Abstract
Over the past decade, microarrays have revolutionized the scientific world as dramatically as the internet has changed everyday life. From the initial applications of DNA microarrays to uncover gene expression patterns that are diagnostic and prognostic of cancer, understanding the interplay between immune responses and disease has been a prime application of this technology. More recent efforts have moved beyond genetic analysis to functional analysis of the molecules involved, including identification of immunodominant antigens and peptides as well as the role of post-translational glycosylation. Here, we focus on recent applications of microarray technology in understanding the detailed chemical biology of immune responses to disease in an effort to guide development of vaccines and other protective therapies.
Introduction
Remarkably, live rotavirus vaccines, which were introduced in 2004, were developed using essentially the same empirical methods for attenuation as the Sabin polio vaccine nearly 50 years earlier: loss of virulence through multiple passages in non-human cells. The vaccine seems to be safe and effective and is desperately needed to prevent the 600 000 deaths annually that are attributed to rotavirus [1]. But its history underscores that, despite the profound role of vaccines in reducing human and animal morbidity and mortality, the field has relied on technological advances in other areas to spur its own development. In particular, the dual advances of genomic sequencing and microarray design have resulted in a renaissance of research in immunity and infectious diseases.
The applications of microarrays span from the bench to the bedside, providing tools that require less effort, expense and sample than other technologies and which are also highly multiplexed, building on established pattern-recognition techniques and statistics [2]. In this review, we do not discuss technical aspects common to all arrays (e.g. statistical analysis and immobilization chemistry) because these are extensively reviewed elsewhere. Instead, we limit ourselves to novel applications in infection and immunity using four variants of array technology: DNA, antigen, peptide–MHC complex (pMHC) and carbohydrate (Figure 1 ). Collectively, these technologies are already advancing our understanding of the interplay between immunity and disease, providing a rational basis for the design of vaccines and agents that interfere with disease progression (Table 1 ).
Figure 1.

Opportunities for arrays in infection and immunity. Aspects of a pathogen that are now accessible to array analysis include the genetic material (transmissible elements and gene variants) and the binding specificity and temporal expression of carbohydrates and lectins involved in, for instance, host cell attachment. The antigenicity of surface exposed and secreted molecules can also be assessed at the genome-wide scale to aid in vaccine and diagnostics development. From the host perspective, arrays can reveal immune cell responses in terms of transcriptional responses, antibody-binding specificity, T cell–pMHC reactivity and the functional consequences of T cell activation.
Table 1.
Recent applications of arrays in infection and immunity
| Array format | Biomarker discovery | Diagnostic profiles | Vaccine candidates | Surveillance/epidemiology | Number of unique elementsa |
|---|---|---|---|---|---|
| DNA | – | [7, 15••, 19, 22, 49] | – | [15••, 20•, 24•, 27•] | 55–22 000 |
| Antigen/ORFeome | [51, 55, 57•] | [52, 54, 56•, 58••, 124] | [50, 125] | – | 22–232 |
| pMHC | [65•, 66] | – | [67••] | – | 7 |
| Glycan | – | [79, 91•, 97, 126] | [80•, 81•, 90, 93, 127] | [95••] | 9–200 |
| Lectin | – | [105] | – | – | 9–21 |
Refers to the number of unique elements (e.g. DNA oligonucleotides) individually produced and immobilized on the array.
Genome arrays for disease surveillance, diagnosis and characterization
Since the days of Pasteur, pathogen identification has been accomplished using a combination of culture and serological techniques, all of which are labor-intensive, require highly trained personnel and incur a delay of hours to days before a conclusion can be reached. Furthermore, many organisms are refractory to culture, whereas serotyping is limited by the availability of specific antisera — a situation especially problematic when emerging or evolving pathogens are considered. Because of the availability of numerous genome sequences and the need to contain rising healthcare costs, DNA microarrays that can simultaneously probe clinical and environmental samples for the presence of conserved viral and bacterial sequences, specific virulence factors and antimicrobial resistance genes, and can even identify point mutations, present a novel alternative (Figure 2 ) [3, 4].
Figure 2.
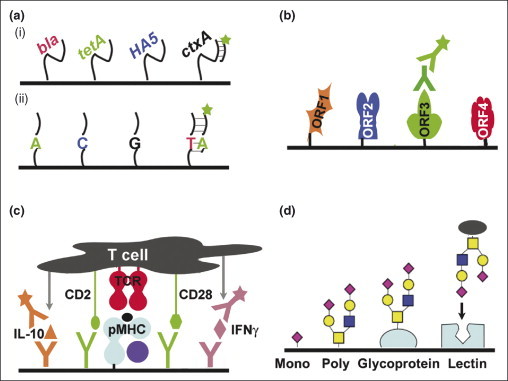
Arrays used to study infection and immunity. (a) DNA arrays. (i) Immobilized DNA sequences can be chosen to represent highly conserved regions from a broad range of organisms, as in the pan Virochip, and used for disease diagnosis. Alternatively, the DNA chosen can be more specialized, detecting clinically relevant genes (e.g. antibiotic resistance and export pump genes, key antigens for serotyping and virulence factors capable of horizontal transmission). Sample nucleic acids are purified, fluorescently labelled and allowed to specifically hybridize with immobilized complementary DNA. (ii) Use of resequencing arrays provides single base pair information, which is useful when tracking genetic drift and single nucleotide polymorphisms. (b) Antigen arrays to monitor humoral immunity. Open reading frames (or peptides) from a genome are expressed recombinantly, purified and immobilized in an array. Serum from an infected, recovered or vaccinated individual is incubated with the array; bound antibodies are detected by subsequent incubation with a fluorescently labeled secondary antibody. These arrays can be used to rapidly characterize the protective immunome of an organism, to identify novel vaccine candidates and to compare vaccine-induced humoral responses with those resulting from natural infection. (c) pMHC arrays to monitor cellular immunity. A variety of soluble peptide–MHC (pMHC) complexes are immobilized with co-stimulatory antibodies to form an array of artificial antigen-presenting cells. Fluorescently labeled T cell populations can be incubated with the array to quantify the fraction specific for a given pMHC. Additionally, antibodies can be co-deposited with the pMHC to capture locally secreted cytokines in a sandwich assay (illustrated here for interleukin 10 and interferon–γ). The nature of the cytokines released reveals the responses of the T cells to stimulation with a particular pMHC. Using secondary antibodies with different fluorescent labels, the assay can be multiplexed to detect multiple cytokines simultaneously. (d) Glycoarrays and lectin arrays. Monosaccharides, oligosaccharides and polysaccharides of varying sequence and structure, in addition to glycoproteins and lectins, are immobilized in an array. Fluorescently labeled proteins, viruses or bacterial cells are applied to the array to assess their carbohydrate-binding specificity; for example, the species specificity of bird and human influenza variants can be attributed to the glycan-binding propensitites of the respective hemagglutinin variants. In the case of lectin arrays, these arrays can rapidly assess the nature of the glycans attached to the protein or cell surface.
