Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) membrane protein is an abundant virion protein, and its interaction with the nucleocapsid protein is crucial for viral assembly and morphogenesis. Although the interacting region in the nucleocapsid protein was mapped to residues 168–208, the interacting region in the membrane protein and the interaction nature are still unclear. In this work, by using yeast two-hybrid and surface plasmon resonance techniques, the residues 197–221 of the membrane protein and the residues 351–422 of the nucleocapsid protein were determined to be involved in their interaction. Sequence analysis revealed that these two fragments are highly charged at neutral pH, suggesting that their interaction may be of ionic nature. Kinetic assays indicated that the endodomain (aa102–221) of the membrane protein interacts with the nucleocapsid protein with high affinity (KD = 0.55 ± 0.04 μM), however, this interaction could be weakened greatly by acidification, higher salt concentration (400 mM NaCl) and divalent cation (50 mM Ca2+), which suggests that electrostatic attraction might play an important role in this interaction. In addition, it is noted that two highly conserved amino acids (L218 and L219) in the membrane protein are not involved in this interaction. Here, we show that electrostatic interactions between the carboxyl termini of SARS-CoV membrane protein and nucleocapsid protein largely mediate the interaction of these two proteins. These results might facilitate therapeutic strategies aiming at the disruption of the association between SARS-CoV membrane and nucleocapsid proteins.
Abbreviations: SARS-CoV, severe acute respiratory syndrome coronavirus; N, the nucleocapsid protein; M, the membrane protein; ME, the endodomain of M protein; SPR, surface plasmon resonance
Keywords: Protein–protein interaction, SARS coronavirus (SARS-CoV), Membrane protein, Nucleocapsid protein, Surface plasmon resonance (SPR), Yeast two-hybrid
1. Introduction
Severe acute respiratory syndrome (SARS) is a novel worldwide infectious disease, which is caused by a newly identified member of the coronaviridae family, SARS coronavirus (SARS-CoV) (Ksiazek et al., 2003, Marra et al., 2003, Rota et al., 2003). In coronavirus, the highly basic nucleocapsid protein (N) binds to RNA to form a helical nucleocapsid, followed by the combination with three other virion proteins, Membrane protein (M), Spike protein (S), and Envelope protein (E). Both the M and E proteins are essential for coronavirus morphogenesis (de Haan, Kuo, Masters, Vennema, & Rottier, 1998; Vennema et al., 1996), and interactions between these two proteins seem to drive the coronavirus envelope assembly, thus producing virus-like particles in the absence of other viral components (Escors, Ortego, & Enjuanes, 2001b). The M protein is able to bind to the nucleocapsid protein in the absence of S and E protein (Narayanan, Maeda, Maeda, & Makino, 2000), and the M protein binds to nucleocapsid with its carboxyl terminus in mouse hepatitis virus (MHV) (Kuo & Masters, 2002; Narayanan et al., 2000) and transmissible gastroenteritis coronavirus (TGEV) (Escors et al., 2001b, Escors et al., 2001c). Moreover, two homologous segments, residues 201–224 of MHV M protein and residues 233–257 of TGEV M protein, were mapped to be involved in the M/N protein–protein interaction (Escors et al., 2001c, Kuo and Masters, 2002). Recently, SARS-CoV M/N interaction in the absence of viral RNA was validated in vivo through the mammalian two-hybrid system, which reinforced the fact that the M/N interaction makes the formation of the crucial structure for coronavirus (He et al., 2004). However, the knowledge about this interaction is still limited and more explorations are needed.
As shown in Fig. 1 , SARS-CoV M protein was predicted to span the membrane three times and display a short ectodomain and a large endodomain on the basis of other coronavirus M proteins (de Haan, Vennema, & Rottier, 2000; Kuo & Masters, 2002; Marra et al., 2003). It is believed that the C-terminal polar tail within the endodomain interacts with N and S proteins (de Haan, Smeets, Vernooij, Vennema, & Rottier, 1999; Opstelten, Raamsman, Wolfs, Horzinek, & Rottier, 1995), which suggests that the large M endodomain (ME) probably makes great sense in SARS-CoV assembly.
Fig. 1.

Diagrammatic representation of SARS-CoV M protein. SARS-CoV M protein is characterized as three domains: a short N-terminal ectodomain, a triple-span transmembrane domain, and a C-terminal endodomain. This result was predicted by Antherpro V5.0 based on the references (de Haan et al., 2000, Kuo and Masters, 2002; Marra et al., 2003).
