Graphical abstract
Abbreviations: IFN, interferon; ISGs, IFNstimulated genes; HCV, hepatitis C virus; IL-10, interleukin-10; PDL-1, programmed death-ligand 1; LCMV, lymphocytic choriomeningitis virus; IFNAR, IFNA receptor; IRFs, IFN regulatory factors; pDCs, plasmacytoid dendritic cells; JAK-STAT, Janus kinase/signal transducer and activator of transcription; TLR, toll-like receptors; IP10, IFNγ-inducible protein 10; PRRs, pattern recognition receptors; MYD88, myeloid differentiation primary response gene 88; HAART, highly active antiretroviral therapy; GALT, gut-associated lymphoid tissue; MIP-3α, macrophage inflammatory protein 3α; CCL20, chemokine (C-C motif) ligand 20; TRAIL, tumor necrosis factor (TNF)-related apoptosis-inducing ligand; DR5, death receptor5; Bak, Bcl-2 homologous antagonist/killer; TF, transmitted founder; IFITMs, IFN-induced transmembrane proteins; DCs, dendritic cells; CPSF6, cleavage and polyadenylation specificity factor subunit 6; cGAS, cyclic guanosine monophosphate-adenosine monophosphate synthasec; IFI16, IFNγ-inducible protein; TREX1, intracellular enzyme 3′ repair exonuclease; NF-κB, nuclear factor kappa-light-chain-enhancer of B cell; RIG1, retinoic acid inducible gene 1 protein; SOCS3, suppressor of cytokine signaling 3; PKR, protein kinase R; 2-5 OAS, 2′-5′-oligoadenylate synthetase; APOBEC, apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3; BST-2, bone marrow stromal antigen 2; Mx2, and myxovirus resistance 2; SAMHD1, sterile alfa motif and histidine/aspartic acid domain containing protein 1; SNP, single nucleotide polymorphism; MSM, men who have sex with men
Keywords: HIV-1, IFN, ISGs, IFNλ, IFNα subtypes, Age, Gender, SNP, Microbiome, Gut, PRR, IFNα receptor
Highlights
-
•
Induction and action of IFN during acute and chronic HIV-1 infection.
-
•
Host and viral factors influencing IFN response in HIV-1 infected patients.
-
•
Type I IFN and IFNα subtypes signatures and their antiviral activity during HIV-1 infection.
-
•
The microbiome and intestinal IFN responses relationship in HIV-1 infection and disease.
Abstract
Type I interferon (IFN) response initially limits HIV-1 spread and may delay disease progression by stimulating several immune system components. Nonetheless, persistent exposure to type I IFN in the chronic phase of HIV-1 infection is associated with desensitization and/or detrimental immune activation, thereby hindering immune recovery and fostering viral persistence. This review provides a basis for understanding the complexity and function of IFN pleiotropic activity in HIV-1 infection. In particular, the dichotomous role of the IFN response in HIV-1 immunopathogenesis will be discussed, highlighting recent advances in the dynamic modulation of IFN production in acute versus chronic infection, expression signatures of IFN subtypes, and viral and host factors affecting the magnitude of IFN response during HIV-1 infection. Lastly, the review gives a forward-looking perspective on the interplay between microbiome compositions and IFN response.
1. Introduction
Despite over 30 years of research, the contribution of type I IFNs to both the control of HIV-1 spread and the initiation of immunologic damage remains controversial [[1], [2]]. As with most viral infections, HIV-1 can efficiently stimulate host antiviral defense responses, but the production of type I IFNs and induction of a large set of ISGs fail to keep the virus entirely in check. Strikingly, and most distressingly for HIV-1-infected patients, prolonged exposure to type I IFN in the chronic phase of infection is associated with desensitization and/or detrimental hyperimmune activation contributing to disease progression [[1], [2]]. This apparently paradoxical action of IFN in HIV-1 infection is also well-established in chronic HCV infection where the majority of patients are unable to clear the virus and develop viral persistence in the face of a rapidly induced ISG response [[3], [4], [5]]. In parallel, when chronically HCV-infected patients with strong ISG expression were treated with IFNα and ribavirin, a virological response was very rare [[3], [4], [5]]. Moreover, when viral infections cannot be cleared, sustained type I IFN signaling seems to assume a predominantly immunosuppressive role, possibly to limit host toxicity and morbidity during persistent infection. Indeed, a direct causal link between type I IFN signaling, immune activation, negative immune regulator expression (e.g. IL-10 and PD-L1), lymphoid tissue disorganization, impaired humoral responses to secondary viral infection, and persistent LCMV has been reported [[6], [7], [8], [9]]. Thus, emerging evidence shows that type I IFN is a common nexus in the pathogenesis of chronic viral diseases.
Interestingly, type I IFNs have been shown to cause immunopathology and/or negative effects even in some acute viral infections (e.g., influenza, severe acute respiratory syndrome-associated coronavirus) and in several bacterial infections [[1], [10], [11], [12]].
Given the growing evidence of type I IFN involvement in viral disease progression, it is not surprising that the possibility to target either type I IFN or the mechanisms leading to IFN production in HIV-1 disease has been considered a promising new therapeutic approach [[13], [14]]. However, the complexity of both the IFN system and HIV-1 immunopathogenesis make the clinical consequences of manipulating type I IFN signaling difficult to predict in vivo. Such therapeutic interventions should therefore be approached with caution in HIV-1 positive subjects [15]. Indeed, several key questions on the controversial role played by type I IFNs in controlling HIV-1 infection remain unanswered despite intense research in this area.
This review attempts to address the above issues summarizing current knowledge of how type I IFN response plays dual roles in HIV-1 replication and disease progression. Emphasis is placed on the importance of type I IFN subtype signature, HIV-1 strategies of IFN evasion, host genetics and the interplay between IFN and the microbiome as the main factors affecting the magnitude of IFN response during HIV-1 infection.
2. IFN system
The IFN system consists of a broad family of cytokines that are key players in the development of both innate and adaptive immune responses. According to their cellular origin and the type of receptors they bind to, IFNs are grouped into three different types (Table 1 ) [5]. Type I IFN comprises multiple species, all of which signal through the same heterodimeric receptor [IFNA receptor (IFNAR)]. In humans, there are several subtypes of IFNα, one IFNβ, one IFNω, one IFNε, and one IFNκ. In particular, excluding the pseudogenes, there are at least 13 distinct IFNα subtypes, which are genetically and structurally very similar. They all lack introns, their protein sequence is highly conserved (75–99% amino acid sequence identity) and they are clustered on the short arm of chromosome 9 [16]. It is unclear what advantage so many different IFNα subtypes provides, but the in vitro antiviral and antiproliferative activities of the IFNα subtypes vary as do their effects on innate and adaptive immunity [16]. The differences among IFNα subtypes may reveal their variation in affinity for the IFNA receptor or variable signaling through complement receptor type 2 [17]. Furthermore, depending on the cell type and the kind of stimulation, the IFNα subtype pattern expressed is regulated by the differential expression of various IFN regulatory factors (IRFs) [16].
Table 1.
Main characteristics of human IFN system (un update).
| IFN | Members | Main Cellular Source | Receptor | Receptor Expression |
|---|---|---|---|---|
| Type I IFN | IFNα1/13, IFNα2,IFNα4, IFNα5, IFNα6, IFNα7, IFNα8, IFNα10, IFNα14, IFNα16,IFNα17, IFNα21 | pDCs, fibroblasts, macrophages | IFNA receptor R1 and R2 | Ubiquitous expression |
| IFNβ | ||||
| IFNε | ||||
| IFNκ | ||||
| IFNω | ||||
| Type II IFN | IFNγ | Natural killer cells,natural killer T cells, Th1 CD4, CD8 cytotoxic T lymphocytes | IFNG receptor R1and R2 | Ubiquitous expression |
| Type III IFN | IFNλ1, IFNλ2, IFNλ3, IFNλ4 | Epithelial cells, hepatocytes, pDCs, myeloid DCs and macrophages | IFNL receptor R1 | Expressed preferentially by epithelial cells |
| IL10 receptor R2 | Ubiquitous expression |
Despite these differences, all IFNα subtypes are mainly produced by pDCs, while fibroblasts are the main source of IFNβ. IFNα and IFNβ are one of the body’s best natural weapons against viruses. After recognition of their specific receptor, all type I IFNs trigger the JAK-STAT pathway leading to the expression of hundreds of ISGs. These ISGs encode proteins that may limit the spread of infection affecting host and/or viral pathways (e.g. retroviral restriction factors). Moreover, both innate and adaptive immune cells respond to type I IFNs by enhancing antigen presentation and chemokine production, increasing antibody production by B cells and amplifying the effector function of T cells [5]. Unlike the multiple type I IFNs, type II IFN includes a single cytokine, IFNγ, mainly secreted by T lymphocytes and NK cells. IFNγ binds to a heterodimeric receptor, IFNGR1/2, and coordinates a wide array of cellular programs through transcriptional regulation of immunologically relevant genes. Type III IFNs are a recently identified class belonging to the IFN system, including four subtypes, IFNλ1-4 [[18], [19]]. Among type III IFNs, IFNλ4 expression and production are controlled by a dinucleotide polymorphism known as IFNL4 rs368234815 (ΔG/TT) located in exon 1 of IFNL4, and IFNλ4 protein can be produced only by individuals carrying the functional genetic variant IFNL4-ΔG allele [20]. IFNλs share features with both type I IFNs and the IL-10 family and display type I IFN-like antiviral activity and induction of classical ISGs [[20], [21]]. These functional similarities result from the activation of a common signaling pathway, although IFNλs engage a specific receptor complex composed of IFNLR1 (also known as IL-28RA) and the accessory chain IL-10R2. They have a more tissue-restricted expression than type I IFN, the liver tissues and epithelial cells being highly sensitive to their action [5].
3. Activation of type I IFN response in acute and chronic HIV-1 infection
HIV-1 infection is usually acquired by sexual mucosal transmission. Studies in simian immunodeficiency virus (SIV) intravaginally infected macaques demonstrate that SIV multiplication is initially confined to the mucosal infection site. Following HIV-1 transmission, there is an eclipse phase of about 10 days during which virus is initially incresead at the mucosal site and in local lymphoid tissues, and then systemic spreading begins [22]. In this context, primate models have demonstrated that cytokine responses, including IFN production, are earlier and stronger in mucosal tissues of the genital tract and lower in systemic lymphoid tissues after vaginal SIV inoculation [23]. In particular, IFNs and other antiviral chemokines seem to be produced locally by pDCs recruited to the mucosal sites within 24 h of SIV exposure [24]. Later SIV spread to lymphoid tissues is associated with strong IFNα and IFNβ production at these sites [25]. In lymph nodes, pDCs are the major but not exclusive producers of IFNα that mediated a transient IFNα response during the acute phase of SIV infection [25]. Indeed, the administration of TLR7 and TLR9 antagonist did not impact the SIV load or the acute IFNα response in plasma and had minimal effects on ISG expression in both blood and lymph node, indicating that other cells may be involved in this process (e.g. monocytes/macrophages and myeloid DC) [[25], [26]]. Similarly, IFNα and IFN mediators (e.g. IP-10) are induced within a few weeks prior to peak viremia during acute HIV-1 infection [27]. Likewise, after SIV infection of rhesus macaques in both peripheral blood and the jejunum, the expression of several IFN-induced restriction factors substantially increased in all CD4 T cell memory subsets at the peak of acute infection [28]. Alongside the induction of type I IFNs and IFN-related pathways, a robust increase in PRRs was recorded during acute SIV infection in both peripheral blood and gut mucosa, coinciding with viral replication [29]. The pronounced IFNα/β response induced during acute HIV-1 infection may also have detrimental effects, contributing to the initial control of viral replication, but also to the immunopathology of the infection [9]. However, the negative effects of type I IFN become more evident during the chronic phase of HIV-1 infection. Indeed, during HIV-1 disease progression, IFNα occurred in serum with increasing regularity and concentration compared to the acute phase and IFNα was correlated with the occurrence of HIV-1 p24 antigen in serum [30]. Moreover, differences in the expression profile of type I IFN pathways of T cells (CD4 and CD8) were recorded from early HIV-1 infection (infected for less than 6 months) and chronic progressors (infected for more than 1 year) [31]. The latter suggests that a stereotypical HIV-1-induced IFN pattern of gene expression in T cells might be established early in the infection and persist for years thereafter. Like HIV-1 infection in humans, pathogenic SIV infection of non-natural host species, such as rhesus macaques, is associated with a robust type I IFN response and ISG expression that persist in SIV-infected macaques even though IFNα-producing pDCs are partially depleted from the blood and/or lymph nodes [[32], [33], [34], [35], [36], [37], [38]]. By contrast, non-pathogenic SIV infections (African green monkey and sooty mangabey) display strong pDC activation and type I IFN production during acute SIV infection but low IFNα activation during chronic infection [[33], [35], [38]]. Intriguingly, pDCs levels capable of sensing SIV and releasing type I IFNs in nonpathogenic SIV infection seems to be decreased during acute infection [39]. Furthermore, SIV-infected sooty mangabeys are not intrinsically resistant to type I IFN signaling so it should be concluded that the mechanisms involved in the capability of these animals to retain low immune activation/inflammation are expected to be multifactorial and not exclusively dependent on IFN/ISG stimulation [40]. In agreement with the results obtained from the primate models of SIV infection, several studies have reported a strong activation of different components associated with the type I IFN response (e.g. IRFs, ISGs, viral DNA sensors, MYD88, IFNAR] during chronic HIV-1 infection [[41], [42], [43], [44], [45]]. High levels of ISGs such as IP-10 were also associated with a more rapid CD4T cell depletion [[46], [47]]. Consistent with the above findings, no significant ISG upregulation has been observed in viremic non-progressors, and global T-cell gene expression profiles of IFN-related genes in nonprogressors appeared similar from those with no HIV-1 infection [[31], [44], [48], [49]]. The notion that IFN pathways are increased during persistent HIV-1 replication is also supported by the observation that a large ISGs group were downregulated in lymph node biopsy samples in post-HAART therapy [50]. Moreover, the interruption of HAART is accompanied by increased mucosal gene expression of IFN-mediated antiviral responses, a profile similar to those found in naive HIV-1 infected patients [51]. Although prolonged antiretroviral treatment can improve GALT immune function, the type I IFN response in gut seems to persist in long-term treated HIV-1-infected patients [52]. This finding could be due to the rapid pDC migration to the intestinal and lymphoid tissues after acute SIV infection, causing a local inflammation with type I IFN production [53].