This concept is elegantly illustrated by the pan-viral DNA microarray (Virochip), which comprises highly conserved 70-mer oligonucleotides from every partially and fully sequenced viral genome in GenBank (as of June 2004). The third generation chip includes 22 000 oligonucleotides, representing ∼277 000 sequences [5, 6]. Patient samples are collected by nasal lavage, the ribonucleic acids are purified, reverse-transcribed, amplified by random-primer PCR, and finally incubated with the array under stringent binding conditions. Using such an array, deRisi and colleagues were able to correctly identify a range of viruses from the RNA of infected tissue culture cells and human samples [5]. More impressively, the array was subsequently used to identify, isolate and even sequence ∼1000 bases of a virus now known as the severe acute respiratory syndrome coronavirus (SARS-CoV) [7, 8, 9]. Originally designed as an experimental research tool, the Virochip also seems to be capable of viral diagnosis in a clinical setting [6, 10].
Although the success of the Virochip is impressive, significant hurdles remain for broad application of this technology, primarily in terms of sample amplification and probe design [11, 12]. Other arrays under development operate at three levels of detail, each of which can be used in surveillance, diagnostic or vaccine development programs to answer the following questions:
Which organisms are present?
Arrays at this level include the Virochip and other microarrays for more specialized detection of viruses of the central nervous system [13] respiratory pathogens [14], and for bacterial strain identification [15••, 16, 17]. Importantly, these chips have demonstrated the ability to detect bacteria in the viable but non-culturable state and are amenable to automation [18].
Which genes are present?
Clinically, the presence of genes or mutations that confer antibiotic resistance influences treatment options [19, 20•, 21], whereas transmissible virulence factors and serotypes can be used for strain identification and to indicate the pathogenic potential of the organism [15••, 22]. Similarly, genes that are differentially present in pathogenic versus commensal or live attenuated strains can guide vaccine development [23].
Which gene variants are present?
Surveillance and molecular epidemiological programs are developing focused arrays to track antigenic drift, anticipate dominant serotypes and monitor the genomes of live attenuated vaccine strains [24•, 25, 26]. For instance, the low-density FluChip can distinguish all influenza A hemagglutinin and neuraminidase subtypes, and tiled resequencing arrays can detect single nucleotide polymorphisms within these genes [24•, 27•, 28, 29].
The complement to specific pathogen hybridization is interrogation of host transcriptional responses [30, 31], primarily using peripheral blood mononuclear cells. The premise is that all coronaviruses will induce similar host responses, and these responses will be distinguishable from those induced by, for instance, bacterial sepsis. Thus, transcriptional profiling could be used to diagnose disease completely independent of any knowledge of the pathogen or even after the pathogen has been cleared from the system. Despite preliminary successes [32, 33, 34], this approach has met with several obstacles in terms of individual heterogeneity and strong stereotyped inflammatory responses mediated by the nuclear factor NFκ-B, which obscure pathogen-specific responses [35, 36, 37].
Antigen arrays to monitor humoral immunity
Once the genes harbored by an organism have been identified, the next step is to probe the host immune responses to the genes products. Which protein antigens are recognized? And which confer lasting immunity? Antibody recognition of a set of antigens can sensitively diagnose disease [38] and an immunodominant antigen can be a candidate target for passive or active vaccination [39]. Antigen arrays are a natural fit for biomarker discovery and complement recent advances in vaccine development.
One advance, termed reverse vaccinology, capitalizes on the available genome sequence of a pathogen that is refractory to traditional vaccine development strategies [40, 41, 42, 43]. In silico techniques identify conserved open reading frames (ORFs) predicted to encode surface exposed or secreted proteins; hundreds of these are cloned in Escherichia coli, expressed and purified in parallel and used to immunize mice [44]. Proteins that either protect mice from subsequent challenge or induce sera that are protective in in vitro assays are pursued. First applied to serotype B Neisseria meningococcus, researchers ultimately queried a metagenomic database using sequence data from eight strains [44, 45]. Five antigens were identified; none of these was broadly protective alone but together they induced bactericidal antibodies in mice against 66 of 85 meningococcal strains [45].
A key element of reverse vaccinology is analysis of immune sera reactivity and it is here that protein antigen arrays are beginning to play a role [46]. Instead of immunizing mice directly, proteins corresponding to the entire ORFeome (or fractionated lysate [47]) are spotted onto a glass slide. Naïve, convalesent or immune sera are applied to the array, followed by a fluorescent secondary antibody (e.g. goat anti-mouse IgG). After washing, the array is scanned and fluorescence intensity is used to indicate the presence of antibodies that recognize the antigen immobilized in that spot (Figure 2). In this way, antigenic proteins can be rapidly recognized, the response of an individual to vaccination or infection monitored, and infections past and present diagnosed. Therapeutic monoclonal antibodies, destined for passive vaccination, can also be documented for cross-reactivity on a broad scale [48].
Such arrays are being developed for multiple diseases, including tuberculosis [49], Yersinia pestis [50], Neisseria meningitidis [51], leprosy [52] and HIV [53], and also for autoimmune diseases [38, 54] and tumor-associated antigens [55]. Early successes include a vaccinia viral array consisting of 185 proteins that were probed using sera from naïve and immunized mice, non-human primates and humans [56•]. Interestingly, the three species did not recognize the same subsets of viral proteins. The array was later used to identify the H3L envelope protein as the immunodominant antigen in the live viral vaccine [57•], perhaps paving the way for a less traumatic subunit vaccine. Similarly, a diagnostic array representing the entire ORFeome of SARS-CoV and portions of five additional coronaviruses was developed and tested using serum from 400 Canadian and 206 Chinese patients [58••]. The array was shown to be at least as sensitive as and more specific than enzyme-linked immunosorbent assay (ELISA) tests for diagnosing SARS, requiring minimal sample processing compared with genome chips.
pMHC arrays to monitor cellular immunity
Cellular responses have always been harder to study than antibody responses: antigen binding, as opposed to the high-affinity binding reaction between two soluble molecules, involves a low affinity tri-molecular interaction that comprises two membrane-bound molecules and a post-translationally processed peptide. From the standpoint of vaccine development or targeted therapies, is it important to identify not only the amino acid sequence corresponding to a key peptide epitope but also the functional T cell responses that result from recognition.
Given these constraints, it is hard to imagine a screening technology that does not involve a cellular readout. Phage and cDNA display technologies, widely used to study antibody–antigen interactions, have been difficult to apply to analysis of T cell receptor (TCR)—pMHC interactions [59, 60, 61]. Non-genetic approaches involve incubating synthetic peptides with antigen-presenting cells and T cells, with stimulatory peptides identified by interleukin 2 release [62]. Computational prediction methods, especially for class I MHC, are also improving but still require experimental validation [63]. For epitopes that have been identified, enzyme-linked immunosorbent spot (ELIspot) and flow cytometry assays using tetramerized pMHC have found wise-spread use to monitor the spatial and temporal presence of cognate T cells [64].