In this study, using yeast two-hybrid and surface plasmon resonance (SPR) techniques, we have fully characterized SARS-CoV ME/N interaction. It is found that the residues 197–221 of M protein and the residues 341–422 of N protein are contributed to this interaction. These two highly polar fragments imply that SARS-CoV ME/N interaction might be of ionic nature. SPR based results indicated that SARS-CoV ME interacts with N protein with a high affinity, and the electrostatic attractive force plays an important role in their interaction. In addition, it is noted that two highly conserved amino acids (L218 and L219) of SARS-CoV M protein are not involved in the M/N interaction. This current report is hoped to contribute some useful information to shedding light on the molecular mechanism of SARS-CoV M/N interaction.
2. Materials and methods
2.1. Chemicals and enzymes
The restriction and modification enzymes in this work were purchased from NEB. The bacterial strains are from Qiagen. The vectors pET15b and pGEX 4T-1, the affinity columns and lower molecular weight (LMW) marker were purchased from Amersham Pharmacia Biotech. Isopropyl β-d-thiogalactoside (IPTG) was purchased from Promega. The MCT peptide (RYRIGNYKLNTDHAGSNDNIALLVQ, the residues 197–221 of SARS-CoV M protein) and the control peptide (AGGAGAGGEA) were synthesized by Sangon (China). All other chemicals were from Sigma in their analytical grade.
2.2. Recombinant vectors construction for yeast two-hybrid assay
The yeast vectors of pGADT7 and pGBKT7 were obtained from Clontech (Palo Alto, CA). The cDNA of SARS-CoV ME was amplified with the following primers: 5′-AATTGAATTCTTTGCTCGTACCCGCTCAATGTG-3′ and 5′-ACCCGGATCCTT ACTGTACTAGCAAAGCAATAT-3′. The amplified products were digested with EcoRI and BamHI then cloned into the pGADT7 vector between these two sites. The cDNA of SARS-CoV N protein was amplified with the following primers: 5′-GGGTGAATTCATGTCTGATAATGGACCCCAAT-3′ and 5′-AATTGGATCCTT ATGCCTGAGTTGAATCAGCAG-3′. The amplified products were digested with EcoRI and BamHI then cloned into pGBKT7 vector between these two sites. To identify the amino acid sequence required for SARS-CoV ME/N interaction, different fragments were prepared by PCR. The primers are listed in Table 1 .
Table 1.
Primers for the truncated fragments and mutants used in yeast two-hybrid
| ΔN1 (sense) | GGTTGAATTCCCAGATGACCAAATTGGCTACTACC |
| ΔN2 (sense) | GGTTGAATTCCAACTTCCTCAAGGAACAACATTGCC |
| ΔN3 (ΔN5) (sense) | AATTGAATTCATGGCTAGCGGAGGTGGTGAAA |
| ΔN4 (sense) | GCGTGAATTCGACCTAATCAGACAAGGAACTGA |
| ΔN5 (anti-sense) | GGCCGGATCCTTAGTTGTCTTTGAATTGTGGAT |
| ΔME1 (anti-sense) | ACGGGGATCCTTAGTTGTATGCAGCAAAACCTG |
| ΔME2 (anti-sense) | ACGGGGATCCTTATGTAGCCACAGTGATCTCTT |
| Mut-ME (anti-sense) | GGCCGGATCCTTACTGTACTAGCAAAGCAATAT |
2.3. Yeast transformation and culture
Transformations were performed according to the manufacturer's protocol. Briefly, 500 ng of plasmid DNA was added to 50 μL of competent cells and mixed with 36 μL lithium acetate and 240 μL 50% PEG3350 and 50 ng single-strain DNA at 30 °C for 30 min followed by heat-shock at 42 °C for 30 min. The mixture was subsequently spread on a drop-out-agar plate in the absence of leucine and tryptophan. The plate was incubated at 30 °C for 48 h for yeast growth. PCR was used to confirm transformation with the target gene. A positive clone was inoculated in the SD medium lacking leucine, typtophan and histidine (SD-LTH) and supplemented with 1 mM 3-amino-1′,2′,4′-triazole (SD-LTH + 3-AT). The medium was shaken at 200 rpm at 30 °C for 96 h before 300 μL of the medium was filled into the well of a 96-well microplate. The absorbance at 595 nm of the medium in the 96-well microplate was measured in a TECAN reader (Switzerland). The growing and measuring courses were repeated three times for each experiment and the absorbance was obtained by average. All the yeast media were prepared according to the standard Protocols Handbook (PT3024-1, Clontech). This is an alternative method based on the growth curve analysis that is reproducible and of equal or greater sensitivity compared with the β-galactosidase method (Diaz-Camino, Risseeuw, Liu, & Crosby, 2003).