4. IFN mechanism of action during HIV-1 infection
A complex and seemingly paradoxical role of type I IFNs in HIV-1 infection has been well documented [[1], [2]]. The pronounced type I IFN response induced during HIV-1 infection may likewise have detrimental effects. Although IFN is produced during HIV-1 infection, it is not able to inhibit early infection and a persistent IFN-mediated immune activation becomes one of the main pathogenic mechanisms of HIV-1 disease. On the contrary, IFNα administration initially upregulated expression of ISGs and prevented systemic infection in a pathogenic SIV rhesus macaque model using intrarectal challenge [15], suggesting a positive action of IFN in controlling early viral infection. However, IFNα treatment also resulted in accelerated CD4T cell depletion and increased viremia, probably due to the activation of IFN-related desensitization mechanisms induced by continued treatment [15]. Hence, type I IFN response during HIV-1 infection, as discussed below, could exert positive and negative effects indicating its dual role in regulating and promoting both HIV-1 infection and immunopathogenesis.
4.1. Anti-HIV activity
Several groups have demonstrated that type I IFNs inhibit HIV-1 replication in human cells cultivated in vitro [[54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73]]. These studies identified multiple mechanisms by which type I IFNs can affect the HIV-1 infectious cycle, determined by the target cells and the timing of IFN addition and virus infection (Table 2 ). In particular, IFNα/β treatment of primary T lymphocytes, monocytes/macrophages, and some T cell lines before HIV-1 infection efficiently suppresses the early steps of the virus life cycle, including HIV-1-induced cell fusion, viral RNA, DNA and protein production (Table 2). However, it appears that the mechanisms by which type I IFNs inhibit HIV-1 replication in T cells and macrophages were not identical. While the principal effect of IFN in infected T cells or primary lymphoblast cultures was on the terminal stage of the HIV-1 life cycle (e.g. assembly and release) and HIV-1 morphogenesis [[66], [68]], IFN in macrophages appears to interrupt an early step of the HIV-1 replication cycle (Table 2). Moreover, monocytes treated with type I IFN a week after HIV-1 infection were not free of the retroviral pathogen: levels of proviral DNA in the IFN-treated and control HIV-infected cells were indistinguishable [63]. Similarly, IFN-induced inhibition of viral protein synthesis could be detected in T cells only when cells were treated with IFNα prior to infection or when IFNα was added up to 10 h postinfection, but not if IFNα was added at the later stages of HIV-1 replication cycle or after HIV-1 infection was already established [67]. IFNα/β also impair later stages of the HIV-1 replication cycle, especially in persistently infected cells cultures, ranging from deregulation of viral protein processing and protein stability, impaired gp120 incorporation and morphogenesis, to altered virion release [[60], [70], [71], [72], [73]]. However, the effects on chronic HIV-1 infection seem to be much less pronounced, and very high concentration of IFNs (in some cases exceeding 1000 International Units/ml) were used in the experiments since the effects were absent or negligible at the doses shown to be effective against the majority of lytic viruses. Moreover, the long-term efficacy of IFNs on HIV-1 spread in T cells, measured by following Gag-expressing cells, seems to be less potently inhibited by IFNα [74]. In particular, a gradual increase in the percentage of HIV-1-infected cells over time was observed in T cells exposed to high HIV-1 doses [74]. Virus emergence was the result of suboptimal suppression of HIV-1 moltiplication and cell-to-cell transfer was only moderately sensitive to type I IFNs [74]. Thus, the molecular nature of HIV-1 inhibition by IFNs appears to be cell type dependent and multiple parameters like the infectious dose, the time of IFN addition and the subtypes of IFNs used in the experiments should be carefully considered when interpreting the results. This situation is exemplified by the observation that other subtypes within type I IFN other than IFNα/β, such as IFNε, can strongly suppress HIV-1 replication at multiple stages of infection [75]. In addition, closely related lentiviruses appeared to be inhibited by type I IFNs at different stages of expression in T cells (Table 2), highlighting the complexity of the phenomenon [71]. All together these limitations emphasize the risk of extrapolating results obtained under in vitro conditions to clinical applications in HIV-1-infected patients, explaining in part why the in vitro anti-HIV potency of IFNs generally contrasts with their poor clinical efficacy.
Table 2.
In vitro type I IFN action against HIV/SIV infection.
| Type I IFN | Cells | Main Results | Ref. |
|---|---|---|---|
| IFNα | Peripheral blood mononuclear cells | IFN has a dose-related suppressive effect on HTLV-III replication | 54 |
| IFNα1, IFNα2 IFNβ, IFN leukocyte | Peripheral blood mononuclear cells | IFN preparations suppress LAV, HTLV-III, and ARV-2 replication as measured by reverse transcriptase (RT) activity by greater than 50%. This suppression was dose dependent and high dosages (500 Units/ml) of IFNα resulted in almost complete suppression of RT activities (77–99%) | 55 |
| IFNα, IFNβ | Peripheral blood mononuclear cells, H9 lymphocytic and monocytoid U937 cell lines | IFNs show similar concentration-dependent antiHIV activity. No reduction in HIV expression are observed when persistently infected H9 cells are treated with high dose of IFN | 56 |
| IFNα2 | Stable cell lines, derived from Vero cells and A3.01 cells, that express IFN gene | The transcription and replication of HIV was completely inhibited by IFN | 57 |
| IFNα | HIV infected H9 lymphocytic cells | HIV-1 replication is inhibited by a maximum of 22% at 1000 Units/ml of IFN | 58 |
| IFNβ | Peripheral blood mononuclear cells | IFN reduces replication of HIV. The effect is most pronounced when high levels of the IFN are employed | 59 |
| IFNα | Promonocytic (U1) and T lymphocytic (ACH-2) cell lines chronically infected with HIV | IFN inhibits the release of RT, viral antigens, the production or release (or both) of whole HIV virions, but has no effect on the amount of cell-associated viral proteins | 60 |
| IFNα, IFNβ | Chronically HIV infected monocytoid U937 cells | The addition of 1000 Units of IFN per ml to HIV-infected U937 cells resulted in some inhibition of virus production | 61 |
| IFNα, IFNβ | Monocyte-derived macrophage | IFN acts to restrict the formation of proviral DNA | 62 |
| IFNα, IFNβ | T cells or monocytes | Levels of RT activity in IFN-treated HIV-infected T cells are half those in control cultures, but the frequency of infected cells or the levels of p24 released in culture fluids are unchanged | 63 |
| Monocytes treated with IFNs at the time of virus challenge showed no evidence of HIV infection: no p24 antigen or RT activity, no viral mRNA, and no proviral DNA. Monocytes treated with IFN 7 days after HIV infection are not free of the retroviral pathogen | |||
| IFNα | Chronically HIV infected Tlymphocytic ACH-2 and promonocytic Ul cell lines | IFN, although effective in suppressing the release of HIV particles, do not inhibit shedding of p24, gag into the culture supernatants | 64 |
| IFNα, IFNβ | HeLa T4 cells | IFNs inhibit syncytium formation induced by HIV-1 envelope glycoprotein and are found to be potent inhibitors of HIV-1 induced cell fusion | 65 |
| IFNα | T-cell, H9, CEM, C3, and Jurkat cell lines | IFN decreases virus production (extracellular RT and p24 antigen levels in the supernatant medium). Chronically infected Jurkat cells treated with IFN appear to be inhibited in growth rate, as virus production decreased with cell number | 66 |
| IFNα | CEM-174 | Pretreatment of cells with 50 to 500 Units of IFN per ml result in a marked reduction in HIV RNA and protein synthesis. IFN-induced inhibition of viral protein synthesis is detected only when cells were treated with IFNα prior to infection or when IFNα are added up to 10 h postinfection, but not if IFNα are added at the later stages of HIV-1 replication cycle or after the HIV-1 infection is already established | 67 |
| IFNα | Peripheral blood mononuclear cell | A marked depletion of envelope glycoprotein (gp120) in HIV virions released from IFN-treated cells | 68 |
| IFNα, IFNβ | Monocytes derived macrophage | Macrophages pretreated with IFNs have a reduced HIV DNA signal while the spliced mRNA signal is essentially abolished. No virus is produced. The addition of IFNs does not affect the levels of HIV spliced transcripts in cells with established productive infection | 69 |
| IFNα, IFNβ | Chronically HIV infected monocytoid U937 cells and CEM cells | IFN treatment induces a specific block on HIV mRNA translation | 70 |
| IFNα, IFNβ | MT4 cells | IFNs block an early step in SIV replication while HIV gene expression was disrupted at a later point. Both the stability and proteolytic processing of HIV specific proteins were altered in IFN-treated cells | 71 |
| IFNα | Monocytes derived macrophage | IFN affect early steps of HIV-1 BaL replication, preceding the completion of viral DNA synthesis | 72 |
| IFNα | Chronically HIV infected monocytoid U937 cells | IFN affects late stages of HIV-1 replication, by inhibiting virus assembly and release, and by reducing the infectivity of shed virions | 73 |
| IFNα1b, IFNα2a, IFNα2b, IFNβ1a | T-cell lines (MT4R5, Jurkat, HUT-78, CEM) and primary CD4 T lymphocytes | IFNs have a limited effect on HIV spread, measured as the appearance of Gag-expressing cells | 74 |
| Cell-to-cell HIV transfer is less sensitive to IFN than infection by cell-free virions | |||
| IFNε | FRT epithelial cancer cell line and Sup-T1 lymphoma line | IFN impairs HIV infection at stages post HIV entry and up to the translation of viral proteins | 75 |
4.2. Detrimental effects
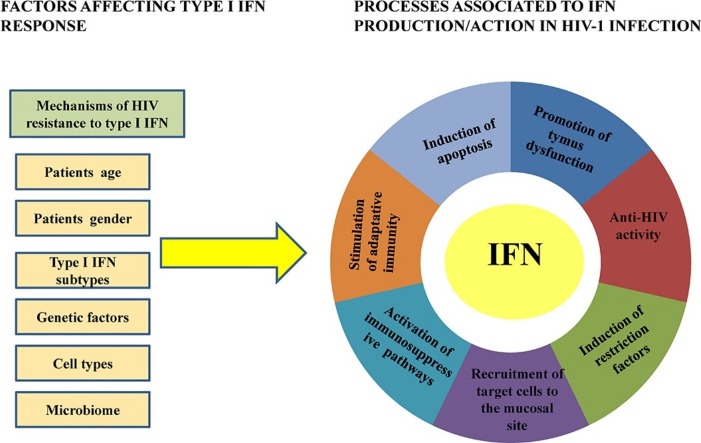
Several mechanisms have been proposed to explain the negative action of type I IFNs in HIV-1-infected patients (Fig. 1 ). First of all, type I IFNs could partly trigger chronic immune activation by inducing ISGs and chemokines able to attract target cells to the initial site of HIV-1 replication [24]. In this regard, the SIV macaca model showed the induction of an endocervical mucosal signaling system involving MIP-3α, also known as CCL20, and the production of IFNα and virus-inhibiting chemokines by pDCs and T cells, which create an environment rich in target cells, CD4T cells and CCR5T cell at the sites of initial infection [24].
Fig. 1.
Processes associated to IFN production/action in HIV-1 infection. Examples of positive and negative effects of IFN reported during HIV-1 infection.
Second, a role for IFNα in TRAIL/DR5-mediated apoptosis and immunopathogenesis has been proposed [76]. In particular, HIV-1-stimulated TRAIL production was IFNα dependent and mediated by STAT-1/2, antibodies against IFNα/β block TRAIL production and apoptosis of HIV-1 exposed CD4T cells, while IFNα produced by HIV-1-exposed pDCs was responsible for TRAIL expression on primary CD4T cells [[41], [76], [77], [78]]. Importantly, increased mRNA expression for IFNα, TRAIL and DR5 was recorded in tonsils of patients with progressive HIV-1 disease compared to tonsils of patients with nonprogressive disease [41]. Upregulated TRAIL expression was also observed in HIV-1 positive patients with high levels of ISGs (e.g. ISG15) and viremia [45]. Besides the relationship between IFNα and TRAIL/DR5-mediated apoptosis, in chronic HIV-1 infection the pro-apoptotic Bak was found increased in CD4T cells and correlated positively with sensitivity to Fas/CD95-mediated apoptosis and negatively with CD4T cell numbers [79]. Moreover, a model in which CD4T cell-intrinsic type I IFN signaling due to microbial exposure was recently proposed to potentiate gut CD4T cells for accelerated HIV-1 by inhibiting CDK4/6, cyclins and c-Myc [80].