The opportunities for arrays in analysis of cellular immunity are threefold: (i) to quantify the fraction of T cells in a population reacting with a given pMHC; (ii) to identify crucial peptide epitopes from candidate sequences; and (iii) to assess T cell responses resulting from recognition of these peptides. Recent reports have devised strategies to achieve these goals by using pMHC arrays [65•, 66, 67••, 68]. In their current form, each feature on the array contains immobilized pMHC molecules, co-stimulatory antibodies and cytokine capture antibodies. Thus, a fluorescent T cell recognizing a particular pMHC will bind to the spot, be activated and secrete cytokines locally. These cytokines are captured and subsequently detected using fluorescent antibodies (Figure 2). Not only can the number of T cells bound to a spot be counted, replicating flow cytometry assays, but cellular responses can also be elucidated based on the cytokines released. Variations in design reveal the detailed consequences of binding to a single pMHC (using multicolor secondary antibodies to quantify levels of a series of cytokines) or to a peptide sequence (using arrays of peptide variants to identify agonist and antagonist peptides).
After the initial report on use of pMHC microarrays [66], Stone et al. used a modified approach to match T cell lines to activating viral epitopes [65•]. In the first clinical application of the technology, Chen et al. used the array to analyze patient T cell responses to a peptide vaccine against melanoma [67••]. Ten patients were immunized, and CD8+ T cell responses to seven pMHC and 26 secreted factors were measured using an array. Interestingly, the investigators were able to detect fractional T cell abundances as low as one cell in 10 000 (0.01%) and they identified a correlation between the functional profile and clinical outcome of the patients.
These early successes indicate that pMHC arrays might be used broadly in vaccine and therapeutic development [68, 69]. For instance, they could identify tumor-associated antigens from a panel of candidates for targeting by TCR therapeutics [70, 71] or could identify viral peptides for inclusion in epitope vaccines. Conversely, the arrays could be used during de-immunization of therapeutic proteins. Although not yet demonstrated, the experiment could also be inverted, using soluble TCRs to capture cells that present specific pMHC for diagnosis or to assess vaccine-induced cellular immunity.
Carbohydrate arrays
Carbohydrates consist of sugar units, including monosaccharides, disaccharides, oligosaccharides and polysaccharides, that can be linked to proteins as linear or branched extensions with varying connectivity. These molecules can profoundly affect protein folding and solubility, pathogen infection and immune system responses [72] and can form the basis of several vaccines (e.g. Haemophilus influenzae type b) [73]. However, because of their intrinsic heterogeneity and non-template-driven biosynthesis, identifying and characterizing the linkage of sugar groups, for example by HPLC of enzymatically released carbohydrates, has been difficult. Recent and ongoing development of glycan arrays has been motivated largely by the Consortium for Functional Glycomics (http://www.functionalglycomics.org/) and has been made possible by several technical breakthroughs, including advances in carbohydrate immobilization [74, 75, 76, 77, 78].
The first arrays consisted of a variety of sugars deposited on a slide and were used to profile the glycan-binding specificity of fluorescently-labeled anti-carbohydrate monoclonal antibodies, lectins and bacterial toxins [79, 80•, 81•, 82, 83, 84, 85, 86, 87, 88]. These might be useful for future development of multivalent toxin inhibitors [89] or peptide mimetics for immunization. Similar arrays have been used to demonstrate potential cross-reactivity between the immune response to an attenuated SARS vaccine and a self-carbohydrate [90]. Moving towards diagnostic glycoarrays, the GloboH hexsaccharide cancer marker and nine analogs were arrayed and used to test monoclonal antibodies and patient sera for GoboH-specific binding [91•].
A major application of these arrays has been to dissect the chemical biology of pathogen–host cell attachment. A rare and potently neutralizing antibody, 2G12, protects against viral challenge in vivo in animal models of HIV infection, by binding terminal Man α1-2Man residues on gp120 [92]. Carbohydrate arrays have been developed to characterize the affinity and structural specificity of 2G12 mannose recognition compared with other mannose-binding or gp120-binding proteins to develop a carbohydrate template for HIV vaccine design [81•, 93, 94]. With influenza A viruses, arrays have probed the basis of species specificity — a crucial aspect when evaluating serotypes for pandemic potential. The virus invades cells by hemagglutinin binding to cell surface sialic acid residues, which vary in structure based on the host species and anatomical location. Binding of hemagglutinin variants recovered from pandemic and circulating strains on a 260-member glycan array demonstrated differences in recognition of carbohydrate linkages (α2-3 or α2-6, characteristic of avian and human viruses, respectively), fucosylation and sulfation. Interestingly, a single amino acid change (Asp255Gly) in the pandemic 1918 H1 was found to switch specificity from exclusively α2-6 to mixed specificity, whereas Asp190Glu conferred complete reversion [95••, 96].
A different approach is the use of these arrays to detect pathogens directly and indirectly. Bacterial glyco-conjugates have been arrayed and interrogated with sera in an effort to determine an individual's prior exposure to the corresponding microbe [76]. Similarly, host carbohydrates and glycoproteins that are used as bacterial receptors have been arrayed, followed by specific capture of bacteria binding those receptors. This approach even allowed for microorganism recovery from arrayed spots for subsequent growth and antibiotic susceptibility testing [97, 98].
Once lectin specificity has been determined (perhaps using comprehensive glycan arrays), the proteins can be employed to generate complementary lectin arrays. Here, the carbohydrate-binding proteins are immobilized and incubated with fluorescently labeled molecules or cells to assess the carbohydrate moieties [99, 100, 101, 102, 103, 104]. Although this format is still in its early stages, one array that contains 21 commercially available lectins was used not only to discriminate between strains of laboratory and pathogenic E. coli bacteria based on whole-cell binding patterns, but also to track the temporal expression of different glycans during the growth cycle [105•]. A limitation is the number of well-characterized lectins and antibodies available, but mechanisms for creating lectin diversity exist in nature and could be applied to the engineering of specific lectin properties [106].
One-third of approved biopharmaceuticals are glycoproteins, and the carbohydrate components of these have long been known to affect functions such as circulating half life, solubility and, for antibodies, complement activity and Fc-receptor binding specificity [107]. These effects have been primary forces in motivating the homogenization of carbohydrates on protein therapeutics expressed in recombinant hosts [108, 109, 110]. The recent discovery that naturally occurring antibody glycoforms vary in their ability to mediate inflammation and cytotoxicity (crucial for autoimmune and cancer treatments, respectively) [111•, 112] has provided additional impetus to control post-translational modifications. Lectin arrays could aid in characterizing, optimizing and monitoring the quality of biologic therapies [99].
Emerging technologies
In addition to concerns common to all microarray applications, advances in several areas are likely to be directly applicable to studies in infection and immunity.
-
1.