2.4. Preparations of the wild type and truncated mutants of SARS-CoV N protein
The recombinant SARS-CoV N protein was prepared according to the reported procedure (Luo et al., 2004). Two truncated fragments of SARS-CoV N, N4 (aa1–340, Scheme 1 ) and N11 (aa283–422, Scheme 1), were cloned, expressed and purified by the similar procedure. The primers for N4 are 5′-AATTGGATCCATGTCTGATAATGGACCCCAATC-3′ and 5′-GGGAGTCGACTTACAATTTAATGGCTCCATGAT-3′. The primers for N11 are 5′-AATTGGATCCACCCAAGGAAATTTCGGGGACCAA -3′ and 5′-GGTTGTCGACTTATGCCTGAGTTGAATCAGCAG-3′.
Scheme 1.

Schematic diagram of the different recombinant domains of SARS-CoV N and M proteins.
2.5. Preparations of the wild type and mutated SARS-CoV ME (L218AL219A)
The plasmid of pMAL-cRI-SARS-CoV-M (Zhang et al., 2003) was kindly provided by Prof. Yang (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, China). The cDNA of SARS-CoV ME was cloned into pET15b vector (PET15b-ME) using pMAL-cRI-SARS-CoV-M as a template. The recombinant plasmid for the mutant (L218AL219A), pET15b-Mut-ME, was constructed by the same protocol using the corresponding primers. All the cloning procedures are standard (Sambrook, Fritsch, & Maniatis, 1989), and the cloned DNA sequences were all sequenced.
The primers for the wild type and the mutant were listed as follows: (sense) 5′-AATTCATATGTTTGCTCGTACCCGCTCA-3′; (anti-sense) 5′-AATTGGATCCTTACTGTACTAGCAAAGGC-3′; (sense) 5′-AATTCATATGTTTGCTCGTACCCGCTCAATGTGG-3′; (anti-sense) 5′-CCGGGGATCCTTACTGTACAGCAGCAGCAATATTGTCGTTGCTACC-3′.
The preparation procedures of the wild type and the mutant of SARS-CoV ME are similar to each other. Here we described the preparation procedure for the wild type as an example. The recombinant plasmid of pET15b-ME was transformed into BL21 (DE3) E. coli bacteria. Clones were grown overnight in LB medium containing 100 mg L−1 ampicillin. Expression of the protein was induced at an OD600 of 0.5–0.7 with addition of isopropyl-β-d-galactoside (IPTG) to a final concentration of 0.2 mM. After induction for 4 h at 18 °C, the cells were harvested by centrifugation for 30 min at 4000 rpm, 4 °C and stored at −80 °C overnight. The frozen cells were thawed, re-suspended in 18 mL of buffer A (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 5 mM imidazole, 1 mM PMSF), and then lysed by sonication for 15 min in ice bath. The lysate was cleared by centrifugation and the supernatant was loaded on a Sepharose Ni-NTA column (Amersham Pharmacia) equilibrated with buffer A. The column was eluted with elution buffer (20 mM Tris–HCl, pH 8.0, 500 mM NaCl, 500 mM imidazole) after being washed by wash buffer (20 mM Tris–HCl, pH 8.0, 500 mM NaCl, 100 mM imidazole). The elution fraction was further applied to a gel filtration column (Hiprep™ 16/60 sephacyl S100) on an FPLC system. The column was pre-equilibrated with HBS buffer (10 mM HEPES, pH 7.4, 150 mM NaCl) before loading the protein sample. ME protein was thus eluted by HBS buffer, and concentrated by Ultrafree4 tube (Milipore Co.) The protein purity was determined by SDS-PAGE and concentration was monitored by measuring the absorbance at 280 nm in HBS buffer using the extinction coefficient of 13075 L mol−1 cm−1.
2.6. Preparation of GST-tagged ME1 (residues 101–196 of SARS-CoV M protein)
The cDNA of ME1 (aa101–196, Scheme 1) was cloned into pGEX 4T-1 vector using BamHI and XholI restrict enzyme sites and was then sequenced. The primers for ME1 are 5′-AATTGAATCCTTTGCTCGTACCCGCTCAATGT-3′ and 5′-GGCCCTCGAGTTACTGTACTAGCAAAGCAATAT-3′. The expression and purification of this recombinant protein by GST affinity column was strictly according to the standard procedures (Amersham Pharmacia Biotech).
2.7. SPR technology based Biacore 3000 analysis
The covalent immobilization of the ligand on CM5 sensor chip was carried out using the standard amine coupled wizard at pH 6.0 in 10 mM sodium acetate. The control flow cell was immobilized with lysozyme at a parallel level as negative reference. Signals obtained from the control flow cell were subtracted from those of the experimental flow cell to reduce the non-specific binding. The response is the direct indication of the amount of the bound analyte. The running and sample buffers were HBS-EP buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% P20), except for the special buffer indicated later in assays. All buffers were filtered and degassed before use. The analyte was injected (120 s, 30 μl s−1) followed by a dissociation phase (120 s). The flow cells were regenerated by 50 mM NaOH after each injection. Each experiment was repeated at least twice. The parameters, association rate constant (k on), dissociation rate constant (k off), and equilibrium dissociation constant (K D), were obtained by fitting to 1:1 Langmuir binding model using a global fitting method from the BIAevaluation 3.1 software. As for the surface competition experiments, SARS-CoV ME (5 μM) was mixed with the MCT peptide (RYRIGNYKLNTDHAGSNDNIALLVQ) in various concentrations before injection. All data were obtained at 25 °C.
