Highlights
► Acylguanidines are a new class of antiviral compounds, targeting diverse proteins of viruses from different families. ► BIT225 is the first acylguanidine in clinical trials against human immunodeficiency virus type 1 and hepatitis C virus. ► This review focuses on the mechanisms of inhibition of viral proteins by acylguanidines.
Abstract
Acylguanidines are a new class of antiviral compounds with the unique ability to target both RNA polymerase and transmembrane proteins of viruses from different families. Importantly, they inhibit proteins which are not targeted by existing antiviral therapies, for example, Vpu of HIV type 1, p7 of hepatitis C virus, E of severe acute respiratory syndrome coronavirus and RNA-dependent RNA polymerase of coxsackievirus B3. BIT225, developed by Biotron Limited, is the first acylguanidine in clinical trials against HIV type 1 and hepatitis C virus. In this article we focus on the mechanisms of inhibition of viral proteins by acylguanidines.
Available treatments for viral infections are limited. Approximately 50 antiviral compounds have been approved for clinical use by the US FDA, with 26 of them targeting HIV. For many important viruses no antiviral treatment is available, and the existing treatments have limited efficacy because of development of virus resistance and side effects. The current strategy of combating the development of drug resistance is combination therapy, using drugs that target different processes in the virus replication cycle, in addition to using different classes of compounds targeting the same process (e.g. nucleoside and non-nucleoside polymerase inhibitors). Although many current drug discovery programs are focused on finding new combination therapeutics, there is also an ongoing search for compounds targeting viruses for which no treatment is currently available. In this article we highlight antiviral actions of acylguanidines, a class of compounds with a remarkable range of inhibitory targets, both cellular and viral. The peculiarity of these compounds is that they can inhibit diverse types of proteins and hence various processes in the virus replication cycle.
Cellular targets of acylguanidines
Acylguanidines are a large class of chemicals containing a carbonylguanidino moiety. The prototypic acylguanidine is amiloride (Fig. 1 ), a potassium-sparing diuretic widely used in medicine, which was first described in 1967 by Cragoe et al. [1]. The primary target of amiloride is the epithelial Na+ channel, which it inhibits at nanomolar concentrations [2]. At micromolar concentrations amiloride inhibits several other cellular ion transporters (e.g. Na+/H+ exchanger), enzymes and receptors [2]. The epithelial Na+ channel is an important target in the treatment of hypertension and cystic fibrosis, and the Na+/H+ exchanger is a central factor in growth and invasive properties of many cancer cells and in cell damage following myocardial infarction. For these reasons numerous acylguanidines with increased potency and specificity towards either the epithelial Na+ channel or the Na+/H+ exchanger compared with amiloride have been synthesised 2, 3, 4.
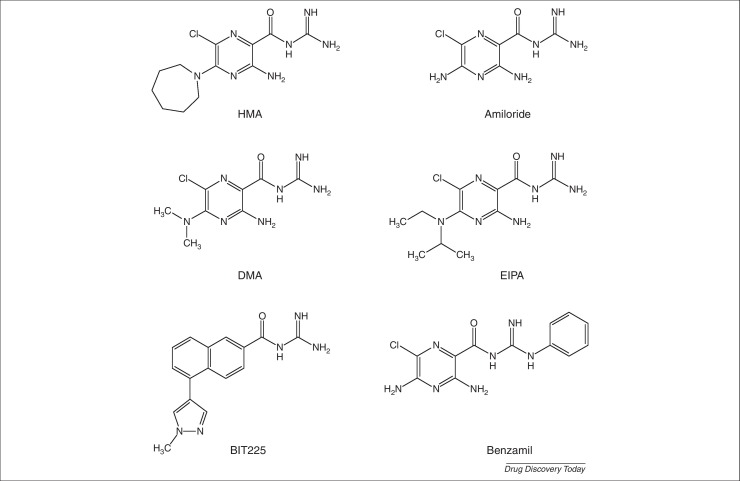
Figure 1.
Structures of acylguanidines inhibiting viral proteins. HMA inhibits Vpu of human immunodeficiency virus type 1, p7 of hepatitis C virus, M protein of dengue virus, E protein of severe acute respiratory syndrome coronavirus and 3Dpol of coxsackievirus B3. DMA inhibits Vpu. BIT225 inhibits Vpu and p7. Amiloride, EIPA and benzamil inhibit 3Dpol.