Third, as reported in other viral infections, highly potent immunosuppressive programs mediated by IL-10 and PD-L1 that reduce T cell responses might be responsible for the harmful effects of IFNα/β also in chronic HIV-1 infection [[1], [81]]
Lastly, a relationship between type I IFNs, thymopoiesis and the HIV-induced immunodeficient state has been observed. Thymus dysfunction is known to characterize HIV-1/SIV infections and contributes to their pathophysiology. In human thymus, thymocytes and medullary pDCs constitutively express IFNα and type I IFNs can suppress thymic output, further limiting CD4T cell recovery [82]. A strong alteration of both chemokines and IFNα subtype transcriptional patterns in SIV-infected thymuses was also recorded, and the IFNα subtypes produced in the infected thymuses inhibited thymocyte proliferation [82]. Moreover, HIV-1 infection of the human thymus results in increased levels of IFNα. These are associated with MHC I upregulation on thymic epithelial cells and subsequent preferential selection of CD4−CD8+ (SP8) thymocytes with a low level of CD8 expression which may contribute to the generalized immunosuppression [83]. CCR5 expression was also induced by IFNα in thymic organ cultures, suggesting that IFNα may paradoxically expand the tropism of R5 HIV-1 and, in so doing, enhancing viral infectivity in the HIV-1 infected patients [84].
5. Factors affecting the type I IFN response during HIV infection
It is well-documented that type I IFNs and/or ISGs might exert deleterious effects during HIV-1 infection [[1], [2]], although the actual relationship between IFN response and HIV-1 infection is still a matter of debate. Indeed, the differential effects of age, gender, host genetics, immune environments and cell type on the overall outcome of type I IFN signaling in HIV-1 infection remain unexplained. Also unknown is the main type I IFN subtypes expressed during acute and chronic HIV infection. Translational studies are needed to establish the magnitude of the effects of microbiome composition on IFN activation in HIV-1-infected individuals. Lastly, it has become important to understand the impact of mechanisms of refractoriness to type I IFN activated during HIV-1 infection. Below, the main factors affecting the type I IFN response during HIV-1 infection are discussed, providing some evidence detailing the role of IFNλ in this picture.
5.1. Mechanisms of HIV resistance to type I IFN
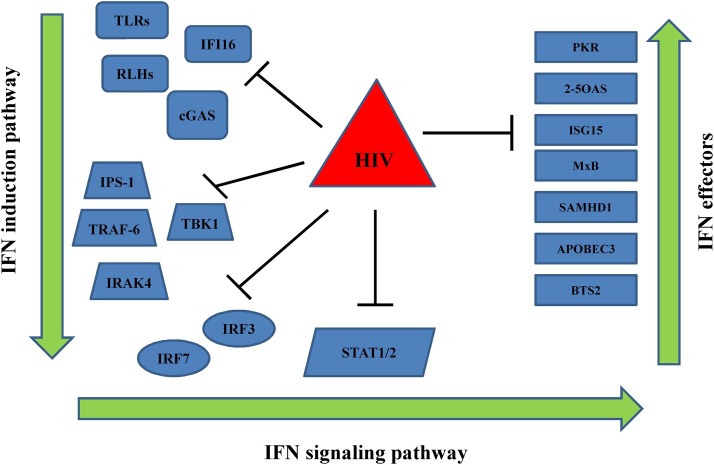
HIV-1 antagonism against type I IFN production and their anti-viral responses strongly support the concept that IFNs have the innate potential to control HIV-1 infection. In vitro type I IFNs can inhibit HIV-1 infection, and many of the ISGs (e.g. retroviral restriction factors) have strong anti-HIV potency [85]. However, HIV-1 is able to replicate in vivo even in the presence of type I IFNs and ISGs, though the mechanisms by which HIV-1 is able to persist remain mysterious. It is known that the inadequacy of HIV-1 transmission across mucosal surfaces is illustrated by a stringent population bottleneck, in which only one or a limited number of variants with improved transmission fitness from the diverse quasispecies of the transmitting donor are able to start the new infection. In this regard, HIV-1 TF viruses seem to be phenotypically distinct. Increased type I IFN resistance is the key feature to elucidate their ability to replicate and spread efficiently despite a potent mucosal innate immune response [[86], [87]]. Moreover, HIV-1 TF viruses are uniformly resistant to IFITMs, a requirement that is lost during chronic infection, in part due to escape mutations acquired in response to autologous neutralizing responses [88]. Besides the demonstration that TF viruses can overcome the antiviral action of IFNα/β and specific ISGs, several in vitro studies have shown that HIV-1 inhibits type I IFN induction, mainly through its interaction with IRF members in its three most important target cells: CD4T cells, macrophages, and DCs [[89], [90], [91]]. Remarkable multiple evasion strategies can be employed by HIV-1 to block type I IFN induction and action (Fig. 2 ). In particular, HIV-1 has evolved to use host cofactors, such as CPSF6 and cyclophilins recuited to its capsid, to cloak its moltiplaction, permitting evasion of PRRs and cGAS, and type I IFN secretion and initiation of an antiviral state [92]. Further defects in innate viral sensing during HIV-1 infection involve the PRRs, cGAS and IFI16 by inducing TREX1 expression and inhibiting NF-κB activation by HIV-1 Vpu [93]. Moreover, an impaired TLRs and RIG1 response can be recorded during HIV-1 infection [93]. HIV-1 can also affect PRR-mediated signaling through the inhibition of several downstream effectors including IRF3 and IRF7 levels and/or activity [[89], [90], [93]]. Following type I IFN production, HIV-1 can also subvert their mechanism of action (Fig. 2). Specifically, HIV-1 can stimulate the production of SOCS3, a molecular inhibitor of IFN signaling, impairing phosphorylation of STAT-1/2 [94]. Moreover, downregulated IFNAR expression and consequent hyporesponsiveness to IFNα in PBMC of HIV-1-infected patients has been recorded, although the specific evasion mechanism employed by HIV-1 is currently undefined [[95], [96]].
Fig. 2.
In vitro impairment of the type IFN response by HIV-1. Black lines indicate countermeasures against the IFN by HIV-1 to overcome IFN induction (left side), IFN signaling (centre) and IFN effectors (right side).
Lastly, several ISGs can be blocked by HIV-1, including well-established antiviral proteins (e.g. PKR, 2–5 OAS, and ISG15) and the recently identified retroviral restriction factors (e.g. APOBEC3, BST-2, and Mx2) [[85], [93]]. Controversial results have been reported on the ability of type I IFNs to directly induce the restriction factor, namely SAMHD1 [[97], [98]]. However, besides the established ability of HIV-2 to subvert SAMHD1 activity through the vpx gene, SAMHD1 can be regulated by type I IFN treatment by phosphorylation at threonine 592 and IRF3 can be responsible for the direct induction of SAMHD1 [[93], [99]]. Hence, it is clear that in order to replicate efficiently, HIV-1 has evolved multiple strategies to circumvent the ‘first line of defense’ embodied by type I IFN. It is probable that most (if not all) antiviral effectors activated by HIV-1 infections are to some degree circumvented by a virus strategy. Nevertheless, the modes of action of most ISGs remain unclear as do the strategies required by HIV-1 to allow it to circumvent particular IFN responses. Determining the mechanisms by which these ISGs function at different steps of the HIV-1 replication cycle and how the virus-cell interplay subsequently reshapes the host defense mechanisms would be interesting to gain a complete understanding of the molecular basis of the HIV-1 host interaction in the immunopathogenesis of HIV-1 disease.
5.2. Host factors
Different host factors, such as age, gender, specific molecular pattern and cell environments could affect the magnitude of type I IFN activation during HIV-1 infection. However, their relative influence in HIV-1 infection remains poorly understood. Despite these limitations, marked sex differences in type I IFN expression have been described in the course of HIV-1 disease (100, 101). In particular, pDCs from women have been shown to exhibit a stronger IFNα response to HIV-1-encoded TLR7 ligands than pDCs from men, causing an higher secondary activation of CD8 T cells [100]. Expression levels of a subset of ISGs, including ISG15, MxA, and CCR5, were also found to be higher in naive HIV-1 infected females than in males after adjusting for HIV-1 RNA levels and to be associated with eleveated levels of immune activation in HIV-1 infection [101].
Few studies have attempted to characterize the influence of patient age on the production of type I IFNs, demonstrating an age-related dysregulation of the capacity to synthesize IFN and ISGs during HIV-1/SIV infection [102].
As far as the genomic study of type I IFNs is concerned, a major problem is the high number of genes (e.g. IFNα, IFNβ, IFNκ, IFNε, IFNω) or “pseudo”-genes (13 different IFNα subtypes), numerous type I IFN signaling pathway components and ISGs [16]. However, several studies have pointed out the importance of SNP related to the type I IFN pathways on the clinical outcome of HIV-1 disease [[103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116], [117]]. Rapid progression of HIV-1 infection, altered viral load and CD4T cell count were associated with TLR9 polymorphisms [[103], [104], [105]]. Further, genetic variations of TLR4 were more common among HIV-1-positive individuals with high peak viral load compared with low/moderate peak viral load [106]. SNPs in other PRR genes (i.e. IFI16) involved in the HIV-1 genome sensor have also been identified [107]. Exhaustive genotyping of the IFNAR1 gene in a French AIDS cohort disclosed an IFNAR1 protein variant associated with AIDS progression or susceptibility to HIV-1 infection [108]. Additionally, SNPs associated with progression from HIV-1 infection to AIDS were identified in two 2′-5′-OAS genes (e.g. OAS2 and OAS3) [109]. In contrast, other SNPs identified in OAS2 and OAS3 genes and in the TRIM5 gene were associated with a slower progression of disease [109]. Several studies have reported controversial results on the influence of retroviral restriction factor SNPs (e.g. APOBEC3, SAMHD1, and BST2) on HIV-1 disease status [[110], [111], [112], [113], [114], [115], [116], [117]]. Although the clinical relevance of these genetic variants during HIV-1 infection has been emphasized, their biological significance has seldom been examined [[45], [118]]. In this regard, two SNPs of IRF-7, both located at intron/extron boundaries, were significantly associated with decreased levels of IFNα production by pDCs in response to HIV-1 [118]. Furthermore, no effects of SNPs located within the 5′UTR region and in the exon 2 sequence on ISG15 expression in HIV-1-infected patients were recorded [45]. Besides the analysis of these genetic variants related to the IFN response in HIV-1-positive patients, few studies have looked at the type III IFNs SNPs so far. IFNλ3/IL28B has attracted much attention from HCV researchers because in 2009 genome-wide association studies identified an association between certain SNPs located in the upstream region of IFNλ3 and both spontaneous and IFN treatment-induced viral clearance in chronic HCV infection [[5], [19]]. Interestingly, the original SNP (rs12979860) identified near IFNλ3 has now been positioned within an intron of the gene encoding the newly discovered IFNλ4 [20]. Importantly, these IFNλ3/4 SNPs have been reported to affect IFNλ and ISG production, particularly in the liver of HCV-infected patients before or after IFNα or DAAs therapy [5].
Discordant results have been obtained on the clinical significance of IFNλ3/4 SNPs in HIV-1 infection [[119], [120], [121], [122], [123], [124], [125]]. On the one hand, IFNλ3/4 SNPs have been associated with unfavorable clinical and immunological status, increased susceptibility to cytomegalovirus retinitis, and protection against HIV-1 disease progression in controllers. On the other, the same SNPs had no effects on clinical outcome or susceptibility to HIV-1 infection.
As far as the biological significance of type III IFN SNPs in HIV-1 infection is concerned and considering that both types I and III IFN share the same JAK-STAT pathway, endogenous ISGs expression in PBMCs from HIV-1-positive patients seems to be independent of the IFNλ4 rs368234815 ΔG/TT dinucleotide polymorphism [126].