3. Results and discussion
3.1. SARS-CoV ME/N interaction region mapping by yeast two-hybrid assays
Recently, He et al. (2004) reported that SARS-CoV M protein directly interacts with SARS-CoV N protein in the absence of viral RNA. In the current work, to obtain more information about this interaction, a yeast two-hybrid technology based system was constructed. The cDNAs of SARS-CoV N and SARS-CoV ME were cloned in-frame into pGBKT7 and pGADT7 vectors. Upon co-transforming the both constructs into AH109, cell growth was observed in stringency nutrition medium (SD-LTH + 1 mM 3-AT) (Fig. 2 B), indicating the SARS-CoV ME/N interaction in vivo, which is in agreement with the reported result (He et al., 2004). Moreover, the fact that the control yeast cells transforming with empty plasmids did not show cell growth (Fig. 2B) suggested that this ME/N interaction is specific in yeast. Accordingly, our yeast two-hybrid system is reliable and could be used for further research.
Fig. 2.

Mapping the domains involved in SARS-CoV ME/N interaction. Schematic description of the truncated fragments (A) and the yeast two-hybrid assay results for SARS-CoV ME/N interactions in their truncated and non-truncated forms (B). The empty vectors pGBKT7 and pGADT7 co-transformed were used as the negative control. Every experiment was repeated for at least three times and the data were obtained by average. The error bars represent standard error of the mean.
To map the involved regions of N protein in the SARS-CoV N/ME interaction, five truncated fragments of N protein were generated (Fig. 2A). These fragments, designated from ΔN1 to ΔN5, were cloned into pGBKT7 vector, and then co-transformed respectively with pGADT7-ME into the AH109 yeast cell. After 96-h growth in stringency nutrition medium (SD-LTH + 1 mM 3-AT) at 30 °C with shaking, 300 μL cells culture was transferred into 96-well microplate. The growth curves of these cells were then evaluated by measuring their absorbance at 595 nm in 96-well microplate. As indicated in Fig. 2B, the yeast cells containing ΔN1–ΔN4 showed viability compared to the empty control, while cells involving ΔN5 did not grow, suggesting that the carboxyl terminus of SARS-CoV N (aa351–422, Fig. 1A) is very important for SARS-CoV M/N interaction. It is noticed that even though ΔN3 and ΔN4 showed somewhat weaker ME-interaction ability in comparison with the full-length N as indicated in Fig. 2B, residues 161–290 of SARS-CoV N could not be considered as the critical region for SARS-CoV M/N interaction, which is not compatible with the reported result that residues 168–208 of SARS-CoV N protein may be critical for SARS-CoV M/N interaction (He et al., 2004).
To explore the binding fragment of SARS-CoV M within SARS-CoV M/N interaction, ΔME1 (aa102–196) and ΔME2 (aa102–171) of SARS-CoV M were also studied by the constructed yeast two-hybrid system (Fig. 2A). These two fragments were cloned into pGADT7 plasmid as pGADT7–ΔME1 and pGADT7–ΔME2, followed by co-transformation separately with pGBKT7–N into AH109. By measuring the cell growth (Fig. 2B) in SD-LTH-3-AT medium, it is found that neither ΔME1 nor ΔME2 showed interaction with SARS-CoV N protein, which suggests that residues 197–221 of SARS-CoV M protein are crucial for SARS-CoV M/N interaction.
3.2. SARS-CoV ME preparation
To further investigate the SARS-CoV M/N interaction in vitro, the expression of the SARS-CoV ME (Scheme 1) in E. coli and purification were carried out. The purity of the His-tagged SARS-CoV ME was confirmed by SDS-PAGE as shown in Fig. 3 . It is noticed that the recombinant ME was prone to expressing in inclusion body, and induction at low temperature (18 °C) might produce more soluble protein. The protein identity was verified by MS spectral analysis. The result of data search using MS/MS raw data of tryptic peptides shows that the sequence coverage of SARS-CoV ME was 63.8% and the total amino acid contained by 10 tryptic peptides were 90 (data not shown), which completely determined the identity of SARS-CoV ME in this work.
Fig. 3.