Viral targets of acylguanidines
Acylguanidines have made their way into antiviral drug development in the past decade. Based on their known activity as cellular ion channel blockers, some amiloride derivatives have been tested against viral proteins possessing ion channel activity. One of these compounds, 5-(N,N-hexamethylene)amiloride (HMA) (Fig. 1), was shown to inhibit ion channel proteins of four viruses belonging to three families: Vpu protein of HIV type 1 (Retroviridae family), p7 protein of hepatitis C virus (HCV) and M protein of dengue virus (Flaviviridae family), and E protein of severe acute respiratory syndrome coronavirus (Coronaviridae family). A subsequent drug development program produced a compound which is currently in clinical trials against two of these viruses. Different studies have demonstrated that a non-ion-channel protein, RNA-dependent RNA polymerase of coxsackievirus B3 (Picornaviridae family), is also inhibited by amiloride derivatives.
Vpu protein of HIV type 1
HIV isolates are grouped into two types, type 1 (HIV-1) and type 2 (HIV-2). The dominant worldwide agent of acquired immunodeficiency syndrome is HIV-1, whereas HIV-2 is restricted to some regions of Western and Central Africa. The introduction of highly active antiretroviral therapy, a treatment paradigm using three or more compounds in combination, has achieved efficient inhibition of virus replication in most patients; however, HIV cannot be cured. The global prevalence of HIV-1 infections has stabilised at 0.8% [5].
HIV-1 encodes nine proteins that are required for its replication and for manipulation of the host; one of them is an accessory protein Vpu, which is absent from HIV-2 [6]. The established functions of Vpu in infected cells are downregulation of tetherin (also known as BST-2, CD317 or HM1.24) and CD4 [7]. Tetherin inhibits virus release by being incorporated into nascent virions and cross-linking the viral and the cellular membranes. CD4 is crucial for virus entry, but later in the replication cycle it can inhibit assembly, release and infectivity of progeny virions by interacting with viral glycoproteins. Two studies have demonstrated that Vpu contributes to the efficient spread of HIV-1 in vivo. Singh et al. infected macaques with chimeric simian-HIV, expressing either wild-type Vpu, or mutated Vpu that did not downregulate CD4 [8]. The animals producing wild-type Vpu maintained significantly higher viral loads than those producing the mutated protein, which suggested that downregulation of CD4 by Vpu contributes to the pathogenicity of HIV-1 [8]. In another study, Sato et al. infected ‘humanised’ mice producing human leukocytes, including CD4+ T cells, with wild-type and Vpu-deficient HIV-1 and showed that the wild-type virus propagated more rapidly than the Vpu-deficient virus [9]. Furthermore, they showed that Vpu increased the release of cell-free virions without affecting cell-to-cell HIV-1 transmission [9].
Vpu is an 80- to 82-amino acid (depending on viral isolate) membrane protein, consisting of an N-terminal transmembrane domain and a C-terminal cytoplasmic domain. It forms pentameric cation-selective ion channels in planar lipid bilayers 10, 11. Studies by Ewart et al. at Biotron Limited showed that the ion channels formed by Vpu or its transmembrane domain are blocked by amiloride derivatives HMA and 5-(N,N-dimethyl)amiloride (DMA) (Fig. 1), but not by amiloride itself [12]. HMA was also shown to inhibit budding of virus-like particles from HeLa cells, suggesting a link between the ion channel activity of Vpu and virus release [12]. Subsequent studies demonstrated that HMA and DMA inhibit HIV-1 replication in cultured human macrophages [13]. An ensuing program of chemical synthesis by the Biotron generated a library of approximately 300 compounds, with a leading compound BIT225 (Fig. 1) [14]. BIT225, at 40 μm concentration, completely blocked the ion channel formed by the Vpu transmembrane domain in planar lipid bilayers, and reduced HIV-1 yield in the supernatant of cultured human macrophages with a 50% effective concentration (EC50) of 2.25 μm and a selectivity index (a ratio between 50% toxic concentration and EC50) of 126 [14]. No reduction in the yield of extracellular HIV-2, which lacks Vpu, was observed under these conditions, demonstrating that Vpu is indeed the antiviral target of BIT225 [14]. Biotron has recently started stage Ib/IIa clinical trials of BIT225, in which 18 HIV-positive, treatment-naive patients will be administered the drug over 10 days, with other eight patients receiving a placebo. The results are expected in the first quarter of 2012 (Biotron: http://www.biotron.com.au).