One approach is to streamline cloning, expression and purification of entire ORFeomes for antigen arrays. Such methods include those borrowed from structural genomics (e.g. tags for purification and immobilization [113]) and those that generate proteins directly from DNA (e.g. spotted viral or cell particles that present the protein of interest on their surface [55, 114], in vitro transcription and translation of spotted transcriptionally active DNA to produce and directly capture expressed protein [56•, 115], and spotted lentiviral arrays to directly transfect overlaid eukaryotic cells [116]).
-
2.
A second important area of development is movement into the array format of technologies that have expanded capabilities compared with most current arrays, which measure binding under near-equilibrium conditions [117]. Newer formats can extract kinetic (e.g. the BIAcore flexchip), force (e.g. Bioforce Nanosciences) [118, 119] or thermodynamic [120] parameters associated with the interaction, permitting rigorous quantitative comparisons and providing mechanistic insight.
-
3.
A third approach involves high-throughput array-based analysis of additional post-translational modifications, such as phosphorylation [121] and lipidation [122, 123].
Concluding remarks
Now that many of the technical hurdles have been addressed, microarrays with new and expanded capabilities (Figure 3 ) can monitor the genome of a pathogen with single nucleotide precision, identify antigens that stimulate both arms of the immune system, and even investigate the role of post-translational modifications. These new arrays are being used to probe the interactions between immunity and disease, and are already resulting in significant discoveries regarding the molecular mechanisms of disease, vaccine development and novel therapeutics. Consistent with the history of infectious diseases and technology, these newer technologies will lead to many more exciting discoveries.
Figure 3.
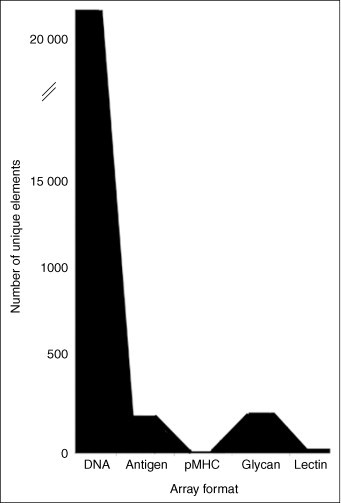
Complexity of arrays. Depending on the ease or difficulty of producing the material used for array elements, different array formats vary greatly in the number of unique molecules that are assessed. These range from as few as seven (for the newcomer, the pMHC array) to >22 000 (for DNA oligonucleotide arrays).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
References
- 1.Glass R.I., Parashar U.D., Bresee J.S., Turcois R., Fischer T.K., Widdowson M.A., Jiang B., Gentsch J.R. Rotavirus vaccines: current prospects and future challenges. Lancet. 2006;368:323–332. doi: 10.1016/S0140-6736(06)68815-6. [DOI] [PubMed] [Google Scholar]
- 2.Royce T.E., Rozowsky J.S., Luscombe N.M., Emanuelsson O., Yu H., Zhu X., Snyder M., Gerstein M.B. Extrapolating traditional DNA microarray statistics to tiling and protein microarray technologies. Methods Enzymol. 2006;411:282–311. doi: 10.1016/S0076-6879(06)11015-0. [DOI] [PubMed] [Google Scholar]
- 3.Dietrich G. DNA microarrays in vaccine research. Curr Opin Mol Ther. 2003;5:575–583. [PubMed] [Google Scholar]
- 4.Bryant P.A., Venter D., Robins-Browne R., Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect Dis. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- 5.Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu C.Y., Rouskin S., Koshy A., Urisman A., Fischer K., Yagi S., Schnurr D., Eckburg P.B., Tompkins L.S., Blackburn B.G. Microarray detection of human Parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin Infect Dis. 2006;43:e71–e76. doi: 10.1086/507896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Urisman A., Liu Y.T., Springer M., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 9.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 10.Urisman A., Molinaro R.J., Fischer N., Plummer S.J., Casey G., Klein E.A., Malathi K., Magi-Galluzzi C., Tubbs R.R., Ganem D. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Urisman A., Fischer K.F., Chiu C.Y., Kistler A.L., Beck S., Wang D., DeRisi J.L. E-predict: a computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol. 2005;6:R78. doi: 10.1186/gb-2005-6-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loy A., Bodrossy L. Highly parallel microbial diagnostics using oligonucleotide microarrays. Clin Chim Acta. 2006;363:106–119. doi: 10.1016/j.cccn.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 13.Conejero-Goldberg C., Wang E., Yi C., Goldberg T.E., Jones-Brando L., Marincola F.M., Webster M.J., Torrey E.F. Infectious pathogen detection arrays: viral detection in cell lines and postmortem brain tissue. Biotechniques. 2005;39:741–751. doi: 10.2144/000112016. [DOI] [PubMed] [Google Scholar]
- 14.Lin B., Wang Z., Vora G.J., Thornton J.A., Schnur J.M., Thach D.C., Blaney K.M., Ligler A.G., Malanoski A.P., Santiago J. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 2006;16:527–535. doi: 10.1101/gr.4337206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Vora G.J., Meador C.E., Bird M.M., Bopp C.A., Andreadis J.D., Stenger D.A. Microarray-based detection of genetic heterogeneity, antimicrobial resistance, and the viable but nonculturable state in human pathogenic Vibrio spp. Proc Natl Acad Sci USA. 2005;102:19109–19114. doi: 10.1073/pnas.0505033102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Uses DNA arrays to monitor genetic composition of viable but non-culturable bacterial strains.
- 16.Chandler D.P., Alferov O., Chernov B., Daly D.S., Golova J., Perov A., Protic M., Robison R., Schipma M., White A. Diagnostic oligonucleotide microarray fingerprinting of Bacillus isolates. J Clin Microbiol. 2006;44:244–250. doi: 10.1128/JCM.44.1.244-250.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burton J.E., Oshota O.J., North E., Hudson M.J., Polyanskaya N., Brehm J., Lloyd G., Silman N.J. Development of a multi-pathogen oligonucleotide microarray for detection of Bacillus anthracis. Mol Cell Probes. 2005;19:349–357. doi: 10.1016/j.mcp.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Malanoski A.P., Lin B., Wang Z., Schnur J.M., Stenger D.A. Automated identification of multiple micro-organisms from resequencing DNA microarrays. Nucleic Acids Res. 2006;34:5300–5311. doi: 10.1093/nar/gkl565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davignon L., Walter E.A., Mueller K.M., Barrozo C.P., Stenger D.A., Lin B. Use of resequencing oligonucleotide microarrays for identification of Streptococcus pyogenes and associated antibiotic resistance determinants. J Clin Microbiol. 2005;43:5690–5695. doi: 10.1128/JCM.43.11.5690-5695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Albert T.J., Dailidiene D., Dailide G., Norton J.E., Kalia A., Richmond T.A., Molla M., Singh J., Green R.D., Berg D.E. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat Methods. 2005;2:951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]; Demonstrates that DNA arrays can be used for comparative genome sequencing.