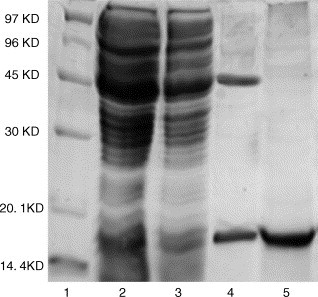
SDS-PAGE of purification for His-tagged SARS-CoV ME. Samples at each purification step were analyzed by SDS-PAGE. Lane 1, molecular mass marker; lane 2, crude lysate before nickel column purification; lane 3, sample flow-through the column; lane 4, ME purified by Ni-NTA resin; lane 5, purified ME after gel filtration.
3.3. SPR based kinetic analysis of SARS-CoV ME/N interaction
In order to perform the kinetic analysis of the SARS-CoV ME/N interaction by Biaocre 3000, the purified SARS-CoV N protein was immobilized on the surface of CM5 sensory chip with 2000 RU (resonance unit), and a series of purified SARS-CoV ME were injected to flow over the immobilized SARS-CoV N protein for kinetic investigation. The flow cell immobilized with lysozyme (2200 RU) was used as the negative reference, and all responses on the experimental flow cell were subtracted to the responses on the control flow cell. The interaction between SARS-CoV ME and the immobilized SARS-CoV N protein were shown as in Fig. 4 A (gray curves). It is found that the binding response increased from zero up to 700 RU with the increasing ME concentrations from 0 to 5 μM, indicating that SARS-CoV ME could bind directly to SARS-CoV N protein. The kinetic parameters for SARS-CoV ME/N interaction (k on and k off) were successfully determined by evaluating the curves according to the 1:1 Langmuir binding model. The evaluated curves were found to superpose the experimental curves very well (Fig. 4A) and the reliability of the fitting model could be also reflected by the small residuals (Fig. 4B). The rate associate (k on) and dissociate (k off) constants for this interaction were evaluated as 4.18 ± 0.32 × 103 M−1 s−1 and 2.23 ± 0.19 × 10−3 s−1, then K D (K D = k off/k on) was calculated as 0.55 ± 0.04 μM, revealing that SARS-CoV ME binds to SARS-CoV N protein with a high affinity. Furthermore, this M/N protein–protein-binding stoichiometry (1:1) for SARS coronavirus seems to be comparable with that for TGEV coronavirus, whose 1:1 binding ratio was accurately estimated (Escors, Camafeita, Ortego, Laude, & Enjuanes, 2001a).
Fig. 4.

Kinetic analysis of SARS-CoV ME/N interaction by SPR. SPR sensorgrams (A) obtained with SARS-CoV ME at different concentrations (marked in the right). The responses in RU (gray line) were recorded as a function of time. They were fitted to 1:1 langmuir binding model and depicted as black solid lines. Superposition of fitted curves (A) to original curves and the small residuals (B) demonstrates the goodness of the fitting.
3.4. The carboxyl terminus of SARS-CoV N protein plays an important role in SARS-CoV M/N interaction
To further evaluate the significance of N protein carboxyl terminus in SARS-CoV N/M interaction, His-tagged N11 (aa283–422, Scheme 1) and N4 (aa1–340, Scheme 1) proteins generated from E. coli system were tested on Biacore 3000. SARS-CoV ME/N11 (N4) interactions were studied with the purified N11 and N4 immobilized on the CM5 sensor chip surface. As shown in Fig. 5 A, 30 μM of SARS-CoV ME showed only 13 RU in response when binding to N4, and 5 μM of SARS-CoV ME demonstrated up to 700 RU when binding to the full-length N (Fig. 4A), this indicated that N4 almost lost the ME-binding capability. By contrast, as shown in Fig. 5B, the immobilized N11 interact with SARS-CoV ME with a high affinity (K D = 0.61 ± 0.06 μM) close to that of SARS-CoV ME/N interaction (K D = 0.55 ± 0.04 μM). These results thus further proved that the carboxyl terminus of SARS-CoV N protein, residues 341–422 in particular, are critical for SARS-CoV M/N interaction. Moreover, our result also indicated that the reported residues 168–208 (He et al., 2004) might not contribute greatly to SARS-CoV M/N interaction, in accordance with the above-mentioned yeast two-hybrid results. In fact, for mouse hepatitis virus (MHV) and TGEV, their M proteins have been believed to bind to the carboxyl terminuses of the N proteins (Kuo and Masters, 2002). Although SARS-CoV N shares only little homology with other coronavirus N protein, there might be a general pattern common to all the coronavirus N proteins: the protein is highly basic overall, and there is a concentration of acidic residues in its carboxyl terminal domain (Laude and Masters, 1995). Thus it is possible that the carboxyl terminus of N protein binding to M protein might be the common feature for coronavirus.
Fig. 5.