5.3. Type I IFN subtypes
12 Despite many indications on a type I IFN signature in HIV-1 infection [[1], [2]], the predominant type I IFN species, and perhaps IFNα subtype, that is upregulated in HIV-1-infected patients has not been conclusively determined. One of oldest studies on the history of IFN and HIV-1 reported an acid-labile human leukocyte IFN in homosexual men with Kaposi’s sarcoma and lymphadenopathy [127]. High levels of IFNα or IFNβ were also detected in sera from HIV-1-infected patients, but other type I IFN subtypes were not analyzed to provide a quantitative comparison [[30], [128], [129]]. One of the first studies on this topic was done by Gendelman et al., examining IFNα, IFNβ, and IFNω expression in HIV-1-infected monocytes [130]. However many years later, IFNα was shown to be the main type I IFN measurable in the peripheral blood of HIV-1-positive subjects [131]. Although the latter work reported elevated IFNα expression in HIV-1 infection, the extent of IFNα subtype upregulation has not been investigated. Some attempts have been made to tackle this issue in recent years. In particular, a preferential upregulation of IFNα subtype 2 expression has been recorded in HIV-1 positive patients [132]. Using a pigtailed macaque SIV model, Zaritsky et al. also demonstrated that IFNα subtype expression and regulation differ between brain, lung, and spleen [133]. Furthermore, in response to oral pathogenic SIV infection, multiple IFNα subtypes (IFNα1/13, IFNα2, IFNα4, IFNα6 and IFNα8) are quickly induced in lymphoid but not at mucosal surfaces of the oral and gastrointestinal tracts [134]. A more recent next-generation sequencing study also analyzed the expression of all IFNα subtypes in HIV-1-exposed pDCs [135]. It was found that IFNα subtype mRNAs from the centromeric half of the IFNA gene cluster were highly expressed in pDCs following HIV-1 exposure. An inverse relationship between IFNα subtype expression and potency was recorded: IFNα8, IFNα6 and IFNα14 were the most potent in blocking HIV-1 infection, while IFNα2 and IFNα1 were both highly expressed but exhibited relatively weak antiviral activity [135]. In agreement, the IFNα14 subtype has been shown to possess potent anti-HIV-1 activity compared to other IFNα subtypes and gene therapy with plasmids encoding IFNβ and IFNα14, but not the commonly used IFNα2, conferred long-term suppression of HIV-1 replication in humanized mouse models [[136], [137]]. Thus the following new concepts have emerged from these studies: i) HIV-1 induces different expression patterns of type I IFN subtypes; ii) the production of individual IFNα subtypes can differ in relation to the cell type and anatomical site analyzed; ii) different IFNα subtypes can mediate distinct anti-HIV-1 effects, indirectly suggesting that different type I IFN preparations might provide different therapeutic outcomes of HIV-1 infection.
5.4. The interplay between the microbiome and the IFN response
It is known that, during the early HIV-1 infection, the initial and main pronounced depletion of CD4T cells generally occurs in the gastrointestinal mucosa [138], where the elevated levels of CCR5-expressing CD4T cells within the gut may permit HIV-1 entry and replication [139]. Chronic HIV-1 infection within the gastrointestinal tract, and the closely related secondary decrease in CD4T cells significantly shape gut physiology, thus leading to a dysregulation of the mucosal immune-epithelial network [140]. In addition to its direct role in changing the gastrointestinal CD4T cell compartment, it is widely established that HIV-1 infection is characterized by gut microbiome compositional and functional changes. Gori et al. were the first to study gut microbiota in the early phases of HIV-1 infection and found an high abundance of Pseudomonas aeruginosa and Candida albicans as well as lower levels in other microbial species, e.g., Bifidobacteria and Lactobacilli, in the fecal microbiota compared to those reported for healthy individuals [141] Nevertheless, the exact microbiome configuration in naive HIV-1-positive individuals, HAART-treated patients and healthy subjects remains still inconsistent [[142], [143]]. Below we first attempt to briefly describe the gut microbiome composition in HIV-1 infected patients and then we discuss on the emerging novel concepts concerning the interaction between gut microbiota and IFN immune response.
5.4.1. Gut microbiota alterations associated with HIV-1 infection
Previous studies indicated that HIV-1 infection has an impact on the intestinal microbiota composition, given that HIV-1 infection was associated with enhanced bacterial populations in the gut that are pro-inflammatory, and so potentially pathogenic, and whose abundance parallel the inflammatory status and immune recovery [[144], [145]]. In particular, despite considerable experimental variability (e.g. types of samples evaluated; treated or not treated subjects, techniques used to characterize microbiota) the main findings of these investigations were the following: first of all, HIV-1 infection was associated with reduced bacterial richness [[144], [145]]. In particular, diminutions in α diversity have been observed in HIV-1-infected subjects compared to uninfected subjects in both mucosal and fecal samples [[144], [145]]. A greater fecal microbial diversity has been recorded in MSM than in those non-MSM; importantly such a reduction in bacterial richness persists even when HIV-1-positive patients were stratified for MSM vs. non-MSM [146]. This decrease in diversity positively correlated with CD4T cell counts and inversely correlated with markers of microbial translocation and monocyte activation. Surprisingly, HAART did not result in significant changes of the gut microbiota towards increased, and hence beneficial, gut microbiome diversity [[144], [147]].
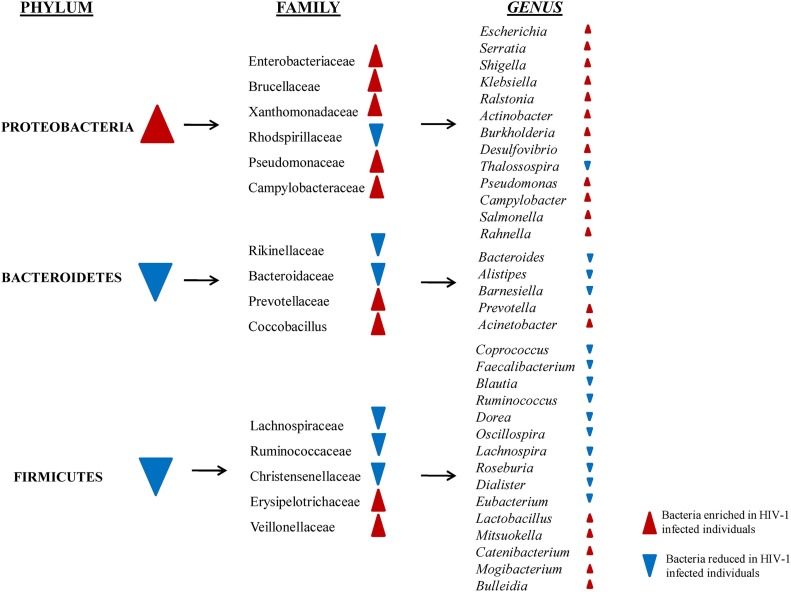
Alterations in bacterial community composition were typically observed in the 3 most dominant phyla detected in the proximal gut: Proteobacteria, Bacteroidetes and Firmicutes (Fig. 3 ).
Fig. 3.
Major findings about gut microbiota compositions in HIV-1 infected patients (adapted from [[144], [145]].
In this context, an overall enrichment of members of the phylum Proteobacteria and decreased abundances of Firmicutes were observed in the mucosal tissues and stool samples of both untreated and treated HIV-1 infected individuals [[144], [145]]. Unexpectedly, a significant decrease in the abundance of Firmicutes was observed in HIV-infected patients using NRTIs + PIs compared to uninfected controls, suggesting that specific HAART regimens are associated with diverse profiles in gut microbiota composition [151].
At the family level, the relative abundance of Prevotellaceae seems to be higher in HIV-1 infected subjects whereas Lachnospiraceae, Christensenellaceae, Ruminococcaceae and Bacteroidaceae were all lower in HIV-1 infected subjects relative to uninfected individuals [148].
Furthermore, when bacteria were evaluated to the genus level, enrichment for Burkholderia, Acinetobacter, Schigella, Klebsiella, Salmonella, Rahnella were observed in HIV-1 infected patients (Fig. 3) [[146], [149]]. In contrast, Lactobacillus, within Firmicutes phylum, were reduced in duodenal tissue from naive HIV-1 positive individuals, and higher Lactobacillus abundance was also found in fecal samples of a cohort of untreated subjects with early HIV-1 infection with higher CD4 counts, lower viral loads and reduced microbial translocation and, interestingly, these associations continued following HAART [[144], [145], [150]].
Altough it is difficult to draw definite conclusions, it appears that HIV-1 positive patients microbiota is characterized by an increase in potentially pathogenic bacteria and, conversely, a decrease in protective bacteria populations. However, it should be underlined that a complete definition of HIV-1 as well as healthy microbiota is still incomplete given that a myriad of factors can influence the composition of gut microbiome [152]. Since the gut microbiota exhibited immune-modulating qualities and considering that the components of the microbiota can directly contribute to CD4T cell depletion in HIV-1 infection and can regulate the gut T helper subset–related immune homeostasis [152], it is tempting to speculate that a better characterization of the complex interactions between microbial dysbioses and the mucosal immune system is critical to improve our understanding of the functional aspects of microbiome as a key or additional driver of HIV-1 disease.
5.4.2. Interaction between IFN response and microbiome
It is known that the cross-talk between the gut microbiome and host is extensive, and involves both innate and adaptive immunity. It is established that HIV-1 infection upsets the delicate equilibrium in the host-microbe interplay through both alterations in the gut microbiome (see above) and interference with the host response mechanisms [152]. In this regard, it has been reported that endogenous type I IFN response induced by signals from the commensal microbiota can affect the local signaling environment to prime the intestinal mucosal immune system to determine later responses to pathogens and commensal organisms [153]. However, it has yet to be determined if the microbiota dysbiosis of HIV-1-infected patients could influence the local and/or systemic innate antiviral immune response including not only type I IFN production but also that associated with IFNλ expression, which is known to use a specific receptor preferentially expressed at the mucosal surfaces. In this context, a remarkable dampening of PRR expression was recently observed in gut mucosal tissues from acute to chronic SIV infection in rhesus macaques [29]. Decreased PRR expression was associated with increased abundance of numerous pathogenic bacterial taxa, including Pasteurellaceae members, Aggregatibacter and Actinobacillus, and the Mycoplasmataceae family, which in turn might be responsible for promoting persistent IFN activation, gut microbiota alterations, and limited viral clearance [29]. Despite a reduced expression of PRR, microbial exposure influenced how HIV-1 altered the gut CD4T cell transcriptome causing a strong upregulation of type I IFN and ISG production [80]. Surprisingly, the induction of multiple antiviral genes after microbial exposure was also associated with an enhanced TF HIV-1 replication [80]. In agreement, our previous study recorded a strong activation of IFNα/β and its receptor in the gut mucosa of HIV-1-infected subjects [52]. Moreover, our preliminary observations suggest a link between microbiota composition and type I IFN signature in HIV-1-positive patients (unpublished data). All these studies emphasize that the dysregulated IFNα/β and ISG levels observed during HIV-1 infection could be at least in part determined by the specific microbiota composition. This hypothesis is supported by the following recent findings. First of all, commensal bacteria can calibrate the activation threshold of innate antiviral immunity [154]. In particular, a tonic signaling by type I IFNs on antigen-presenting cells is required for efficient viral recognition and generation of adaptive immunity, and this signal is provided by commensal bacteria in the steady state. Second, type I IFN receptor functions on intestinal epithelial cells to restrain Paneth and goblet cells and to shape the microbial composition [155]. Third, the bacterial microbiome can foster enteric viral persistence (e.g. norovirus) in a manner counteracted by specific components of the innate immune system (e.g. type III IFNs) [156, nice].
Besides the ability of the microbiota to modulate the IFN response through as yet unclear mechanisms, specific components of the enteric microbiota (i.e. microbial metabolite desaminotyrosine) have been shown to exert distal effects on responses to lethal viral infections through modulation of type I IFN [157]. Thus, bacterial products can modulate different components of the innate immune system to potentially facilitate responses to microbial pathogens. Further work is required to define specific mechanisms by which bacteria and/or their metabolites as well as other microorganisms (e.g. virome and mycome) might impact IFN signaling and anti-HIV-1 immunity not only in the intestine but also in other anatomical sites.
6. Conclusions
It is well-established that type I IFNs have direct and/or indirect (through the induction of several mediators) beneficial effects on innate and adaptive immune cells during viral infection. It is now known, however, that while low levels of type I IFNs may be required at an early stage to initiate cell-mediated immune responses, high concentrations may lead to the production of immunosuppressive molecules. Type I IFN-induced virus protection, immunopathology or autoimmunity also seem to be a matter of kinetics other than magnitude. In particular, delayed type I IFN production has been implicated in the promotion of viral disease [12]. It is tempting to speculate that when viral infections cannot be cleared persistent type I IFN activation assumes a principally immunosuppressive role, probably to reduce host toxicity during chronic infection. Then, type I IFN-induced negative regulatory pathways represent one of the major drivers of persistent inflammation/immune activation and disease progression in chronic viral infections such as those caused by HIV-1 or HCV [[1], [2], [5]]. However, the role of type I IFNs in chronic virus infections is complex, often leading to different outcomes depending on the timing, host cellular environment present, the cumulative amounts of type I IFN components, and the specific IFN subtypes mediating the biological effects. This suggests that during viral diseases a balanced network of type I IFNs does exist to provide protection with minimum damage to the host, but is still largely unknown. Indeed, although we have wide knowledge of the variety of signaling pathways activated by the IFN system, the links between specific pathways and distinct outcomes are not well-understood, especially in a complex viral disease such as HIV-1 infection. In this regard, the observations that an elevated type I IFN signature can be deleterious in chronic HIV-1 infection together with the difficulties encountered in the in vivo manipulation of type I IFN signaling [15] should prompt us to reflect on whether we have properly investigated the therapeutic potential of IFNα in this clinical setting. For instance, since its first use in the early 1980s as a promising therapy for HIV-1, we started IFNα therapy in HIV-1-infected patients regardless of whether the pre-existing elevated type I IFN signature could render the patient intrinsically resistant to IFN therapy. Therefore, as “veterans” of the lesson on ISG-mediated resistance to IFN therapy in HCV infection [5], are we again wrong to evaluate, but above all compare, the anti-HIV-1 or SIV effects of IFNα without first assessing the endogenous levels of type I IFN activation? Moreover, how can we indifferently target the type I IFN pathways without taking into account the individuality and variation in IFN gene expression patterns (e.g. type I IFN subtypes, ISGs, etc.)? Such an achievement would be an important advancement and as such should be thoroughly explored in HIV-1 infection. Many other factors should also be considered when interpreting the data on activation of the type I IFN response in HIV-1 infection and several other issues have yet to be addressed. For example, most studies evaluating the type I IFN response in SIV/HIV-1 infection have analyzed the ISG response. Although ISG production may somehow reflect the direct activation of a type I IFN response, we cannot disregard that ISGs can also be induced by IFN-independent mechanisms (e.g. directly by viruses) and by other types of IFNs/cytokines (e.g. IFNλs). Analysis of the IFN response in HIV-1/SIV infection has also been generally performed with classical or innovative molecular techniques, hiding the real contribution of protein expression. Epigenetic regulation of IFN genes can be an additional confounding factor for the correct interpretation of the role of IFN/ISGs in HIV-1/SIV infection. Lastly, the real contribution of other IFN types or subtypes (e.g. IFNλ, IFNε, IFNα) in the natural history of HIV-1 infection, and how the microbiome can regulate the rate of types I–III IFN activation are currently unknown. Hence, it is essential to understand IFN’s mechanism of induction and action in more detail by studying the factors affecting the kinetics and magnitude of the IFN response during acute and chronic viral infections. This information should help us provide additional insights into the type I IFN paradox in HIV-1 infection and will be essential to harness these powerful products of nature for our benefit.
