adding IL-12 and IL-23 [84].
6. Chemokines
Not many studies have focused their study objectives on discovering relationship between chemokines and DUBs. Some discovered interactions are listed in Table 2.
7. Cytokine-inducible DUBs
We have discussed how DUBs induce cytokine production, signaling and effects. Compared to DUB’s effects on cytokines, cytokine-inducible DUBs are far less studied due to the difficult nature of planning such studies. However, this information can be as important as DUB-induced cytokines. We will now discuss some known examples of cytokine-inducible DUBs, as listed in Table 3 .
Table 3.
Cytokine-inducible DUBs and their effects.
| DUB | Cytokine | Effects (cell line/organism) | References |
|---|---|---|---|
| DUB-1 | IL-3 | increases (Ba/F3) | [85] |
| IL-5 | increases (Ba/F3) | [87] | |
| GM-CSF | |||
| DUB-1A | IL-3 | increases (Ba/F3) | [88] |
| DUB-2 | IL-2 | increases (CTLL) | [89] |
| DUB-2A | CSF3 | increases (myeloid 32D) | [92] |
| DUB-2A | IL-4 | increases (Raji) | [94] |
| IL-6 | increases (U937) | [94] | |
| Otud-6B | IL-3 | increases (Ba/F3) | [96] |
| IL-4 | |||
| IL-13 | |||
| GM-CSF | |||
| USP18 | IFN-β | increases (THP-1, THP-1 derived macrophage) | [42] |
| USP48 | TNFα | + (beas2B) | [71] |
| A20 | TNFα | increases (HUVEC) | [98] |
| Cezanne | TNFα | increases (HEK293, HUVEC) | [68] |
Increase = inc. in production.
+ = induce positive effect.
7.1. DUB-1
DUB-1 is one of the early identified cytokine-inducible DUBs. Studying the sequence of DUB-1 gene, unveiled that it contained a IL-3 inducible enhancer in Ba/F3 murine lymphocyte cell line [85]. The timing of IL-3 induced DUB-1 mRNA increase was identified as early G1 phase [85]. Moreover, when DUB-1 was constitutively expressed, majority of Ba/F3 cells were arrested in G1 phase of the cell cycle, which was DUB activity dependent [85]. Induction of DUB-1 was dependent on viable JAK2 and Raf-1, but not STAT5, suggesting that DUB-1 expression is dependent on two pathways: JAK2 and Ras/Raf-1/MAPK pathway [86]. IL-5 and granulocyte-macrophage colony-stimulating factor (GM-CSF) also induced DUB-1 transcription, which supported that β common (βc) subunit plays a part [87].
DUB-1A decreased in expression when JAK2 was suppressed, also suggesting DUB-1A to be affected by IL-3 and JAK2 pathway [88].
7.2. DUB-2
DUB-2 is similar to DUB-1 in its sequence of amino acids and is also induced by JAK2/STAT5 pathway [89]. However, unlike DUB-1, DUB-2 was induced only by IL-2 in T cells, but not by IL-3 [90]. DUB-2 also showed increase in JAK/STAT signaling pathway products by decreasing IL-2 induced dephosphorylation of STAT5 [89]. Also, DUB-2 decreased apoptosis in Ba/F3 cells upon withdrawal of cytokines [89]. The mechanism by which DUB-2 achieves these effects needs to be further studied.
In myeloid 32D cells, DUB-2 stabilized CSF3R and increased its signaling activity by decreasing lysosomal degradation of CSF3R by deubiquitinating it, leading to prolongation of STAT5 phosphorylation in CSF3 signaling pathway [91].
On the other hand, DUB-2A is a DUB expressed in hematopoietic cells, such as B and T cells [92]. Unlike DUB-2, which was more expressed by IL-2, DUB-2A was further expressed upon exposure to IL-3 [92]. Although similar to DUB-1 induction by IL-3 [85], DUB-2A induction by colony-stimulating factor 3 (CSF3) in myeloid 32D cells did not require Erk [91].