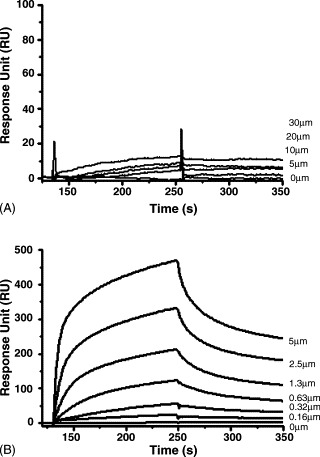
Residues 341–422 of SARS-CoV N protein play an important role in SARS-CoV M/N interaction. The curves for SARS-CoV ME binding to immobilized N4 (aa1–340, Scheme 1) (A) and N11 (aa283–422, Scheme 1) (B) at various concentrations. The concentration for each injection was marked in the right. The immobilized level for N4 and N11 are 1500 and 1200 RU.
3.5. Residues 197–221 of M is crucial for SARS-CoV M/N interaction
To further verify our above results that residues 197–221 of SARS-CoV M are crucial for SARS-CoV M/N interaction, GST-tagged ME1 (aa101–196, Scheme 1) was expressed and purified. Then the interaction between ME1 and the immobilized SARS-CoV N was studied on Biacore 3000. The results showed that SARS-CoV ME1/N interaction is very weak (Fig. 6 A). Together with the above-mentioned fact that SARS-CoV ME bind stongly to SARS-CoV N protein (Fig. 4A), it suggested that residues 197–221 of SARS-CoV M protein were involved in SARS-CoV M/N interaction. To further confirm the contribution of these 25 residues to this interaction, the MCT peptide (RYRIGNYKLNTDHAGSNDNIALLVQ, residues 197–221 of SARS-CoV M) interaction with the immobilized SARS-CoV N protein was also quantitatively investigated on Biacore 3000. Fig. 6B clearly showed that the MCT peptide could bind to the immobilized SARS-CoV N protein, and the related kinetic parameters could be determined by 1:1 Langmuir binding model as k on = 509 ± 42 M−1 s−1, k off = 0.0394 ± 0.002 s−1 and K D = 75 ± 5 μM. Moreover, the inhibition of the MCT peptide against SARS-CoV ME/N interaction was also studied. In the surface competition assays, 5 μM SARS-CoV ME protein was mixed with different concentrations of the MCT peptide before injection. As shown in Fig. 6C, the responses for the mixtures decreased with the increase of the MCT peptide, which thereby suggested that the MCT peptide could inhibit SARS-CoV ME protein binding to the immobilized N protein. In addition, as shown in Fig. 6D, the fact that the control peptide (AGGAGAGGEA) did not interact with the immobilized SARS-CoV N and rendered no effects on SARS-CoV ME/N interaction suggested the specificity of SARS-CoV N/MCT peptide interaction.
Fig. 6.

Residues 197–221 of M protein are crucial for SARS-CoV M/N interaction. (A) The curves of ME1 (aa101–196, Scheme 1) binding to the immobilized SARS-CoV N protein. (B) The curves for the MCT peptide (RYRIGNYKLNTDHAGSNDNIALLVQ, residues 197–221 in SARS-CoV M protein) interacting with the immobilized N protein. (C) Inhibition of SARS-CoV ME/N interaction by the MCT peptide. The concentration of SARS-CoV ME was fixed as 5 μM and the MCT peptide concentration was labelled on the right. (D) The control peptide (AGGAGAGGEA) showed no binding to the immobilized SARS-CoV N protein and no inhibition against SARS-CoV ME/N interaction.
Accordingly, all these above data obviously revealed that residues 197–221 of SARS-CoV M protein might be crucial for SARS-CoV M/N interaction. This is similar to the reported result that the 16 residues (aa237–252) were responsible for M protein binding to the nucleocapsid protein in TGEV (Escors et al., 2001c). The result that SARS-CoV M protein specifically interacts with N protein via the carboxyl terminus is not uncommon, since some viral M proteins usually interact either with an internal matrix protein or nucleocapsid according to their C-terminuses, such as in retrovirus, rhabdovirus, orthomyxovirus, and alphavirus (Escors et al., 2001c).
3.6. The electrostatic attraction plays an important role in SARS-CoV M/N interaction
As discussed above, SARS-CoV M interacts with SARS-CoV N protein mostly through their carboxyl termini. By sequence analysis, it was found that these two fragments are hydrophilic and highly charged at neutral pH. This fact thus suggests that SARS-CoV M/N interaction might be of ionic nature.
To verify this assumption, using Biacore3000 system the interactions between SARS-CoV ME protein and the immobilized SARS-CoV N protein were investigated firstly in three different pHs (7.4, 5.4 and 9.6). As indicated in Fig. 7 A, the binding response for SARS-CoV ME (5 μM) at pH 5.4 or pH 9.6 is lower than that at pH 7.4. This suggested that both the acidification and basification could weaken the ME/N interaction, especially the decrease due to acidification is more apparent. Quantitatively kinetic studies revealed that the equilibrium dissociation constants (K D) for SARS-CoV ME/N interaction at pH 5.4 (21.2 μM) and pH 9.6 (0.85 μM) were greater than that at pH 7.4 (0.55 μM), which thus suggests that SARS-CoV M/N interaction might be of ionic nature.