Conflict of interest
None.
Biographies

Carolina Scagnolari is an Associate Professor of Virology at the Sapienza University in Rome. After receiving her Ph.D. and Microbiology and Virology post graduated school degrees, in 2010, she joined the Sapienza University of Rome as Assistant Professor of Virology. Currently, she is the head of antiviral innate immunity research Unit at the Department of Molecular Medicine, Laboratory of Virology affiliated to Istituto Pasteur Italia-Fondazione Cenci Bolognetti. Scagnolari's career has been devoted to viral infectious diseases focusing principally on the role of type I and III interferons in the control of acute and chronic viral infections, mostly those caused by respiratory viruses, HPV, HCV, and HIV. She received the “Sivim” (2004) and “ICAR-CROI” (2014, 2015) awards, the special Jury Price CROI/2014 and several research grants from Sapienza University and Institutes of Pasteur International Network. She is author/co-author of about 90 papers appeared on international journals in the field of Virology and Immunology.

Guido Antonelli is full Professor of Virology at the Sapienza University in Rome. He is also head of the Microbiology unit and the Department of Diagnostic Services at the “Sapienza” University Hospital “Policlinico Umberto I” in Rome. He took a position at Institute of Virology/Sapienza University (1989) and then at the Department of Biomedicine/University of Pisa (1992); he worked also with the Department of Microbiology, University of Texas Medical Branch (Galveston, Texas, USA) and the Clinical Research Center, Division of Immunological Medicine (Harrow, London, UK). His interest was initially focused on the in vivo and in vitro mechanism of production and action of interferons. His group’s research currently focuses on pathogenesis, control, and diagnosis of viral infections, particularly HIV, HCV, respiratory viruses, and HPV. He participates in several national and international research projects and few years ago served as president of SIVIM (Italian Society for Medical Virology). He has authored and co-authored more than 250 peer-reviewed articles and is now serving as co-editor or member of the editorial board of several peer-reviewed Journals.
References
- 1.McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doyle T., Goujon C., Malim M.H. HIV-1 and interferons: who's interfering with whom? Nat. Rev. Microbiol. 2015;13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heim M.H., Thimme R. Innate and adaptive immune responses in HCV infections. J. Hepatol. 2014;61:S14–25. doi: 10.1016/j.jhep.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 4.Antonelli G., Scagnolari C., Moschella F., Proietti E. Twenty-five years of type I interferon-based treatment: a critical analysis of its therapeutic use. Cytokine Growth Factor Rev. 2015;26:121–131. doi: 10.1016/j.cytogfr.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scagnolari C., Monteleone K., Cacciotti G., Antonelli G. Role of interferons in chronic hepatitis C infection. Curr. Drug Targets. 2017;18:844–850. doi: 10.2174/1389450117666160201112632. [DOI] [PubMed] [Google Scholar]
- 6.Teijaro J.R., Ng C., Lee A.M., Sullivan B.M., Sheehan K.C., Welch M., Schreiber R.D., dela Torre J.C., Oldstone M.B. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson E.B., Yamada D.H., Elsaesser H., Herskovitz J., Deng J., Cheng G., Aronow B.J., Karp C.L., Brooks D.G. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teijaro J.R. Too much of a good thing: sustained type 1 interferon signaling limits humoral responses to secondary viral infection. Eur. J. Immunol. 2016;46:300–302. doi: 10.1002/eji.201546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honke N., Shaabani N., Merches K., Gassa A., Kraft A., Ehrhardt K., Häussinger D., Löhning M., Dittmer U., Hengel H., Recher M., Lang P.A., Lang K.S. Immunoactivation induced by chronic viral infection inhibits viral replication and drives immunosuppression through sustained IFN-I responses. Eur. J. Immunol. 2016;46:372–380. doi: 10.1002/eji.201545765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baskin C.R., Bielefeldt-Ohmann H., Tumpey T.M., Sabourin P.J., Long J.P., García A., Sastre A.E., Tolnay R., Albrecht J.A., Pyles P.H., Olson L.D., Aicher E.R., Rosenzweig K., Murali-Krishna E.A., Clark M.S., Kotur J.L., Fornek S., Proll R.E., Palermo C.L., Sabourin M.G. Katze Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson S., Crotta S., McCabe T.M., Wack A. Pathogenic potential of interferon αβ in acute influenza infection. Nat. Commun. 2014;5:3864. doi: 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-Infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng L., Ma J., Li J., Li D., Li G., Li F., Zhang Q., Yu H., Yasui F., Ye C., Tsao L.C., Hu Z., Su L., Zhang L. Blocking type I interferon signaling enhances T cell recovery and reduces HIV-1 reservoirs. J. Clin. Invest. 2017;127:269–279. doi: 10.1172/JCI90745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhen A., Rezek V., Youn C., Lam B., Chang N., Rick J., Carrillo M., Martin H., Kasparian S., Syed P., Rice N., Brooks D.G., Kitchen S.G. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J. Clin. Invest. 2017;127:260–268. doi: 10.1172/JCI89488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandler N.G., Bosinger S.E., Estes J.D., Zhu R.T., Tharp G.K., Boritz E., Levin D., Wijeyesinghe S., Makamdop K.N., del Prete G.Q., Hill B.J., Timmer J.K., Reiss E., Yarden G., Darko S., Contijoch E., Todd J.P., Silvestri G., Nason M., Norgren R.B., Jr., Keele B.F., Rao S., Langer J.A., Lifson J.D., Schreiber G., Douek D.C. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbert K., Schlaak J.F., Yang D., Dittmer U. IFN-α subtypes: distinct biological activities in anti-viral therapy. Br. J. Pharmacol. 2013;168:048–58. doi: 10.1111/bph.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asokan R., Hua J., Young K.A., Gould H.J., Hannan J.P., Kraus D.M., Szakonyi G., Grundy G.J., Chen X.S., Crow M.K., Holers V.M. Characterization of human complement receptor type 2 (CR2/CD21) as a receptor for IFN-alpha: a potential role in systemic lupus erythematosus. J. Immunol. 2006;177:383–394. doi: 10.4049/jimmunol.177.1.383. [DOI] [PubMed] [Google Scholar]
- 18.Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 19.Riva E., Scagnolari C., Turriziani O., Antonelli G. Hepatitis C virus and interferon type III (interferon-λ3/interleukin-28 B and interferon-λ4): genetic basis of susceptibility to infection and response to antiviral treatment. Clin. Microbiol. Infect. 2014;20:1237–1245. doi: 10.1111/1469-0691.12797. [DOI] [PubMed] [Google Scholar]
- 20.Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., Chen S., Brand N., Tarway M., Liu L., Sheikh F., Astemborski J., Bonkovsky H.L., Edlin B.R., Howell C.D., Morgan T.R., Thomas D.L., Rehermann B., Donnelly R.P., O'Brien T.R. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uzé G., Monneron D. L-28 and IL-29: newcomers to the interferon family. Biochimie. 2007;89:729–734. doi: 10.1016/j.biochi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Haase A.T. Targeting early infection to prevent HIV-1 mucosal transmission. Nature. 2010;464:217–223. doi: 10.1038/nature08757. [DOI] [PubMed] [Google Scholar]
- 23.Abel K., Rocke D.M., Chohan B., Fritts L., Miller C.J. Temporal and anatomic relationship between virus replication and cytokine gene expression after vaginal simian immunodeficiency virus infection. J. Virol. 2005;79:12164–12172. doi: 10.1128/JVI.79.19.12164-12172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q., Estes J.D., Schlievert P.M., Duan L., Brosnahan A.J., Southern P.J., Reilly C.S., Peterson M.L., Schultz-Darken N., Brunner K.G., Nephew K.R., Pambuccian S., Lifson J.D., Carlis J.V., Haase A.T. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;10:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kader M., Smith A.P., Guiducci C., Wonderlich E.R., Normolle D., Watkins S.C., Barrat F.J., Barratt-Boyes S.M. Blocking TLR7- and TLR9-mediated IFN-α production by plasmacytoid dendritic cells does not diminish immune activation in early SIV infection. PLoS Pathog. 2013;9:e1003530. doi: 10.1371/journal.ppat.1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wonderlich E.R., Wijewardana V., Liu X., Barratt-Boyes S.M. Virus-encoded TLR ligands reveal divergent functional responses of mononuclear phagocytes in pathogenic simian immunodeficiency virus infection. J. Immunol. 2013;190:2188–2198. doi: 10.4049/jimmunol.1201645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stacey A.R., Norris P.J., Qin L., Haygreen E.A., Taylor E., Heitman J., Lebedeva M., DeCamp A., Li D., Grove D., Self S.G., Borrow P. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahmberg A.R., Rajakumar P.A., Billingsley J.M., Johnson R.P. Dynamic modulation of expression of lentiviral restriction factors in primary CD4(+) t cells following simian immunodeficiency virus infection. J. Virol. 2017;91 doi: 10.1128/JVI.02189-16. e02189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glavan T.W., Gaulke C.A., Santos Rocha C., Sankaran-Walters S., Hirao L.A., Raffatellu M., Jiang G., Bäumler A.J., Goulart L.R., Dandekar S. Gut immune dysfunction through impaired innate pattern recognition receptor expression and gut microbiota dysbiosis in chronic SIV infection. Mucosal Immunol. 2016;9:677–688. doi: 10.1038/mi.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Sydow M., Sonnerborg A., Gaines H., Strannegard O. Interferon-alpha and tumor necrosis factor-alpha in serum of patients in various stages of HIV-1 infection. AIDS Res. Hum. Retrovir. 1991;7:375–380. doi: 10.1089/aid.1991.7.375. [DOI] [PubMed] [Google Scholar]
- 31.Hyrcza M.D., Kovacs C., Loutfy M., Halpenny R., Heisler L., Yang S., Wilkins O., Ostrowski M., Der S.D. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J. Virol. 2007;81:3477–3486. doi: 10.1128/JVI.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosinger S.E., Hosiawa K.A., Cameron M.J., Persad D., Ran L. Gene expression profiling of host response in models of acute HIV infection. J. Immunol. 2004;173:6858–6863. doi: 10.4049/jimmunol.173.11.6858. [DOI] [PubMed] [Google Scholar]
- 33.Bosinger S.E., Li Q., Gordon S.N., Klatt N.R., Duan L., Xu L., Francella N., Sidahmed A., Smith A.J., Cramer E.M., Zeng M., Masopust D., Carlis J.V., Ran L., Vanderford T.H., Paiardini M., Isett R.B., Baldwin D.A., Else J.G., Staprans S.I., Silvestri G., Haase A.T., Kelvin D.J. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J. Clin. Invest. 2009;119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotger M., Dalmau J., Rauch A., McLaren P., Bosinger S.E., Martinez R., Sandler N.G., Roque A., Liebner J., Battegay M., Bernasconi E., Descombes P., Erkizia I., Fellay J., Hirschel B., Miró J.M., Palou E., Hoffmann M., Massanella M., Blanco J., Woods M., Günthard H.F., de Bakker P., Douek D.C., Silvestri G., Martinez-Picado J., Telenti A. Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. J. Clin. Invest. 2011;121:2391–2400. doi: 10.1172/JCI45235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacquelin B., Mayau V., Targat B., Liovat A.S., Kunkel D., Petitjean G., Dillies M.A., Roques P., Butor C., Silvestri G., Giavedoni L.D., Lebon P., Barré-Sinoussi F., Benecke A., Müller-Trutwin M.C. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J. Clin. Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abel K., Alegria-Hartman M.J., Rothaeusler K., Marthas M., Miller C.J. The relationship between simian immunodeficiency virus RNA levels and the mRNA levels of alpha/beta interferons (IFN-alpha/beta) and IFN-alpha/beta-inducible Mx in lymphoid tissues of rhesus macaques during acute and chronic infection. J. Virol. 2002;76:8433–8445. doi: 10.1128/JVI.76.16.8433-8445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaefer T.M., Fuller C.L., Basu S., Fallert B.A., Poveda S.L., Sanghavi S.K., Choi Y.K., Kirschner D.E., Feingold E., Reinhart T.A. Increased expression of interferon-inducible genes in macaque lung tissues during simian immunodeficiency virus infection. Microbes Infect. 