- 21.Denkin S., Volokhov D., Chizhikov V., Zhang Y. Microarray-based pncA genotyping of pyrazinamide-resistant strains of Mycobacterium tuberculosis. J Med Microbiol. 2005;54:1127–1131. doi: 10.1099/jmm.0.46129-0. [DOI] [PubMed] [Google Scholar]
- 22.Palaniappan R.U., Zhang Y., Chiu D., Torres A., Debroy C., Whittam T.S., Chang Y.F. Differentiation of Escherichia coli pathotypes by oligonucleotide spotted array. J Clin Microbiol. 2006;44:1495–1501. doi: 10.1128/JCM.44.4.1495-1501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behr M.A., Wilson M.A., Gill W.P., Salamon H., Schoolnik G.K., Rane S., Small P.M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 24•.Townsend M.B., Dawson E.D., Mehlmann M., Smagala J.A., Dankbar D.M., Moore C.L., Smith C.B., Cox N.J., Kuchta R.D., Rowlen K.L. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J Clin Microbiol. 2006;44:2863–2871. doi: 10.1128/JCM.00134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes a low-density DNA array that can distinguish rapidly between circulating influenza A and B subtypes
- 25.Beres S.B., Richter E.W., Nagiec M.J., Sumby P., Porcella S.F., DeLeo F.R., Musser J.M. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A streptococcus. Proc Natl Acad Sci USA. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neverov A.A., Riddell M.A., Moss W.J., Volokhov D.V., Rota P.A., Lowe L.E., Chibo D., Smit S.B., Griffin D.E., Chumakov K.M. Genotyping of measles virus in clinical specimens on the basis of oligonucleotide microarray hybridization patterns. J Clin Microbiol. 2006;44:3752–3759. doi: 10.1128/JCM.00998-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Wang Z., Daum L.T., Vora G.J., Metzgar D., Walter E.A., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. Identifying influenza viruses with resequencing microarrays. Emerg Infect Dis. 2006;12:638–646. doi: 10.3201/eid1204.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that DNA resequencing arrays can be used in viral surveillance programs.
- 28.Sulaiman Irshad M., Liu X., Frace M., Sulaiman N., Olsen-Rasmussen M., Neuhaus E., Rota P.A., Wohlhueter R.M. Evaluation of Affymetrix severe acute respiratory syndrome resequencing genechips in characterization of the genomes of two strains of coronavirus infecting humans. Appl Environ Microbiol. 2006;72:207–211. doi: 10.1128/AEM.72.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lodes M.J., Suciu D., Elliott M., Stover A.G., Ross M., Caraballo M., Dix K., Crye J., Webby R.J., Lyon W.J. Use of semiconductor-based oligonucleotide microarrays for influenza A virus subtype identification and sequencing. J Clin Microbiol. 2006;44:1209–1218. doi: 10.1128/JCM.44.4.1209-1218.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M., Popper S.J., Rubins K.H., Relman D.A. Early days: genomics and human responses to infection. Curr Opin Microbiol. 2006;9:312–319. doi: 10.1016/j.mib.2006.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenner R.G., Young R.A. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 32.Huang Q., Liu D., Majewski P., Schulte L.C., Korn J.M., Young R.A., Lander E.S., Hacohen N. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 33.Boldrick J.C., Alizadeh A.A., Diehn M., Dudoit S., Liu C.L., Belcher C.E., Botstein D., Staudt L.M., Brown P.O., Relman D.A. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci USA. 2002;99:972–977. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubins K.H., Hensley L.E., Jahrling P.B., Whitney A.R., Geisbert T.W., Huggins J.W., Owen A., Leduc J.W., Brown P.O., Relman D.A. The host response to smallpox: analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc Natl Acad Sci USA. 2004;101:15190–15195. doi: 10.1073/pnas.0405759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitney A.R., Diehn M., Popper S.J., Alizadeh A.A., Boldrick J.C., Relman D.A., Brown P.O. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCaffrey R.L., Fawcett P., O’Riordan M., Lee K.D., Havell E.A., Brown P.O., Portnoy D.A. A specific gene expression program triggered by gram-positive bacteria in the cytosol. Proc Natl Acad Sci USA. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thach D.C., Agan B.K., Olsen C., Diao J., Lin B., Gomez J., Jesse M., Jenkins M., Rowley R., Hanson E. Surveillance of transcriptomes in basic military trainees with normal, febrile respiratory illness, and convalescent phenotypes. Genes Immun. 2005;6:588–595. doi: 10.1038/sj.gene.6364244. [DOI] [PubMed] [Google Scholar]
- 38.Robinson W.H., DiGennaro C., Hueber W., Haab B.B., Kamachi M., Dean E.J., Fournel S., Fong D., Genovese M.C., de Vegvar H.E. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 39.Maynard J.A., Maassen C.B., Leppla S.H., Brasky K., Patterson J.L., Iverson B.L., Georgiou G. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 40.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S. Genome analysis of multiple pathogenic isolates of streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maione D., Margarit I., Rinaudo C.D., Masignani V., Mora M., Scarselli M., Tettelin H., Brettoni C., Iacobini E.T., Rosini R. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stranger-Jones Y.K., Bae T., Schneewind O. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci USA. 2006;103:16942–16947. doi: 10.1073/pnas.0606863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scarselli M., Giuliani M.M., Adu-Bobie J., Pizza M., Rappuoli R. The impact of genomics on vaccine design. Trends Biotechnol. 2005;23:84–91. doi: 10.1016/j.tibtech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Pizza M., Scarlato V., Masignani V., Giuliani M.M., Arico B., Comanducci M., Jennings G.T., Baldi L., Bartolini E., Capecchi B. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 45.Giuliani M.M., Adu-Bobie J., Comanducci M., Arico B., Savino S., Santini L., Brunelli B., Bambini S., Biolchi A., Capecchi B. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci USA. 2006;103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bacarese-Hamilton T., Gray J., Crisanti A. Protein microarray technology for unraveling the antibody specificity repertoire against microbial proteomes. Curr Opin Mol Ther. 2003;5:278–284. [PubMed] [Google Scholar]
- 47.Sartain M.J., Slayden R.A., Singh K.K., Laal S., Belisle J.T. Disease state differentiation and identification of tuberculosis biomarkers via native antigen array profiling. Mol Cell Proteomics. 2006;5:2102–2113. doi: 10.1074/mcp.M600089-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Michaud G.A., Salcius M., Zhou F., Bangham R., Bonin J., Guo H., Snyder M., Predki P.F., Schweitzer B.I. Analyzing antibody specificity with whole proteome microarrays. Nat Biotechnol. 2003;21:1509–1512. doi: 10.1038/nbt910. [DOI] [PubMed] [Google Scholar]
- 49.Tong M., Jacobi C.E., van de Rijke F.M., Kuijper S., van de Werken S., Lowary T.L., Hokke C.H., Appelmelk B.J., Nagelkerke N.J., Tanke H.J. A multiplexed and miniaturized serological tuberculosis assay identifies antigens that discriminate maximally between TB and non-TB sera. J Immunol Methods. 2005;301:154–163. doi: 10.1016/j.jim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Li B., Jiang L., Song Q., Yang J., Chen Z., Guo Z., Zhou D., Du Z., Song Y., Wang J. Protein microarray for profiling antibody responses to Yersinia pestis live vaccine. Infect Immun. 2005;73:3734–3739. doi: 10.1128/IAI.73.6.3734-3739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steller S., Angenendt P., Cahill D.J., Heuberger S., Lehrach H., Kreutzberger J. Bacterial protein microarrays for identification of new potential diagnostic markers for Neisseria meningitides infections. Proteomics. 2005;5:2048–2055. doi: 10.1002/pmic.200401097. [DOI] [PubMed] [Google Scholar]