7.3. DUB-3
DUB-3 (also known as USP17) is a cytokine-inducible human DUB that was found to deubiquitinate SDS3 and block proliferation in HeLa cells [93]. In mRNA level, DUB-3 expression increased in Raji cells when treated with IL-4 and in U937 cells when treated with IL-6 [94]. DUB-3 also influenced the Ras/MEK/ERK signaling pathway and deubiquitinated Ras converting enzyme 1 (RCE1), decreasing proliferation of cells [95].
7.4. Otud-6B
Otud-6B, a DUB from OTU family, was upregulated in a mouse pro-B cells, Ba/F3 cells, upon cytokine stimulation [96]. Stimulation with IL-3, IL-4, IL-13 or GM-CSF resulted in a dose-dependent increase of Otud-6B mRNA in the first 0 to 2 h of stimulation, but quick decrease was observed from 4 to 6 h [96]. Overexpressing Otud-6B in Ba/F3 cells resulted in downregulation of proliferation and increased the frequency of apoptosis [96].
7.5. USP18
USP18 has been known to be induced by viral infection, genotoxic stress or interferon [97]. Consistent with this, USP18’s mRNA level increased in THP-1 cells and THP-1-derived macrophages upon exposure to IFN-β [42]. Furthermore, TLR ligands, LPS, Pam3CSK4 and CL097 all gave the same result of increased mRNA level of USP18, supporting that TLR-induced signaling pathway induces USP18 expression [42].
7.6. USP48
Exposure to TNFα caused GSK3β-mediated phosphorylation of USP48, leading to deubiquitination of TRAF2 in beas2B cells, increasing JNK signaling upon TNF-α-induction [71].
7.7. A20
A20 was identified since 1990 as a TNF-α-induced DUB in human umbilical vein endothelial cells (HUVEC) [98].
7.8. Cezanne
mRNA of Cezanne quickly increased in HEK293 and HUVEC cells upon exposure to TNF-α, but not upon shear stress [68].
8. Conclusion
We have discussed some known cases of DUB-regulated cytokines and cytokine-inducible DUBs. Cytokines are intertwined in numerous cellular processes and DUBs are closely related to cytokines. This review dealt with many DUBs, but less than half of all known DUBs are discussed. Moreover, we cannot say for certain that all the functions of the DUBs discussed here are discovered. This grants us to further investigate the molecular mechanism and their effects. Studying DUBs and their effects could enlighten us with a novel therapeutic approach to various diseases, including but not limited to immunological diseases and cancer.
Acknowledgement
We would like to thank members of Baek’s laboratory for their critical comments This study was funded by the Korea Ministry of Environment (MOE) as ‘the Environmental Health Action Program (2016001360008)’.
Biographies

Bean Woo obtained a bachelor’s degree in music composition at Emory university in 2015. The following year, he started MD studies in 2017 at the University of Alabama at Birmingham School of Medicine with a special interest in hematology and oncology. His research interest includes interaction between hematological disease processes and cytokines.

Kwang-Hyun Baek is a full professor of Department of Biomedical Science and a director of Cell & Gene Therapy Research Institute at CHA University in Korea. He received his Ph.D. from the department of Zoology and Genetics at Iowa State University in US in 1995 and worked as a postdoctoral research fellow in the division of Genetics at Brigham and Women’s Hospital and in the department of Pediatric Oncology at Dana-Farber Cancer Institute, Harvard Medical School. He has been serving as an editorial board member of more than a dozen of international journals. Current research and clinical interests are in the molecular genetics of ubiquitination and deubiquitination systems relevant to various cancers. He first coined terms ubiquitomics and deubiquitomics in the field of the ubiquitin-proteasome system. In addition, he is also interested in genomics and proteomics during stem cell proliferation, differentiation, and reprogramming.