The mechanism of action of acylguanidines against HIV-1 has not been elucidated. The prevailing hypothesis is that it is because of the inhibition of the ion channel activity of Vpu. However, currently there is no proof that the ion channel activity of Vpu has a role in the virus replication cycle. Published data suggest that it is not required for downregulation of CD4 or tetherin as Vpu mutants without ion channel function retain their ability to downregulate CD4 and tetherin 11, 15, 16. Alternatively, acylguanidines could block Vpu binding to its cellular targets. However, Kuhl et al. have recently demonstrated that BIT225 does not interfere with Vpu-tetherin interaction [17]. Moreover, the compound inhibited HIV-1 yield in CD4+ T-cell lines without affecting tetherin modulation [17]. These data suggest that the antiviral action of acylguanidines is not because of the inhibition of tetherin downregulation by Vpu. A potential effect of BIT225 on CD4 downregulation remains to be investigated. Also, recent studies identified new cellular targets of Vpu, SLAMF6 and CD1d 9, 18, 19. Further work is required to determine which functions of Vpu in infected cells are inhibited by acylguanidines, and whether the inhibition occurs through the blockade of its ion channel activity or through an alternative mechanism.
p7 protein of HCV
HCV belongs to the genus Hepaciviruses of the Flaviviridae family. The other two genera of this family are Flaviviruses (e.g. dengue virus) and Pestiviruses (e.g. bovine viral diarrhoea virus). An estimated 2.35% of the world's population is chronically infected with HCV, which can lead to life-threatening liver disease, including hepatocellular carcinoma [20]. The current therapy with pegylated interferon α and ribavirin is effective in approximately 50% of cases [21].
The HCV genome encodes a polyprotein of approximately 3010 amino acids, which is processed by host and viral proteases into at least ten polypeptides: core (capsid) protein, two envelope proteins, p7 and seven non-structural proteins [22]. HCV p7 is a 63-amino acid membrane protein comprising two transmembrane domains connected by a basic cytoplasmic loop [23]. It forms hexa- and heptameric cation-selective channels in phospholipid membranes 24, 25, 26, 27, and is required, together with one of the non-structural proteins, NS2, for effective assembly and release of infectious virus 28, 29. In vivo, p7 is essential for HCV infectivity in chimpanzees [30].
Both ion channel and non-ion-channel functions of p7 are required for production of infectious virus. HCV assembly is believed to occur at the endoplasmic reticulum (ER) where viral envelope proteins accumulate 31, 32. Recent studies of Boson et al. demonstrated that co-localisation of the core protein with the envelope proteins at the ER is required for efficient virion assembly, and that p7 and NS2 are the strain-specific factors governing intracellular localisation of core protein in infected cells [33]. Although the ion channel activity of p7 appears unlikely to be involved in the core protein localisation 33, 34, it is crucial for intracellular virus viability. Extracellular HCV virions are acid-resistant, whereas intracellular virus particles have greatly increased acid sensitivity [35]. Proton channel activity of p7 protects intracellular virus particles by preventing the obligate acidification of intracellular compartments [35]. Mutations in p7 abrogating its ion channel activity resulted in complete loss of infectious virus production, which could be partially restored by preventing acidification with the inhibitor bafilomycin A1 [35].
Several compounds inhibiting HCV p7 ion channel in lipid bilayers have been identified, including HMA and BIT225 25, 26, 27, 36. In Madin-Darby bovine kidney (MDBK) cells, BIT225 inhibited bovine viral diarrhoea virus, a pestivirus used as HCV model, with an EC50 of 314 nM and a selectivity index of approximately 40, and showed synergy with currently used anti-HCV drugs, interferon α and ribavirin [36]. The current hypothesis regarding the mechanism of antiviral activity of BIT225 is that it is related to the inhibition of p7 ion channel activity, which remains to be confirmed. The compound has recently completed stage IIa clinical trials on 24 hepatitis C patients who were randomly assigned to receive either 400 mg or 200 mg BIT225, or placebo (ratio of 1:1:1), for the first 28 days of their standard treatment with interferon and ribavirin. Preliminary analysis of trial data showed approximately tenfold greater virus reduction in the blood of patients receiving 400 mg dose of BIT225 compared to those receiving interferon and ribavirin only (Biotron: http://www.biotron.com.au).