- 52.Groathouse N.A., Amin A., Marques M.A., Spencer J.S., Gelber R., Knudson D.L., Belisle J.T., Brennan P.J., Slayden R.A. Use of protein microarrays to define the humoral immune response in leprosy patients and identification of disease-state-specific antigenic profiles. Infect Immun. 2006;74:6458–6466. doi: 10.1128/IAI.00041-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neuman de Vegvar H.E., Amara R.R., Steinman L., Utz P.J., Robinson H.L., Robinson W.H. Microarray profiling of antibody responses against simian–human immunodeficiency virus: postchallenge convergence of reactivities independent of host histocompatibility type and vaccine regimen. J Virol. 2003;77:11125–11138. doi: 10.1128/JVI.77.20.11125-11138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quintana F.J., Hagedorn P.H., Elizur G., Merbl Y., Domany E., Cohen I.R. Functional immunomics: microarray analysis of IgG autoantibody repertoires predicts the future response of mice to induced diabetes. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14615–14621. doi: 10.1073/pnas.0404848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Yu J., Sreekumar A., Varambally S., Shen R., Giacherio D., Mehra R., Montie J.E., Pienta K.J., Sanda M.G. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 56•.Davies D.H., Liang X., Hernandez J.E., Randall A., Hirst S., Mu Y., Romero K.M., Nguyen T.T., Kalantari-Dehaghi M., Crotty S. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with [57•], the authors express the entire vaccinia ORFeome recombinantly, using the products to create an antigen array. Serum antibody responses from three species, including humans, are incubated with the array to identify immunodominant antigens.
- 57•.Davies D.H., McCausland M.M., Valdez C., Huynh D., Hernandez J.E., Mu Y., Hirst S., Villarreal L., Felgner P.L., Crotty S. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J Virol. 2005;79:11724–11733. doi: 10.1128/JVI.79.18.11724-11733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; In conjunction with [56•], the authors identify a specific gene product, H3L envelope protein, as an immunodominant antigen in the vaccine.
- 58••.Zhu H., Hu S., Jona G., Zhu X., Kreiswirth N., Willey B.M., Mazzulli T., Liu G., Song Q., Chen P. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc Natl Acad Sci USA. 2006;103:4011–4016. doi: 10.1073/pnas.0510921103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Validates an antigen array for diagnosing SARS-CoV infection using a large number of clinical samples.
- 59.Boder E.T., Bill J.R., Nields A.W., Marrack P.C., Kappler J.W. Yeast surface display of a noncovalent MHC class II heterodimer complexed with antigenic peptide. Biotechnol Bioeng. 2005;92:485–491. doi: 10.1002/bit.20616. [DOI] [PubMed] [Google Scholar]
- 60.Esteban O., Zhao H. Directed evolution of soluble single-chain human class II MHC molecules. J Mol Biol. 2004;340:81–95. doi: 10.1016/j.jmb.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y., Rubtsov A., Heiser R., White J., Crawford F., Marrack P., Kappler J.W. Using a baculovirus display library to identify MHC class I mimotopes. Proc Natl Acad Sci USA. 2005;102:2476–2481. doi: 10.1073/pnas.0409798102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maynard J., Petersson K., Wilson D.H., Adams E.J., Blondelle S.E., Boulanger M.J., Wilson D.B., Garcia K.C. Structure of an autoimmune T cell receptor complexed with class II peptide–MHC: insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 63.De Groot A.S., McMurry J., Marcon L., Franco J., Rivera D., Kutzler M., Weiner D., Martin B. Developing an epitope-driven tuberculosis (TB) vaccine. Vaccine. 2005;23:2121–2131. doi: 10.1016/j.vaccine.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 64.Altman J.D., Moss P.A., Goulder P.J., Barouch D.H., McHeyzer-Williams M.G., Bell J.I., McMichael A.J., Davis M.M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 65•.Stone J.D., Demkowicz W.E., Jr., Stern L.J. HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays. Proc Natl Acad Sci USA. 2005;102:3744–3749. doi: 10.1073/pnas.0407019102. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first paper to use arrays to examine T cell activation.
- 66.Soen Y., Chen D.S., Kraft D.L., Davis M.M., Brown P.O. Detection and characterization of cellular immune responses using peptide–MHC microarrays. PLoS Biol. 2003;1:E65. doi: 10.1371/journal.pbio.0000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Chen D.S., Soen Y., Stuge T.B., Lee P.P., Weber J.S., Brown P.O., Davis M.M. Marked differences in human melanoma antigen-specific T cell responsiveness after vaccination using a functional microarray. PLoS Med. 2005;2:e265. doi: 10.1371/journal.pmed.0020265. [DOI] [PMC free article] [PubMed] [Google Scholar]; OnlythethirdreporttoemploypMHCarraysThearrayswereusedtomeasurepatientresponsestoamelanomapeptidevaccineintermsofpMHCrecognitionandcytokinesecretion
- 68.Deviren G., Gupta K., Paulaitis M.E., Schneck J.P. Detection of antigen-specific T cells on p/MHC microarrays. J Mol Recognit. 2007;20:32–38. doi: 10.1002/jmr.805. [DOI] [PubMed] [Google Scholar]
- 69.Chen D.S., Davis M.M. Molecular and functional analysis using live cell microarrays. Curr Opin Chem Biol. 2006;10:28–34. doi: 10.1016/j.cbpa.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 70.Laugel B., Price D.A., Sewell A.K. On the path to TCR-directed therapeutics. Nat Biotechnol. 2006;24:615–616. doi: 10.1038/nbt0606-615b. [DOI] [PubMed] [Google Scholar]
- 71.Morgan R.A., Dudley M.E., Wunderlich J.R., Hughes M.S., Yang J.C., Sherry R.M., Royal R.E., Topalian S.L., Kammula U.S., Restifo N.P. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daniels M.A., Hogquist K.A., Jameson S.C. Sweet ‘n’ sour: the impact of differential glycosylation on T cell responses. Nat Immunol. 2002;3:903–910. doi: 10.1038/ni1002-903. [DOI] [PubMed] [Google Scholar]
- 73.Mazmanian S.K., Kasper D.L. The love–hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 74.Xia B., Kawar Z.S., Ju T., Alvarez R.A., Sachdev G.P., Cummings R.D. Versatile fluorescent derivatization of glycans for glycomic analysis. Nat Methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 75.Aharoni A., Thieme K., Chiu C.P., Buchini S., Lairson L.L., Chen H., Strynadka N.C., Wakarchuk W.W., Withers S.G. High-throughput screening methodology for the directed evolution of glycosyltransferases. Nat Methods. 2006;3:609–614. doi: 10.1038/nmeth899. [DOI] [PubMed] [Google Scholar]
- 76.ang D., Liu S., Trummer B.J., Deng C., Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 77.Fukui S., Feizi T., Galustian C., Lawson A.M., Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate–protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 78.Feizi T., Fazio F., Chai W., Wong C.H. Carbohydrate microarrays — a new set of technologies at the frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–645. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 79.Ngundi M.M., Taitt C.R., McMurry S.A., Kahne D., Ligler F.S. Detection of bacterial toxins with monosaccharide arrays. Biosens Bioelectron. 2006;21:1195–1201. doi: 10.1016/j.bios.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80•.Manimala J.C., Li Z., Jain A., VedBrat S., Gildersleeve J.C. Carbohydrate array analysis of anti-Tn antibodies and lectins reveals unexpected specificities: implications for diagnostic and vaccine development. ChemBioChem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]; Uses a glycoprotein array to evaluate the potential for using the Tn carbohydrate antigen as a biomarker.