Fig. 7.

Influence of pH and ionic strength on SARS-CoV ME/N interaction studied by SPR. (A) The curves of 5 μM SARS-CoV ME interacting with the immobilized N at different pH: 10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% P20; 10 mM Tris–HCl, pH 9.6, 150 mM NaCl, 3 mM EDTA, 0.005% P20; and 10 mM sodium acetate, pH 5.4, 150 mM NaCl, 3 mM EDTA, 0.005% P20. (B) The curves of 5 μM SARS-CoV ME interacting with immobilized N in different salts: 10 mM HEPES, pH 7.4, 150 mM NaCl; 10 mM HEPES, pH 7.4, 400 mM NaCl; and 10 mM HEPES, pH 7.4, 50 mM CaCl2.
As the acidification and basification might alter the conformations of the interacting regions, the interactions between SARS-CoV ME protein and the immobilized SARS-CoV N protein were further studied at different salt concentrations on Biacore 3000 system. As shown in Fig. 7B, when NaCl concentration was increased from 150 to 400 mM, the binding response for SARS-CoV ME binding to the immobilized SARS-CoV N decreased from 700 to 300 RU. Kinetic analysis further indicated that the binding affinity for SARS-CoV ME/N decreased by two orders of magnitude, with the K D value from 0.55 μM increased to 49.2 μM. This indicated that SARS-CoV M/N interaction was relative to ionic strength, and electrostatic force might play an important role in SARS-CoV M/N interaction. To further explore the electrostatic attractive or repulsive force involved in this interaction, we replaced 400 mM NaCl (monovalent cation) by 50 mM CaCl2 (divalent cation) for maintaining the similar ionic strength to repeat the SPR experiment. Interestingly, in the presence of 50 mM Ca2+, SARS-CoV ME almost showed a similar binding response (260 RU) to that in 400 mM NaCl as shown in Fig. 7B. All facts suggested that SARS-CoV M/N interaction was dominated by electrostatic attraction.
3.7. Mutation of L218A and L219A has no impact on SARS-CoV M/N interaction
As reported for mouse hepatitis virus (MHV) (Kuo and Masters, 2002), the lost electrostatic interaction could be replaced by a hydrophobic interaction between the amino acids L225 and L226 of the M and N proteins in the absence of R227 in the M protein. By multiple sequence alignment (Fig. 8 A), we found that in the C-terminus of SARS-CoV M, there was no ‘LLR(K)’ or ‘R(K)LL’ motif but a conserved “LL”. We thereby suspected that L218 and L219 of M protein might also contribute to SARS-CoV M/N interaction. To validate this, we studied the interaction between the double-site mutated SARS-CoV ME (L218L219–A218A219) and the immobilized SARS-CoV N protein on Biacore 3000. As shown in Fig. 8B, the fitted K D value of 0.58 μM for the mutated SARS-CoV ME binding to the immobilized SARS-CoV N protein is very similar to that for the wild type SARS-CoV ME binding to SARS-CoV N (K D = 0.55 μM) (Fig. 4A). Furthermore, as indicated by the yeast two-hybrid assay (Fig. 8C), the double-site mutated SARS-CoV ME showed the similar N protein binding ability to the wild type SARS-CoV ME protein. Therefore, these results indicated that L218 and L219 might be not important in SARS-CoV M/N interaction.
Fig. 8.

L218L219 of the M protein renders no function in SARS-CoV M/N interaction. (A) Alignment of the C-terminuses of SARS-CoV M with those from other coronaviruses. The (*) indicates that amino acids in this row are identical; (.), similar; (:), more similar. The M protein sequences are from: transmissible gastroenteritis virus strain PUR46-MAD (TGEV-PUR46-MAD), canine coronavirus strain HF3 (CCoV-HF3), feline infectious speritionitis virus strain WSU 79-1146 (FIPV-WSU-79-1146), human coronavirus strain OC 4 (HCoV-OC4), SARS coronavirus stain BJ01 (SARS-CoV-BJ01) and murine hepatitis virus strain TY (MHV-TY). (B) The interaction curves for the L218AL219A-mutated ME binding to the immobilized SARS-CoV N protein. The protein concentrations were labeled on the right. (C) Yeast two-hybrid assays for interaction of the L218AL219-mutated ME and SARS-CoV N protein. The vales were obtained by average of the data from three repeated experiments.