M protein of dengue virus
Dengue is the most widespread mosquito-borne viral disease. Each year, there are ∼50 million dengue infections and ∼500,000 individuals are hospitalised with dengue haemorrhagic fever, mainly in Southeast Asia, the Pacific and the Americas [37]. At present there is no antiviral treatment for dengue.
Dengue virus genome encodes a polyprotein that is cleaved into three structural proteins (C, prM and E) and seven non-structural proteins [38]. prM is a 166-amino acid membrane protein having a crucial role in virus assembly and maturation. Immature virus particles assemble at the ER, where viral nucleocapsid formed by protein C and the viral genome becomes enveloped by lipid membrane containing prM-E heterodimers. These virus particles are transported through the secretory pathway to the trans-Golgi network, where acidic environment induces a conformational change followed by cleavage of prM into 91-amino acid pr and 75-amino acid M [39]. The resulting mature virions contain M, whereas pr remains attached to the virions until release into extracellular environment, thus protecting E from fusion with intracellular membranes [39]. The function of M is unknown.
Premkumar et al. have shown that a peptide corresponding to the C-terminal transmembrane domain of prM/M has an ion channel activity in planar lipid bilayers, which is inhibited by HMA [40]. By contrast, no ion channel activity was observed when prM or M were expressed in Xenopus oocytes, despite the proteins’ localisation on the oocyte surface [41]. Thus it is not clear whether prM or M form ion channels, and an antiviral activity of HMA against Dengue viruses has not yet been shown.
E protein of severe acute respiratory syndrome coronavirus
The Coronaviridae cause respiratory diseases in humans and a variety of severe diseases in animals. Severe acute respiratory syndrome coronavirus (SARS-CoV) jumped the animal–human host barrier and infected 8096 people in 2002–2003, causing 774 deaths (WHO: http://www.who.int/csr/sars/country/table2004_04_21/en/index.html).
SARS-CoV genome encodes four structural proteins (spike, membrane, envelope and nucleocapsid), in addition to 16 non-structural and eight accessory proteins [42]. The 76-amino acid envelope protein (E) is embedded in virus lipid envelope together with the spike and membrane proteins. The cell culture titre of SARS-CoV lacking the E gene was 20- to 200-fold lower compared to the wild-type (depending on cell line), with virus assembly likely to be inhibited [43]. The mutant virus was also significantly attenuated in animal models 43, 44.
The E protein is located mainly in the ER-Golgi intermediate compartment when expressed alone or during SARS-CoV infection [45]. It comprises ∼25-amino acid transmembrane domain, lumenal N-terminal domain and cytoplasmic C-terminal domain 45, 46. Pentameric cation-selective ion channels are formed by E protein or its transmembrane domain in lipid bilayers 47, 48, 49. By using NMR, HMA was shown to bind at two sites inside the lumen of the channel formed by the E transmembrane domain, with no specific interaction observed for amiloride [49]. No data on the effect of the compounds on SARS-CoV replication are available yet; however, HMA, but not amiloride, inhibited replication of two other coronaviruses, human coronavirus 229E and mouse hepatitis virus, in cell culture, with EC50 of 1.34 μm and 3.94 μm, respectively [50]. The compound had no effect on replication of E-deleted mouse hepatitis virus, confirming that E protein is its target [50].
RNA-dependent RNA polymerase of coxsackievirus B3
The Picornaviridae are a family of positive-sense RNA viruses, which contains numerous human pathogens causing poliomyelitis, myocarditis, meningitis, hepatitis, common cold and other diseases. Coxsackievirus B3 (CVB3) is responsible for 14–32% of human myocarditis cases [51]. No antiviral treatment is currently available for picornaviral infections.
The genomic RNA contains a viral peptide, VPg, covalently linked to the 5′ end. RNA replication occurs through synthesis of a complementary RNA strand catalysed by viral RNA-dependent RNA polymerase, 3Dpol, with VPg serving as a primer 52, 53, 54.