- 81•.Calarese D.A., Lee H.K., Huang C.Y., Best M.D., Astronomo R.D., Stanfield R.L., Katinger H., Burton D.R., Wong C.H., Wilson I.A. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc Natl Acad Sci USA. 2005;102:13372–13377. doi: 10.1073/pnas.0505763102. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors use a specially designed carbohydrate array to detail the carbohydrate-binding specificity of a potent HIV-neutralizing antibody.
- 82.Gama C.I., Tully S.E., Sotogaku N., Clark P.M., Rawat M., Vaidehi N., Goddard W.A., 3rd, Nishi A., Hsieh-Wilson L.C. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 83.Bochner B.S., Alvarez R.A., Mehta P., Bovin N.V., Blixt O., White J.R., Schnaar R.L. Glycan array screening reveals a candidate ligand for Siglec-8. J Biol Chem. 2005;280:4307–4312. doi: 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- 84.Blixt O., Head S., Mondala T., Scanlan C., Huflejt M.E., Alvarez R., Bryan M.C., Fazio F., Calarese D., Stevens J. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwarz M., Spector L., Gargir A., Shtevi A., Gortler M., Altstock R.T., Dukler A.A., Dotan N. A new kind of carbohydrate array, its use for profiling antiglycan antibodies, and the discovery of a novel human cellulose-binding antibody. Glycobiology. 2003;13:749–754. doi: 10.1093/glycob/cwg091. [DOI] [PubMed] [Google Scholar]
- 86.Guo Y., Feinberg H., Conroy E., Mitchell D.A., Alvarez R., Blixt O., Taylor M.E., Weis W.I., Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat Struct Mol Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- 87.Patwa T.H., Zhao J., Anderson M.A., Simeone D.M., Lubman D.M. Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem. 2006;78:6411–6421. doi: 10.1021/ac060726z. [DOI] [PubMed] [Google Scholar]
- 88.Coombs P.J., Taylor M.E., Drickamer K. Two categories of mammalian galactose-binding receptors distinguished by glycan array profiling. Glycobiology. 2006;16:1C–7C. doi: 10.1093/glycob/cwj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ngundi M.M., Taitt C.R., McMurry S.A., Kahne D., Ligler F.S. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature. 2000;403:669–672. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 90.Wang D., Lu J. Glycan arrays lead to the discovery of autoimmunogenic activity of SARS-CoV. Physiol Genomics. 2004;18:245–248. doi: 10.1152/physiolgenomics.00102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91•.Huang C.Y., Thayer D.A., Chang A.Y., Best M.D., Hoffmann J., Head S., Wong C.H. Carbohydrate microarray for profiling the antibodies interacting with Globo H tumor antigen. Proc Natl Acad Sci USA. 2006;103:15–20. doi: 10.1073/pnas.0509693102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstratesthepotentialforcarbohydratearraystobeusedascancerdiagnostics
- 92.Burton D.R., Desrosiers R.C., Doms R.W., Koff W.C., Kwong P.D., Moore J.P., Nabel G.J., Sodroski J., Wilson I.A., Wyatt R.T. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 93.Adams E.W., Ratner D.M., Bokesch H.R., McMahon J.B., O’Keefe B.R., Seeberger P.H. Oligosaccharide and glycoprotein microarrays as tools in HIV glycobiology: glycan-dependent gp120/protein interactions. Chem Biol. 2004;11:875–881. doi: 10.1016/j.chembiol.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 94.Pashov A.D., Plaxco J., Kaveri S.V., Monzavi-Karbassi B., Harn D., Kieber-Emmons T. Multiple antigenic mimotopes of HIV carbohydrate antigens: relating structure and antigenicity. J Biol Chem. 2006;281:29675–29683. doi: 10.1074/jbc.M604137200. [DOI] [PubMed] [Google Scholar]
- 95••.Stevens J., Blixt O., Tumpey T.M., Taubenberger J.K., Paulson J.C., Wilson I.A. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]; Together with [96], uses a 200-member glycan array plus hemagglutinin variants and site-directed mutants to detail the fine biochemical differences that result in avian versions of human influenza viruses.
- 96.Stevens J., Blixt O., Glaser L., Taubenberger J.K., Palese P., Paulson J.C., Wilson I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 97.Disney M.D., Seeberger P.H. The use of carbohydrate microarrays to study carbohydrate–cell interactions and to detect pathogens. Chem Biol. 2004;11:1701–1707. doi: 10.1016/j.chembiol.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 98.Walz A., Odenbreit S., Mahdavi J., Boren T., Ruhl S. Identification and characterization of binding properties of Helicobacter pylori by glycoconjugate arrays. Glycobiology. 2005;15:700–708. doi: 10.1093/glycob/cwi049. [DOI] [PubMed] [Google Scholar]
- 99.Rosenfeld R., Bangio H., Gerwig G.J., Rosenberg R., Aloni R., Cohen Y., Amor Y., Plaschkes I., Kamerling J.P., Maya R.B. A lectin array-based methodology for the analysis of protein glycosylation. J Biochem Biophys Methods. 2007;10:415–426. doi: 10.1016/j.jbbm.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 100.Angeloni S., Ridet J.L., Kusy N., Gao H., Crevoisier F., Guinchard S., Kochhar S., Sigrist H., Sprenger N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 101.Koshi Y., Nakata E., Yamane H., Hamachi I. A fluorescent lectin array using supramolecular hydrogel for simple detection and pattern profiling for various glycoconjugates. J Am Chem Soc. 2006;128:10413–10422. doi: 10.1021/ja0613963. [DOI] [PubMed] [Google Scholar]
- 102.Kuno A., Uchiyama N., Koseki-Kuno S., Ebe Y., Takashima S., Yamada M., Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 103.Pilobello K.T., Krishnamoorthy L., Slawek D., Mahal L.K. Development of a lectin microarray for the rapid analysis of protein glycopatterns. ChemBioChem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 104.Zheng T., Peelen D., Smith L.M. Lectin arrays for profiling cell surface carbohydrate expression. J Am Chem Soc. 2005;127:9982–9983. doi: 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- 105•.Hsu K.L., Pilobello K.T., Mahal L.K. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat Chem Biol. 2006;2:153–157. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]; Constructs and validates a large lectin array, and uses it to distinguish between bacterial strains and to follow temporal changes in bacterial surface glycans.