Summarily, in our current work, the residues 197–221 of SARS-CoV M and the residues 341–422 of SARS-CoV N protein were proved to be crucial for SARS-CoV M/N interaction by yeast two-hybrid and SPR assays. SPR kinetic assays revealed that the endodomain (aa102–221) of SARS-CoV M interacts with SARS-CoV N protein with high affinity. This binding affinity, however, could be weakened greatly by acidification, high concentration salt (400 mM NaCl) and divalent cation (50 mM Ca2+), thus suggesting that the electrostatic attraction might play an important role in SARS-CoV M/N interaction. These results clearly showed that SARS-CoV M protein interacts with N protein mostly through their carboxyl termini by electrostatic attraction. According to our results, the residues 168–208 of SARS-CoV N might be involved in but not contribute greatly to SARS-CoV M/N interaction, which seems to be not compatible with the reported result that these residues might be critical for SARS-CoV M/N interaction (He et al., 2004). While we are preparing this manuscript, Fang et al. published the result that residues 194–205 of SARS-CoV M were involved in SARS-CoV M/N interaction by GST pull-down method (Fang et al., 2005), this region is within our proved residues 197–221 in SARS-CoV M responsible for binding to SARS-CoV N protein. As discussed above, the electrostatic attraction might play an important role in SARS-CoV M/N interaction, it is thereby suggested that the three basic amino acids, R197, R199 and K204 in the overlapping sequence (aa197–205, RYRIGNYKL) of SARS-CoV M might be responsible for its binding to SARS-CoV N protein. In addition, it is noted that the two highly conserved amino acids (L218 and L219) of SARS-CoV M protein are not involved in SARS-CoV M/N interaction. Our current work will hopefully help understanding the molecular mechanism of SARS-CoV M/N interaction and provide valuable clues for mutagenic studies to disrupt virion assembly.
Acknowledgements
This work was supported by the State Key Program of Basic Research of China (grants 2002CB512802, 2002CB512807, 2004CB58905), the National Natural Science Foundation of China (grants 20372069, 20472095), Shanghai Basic Research Project from the Shanghai Science and Technology Commission (grants 02DJ14070, 03DZ19228 and 03DZ19212), the 863 Hi-Tech Program (grants 2002AA233011, 2005AA235030), Sino-European Project on SARS Diagnostics and Antivirals (Proposal/Contract no.: 003831).
Contributor Information
Xu Shen, Email: xshen@mail.shcnc.ac.cn.
Hualiang Jiang, Email: hljiang@mail.shcnc.ac.cn.
References
- de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. Journal of Virology. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Smeets M., Vernooij F., Vennema H., Rottier P.J. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. Journal of Virology. 1999;73:7441–7452. doi: 10.1128/jvi.73.9.7441-7452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. Journal of Virology. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Camino C., Risseeuw E.P., Liu E., Crosby W.L. A high-throughput system for two-hybrid screening based on growth curve analysis in microtiter plates. Analytical Biochemisty. 2003;316:171–174. doi: 10.1016/s0003-2697(02)00706-6. [DOI] [PubMed] [Google Scholar]
- Escors D., Camafeita E., Ortego J., Laude H., Enjuanes L. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. Journal of Virology. 2001;75:12228–12240. doi: 10.1128/JVI.75.24.12228-12240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Ortego J., Enjuanes L. The membrane M protein of the transmissible gastroenteritis coronavirus binds to the internal core through the carboxy-terminus. Advances in Experimental Medicine and Biology. 2001;494:589–593. doi: 10.1007/978-1-4615-1325-4_87. [DOI] [PubMed] [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. Journal of Virology. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Ye L., Timani K.A., Li S., Zen Y., Zhao M. Peptide domain involved in the interaction between membrane protein and nucleocapsid protein of SARS-associated coronavirus. Journal of Biochemistry and Molecular Biology. 2005;38:381–385. doi: 10.5483/bmbrep.2005.38.4.381. [DOI] [PubMed] [Google Scholar]
- He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F. Characterization of protein–protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Research. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. The New England Journal of Medicine. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. Journal of Virology. 2002;76:4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Masters P.S. Plenmum Press; New York: 1995. The Coronavirus Nucleocapsid Protein. [Google Scholar]
- Luo H.B., Ye F., Sun T., Yue L.D., Peng S.Y., Chen J. In vitro biochemical and thermodynamic charaterization of nucleocapsid of SARS. Biophysical Chemistry. 2004;112:5–25. doi: 10.1016/j.bpc.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. Journal of Virology. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opstelten D.J., Raamsman M.J., Wolfs K., Horzinek M.C., Rottier P.J. Envelope glycoprotein interactions in coronavirus assembly. Journal of Cell Biology. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular Cloning—A Laboratory Manual. [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO Journal. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.L., Wang J.R., Zhang Y., Chen M.L., Zhang W., Yang S. Expression, purification and identification of recombinant SARS coronavirus membrane protein. Acta Biochimica et Biophisica Sinica. 2003;35:1140–1144. [PubMed] [Google Scholar]