2006;8:1839–1850. doi: 10.1016/j.micinf.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 38.Mir K.D., Gasper M.A., Sundaravaradan V., Sodora D.L. SIV infection in natural hosts: resolution of immune activation during the acute-to-chronic transition phase. Microbes Infect. 2011;13:14–24. doi: 10.1016/j.micinf.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jochems S.P., Petitjean G., Kunkel D., Liovat A.S., Ploquin M.J., Barré-Sinoussi F., Lebon P., Jacquelin B., Müller-Trutwin M.C. Modulation of type I interferon-associated viral sensing during acute simian immunodeficiency virus infection in African green monkeys. J. Virol. 2015;89:751–762. doi: 10.1128/JVI.02430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanderford T.H., Slichter C., Rogers K.A., Lawson B.O., Obaede R., Else J., Villinger F., Bosinger S.E., Silvestri G. Treatment of SIV-infected sooty mangabeys with a type-I IFN agonist results in decreased virus replication without inducing hyperimmune activation. Blood. 2012;119:5750–5757. doi: 10.1182/blood-2012-02-411496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbeuval J.P., Nilsson J., Boasso A., Hardy A.W., Kruhlak M.J., Anderson S.A., Dolan M.J., Dy M., Andersson J., Shearer G.M. Differential expression of IFN-alpha and TRAIL/DR5 in lymphoid tissue of progressor versus nonprogressor HIV-1-infected patients. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7000–7005. doi: 10.1073/pnas.0600363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nissen S.K., Højen J.F., Andersen K.L., Kofod-Olsen E., Berg R.K., Paludan S.R., Østergaard L., Jakobsen M.R., Tolstrup M., Mogensen T.H. Innate DNA sensing is impaired in HIV patients and IFI16 expression correlates with chronic immune activation. Clin. Exp. Immunol. 2014;177:295–309. doi: 10.1111/cei.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Killian M.S., Fujimura S.H., Sudhagoni R.G. Brief report: increased expression of the type I interferon receptor on CD4+ t lymphocytes in HIV-1-Infected individuals. J. Acquir. Immune Defic. Syndr. 2017;74:473–478. doi: 10.1097/QAI.0000000000001280. [DOI] [PubMed] [Google Scholar]
- 44.Rotger M., Dang K.K., Fellay J., Heinzen E.L., Feng S., Descombes P., Shianna K.V., Ge D., Günthard H.F., Goldstein D.B., Telenti A. Swiss HIV cohort study; center for HIV/AIDS vaccine immunology. genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000781. (pp. e1000781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scagnolari C., Monteleone K., Selvaggi C., Pierangeli A., D'Ettorre G., Mezzaroma I., Turriziani O., Gentile M., Vullo V., Antonelli G. ISG15 expression correlates with HIV-1 viral load and with factors regulating T cell response. Immunobiology. 2016;221:282–290. doi: 10.1016/j.imbio.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Sedaghat A.R., German J., Teslovich T.M., Cofrancesco J., Jr., Jie C.C., Talbot C.C., Jr, Siliciano R.F. Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J. Virol. 2008;82:1870–1883. doi: 10.1128/JVI.02228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liovat A.S., Rey-Cuille M.A., Lecuroux C., Jacquelin B., Girault I., Petitjean G., Zitoun Y., Venet A., Barré-Sinoussi F., Lebon P., Meyer L., Sinet M., Müller-Trutwin M. Acute plasma biomarkers of T cell activation set-point levels and of disease progression in HIV-1 infection. PLoS One. 2012;7:e46143. doi: 10.1371/journal.pone.0046143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdel-Mohsen M., Raposo R.A., Deng X., Li M., Liegler T., Sinclair E., Salama M.S., Ghanem Hel D., Hoh R., Wong J.K., David M., Nixon D.F., Deeks S.G., Pillai S.K. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology. 2013;10:106. doi: 10.1186/1742-4690-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Masson A., Kirilovsky A., Zoorob R., Avettand-Fenoel V., Morin V., Oudin A., Descours B., Rouzioux C., Autran B. Blimp-1 overexpression is associated with low HIV-1 reservoir and transcription levels in central memory CD4+ T cells from elite controllers. AIDS. 2014;28:1567–1577. doi: 10.1097/QAD.0000000000000295. [DOI] [PubMed] [Google Scholar]
- 50.Li Q., Schacker T., Carlis J., Beilman G., Nguyen P., Haase A.T. Functional genomic analysis of the response of HIV-1-infected lymphatic tissue to antiretroviral therapy. J. Infect. Dis. 2004;189:572–582. doi: 10.1086/381396. [DOI] [PubMed] [Google Scholar]
- 51.Lerner P., Guadalupe M., Donovan R., Hung J., Flamm J., Prindiville T., Sankaran-Walters S., Syvane M., Wong J.K., George M.D., Dandekar S. The gut mucosal viral reservoir in HIV-infected patients is not the major source of rebound plasma viremia following interruption of highly active antiretroviral therapy. J. Virol. 2011;85:4772–4782. doi: 10.1128/JVI.02409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.d' Ettorre G., Ceccarelli G., Andreotti M., Selvaggi C., Giustini N., Serafino S., Schietroma I., Nunnari G., Antonelli G., Vullo V., Scagnolari C. Analysis of Th17 and Tc17 frequencies and antiviral defenses in gut-associated lymphoid tissue of chronic HIV-1 positive patients. Mediators Inflamm. 2015;2015:395484. doi: 10.1155/2015/395484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwa S., Kannanganat S., Nigam P., Siddiqui M., Shetty R.D., Armstrong W., Ansari A., Bosinger S.E., Silvestri G., Amara R.R. Plasmacytoid dendritic cells are recruited to the colorectum and contribute to immune activation during pathogenic SIV infection in rhesus macaques. Blood. 2011;118:2763–2773. doi: 10.1182/blood-2011-02-339515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho D.D., Hartshorn K.L., Rota T.R., Andrews C.A., Kaplan J.C., Schooley R.T., Hirsch M.S. Recombinant human interferon alfa-A suppresses HTLV-III replication in vitro. Lancet. 1985;1:602–604. doi: 10.1016/s0140-6736(85)92144-0. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto J.K., Barré-Sinoussi F., Bolton V., Pedersen N.C., Gardner M.B. Human alpha- and beta-interferon but not gamma- suppress the in vitro replication of LAV HTLV-III, and ARV-2. J. Interferon Res. 1986;6:143–152. doi: 10.1089/jir.1986.6.143. [DOI] [PubMed] [Google Scholar]
- 56.Hartshorn K.L., Neumeyer D., Vogt M.W., Schooley R.T., Hirsch M.S. Activity of interferons alpha, beta, and gamma against human immunodeficiencyvirus replication in vitro. AIDS Res. Hum. Retroviruses. 1987;3:125–133. doi: 10.1089/aid.1987.3.125. [DOI] [PubMed] [Google Scholar]
- 57.Bednarik D.P., Mosca J.D., Raj N.B., Pitha P.M. Inhibition of human immunodeficiency virus (HIV) replication by HIV-trans-activated alpha 2-interferon. Proc. Natl. Acad. Sci. U. S. A. 1989;86:4958–4962. doi: 10.1073/pnas.86.13.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crespi M. The effect of interferon on cells persistently infected with HIV-1. AIDS. 1989;3:27–31. [PubMed] [Google Scholar]
- 59.Michaelis B., Levy J.A. HIV replication can be blocked by recombinant human interferon beta. AIDS. 1989;3:27–31. [PubMed] [Google Scholar]
- 60.Poli G., Orenstein J.M., Kinter A., Folks T.M., Fauci A.S. Interferon alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science. 1989;244:575–577. doi: 10.1126/science.2470148. [DOI] [PubMed] [Google Scholar]
- 61.Locardi C., Petrini C., Boccoli G., Testa U., Dieffenbach C., Buttò S., Belardelli F. Increased human immunodeficiency virus (HIV) expression in chronically infected U937 cells upon in vitro differentiation by hydroxyvitamin D3: roles of interferon and tumor necrosis factor in regulation of HIV production. J. Virol. 1990;64:5874–5882. doi: 10.1128/jvi.64.12.5874-5882.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kornbluth R.S., Oh P.S., Munis J.R., Cleveland P.H., Richman D.D. The role of interferons in the control of HIV replication in macrophages. Clin. Immunol. Immunopathol. 1990;54:200–219. doi: 10.1016/0090-1229(90)90082-2. [DOI] [PubMed] [Google Scholar]
- 63.Gendelman H.E., Baca L.M., Turpin J., Kalter D.C., Hansen B., Orenstein J.M., Dieffenbach C.W., Friedman R.M., Meltzer M.S. Regulation of HIV replication in infected monocytes by IFN-alpha. Mechanisms for viral restriction. J. Immunol. 1990;145:2669–2676. [PubMed] [Google Scholar]
- 64.Fernie B.F., Poli G., Fauci A.S. Alpha interferon suppresses virion but not soluble human immunodeficiency virus antigen production in chronically infected T-lymphocytic cells. J. Virol. 1991;65:3968–3971. doi: 10.1128/jvi.65.7.3968-3971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wells D.E., Chatterjee S., Mulligan M.J., Compans R.W. Inhibition of human immunodeficiency virus type 1-induced cell fusion byrecombinant human interferons. J. Virol. 1991;65:6325–6330. doi: 10.1128/jvi.65.11.6325-6330.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith M.S., Thresher R.J., Pagano J.S. Inhibition of human immunodeficiency virus type 1 morphogenesis in T cells by alpha interferon. Antimicrob. Agents Chemother. 1991;35:62–67. doi: 10.1128/aac.35.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shirazi Y., Pitha P.M. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 1992;66:1321–1328. doi: 10.1128/jvi.66.3.1321-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansen B.D., Nara P.L., Maheshwari R.K., Sidhu G.S., Bernbaum J.G., Hoekzema D., Meltzer M.S., Gendelman H.E. Loss of infectivity by progeny virus from alpha interferon-treated humanimmunodeficiency virus type 1-infected T cells is associated with defectiveassembly of envelope gp120. J. Virol. 1992;66:7543–7548. doi: 10.1128/jvi.66.12.7543-7548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meylan P.R., Guatelli J.C., Munis J.R., Richman D.D., Kornbluth R.S. Mechanisms for the inhibition of HIV replication by interferons-alpha –beta, and –gamma in primary human macrophages. Virology. 1993;193:138–148. doi: 10.1006/viro.1993.1110. [DOI] [PubMed] [Google Scholar]
- 70.Coccia E.M., Krust B., Hovanessian A.G. Specific inhibition of viral protein synthesis in HIV-infected cells in response to interferon treatment. J. Biol. Chem. 1994;269:23087–23094. [PubMed] [Google Scholar]
- 71.Agy M.B., Acker R.L., Sherbert C.H., Katze M.G. Interferon treatment inhibits virus replication in HIV-1- and SIV-infected CD4+T-cell lines by distinct mechanisms: evidence for decreased stability andaberrant processing of HIV-1 proteins. Virology. 1995;214:379–386. doi: 10.1006/viro.1995.0047. [DOI] [PubMed] [Google Scholar]
- 72.Capobianchi M.R., Abbate I., Antonelli G., Turriziani O., Dolei A., Dianzani F. Inhibition of HIV type 1 BaL replication by MIP-1alpha, MIP-1beta, and RANTES in macrophages. AIDS Res. Hum. Retroviruses. 1998;14(February (3)):233–240. doi: 10.1089/aid.1998.14.233. [DOI] [PubMed] [Google Scholar]
- 73.Dianzani F., Castilletti C., Gentile M., Gelderblom H.R., Frezza F., Capobianchi M.R. Effects of IFN alpha on late stages of HIV-1 replication cycle. Biochimie. 1998;80:745–754. doi: 10.1016/s0300-9084(99)80028-5. [DOI] [PubMed] [Google Scholar]
- 74.Vendrame D., Sourisseau M., Perrin V., Schwartz O., Mammano F. Partial inhibition of human immunodeficiency virus replication by type I interferons: impact of cell-to-cell viral transfer. J. Virol. 2009;83:10527–10537. doi: 10.1128/JVI.01235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia-Minambres A., Eid S.G., Mangan N.E., Pade C., Lim S.S., Matthews A.Y., de Weerd N.A., Hertzog P.J., Mak J. Interferon epsilon promotes HIV restriction at multiple steps of viral replication. Immunol. Cell Biol. 2017;95:478–483. doi: 10.1038/icb.2016.123. [DOI] [PubMed] [Google Scholar]