References
- 1.Streich F.C., Jr., Lima C.D. Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 2014;43:357–379. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon Y.T., Ciechanover A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 2017;42(11):873–886. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luise C., Capra M., Donzelli M., Mazzarol G., Jodice M.G., Nuciforo P., Viale G., Di Fiore P.P., Confalonieri S. An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdul Rehman S.A., Kristariyanto Y.A., Choi S.Y., Nkosi P.J., Weidlich S., Labib K., Hofmann K., Kulathu Y. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell. 2016;63(1):146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q., Wu Y., Qin Y., Hu J., Xie W., Qin F.X., Cui J. Broad and diverse mechanisms used by deubiquitinase family members in regulating the type I interferon signaling pathway during antiviral responses. Sci. Adv. 2018;4(5) doi: 10.1126/sciadv.aar2824. eaar2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumari N., Jaynes P.W., Saei A., Iyengar P.V., Richard J.L.C., Eichhorn P.J.A. The roles of ubiquitin modifying enzymes in neoplastic disease. Biochim. Biophys. Acta Rev. Cancer. 2017;1868(2):456–483. doi: 10.1016/j.bbcan.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Kumari P., Kumar H. Viral deubiquitinases: role in evasion of anti-viral innate immunity. Crit. Rev. Microbiol. 2018;44(3):304–317. doi: 10.1080/1040841X.2017.1368999. [DOI] [PubMed] [Google Scholar]
- 11.Snell L.M., McGaha T.L., Brooks D.G. Type I interferon in chronic virus infection and cancer. Trends Immunol. 2017;38(8):542–557. doi: 10.1016/j.it.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita T., Onoguchi K., Onomoto K., Hirai R., Yoneyama M. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie. 2007;89(6-7):754–760. doi: 10.1016/j.biochi.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Cheng G., Zhong J., Chung J., Chisari F.V. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104(21):9035–9040. doi: 10.1073/pnas.0703285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneyama M., Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227(1):54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 15.Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J.U. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H., Wang D., Zhong H., Luo R., Shang M., Liu D., Chen H., Fang L., Xiao S. Ubiquitin-specific protease 15 negatively regulates virus-induced type I interferon signaling via catalytically-dependent and -independent mechanisms. Sci. Rep. 2015;5(11220):11220. doi: 10.1038/srep11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gredmark S., Schlieker C., Quesada V., Spooner E., Ploegh H.L. A functional ubiquitin-specific protease embedded in the large tegument protein (ORF64) of murine gammaherpesvirus 68 is active during the course of infection. J. Virol. 2007;81(19):10300–10309. doi: 10.1128/JVI.01149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inn K.S., Lee S.H., Rathbun J.Y., Wong L.Y., Toth Z., Machida K., Ou J.H., Jung J.U. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 2011;85(20):10899–10904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun C., Schattgen S.A., Pisitkun P., Jorgensen J.P., Hilterbrand A.T., Wang L.J., West J.A., Hansen K., Horan K.A., Jakobsen M.R., O’Hare P., Adler H., Sun R., Ploegh H.L., Damania B., Upton J.W., Fitzgerald K.A., Paludan S.R. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J. Immunol. 2015;194(4):1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong H., Wang D., Fang L., Zhang H., Luo R., Shang M., Ouyang C., Ouyang H., Chen H., Xiao S. Ubiquitin-specific proteases 25 negatively regulates virus-induced type I interferon signaling. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin D., Zhang M., Zhang M.X., Ren Y., Jin J., Zhao Q., Pan Z., Wu M., Shu H.B., Dong C., Zhong B. Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc. Natl. Acad. Sci. U. S. A. 2015;112(36):11324–11329. doi: 10.1073/pnas.1509968112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong B., Liu X., Wang X., Liu X., Li H., Darnay B.G., Lin X., Sun S.C., Dong C. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci. Signal. 2013;6(275):ra35. doi: 10.1126/scisignal.2003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong B., Liu X., Wang X., Chang S.H., Liu X., Wang A., Reynolds J.M., Dong C. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nat. Immunol. 2012;13(11):1110–1117. doi: 10.1038/ni.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan Y., Mao R., Yu Y., Liu S., Shi Z., Cheng J., Zhang H., An L., Zhao Y., Xu X., Chen Z., Kogiso M., Zhang D., Zhang H., Zhang P., Jung J.U., Li X., Xu G., Yang J. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J. Exp. Med. 2014;211(2):313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui J., Song Y., Li Y., Zhu Q., Tan P., Qin Y., Wang H.Y., Wang R.F. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 2014;24(4):400–416. doi: 10.1038/cr.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imaizumi T., Hayakari R., Watanabe S., Aizawa T., Matsumiya T., Yoshida H., Tsuruga K., Kawaguchi S., Tanaka H. Cylindromatosis (CYLD), a deubiquitinase, attenuates inflammatory signaling pathways by activating toll-like receptor 3 in human mesangial cells. Kidney Blood Press. Res. 2017;42(5):942–950. doi: 10.1159/000485084. [DOI] [PubMed] [Google Scholar]
- 27.Friedman C.S., O’Donnell M.A., Legarda-Addison D., Ng A., Cardenas W.B., Yount J.S., Moran T.M., Basler C.F., Komuro A., Horvath C.M., Xavier R., Ting A.T. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9(9):930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid U., Stenzel W., Koschel J., Raptaki M., Wang X., Naumann M., Matuschewski K., Schluter D., Nishanth G. The deubiquitinating enzyme cylindromatosis dampens CD8(+) T cell responses and is a critical factor for experimental cerebral malaria and blood-brain barrier damage. Front. Immunol. 2017;8:27. doi: 10.3389/fimmu.2017.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brummelkamp T.R., Nijman S.M., Dirac A.M., Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424(6950):797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 30.Boone D.L., Turer E.E., Lee E.G., Ahmad R.C., Wheeler M.T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 2004;5(10):1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 31.Hrdinka M., Fiil B.K., Zucca M., Leske D., Bagola K., Yabal M., Elliott P.R., Damgaard R.B., Komander D., Jost P.J., Gyrd-Hansen M. CYLD limits Lys63- and Met1-linked ubiquitin at receptor complexes to regulate innate immune signaling. Cell Rep. 2016;14(12):2846–2858. doi: 10.1016/j.celrep.2016.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79(7):4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289(50):34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450-451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H., Baker S.C., Feng L., Chen Z. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94(Pt 7):1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu X., Tian J., Kang H., Guo D., Liu J., Liu D., Jiang Q., Li Z., Qu J., Qu L. Transmissible gastroenteritis virus papain-like protease 1 antagonizes production of Interferon-beta through its deubiquitinase activity. Biomed Res. Int. 2017;2017 doi: 10.1155/2017/7089091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X., Chen X., Bian G., Tu J., Xing Y., Wang Y., Chen Z. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J. Gen. Virol. 2014;95(Pt 3):614–626. doi: 10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 38.Matthews K., Schafer A., Pham A., Frieman M. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol. J. 2014;11:209. doi: 10.1186/s12985-014-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Kasteren P.B., Bailey-Elkin B.A., James T.W., Ninaber D.K., Beugeling C., Khajehpour M., Snijder E.J., Mark B.L., Kikkert M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110(9):E838–47. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishikawa Y., Matsuzaki Y., Kimura K., Rokunohe A., Nakano H., Sawamura D. Modulation of stimulator of interferon genes (STING) expression by interferon-gamma in human keratinocytes. Biochem. Genet. 2018;56(1-2):93–102. doi: 10.1007/s10528-017-9832-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhang M., Zhang M.X., Zhang Q., Zhu G.F., Yuan L., Zhang D.E., Zhu Q., Yao J., Shu H.B., Zhong B. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Res. 2016;26(12):1302–1319. doi: 10.1038/cr.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z., Xian H., Hu J., Tian S., Qin Y., Wang R.F., Cui J. USP18 negatively regulates NF-kappaB signaling by targeting TAK1 and NEMO for deubiquitination through distinct mechanisms. Sci. Rep. 2015;5:12738. doi: 10.1038/srep12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X., Li H., Zhong B., Blonska M., Gorjestani S., Yan M., Tian Q., Zhang D.E., Lin X., Dong C. USP18 inhibits NF-kappaB and NFAT activation during Th17 differentiation by deubiquitinating the TAK1-TAB1 complex. J. Exp. Med. 2013;210(8):1575–1590. doi: 10.1084/jem.20122327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malakhova O.A., Kim K.I., Luo J.K., Zou W., Kumar K.G., Fuchs S.Y., Shuai K., Zhang D.E. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25(11):2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou W., Kim J.H., Handidu A., Li X., Kim K.I., Yan M., Li J., Zhang D.E. Microarray analysis reveals that Type I interferon strongly increases the expression of immune-response related genes in Ubp43 (Usp18) deficient macrophages. Biochem. Biophys. Res. Commun. 2007;356(1):193–199. doi: 10.1016/j.bbrc.2007.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilmes S., Beutel O., Li Z., Francois-Newton V., Richter C.P., Janning D., Kroll C., Hanhart P., Hotte K., You C., Uze G., Pellegrini S., Piehler J. Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J. Cell Biol. 2015;209(4):579–593. doi: 10.1083/jcb.201412049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlieker C., Korbel G.A., Kattenhorn L.M., Ploegh H.L. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 2005;79(24):15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S., Wang K., Li J., Zheng C. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J. Virol. 2013;87(21):11851–11860. doi: 10.1128/JVI.01211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye R., Su C., Xu H., Zheng C. Herpes simplex virus 1 ubiquitin-specific protease UL36 abrogates NF-kappaB activation in DNA sensing signal pathway. J. Virol. 2017;91(5) doi: 10.1128/JVI.02417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng D., Chen G., Guo B., Cheng G., Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18(11):1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang G., Chen G., Zheng D., Cheng G., Tang H. PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang J., Saad Y., Lei T., Wang J., Qi D., Yang Q., Kolattukudy P.E., Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 2010;207(13):2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X., Zhao Q., Xie Q., Xing Y., Chen Z. MCPIP1 negatively regulate cellular antiviral innate immune responses through DUB and disruption of TRAF3-TBK1-IKKepsilon complex. Biochem. Biophys. Res. Commun. 2018;503(2):830–836. doi: 10.1016/j.bbrc.2018.06.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ning S., Pagano J.S. The A20 deubiquitinase activity negatively regulates LMP1 activation of IRF7. J. Virol. 2010;84(12):6130–6138. doi: 10.1128/JVI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Wu L., Hong K., Pagano J.S. Intracellular signaling molecules activated by Epstein-barr virus for induction of interferon regulatory factor 7. J. Virol. 2001;75(24):12393–12401. doi: 10.1128/JVI.75.24.12393-12401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin R., Yang L., Nakhaei P., Sun Q., Sharif-Askari E., Julkunen I., Hiscott J. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J. Biol. Chem. 2006;281(4):2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 58.van Gent M., Braem S.G., de Jong A., Delagic N., Peeters J.G., Boer I.G., Moynagh P.N., Kremmer E., Wiertz E.J., Ovaa H., Griffin B.D., Ressing M.E. Epstein-Barr virus large tegument protein BPLF1 contributes to innate immune evasion through interference with toll-like receptor signaling. PLoS Pathog. 2014;10(2) doi: 10.1371/journal.ppat.1003960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng H., Gupta V., Patterson-Fortin J., Bhattacharya S., Katlinski K., Wu J., Varghese B., Carbone C.J., Aressy B., Fuchs S.Y., Greenberg R.A. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 2013;5(1):180–193. doi: 10.1016/j.celrep.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang X., Yu M., Ou Y., Cao Y., Yao Y., Cai P., Zhang F. Downregulation of USP4 promotes activation of microglia and subsequent neuronal inflammation in rat spinal cord after injury. Neurochem. Res. 2017;42(11):3245–3253. doi: 10.1007/s11064-017-2361-2. [DOI] [PubMed] [Google Scholar]
- 61.Xiao N., Li H., Luo J., Wang R., Chen H., Chen J., Wang P. Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFalpha-induced cancer cell migration. Biochem. J. 2012;441(3):979–986. doi: 10.1042/BJ20111358. [DOI] [PubMed] [Google Scholar]
- 62.Fan Y.H., Yu Y., Mao R.F., Tan X.J., Xu G.F., Zhang H., Lu X.B., Fu S.B., Yang J. USP4 targets TAK1 to downregulate TNFalpha-induced NF-kappaB activation. Cell Death Differ. 2011;18(10):1547–1560. doi: 10.1038/cdd.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 2005;7(8):758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Azevedo C.S., Guido B.C., Pereira J.L., Nolasco D.O., Correa R., Magalhaes K.G., Motta F.N., Santana J.M., Grellier P., Bastos I.M. Revealing a novel otubain-like enzyme from Leishmania infantum with deubiquitinating activity toward K48-linked substrate. Front. Chem. 2017;5:13. doi: 10.3389/fchem.2017.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shembade N., Ma A., Harhaj E.W. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327(5969):1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu T.T., Onizawa M., Hammer G.E., Turer E.E., Yin Q., Damko E., Agelidis A., Shifrin N., Advincula R., Barrera J., Malynn B.A., Wu H., Ma A. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity. 2013;38(5):896–905. doi: 10.1016/j.immuni.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Carbonell R., Wong J., Kim J.Y., Close L.A., Boland B.S., Wong T.L., Harris P.A., Ho S.B., Das S., Ernst P.B., Sasik R., Sandborn W.J., Bertin J., Gough P.J., Chang J.T., Kelliher M., Boone D., Guma M., Karin M. Elevated A20 promotes TNF-induced and RIPK1-dependent intestinal epithelial cell death. Proc. Natl. Acad. Sci. U. S. A. 2018;115(39):E9192–200. doi: 10.1073/pnas.1810584115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Enesa K., Zakkar M., Chaudhury H., Luong le A., Rawlinson L., Mason J.C., Haskard D.O., Dean J.L., Evans P.C. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J. Biol. Chem. 2008;283(11):7036–7045. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- 69.Ji Y., Cao L., Zeng L., Zhang Z., Xiao Q., Guan P., Chen S., Chen Y., Wang M., Guo D. The N-terminal ubiquitin-associated domain of Cezanne is crucial for its function to suppress NF-kappaB pathway. J. Cell. Biochem. 2018;119(2):1979–1991. doi: 10.1002/jcb.26359. [DOI] [PubMed] [Google Scholar]