- 106.McMahon S.A., Miller J.L., Lawton J.A., Kerkow D.E., Hodes A., Marti-Renom M.A., Doulatov S., Narayanan E., Sali A., Miller J.F. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol. 2005;12:886–892. doi: 10.1038/nsmb992. [DOI] [PubMed] [Google Scholar]
- 107.Walsh G., Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 108.Weikert S., Papac D., Briggs J., Cowfer D., Tom S., Gawlitzek M., Lofgren J., Mehta S., Chisholm V., Modi N. Engineering Chinese hamster ovary cells to maximize sialic acid content of recombinant glycoproteins. Nat Biotechnol. 1999;17:1116–1121. doi: 10.1038/15104. [DOI] [PubMed] [Google Scholar]
- 109.Hamilton S.R., Davidson R.C., Sethuraman N., Nett J.H., Jiang Y., Rios S., Bobrowicz P., Stadheim T.A., Li H., Choi B.K. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313:1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- 110.Cox K.M., Sterling J.D., Regan J.T., Gasdaska J.R., Frantz K.K., Peele C.G., Black A., Passmore D., Moldovan-Loomis C., Srinivasan M. Glycan optimization of a human monoclonal antibody in the aquatic plant Lemna minor. Nat Biotechnol. 2006;24:1591–1597. doi: 10.1038/nbt1260. [DOI] [PubMed] [Google Scholar]
- 111•.Kaneko Y., Nimmerjahn F., Ravetch J.V. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]; Suggests that differential sialylation of the antibody Fc region profoundly affects its ability to mediate pro-inflammatory or anti-inflammatory activities.
- 112.Raju T.S., Scallon B.J. Glycosylation in the Fc domain of IgG increases resistance to proteolytic cleavage by papain. Biochem Biophys Res Commun. 2006;341:797–803. doi: 10.1016/j.bbrc.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 113.Gray J.J. The interaction of proteins with solid surfaces. Curr Opin Struct Biol. 2004;14:110–115. doi: 10.1016/j.sbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 114.Oh S.H., Lee S.H., Kenrick S.A., Daugherty P.S., Soh H.T. Microfluidic protein detection through genetically engineered bacterial cells. J Proteome Res. 2006;5:3433–3437. doi: 10.1021/pr060193a. [DOI] [PubMed] [Google Scholar]
- 115.Ramachandran N., Hainsworth E., Bhullar B., Eisenstein S., Rosen B., Lau A.Y., Walter J.C., LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 116.Bailey S.N., Ali S.M., Carpenter A.E., Higgins C.O., Sabatini D.M. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat Methods. 2006;3:117–122. doi: 10.1038/nmeth848. [DOI] [PubMed] [Google Scholar]
- 117.Ramachandran N., Larson D.N., Stark P.R., Hainsworth E., LaBaer J. Emerging tools for real-time label-free detection of interactions on functional protein microarrays. FEBS J. 2005;272:5412–5425. doi: 10.1111/j.1742-4658.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 118.Blank K., Mai T., Gilbert I., Schiffmann S., Rankl J., Zivin R., Tackney C., Nicolaus T., Spinnler K., Oesterhelt F. A force-based protein biochip. Proc Natl Acad Sci USA. 2003;100:11356–11360. doi: 10.1073/pnas.1934928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dhayal B., Henne W.A., Doorneweerd D.D., Reifenberger R.G., Low P.S. Detection of Bacillus subtilis spores using peptide-functionalized cantilever arrays. J Am Chem Soc. 2006;128:3716–3721. doi: 10.1021/ja0570887. [DOI] [PubMed] [Google Scholar]
- 120.Torres F.E., Kuhn P., De Bruyker D., Bell A.G., Wolkin M.V., Peeters E., Williamson J.R., Anderson G.B., Schmitz G.P., Recht M.I. Enthalpy arrays. Proc Natl Acad Sci USA. 2004;101:9517–9522. doi: 10.1073/pnas.0403573101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zarling A.L., Polefrone J.M., Evans A.M., Mikesh L.M., Shabanowitz J., Lewis S.T., Engelhard V.H., Hunt D.F. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc Natl Acad Sci USA. 2006;103:14889–14894. doi: 10.1073/pnas.0604045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thirumalapura N.R., Morton R.J., Ramachandran A., Malayer J.R. Lipopolysaccharide microarrays for the detection of antibodies. J Immunol Methods. 2005;298:73–81. doi: 10.1016/j.jim.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 123.Kanter J.L., Narayana S., Ho P.P., Catz I., Warren K.G., Sobel R.A., Steinman L., Robinson W.H. Lipid microarrays identify key mediators of autoimmune brain inflammation. Nat Med. 2006;12:138–143. doi: 10.1038/nm1344. [DOI] [PubMed] [Google Scholar]
- 124.Bacarese-Hamilton T., Bistoni F., Crisanti A. Protein microarrays: from serodiagnosis to whole proteome scale analysis of the immune response against pathogenic microorganisms. Biotechniques. 2002;33(Suppl):24–29. [PubMed] [Google Scholar]
- 125.Qiu M., Shi Y., Guo Z., Chen Z., He R., Chen R., Zhou D., Dai E., Wang X., Si B. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7:882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dotan N., Altstock R.T., Schwarz M., Dukler A. Anti-glycan antibodies as biomarkers for diagnosis and prognosis. Lupus. 2006;15:442–450. doi: 10.1191/0961203306lu2331oa. [DOI] [PubMed] [Google Scholar]
- 127.Wang L., Ni J., Singh S., Li H. Binding of high-mannose-type oligosaccharaides and synthetic oligomannose clusters to human antibody 2G12: implications for HIV-1 vaccine design. Chem Biol. 2004;11:127–134. doi: 10.1016/j.chembiol.2003.12.020. [DOI] [PubMed] [Google Scholar]