- 76.Herbeuval J.P., Shearer G.M. HIV-1 immunopathogenesis: how good interferon turns bad. Clin. Immunol. 2007;123:121–128. doi: 10.1016/j.clim.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fonteneau J.F., Larsson M., Beignon A.S., McKenna K., Dasilva I., Amara A., Liu Y.J., Lifson J.D., Littman D.R., Bhardwaj N. Human immunodeficiency virus type 1 activates plasmacytoid dendritic cells and concomitantly induces the bystander maturation of myeloid dendritic cells. J. Virol. 2004;78:5223–5232. doi: 10.1128/JVI.78.10.5223-5232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Herbeuval J.P., Hardy A.W., Boasso A., Anderson S.A., Dolan M.J., Dy M., Shearer G.M. Regulation of TNF-related apoptosis-inducing ligand on primary CD4+ T cells by HIV-1: role of type I IFN-producing plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13974–13979. doi: 10.1073/pnas.0505251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fraietta J.A., Mueller Y.M., Yang G., Boesteanu A.C., Gracias D.T., Do D.H., Hope J.L., Kathuria N., McGettigan S.E., Lewis M.G., Giavedoni L.D., Jacobson J.M., Katsikis P.D. Type I interferon upregulates Bak and contributes to T cell loss during human immunodeficiency virus (HIV) infection. PLoS Pathog. 2013;9:e1003658. doi: 10.1371/journal.ppat.1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoder A.C., Guo K., Dillon S.M., Phang T., Lee E.J., Harper M.S., Helm K., Kappes J.C., Ochsenbauer C., McCarter M.D., Wilson C.C., Santiago M.L. The transcriptome of HIV-1 infected intestinal CD4+ T cells exposed to enteric bacteria. PLoS Pathog. 2017;13:e1006226. doi: 10.1371/journal.ppat.1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cunningham C.R., Champhekar A., Tullius M.V., Dillon B.J., Zhen A., de la Fuente J.R., Herskovitz J., Elsaesser H., Snell L.M., Wilson E.B., de la Torre J.C., Kitchen S.G., Horwitz M.A., Bensinger S.J., Smale S.T., Brooks D.G. Type I and type II interferon coordinately regulate suppressive dendritic cell fate and function during viral persistence. PLoS Pathog. 2016;12:e1005356. doi: 10.1371/journal.ppat.1005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dutrieux J., Fabre-Mersseman V., Charmeteau-De Muylder B., Rancez M., Ponte R., Rozlan S., Figueiredo-Morgado S., Bernard A., Beq S., Couëdel-Courteille A., Cheynier R. Modified interferon-α subtypes production and chemokine networks in the thymus during acute simian immunodeficiency virus infection, impact on thymopoiesis. AIDS. 2014;28:1101–1113. doi: 10.1097/QAD.0000000000000249. [DOI] [PubMed] [Google Scholar]
- 83.Keir M.E., Rosenberg M.G., Sandberg J.K., Jordan K.A., Wiznia A., Nixon D.F., Stoddart C.A., McCune J.M. Generation of CD3+CD8 low thymocytes in the HIV type 1-infected thymus. J. Immunol. 2002;169:2788–2796. doi: 10.4049/jimmunol.169.5.2788. [DOI] [PubMed] [Google Scholar]
- 84.Stoddart C.A., Keir M.E., McCune J.M. IFN-alpha-induced upregulation of CCR5 leads to expanded HIV tropism in vivo. PLoS Pathog. 2010;6:e1000766. doi: 10.1371/journal.ppat.1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Montfoort N., Olagnier D., Hiscott J. Unmasking immune sensing of retroviruses: interplay between innate sensors and host effectors. Cytokine Growth Factor Rev. 2014;25:657–668. doi: 10.1016/j.cytogfr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Fenton-May A.E., Dibben O., Emmerich T., Ding H., Pfafferott K., Aasa-Chapman M.M., Pellegrino P., Williams I., Cohen M.S., Gao F., Shaw G.M., Hahn B.H., Ochsenbauer C., Kappes J.C., Borrow P. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology. 2013;10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iyer S.S., Bibollet-Ruche F., Sherrill-Mix S., Learn G.H., Plenderleith L., Smith A.G., Barbian H.J., Russell R.M., Gondim M.V., Bahari C.Y., Shaw C.M., Li Y., Decker T., Haynes B.F., Shaw G.M., Sharp P.M., Borrow P., Hahn B.H. Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E590–E599. doi: 10.1073/pnas.1620144114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Foster T.L., Wilson H., Iyer S.S., Coss K., Doores K., Smith S., Kellam P., Finzi A., Borrow P., Hahn B.H., Neil S.J.D. Resistance of transmitted founder HIV-1 to IFITM-Mediated restriction. Cell Host Microbe. 2016;20:429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Okumura A., Alce T., Lubyova B., Ezelle H., Strebel K., Pitha P.M. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology. 2008;373:85–97. doi: 10.1016/j.virol.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doehle B.P., Hladik F., McNevin J.P., McElrath M.J., Gale M., Jr. Human immunodeficiency virus type 1 mediates global disruption of innateantiviral signaling and immune defenses within infected cells. J. Virol. 2009;83:10395–10405. doi: 10.1128/JVI.00849-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harman A.N., Lai J., Turville S., Samarajiwa S., Gray L., Marsden V., Mercier S.K., Jones K., Nasr N., Rustagi A., Cumming H., Donaghy H., Mak J., Gale M., Jr., Churchill M., Hertzog P., Cunningham A.L. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood. 2011;118:298–308. doi: 10.1182/blood-2010-07-297721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rasaiyaah J., Tan C.P., Fletcher A.J., Price A.J., Blondeau C., Hilditch L., Jacques D., Selwood D.L., James L.C., Noursadeghi M., Towers G.J. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sandstrom T.S., Ranganath N., Angel J.B. Impairment of the type I interferon response by HIV-1: Potential targets for HIV eradication. Cytokine Growth Factor Rev. 2017;37:1–16. doi: 10.1016/j.cytogfr.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 94.Akhtar L.N., Qin H., Muldowney M.T., Yanagisawa L.L., Kutsch O., Clements J.E., Benveniste E.N. Suppressor of cytokine signaling 3 inhibits antiviral IFN-beta signaling to enhance HIV-1 replication in macrophages. J. Immunol. 2010;185:2393–2404. doi: 10.4049/jimmunol.0903563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lau A.S., Read S.E., Williams B.R. Downregulation of interferon alpha but not gamma receptor expression in vivo in the acquired immunodeficiency syndrome. J. Clin. Invest. 1988;82:1415–1421. doi: 10.1172/JCI113746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hardy G.A., Sieg S.F., Rodriguez B., Jiang W., Asaad R., Lederman M.M., Harding C.V. Desensitization to type I interferon in HIV-1 infection correlates with markers of immune activation and disease progression. Blood. 2009;113:5497–5505. doi: 10.1182/blood-2008-11-190231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berger A., Sommer A.F., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L.C., Fernandez-Sesma A., Rutsch F., Simon V., König R., Flory E. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goujon C., Schaller T., Galão R.P., Amie S.M., Kim B., Olivieri K., Neil S.J., Malim M.H. Evidence for IFNalpha-induced, SAMHD1-independent inhibitors of early HIV-1 infection. Retrovirology. 2013;10:23. doi: 10.1186/1742-4690-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang S., Zhan Y., Zhou Y., Jiang Y., Zheng X., Yu L., Tong W., Gao F., Li L., Huang Q., Ma Z., Tong G. Interferon regulatory factor 3 is a key regulation factor for inducing the expression of SAMHD1 in antiviral innate immunity. Sci. Rep. 2016;6:29665. doi: 10.1038/srep29665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meier A., Chang J.J., Chan E.S., Pollard R.B., Sidhu H.K., Kulkarni S., Wen T.F., Lindsay R.J., Orellana L., Mildvan D., Bazner S., Streeck H., Alter G., Lifson J.D., Carrington M., Bosch R.J., Robbins G.K., Altfeld M. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat. Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chang J.J., Woods M., Lindsay R.J., Doyle E.H., Griesbeck M., Chan E.S., Robbins G.K., Bosch R.J., Altfeld M. Higher expression of several interferon-stimulated genes in HIV-1-infected females after adjusting for the level of viral replication. J. Infect. Dis. 2013;208:830–838. doi: 10.1093/infdis/jit262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng H.Y., Zhang M.X., Chen M., Jiang J., Song J.H., Lian X.D., Tian R.R., Zhang X.L., Zhang L.T., Pang W., Zhang G.H., Zheng Y.T. Accelerated disease progression and robust innate host response in aged SIVmac239-infected Chinese rhesus macaques is associated with enhanced immunosenescence. Sci. Rep. 2017;7:37. doi: 10.1038/s41598-017-00084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bochud P.Y., Hersberger M., Taffé P., Bochud M., Stein C.M., Rodrigues S.D., Calandra T., Francioli P., Telenti A., Speck R.F., Aderem;;1 A. Swiss HIV cohort study. polymorphisms in toll-like receptor 9 influence the clinical course of HIV-1 infection. AIDS. 2007;21:441–446. doi: 10.1097/QAD.0b013e328012b8ac. [DOI] [PubMed] [Google Scholar]
- 104.Soriano-Sarabia N., Vallejo A., Ramirez-Lorca R., Rodriguez M.D., Salinas A., Pulido I. Influence of the toll-like receptor 9 1635A/G polymorphism on the CD4 count HIV viral load, and clinical progression. J. Acquir. Immune Defic. Syndr. 2008;49:128–135. doi: 10.1097/QAI.0b013e318184fb41. [DOI] [PubMed] [Google Scholar]
- 105.Said E.A., Al-Yafei F., Zadjali F., Hasson S.S., Al-Balushi M.S., Al-Mahruqi S., Koh C.Y., Al-Naamani K., Al-Busaidi J.Z., Idris M.A., Balkhair A., Al-Jabri A.A. Association of single-nucleotide polymorphisms in TLR7 (Gln11Leu) and TLR9 (1635A/G) with a higher CD4T cell count during HIV infection. Immunol. Lett. 2014;160:58–64. doi: 10.1016/j.imlet.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 106.Pine S.O., McElrath M.J., Bochud P.Y. Polymorphisms in toll-like receptor 4 and toll-like receptor 9 influence viral load in a seroincident cohort of HIV-1-infected individuals. AIDS. 2009;23:2387–2395. doi: 10.1097/QAD.0b013e328330b489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Booiman T., Kootstra N.A. Polymorphism in IFI16 affects CD4(+) T-cell counts in HIV-1 infection. Int. J. Immunogenet. 2014;41:518–520. doi: 10.1111/iji.12157. [DOI] [PubMed] [Google Scholar]
- 108.Diop G., Hirtzig T., Do H., Coulonges C., Vasilescu A., Labib T., Spadoni J.L., Therwath A., Lathrop M., Matsuda F., Zagury J.F. Exhaustive genotyping of the interferon alpha receptor 1 (IFNAR1) gene and association of an IFNAR1 protein variant with AIDS progression or susceptibility to HIV-1 infection in a French AIDS cohort. Biomed. Pharmacother. 2006;60:569–577. doi: 10.1016/j.biopha.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 109.Anokhin V.V., Bakhteeva L.B., Khasanova G.R., Khaiboullina S.F., Martynova E.V., Tillett R.L., Schlauch K.A., Lombardi V.C., Rizvanov A.A. Previously unidentified single nucleotide polymorphisms in HIV/AIDS cases associate with clinical parameters and disease progression. BioMed Res. Int. 2016;2016 doi: 10.1155/2016/2742648. (2742648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Coon S., Wang D., Wu L. Polymorphisms of the SAMHD1 gene are not associated with the infection and natural control of HIV type 1 in Europeans and African-Americans. AIDS Res. Hum. Retroviruses. 2012;28:1565–1573. doi: 10.1089/aid.2012.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li Q., Qiao Y., Zhang G., He N., Zhang X., Jia X., Sun H., Wang C., Xu L. Association of single nucleotide polymorphisms of APOBEC3G with susceptibility to HIV-1 infection and disease progression among men engaging in homosexual activity in northern China. Arch. Virol. 2017;162:259–268. doi: 10.1007/s00705-016-3080-8. [DOI] [PubMed] [Google Scholar]
- 112.Mhandire K., Duri K., Mhandire D., Musarurwa C., Stray-Pedersen B., Dandara C. Evaluating the contribution of APOBEC3G haplotypes: on influencing HIV infection in a Zimbabwean paediatric population. S. Afr. Med. J. 2016;106:S119–23. doi: 10.7196/SAMJ.2016.v106i6.11013. [DOI] [PubMed] [Google Scholar]
- 113.Compaore T.R., Soubeiga S.T., Ouattara A.K., Obiri-Yeboah D., Tchelougou D., Maiga M., Assih M., Bisseye C., Bakouan D., Compaore I.P., Dembele A., Martinson J., Simpore J. APOBEC3G variants and protection against HIV-1 infection in Burkina Faso. PLoS One. 2016;11:e0146386. doi: 10.1371/journal.pone.0146386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Naruse T.K., Sakurai D., Ohtani H., Sharma G., Sharma S.K., Vajpayee M., Mehra N.K., Kaur G., Kimura A. APOBEC3H polymorphisms and susceptibility to HIV-1 infection in an Indian population. J. Hum. Genet. 2016;61:263–265. doi: 10.1038/jhg.2015.136. [DOI] [PubMed] [Google Scholar]
- 115.Singh K.K., Wang Y., Gray K.P., Farhad M., Brummel S., Fenton T., Trout R., Spector S.A. Genetic variants in the host restriction factor APOBEC3G are associated with HIV-1-related disease progression and central nervous system impairment in children. J. Acquir. Immune Defic. Syndr. 2013;62:197–203. doi: 10.1097/QAI.0b013e31827ab612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bizinoto M.C., Leal E., Diaz R.S., Janini L.M. Loci polymorphisms of the APOBEC3G gene in HIV type 1-infected Brazilians. AIDS Res. Hum. Retroviruses. 2011;27:137–141. doi: 10.1089/aid.2010.0146. [DOI] [PubMed] [Google Scholar]