- 70.Luong le A., Fragiadaki M., Smith J., Boyle J., Lutz J., Dean J.L., Harten S., Ashcroft M., Walmsley S.R., Haskard D.O., Maxwell P.H., Walczak H., Pusey C., Evans P.C. Cezanne regulates inflammatory responses to hypoxia in endothelial cells by targeting TRAF6 for deubiquitination. Circ. Res. 2013;112(12):1583–1591. doi: 10.1161/CIRCRESAHA.111.300119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S., Wang D., Zhao J., Weathington N.M., Shang D., Zhao Y. The deubiquitinating enzyme USP48 stabilizes TRAF2 and reduces E-cadherin-mediated adherens junctions. FASEB J. 2018;32(1):230–242. doi: 10.1096/fj.201700415RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolluri K.K., Alifrangis C., Kumar N., Ishii Y., Price S., Michaut M., Williams S., Barthorpe S., Lightfoot H., Busacca S., Sharkey A., Yuan Z., Sage E.K., Vallath S., Le Quesne J., Tice D.A., Alrifai D., von Karstedt S., Montinaro A., Guppy N., Waller D.A., Nakas A., Good R., Holmes A., Walczak H., Fennell D.A., Garnett M., Iorio F., Wessels L., McDermott U., Janes S.M. Loss of functional BAP1 augments sensitivity to TRAIL in cancer cells. Elife. 2018;7 doi: 10.7554/eLife.30224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheuermann J.C., de Ayala Alonso A.G., Oktaba K., Ly-Hartig N., McGinty R.K., Fraterman S., Wilm M., Muir T.W., Muller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465(7295):243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leznicki P., Natarajan J., Bader G., Spevak W., Schlattl A., Abdul Rehman S.A., Pathak D., Weidlich S., Zoephel A., Bordone M.C., Barbosa-Morais N.L., Boehmelt G., Kulathu Y. Expansion of DUB functionality generated by alternative isoforms - USP35, a case study. J. Cell. Sci. 2018;131(10) doi: 10.1242/jcs.212753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.D’Arcy P., Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int. J. Biochem. Cell Biol. 2012;44(11):1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 76.Sarhan D., Wennerberg E., D’Arcy P., Gurajada D., Linder S., Lundqvist A. A novel inhibitor of proteasome deubiquitinating activity renders tumor cells sensitive to TRAIL-mediated apoptosis by natural killer cells and T cells. Cancer Immunol. Immunother. 2013;62(8):1359–1368. doi: 10.1007/s00262-013-1439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh Y.T., Deng L., Deng J., Sun S.Y. The proteasome deubiquitinase inhibitor b-AP15 enhances DR5 activation-induced apoptosis through stabilizing DR5. Sci. Rep. 2017;7(1):8027. doi: 10.1038/s41598-017-08424-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kropp K.N., Maurer S., Rothfelder K., Schmied B.J., Clar K.L., Schmidt M., Strunz B., Kopp H.G., Steinle A., Grunebach F., Rittig S.M., Salih H.R., Dorfel D. The novel deubiquitinase inhibitor b-AP15 induces direct and NK cell-mediated antitumor effects in human mantle cell lymphoma. Cancer Immunol. Immunother. 2018;67(6):935–947. doi: 10.1007/s00262-018-2151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oh Y.T., Qian G., Deng J., Sun S.Y. Monocyte chemotactic protein-induced protein-1 enhances DR5 degradation and negatively regulates DR5 activation-induced apoptosis through its deubiquitinase function. Oncogene. 2018;37(25):3415–3425. doi: 10.1038/s41388-018-0200-9. [DOI] [PubMed] [Google Scholar]
- 80.Lopez-Castejon G., Luheshi N.M., Compan V., High S., Whitehead R.C., Flitsch S., Kirov A., Prudovsky I., Swanton E., Brough D. Deubiquitinases regulate the activity of caspase-1 and interleukin-1beta secretion via assembly of the inflammasome. J. Biol. Chem. 2013;288(4):2721–2733. doi: 10.1074/jbc.M112.422238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dufner A., Kisser A., Niendorf S., Basters A., Reissig S., Schonle A., Aichem A., Kurz T., Schlosser A., Yablonski D., Groettrup M., Buch T., Waisman A., Schamel W.W., Prinz M., Knobeloch K.P. The ubiquitin-specific protease USP8 is critical for the development and homeostasis of T cells. Nat. Immunol. 2015;16(9):950–960. doi: 10.1038/ni.3230. [DOI] [PubMed] [Google Scholar]