- 117.Kamada A.J., Bianco A.M., Zupin L., Girardelli M., Matte M.C., Medeiros R.M., Almeida S.E., Rocha M.M., Segat L., Chies J.A., Kuhn L., Crovella S. Protective role of BST2 polymorphisms in mother-to-Child transmission of HIV-1 and adult AIDS progression. J. Acquir. Immune Defic. Syndr. 2016;72:237–241. doi: 10.1097/QAI.0000000000000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chang J.J., Lindsay R.J., Kulkarni S., Lifson J.D., Carrington M., Altfeld M. Polymorphisms in IR reduce IFNα responses of pDCs to HIV-1. AIDS. 2011;25:715–717. doi: 10.1097/QAD.0b013e328343c186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martin M.P., Qi Y., Goedert J.J., Hussain S.K., Kirk G.D., Hoots W.K., Buchbinder S., Carrington M., Thio C.L. IL28B polymorphism does not determine outcomes of hepatitis B virus or HIV infection. J. Infect. Dis. 2010;202:1749–1753. doi: 10.1086/657146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rallon N.I., Restrepo C., Naggie S., Lopez M., Del Romero J., Goldstein D., McHutchison J., Soriano V., Benito J.M. Interleukin-28B gene polymorphisms do not influence the susceptibility to HIV infection or CD4 cell decline. AIDS. 2011;25:269–271. doi: 10.1097/QAD.0b013e328341b84e. [DOI] [PubMed] [Google Scholar]
- 121.Bibert A., Taffé P., Manuel E., Bernasconi H., Günthard M., Hoffmann L., Kaiser M., Osthoff M., Bochud P.Y. Swiss HIV cohort study. the IFNL3/4 éG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS. 2014;28:1885–1889. doi: 10.1097/QAD.0000000000000379. [DOI] [PubMed] [Google Scholar]
- 122.Parczewski M., Bander D., Leszczyszyn-Pynka M., Urbańska A., Socha Ł., Boroń-Kaczmarska A. IL28B CC genotype is associated with higher all-cause mortality in antiretroviral-treated HIV-infected patients. AIDS Res. Hum. Retroviruses. 2012;28:1640–1646. doi: 10.1089/AID.2011.0354. [DOI] [PubMed] [Google Scholar]
- 123.Machmach K., Abad-Molina C., Romero-Sánchez M.C., Dominguez-Molina B., Moyano M., Rodriguez M.M., Rafii-El-Idrissi Benhnia M., Jimenez-Mejias M.E., Vidal F., Muñoz-Fernández M.A., Genebat M., Viciana P., González-Escribano M.F., Leal M., Ruiz-Mateos E. IFNL4 ss469415590 polymorphism is associated with unfavourable clinical and immunological status in HIV-infected individuals. Clin. Microbiol. Infect. 2015;21:289. doi: 10.1016/j.cmi.2014.10.012. (e1-4) [DOI] [PubMed] [Google Scholar]
- 124.Real L.M., Herrero R., Rivero-Juárez A., Camacho Á., Macías J., Vic S., Soriano V., Viedma S., Guardiola J.M., Fibla J., Rivero A., Pineda J.A., Caruz A. IFNL4 rs368234815 polymorphism is associated with innate resistance to HIV-1 infection. AIDS. 2015;29:1895–1897. doi: 10.1097/QAD.0000000000000773. [DOI] [PubMed] [Google Scholar]
- 125.Dominguez-Molina B., Tarancon-Diez L., Hua S., Abad-Molina C., Rodriguez-Gallego E., Machmach K., Vidal F., Tural C., Moreno S., Goñi J.M., Ramírez de Arellano E., Del Val M., Gonzalez-Escribano M.F., Del Romero J., Rodriguez C., Capa L., Viciana P., Alcamí J., Yu X.G., Walker B.D., Leal M., Lichterfeld M., Ruiz-Mateos E. ECRIS integrated in the Spanish AIDS Research Network. HLA-B*57 and IFNL4-related polymorphisms are associated with protection againstHIV-1 disease progression in controllers. Clin. Infect. Dis. 2017;64:621–628. doi: 10.1093/cid/ciw833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Monteleone K., Scheri G.C., Statzu M., Selvaggi C., Falasca F., Giustini N., Mezzaroma I., Turriziani O., d'Ettorre G., Antonelli G., Scagnolari C. IFN-stimulated gene expression is independent of the IFNL4 genotype in chroniHIV-1 infection. Arch. Virol. 2016;161:3263–3268. doi: 10.1007/s00705-016-3016-3. [DOI] [PubMed] [Google Scholar]
- 127.DeStefano E., Friedman R.M., Friedman-Kien A.E., Goedert J.J., Henriksen D., Preble O.T., Sonnabend J.A., Vilcek J. Acid-labile human leukocyte interferon in homosexual men with Kaposi's sarcoma and lymphadenopathy. J. Infect. Dis. 1982;146:451–459. doi: 10.1093/infdis/146.4.451. [DOI] [PubMed] [Google Scholar]
- 128.Voth R., Rossol S., Klein K., Hess G., Schütt K.H., Schröder H.C., Meyer zum Büschenfelde K.H., Müller W.E. Differential gene expression of IFN-alpha and tumor necrosis factor-alpha in peripheral blood mononuclear cells from patients with AIDS related complex and AIDS. J. Exp. Med. 1990;172:1433–1442. [PubMed] [Google Scholar]
- 129.Vyakarnam A., Matear P., Meager A., Kelly G., Stanley B., Weller I., Beverley P. Altered production of tumour necrosis factors alpha and beta and interferon gamma by HIV-infected individuals. Clin. Exp. Immunol. 1991;84:109–115. doi: 10.1111/j.1365-2249.1991.tb08132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gendelman H.E., Friedman R.M., Joe S., Baca L.M., Turpin J.A., Dveksler G., Meltzer M.S., Dieffenbach C. A selective defect of interferon alpha production in human immunodeficiency virus-infected monocytes. J. Exp. Med. 1990;172:1433–1442. doi: 10.1084/jem.172.5.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hardy G.A., Sieg S., Rodriguez B., Anthony D., Asaad R., Jiang W., Mudd J., Schacker T., Funderburg N.T., Pilch-Cooper H.A., Debernardo R., Rabin R.L., Lederman M.M., Harding C.V. Interferon-α is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS One. 2013;8:e56527. doi: 10.1371/journal.pone.0056527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lehmann C., Taubert D., Jung N., Fatkenheuer G., van Lunzen J., Hartmann P., Romerio F. Preferential upregulation of interferon-alpha subtype 2 expression in HIV-1 patients. AIDS Res. Hum. Retroviruses. 2009;25:577–581. doi: 10.1089/aid.2008.0238. [DOI] [PubMed] [Google Scholar]
- 133.Zaritsky L.A., Dery A., Leong W.Y., Gama L., Clements J.E. Tissue-specific interferon alpha subtype response to SIV infection in brain, spleen, and lung. J. Interferon Cytokine Res. 2013;33:24–33. doi: 10.1089/jir.2012.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Easlick J., Szubin R., Lantz S., Baumgarth N., Abel K. The early interferon alpha subtype response in infant macaques infected orally with SIV. J. Acquir. Immune Defic. Syndr. 2010;55:14–28. doi: 10.1097/QAI.0b013e3181e696ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Harper M.S., Guo K., Gibbert K., Lee E.J., Dillon S.M., Barrett B.S., McCarter M.D., Hasenkrug K.J., Dittmer U., Wilson C.C., Santiago M.L. Interferon-alpha subtypes in an ex vivo model of acute HIV-1 infection: expression, potency and effector mechanisms. PLoS Pathog. 2015;11:e1005254. doi: 10.1371/journal.ppat.1005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lavender K.J., Gibbert K., Peterson K.E., Van Dis E., Francois S., Woods T., Messer R.J., Gawanbacht A., Müller J.A., Münch J., Phillips K., Race B., Harper M.S., Guo K., Lee E.J., Trilling M., Hengel H., Piehler J., Verheyen J., Wilson C.C., Santiago M.L., Hasenkrug K.J., Dittmer U. Interferon alpha subtype-specific suppression of HIV-1 infection in vivo. J. Virol. 2016;90:6001–6013. doi: 10.1128/JVI.00451-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Abraham S., Choi J.G., Ortega N.M., Zhang J., Shankar P., Swamy N.M. Gene therapy with plasmids encoding IFN-β or IFN-α14 confers long-term resistance to HIV-1 in humanized mice. Oncotarget. 2016;7:78412–78420. doi: 10.18632/oncotarget.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brenchley J.M., Schacker T.W., Ruff L.E., Price D.A., Taylor J.H., Beilman G.J., Nguyen P.L., Khoruts A., Larson M., Haase A.T., Douek D.C. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Parmentier M. CCR5 and HIV infection, a view from brussels. Front. Immunol. 2015;6:295. doi: 10.3389/fimmu.2015.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Klatt N.R., Funderburg N.T., Brenchley J.M. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21:6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gori A., Tincati C., Rizzardini G., Torti C., Quirino T., Haarman M., Ben Amor K., v an Schaik J., Vriesema A., Knol J., Marchetti G., Welling G., Clerici M. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J. Clin. Microbiol. 2008;46:757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zilberman-Schapira G., Zmora N., Itav S., Bashiardes S., Elinav H., Elinav E. The gut microbiome in human immunodeficiency virus infection. BMC Med. 2016;14:83. doi: 10.1186/s12916-016-0625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dubourg G., Surenaud M., Lévy Y., Hüe S., Raoult D. Microbiome of HIV-infected people. Microb. Pathog. 2017;106:85–93. doi: 10.1016/j.micpath.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 144.Dillon S.M., Frank D.N., Wilson C.C. The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS. 2016;30:2737–2751. doi: 10.1097/QAD.0000000000001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ribeiro A.B.D.T.M., Heimesaat M.M., Bereswill S. Changes of the intestinal microbiome-host homeostasis in HIV-infected individuals- a focus on the bacterial gut microbiome. Eu.r J. Microbiol. Immunol. (Bp) 2017;7:158–167. doi: 10.1556/1886.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Noguera-Julian M., Rocafort M., Guillén Y., Rivera J., Casadellà M., Nowak P., Hildebrand F., Zeller G., Parera M. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nowak P., Troseid M., Avershina E., Barqasho B., Neogi U., Holm K., Hov J.R., Noyan K., Vesterbacka J., Svärd J., Rudi K., Sönnerborg A. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 148.Dillon S.M., Lee E.J., Kotter C.V., Austin G.L., Dong Z., Hecht D.K. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vujkovic-Cvijin I., Dunham R.M., Iwai S., Maher M.C., Albright R.G., Broadhurst M.J., Hernandez R.D., Lederman M.M., Huang Y., Somsouk M., Deeks S.G., Hunt P.W., Lynch S.V., McCune J.M. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006438. (pp. 193ra91) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pérez-Santiago J., Gianella S., Massanella M., Spina C.A., Karris M.Y., Var S.R., Patel D., Jordan P.S., Young J.A., Little S.J., Richman D.D., Smith D.M. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS. 2013:1921–1931. doi: 10.1097/qad.0b013e3283611816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Villanueva-Millán M.J., Pérez-Matute P., Recio-Fernández E., Lezana Rosales J.M., Oteo J.A. Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J. Int. AIDS Soc. 2017;20:21526. doi: 10.7448/IAS.20.1.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu J., Williams B., Frank D., Dillon S.M., Wilson C.C., Landay A.L. Inside out: HIV, the gut microbiome, and the mucosal immune system. J. Immunol. 2017;198:605–614. doi: 10.4049/jimmunol.1601355. [DOI] [PubMed] [Google Scholar]
- 153.Giles E.M., Stagg A.J. Type 1 interferon in the human intestine-A Co-ordinator of the immune response to the microbiota. Inflamm. Bowel Dis. 2017;23:524–533. doi: 10.1097/MIB.0000000000001078. [DOI] [PubMed] [Google Scholar]
- 154.Abt M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J., Wherry E.J., Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tschurtschenthaler M., Wang J., Fricke C., Fritz T.M., Niederreiter L., Adolph T.E., Sarcevic E., Künzel S., Offner F.A., Kalinke U., Baines J.F., Tilg H., Kaser A. Type I interferon signalling in the intestinal epithelium affects Paneth cells, microbial ecology and epithelial regeneration. Gut. 2014;63:1921–1931. doi: 10.1136/gutjnl-2013-305863. [DOI] [PubMed] [Google Scholar]
- 156.Baldridge M.T., Nice T.J., McCune B.T., Yokoyama C.C., Kambal A., Wheadon M., Diamond M.S., Ivanova Y., Artyomov M., Virgin H.W. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Steed A.L., Christophi G.P., Kaiko G.E., Sun L., Goodwin V.M., Jain U., Esaulova E., Artyomov M.N., Morales D.J., Holtzman M.J., Boon A.C.M., Lenschow D.J., Stappenbeck. Virgin T.S.H.W. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 2017;357:498–502. doi: 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]