- 82.Damgaard R.B., Walker J.A., Marco-Casanova P., Morgan N.V., Titheradge H.L., Elliott P.R., McHale D., Maher E.R., McKenzie A.N.J., Komander D. The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell. 2016;166(5):1215–1230. doi: 10.1016/j.cell.2016.07.019. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keusekotten K., Elliott P.R., Glockner L., Fiil B.K., Damgaard R.B., Kulathu Y., Wauer T., Hospenthal M.K., Gyrd-Hansen M., Krappmann D., Hofmann K., Komander D. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153(6):1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jin J., Xie X., Xiao Y., Hu H., Zou Q., Cheng X., Sun S.C. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat. Immunol. 2016;17(3):259–268. doi: 10.1038/ni.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu Y., Carroll M., Papa F.R., Hochstrasser M., D’Andrea A.D. DUB-1, a deubiquitinating enzyme with growth-suppressing activity. Proc. Natl. Acad. Sci. U. S. A. 1996;93(8):3275–3279. doi: 10.1073/pnas.93.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jaster R., Zhu Y., Pless M. JAK2 is required for induction of the murine DUB-1 gene. Mol. Cell. Biol. 1997;17:3364–3372. doi: 10.1128/mcb.17.6.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu Y., Pless M., Inhorn R., Mathey-Prevot B., D’Andrea A.D. The murine DUB-1 gene is specifically induced by the betac subunit of interleukin-3 receptor. Mol. Cell. Biol. 1996;16(9):4808–4817. doi: 10.1128/mcb.16.9.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baek K.H., Kim M.S., Kim Y.S., Shin J.M., Choi H.K. DUB-1A, a novel deubiquitinating enzyme subfamily member, is polyubiquitinated and cytokine-inducible in B-lymphocytes. J. Biol. Chem. 2004;279(4):2368–2376. doi: 10.1074/jbc.M304774200. [DOI] [PubMed] [Google Scholar]
- 89.Migone T.S., Humbert M., Rascle A., Sanden D., D’Andrea A., Johnston J.A. The deubiquitinating enzyme DUB-2 prolongs cytokine-induced signal transducers and activators of transcription activation and suppresses apoptosis following cytokine withdrawal. Blood. 2001;98(6):1935–1941. doi: 10.1182/blood.v98.6.1935. [DOI] [PubMed] [Google Scholar]
- 90.Zhu Y., Lambert K., Corless C., Copeland N.G., Gilbert D.J., Jenkins N.A., D’Andrea A.D. DUB-2 is a member of a novel family of cytokine-inducible deubiquitinating enzymes. J. Biol. Chem. 1997;272(1):51–57. doi: 10.1074/jbc.272.1.51. [DOI] [PubMed] [Google Scholar]
- 91.Meenhuis A., Verwijmeren C., Roovers O., Touw I.P. The deubiquitinating enzyme DUB2A enhances CSF3 signalling by attenuating lysosomal routing of the CSF3 receptor. Biochem. J. 2011;434(2):343–351. doi: 10.1042/BJ20101628. [DOI] [PubMed] [Google Scholar]
- 92.Baek K.H., Mondoux M.A., Jaster R., Fire-Levin E., D’Andrea A.D. DUB-2A, a new member of the DUB subfamily of hematopoietic deubiquitinating enzymes. Blood. 2001;98(3):636–642. doi: 10.1182/blood.v98.3.636. [DOI] [PubMed] [Google Scholar]
- 93.Ramakrishna S., Suresh B., Lee E.J., Lee H.J., Ahn W.S., Baek K.H. Lys-63-specific deubiquitination of SDS3 by USP17 regulates HDAC activity. J. Biol. Chem. 2011;286(12):10505–10514. doi: 10.1074/jbc.M110.162321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burrows J.F., McGrattan M.J., Rascle A., Humbert M., Baek K.H., Johnston J.A. DUB-3, a cytokine-inducible deubiquitinating enzyme that blocks proliferation. J. Biol. Chem. 2004;279(14):13993–14000. doi: 10.1074/jbc.M311291200. [DOI] [PubMed] [Google Scholar]
- 95.Burrows J.F., Kelvin A.A., McFarlane C., Burden R.E., McGrattan M.J., De la Vega M., Govender U., Quinn D.J., Dib K., Gadina M., Scott C.J., Johnston J.A. USP17 regulates Ras activation and cell proliferation by blocking RCE1 activity. J. Biol. Chem. 2009;284(14):9587–9595. doi: 10.1074/jbc.M807216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu Z., Zheng Y., Zhu Y., Kong X., Hu L. Evidence for OTUD-6B participation in B lymphocytes cell cycle after cytokine stimulation. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0014514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malakhov M.P., Malakhova O.A., Kim K.I., Ritchie K.J., Zhang D.E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2002;277(12):9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 98.Opipari A.W., Jr., Boguski M.S., Dixit V.M. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J. Biol. Chem. 1990;265(25):14705–14708. [PubMed] [Google Scholar]