Harrison et al. have shown that amiloride and its derivatives 5-(N-ethyl-N-isopropyl) amiloride (EIPA, Fig. 1), HMA and benzamil (Fig. 1) inhibit CVB3 replication in cell culture [55]. The antiviral effect of amiloride and EIPA was because of the inhibition of CVB3 RNA replication, with two individual amino acid substitutions in 3Dpol conferring resistance of the virus to the compounds [55]. Subsequent in vitro studies demonstrated that amiloride and EIPA inhibited enzymatic activity of 3Dpol, affecting both VPg-primed initiation and elongation steps of RNA synthesis [56]. Amiloride acted as a competitive inhibitor, competing with incoming nucleotide and Mg2+ for binding to the active site of the enzyme [56]. There was, however, a marked difference in the rank order of the compounds between inhibition of 3Dpol in vitro (EIPA > amiloride > benzamil > HMA, with EC50 of 11 μm, 20 μm, 52 μm and 136 μm, respectively, in an RNA elongation assay) and inhibition of virus replication in cell culture (EIPA = HMA > benzamil > amiloride, with EC50 of 2 μm, 2 μm, 10 μm and 60 μm, respectively) (55, 56 and E.V. Gazina, unpublished). There are two potential reasons for this discrepancy. Although 3Dpol is the only target of amiloride and EIPA, additional targets cannot be ruled out for benzamil and HMA. For example, CVB3 protein 2B, a transmembrane protein similar to the viral ion channel proteins described above 57, 58, might be inhibited by HMA and benzamil. Alternatively, there may be large differences in intracellular accumulation between the compounds.
Concluding remarks
HMA is unique among the antiviral compounds in its ability to inhibit different types of proteins produced by viruses from different families. Importantly, it inhibits viruses for which no antiviral treatment is currently available (i.e. SARS-CoV and CVB3) or targets proteins, which are not targeted by existing antiviral therapies (i.e. HIV-1 Vpu and HCV p7). Unfortunately, HMA has a much higher potency as a blocker of cellular Na+/H+ exchanger [2] than as inhibitor of viral proteins, which rules it out as an antiviral drug. However, the development of BIT225, which is sufficiently potent and specific against HIV-1 and HCV to progress to clinical trials, clearly demonstrates that amiloride derivatives are very useful as a starting point in antiviral drug development. Structure–activity studies on SARS-CoV E and CVB3 3Dpol may also produce potent and specific antiviral drugs in the future, whereas antiviral activity of acylguanidines against Dengue viruses is yet to be demonstrated.
Currently it is not clear whether the antiviral action of BIT225 and HMA against HIV-1, HCV and SARS-CoV is because of the blockade of ion channels formed by Vpu, p7 and E, respectively, or because of inhibition of other functions of these proteins. Further work is required to elucidate the mechanism(s) of activity of acylguanidines against viral transmembrane proteins.
The competitive inhibition of a viral enzyme, CVB3 3Dpol, by amiloride and its derivatives is not surprising in the light of the previous findings that amiloride acts as a competitive inhibitor of several cellular protein kinases, cholinesterases and a serine protease 2, 59, 60. Other viral enzymes may, therefore, be added to the list of proteins inhibited by these compounds in the future.
References
- 1.Cragoe E.J., Jr. Pyrazine diuretics. II. N-amidino-3-amino-5-substituted 6-halopyrazinecarboxamides. J. Med. Chem. 1967;10:66–75. doi: 10.1021/jm00313a014. [DOI] [PubMed] [Google Scholar]
- 2.Kleyman T.R., Cragoe E.J., Jr. Amiloride and its analogs as tools in the study of ion transport. J. Membr. Biol. 1988;105:1–21. doi: 10.1007/BF01871102. [DOI] [PubMed] [Google Scholar]
- 3.Masereel B. An overview of inhibitors of Na(+)/H(+) exchanger. Eur. J. Med. Chem. 2003;38:547–554. doi: 10.1016/s0223-5234(03)00100-4. [DOI] [PubMed] [Google Scholar]
- 4.Lee S. (5-Arylfuran-2-ylcarbonyl)guanidines as cardioprotectives through the inhibition of Na+/H+ exchanger isoform-1. J. Med. Chem. 2005;48:2882–2891. doi: 10.1021/jm0492305. [DOI] [PubMed] [Google Scholar]
- 5.Kilmarx P.H. Global epidemiology of HIV. Curr. Opin. HIV AIDS. 2009;4:240–246. doi: 10.1097/COH.0b013e32832c06db. [DOI] [PubMed] [Google Scholar]
- 6.Fanales-Belasio E. HIV virology and pathogenetic mechanisms of infection: a brief overview. Ann. Ist. Super. Sanita. 2010;46:5–14. doi: 10.4415/ANN_10_01_02. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz A. The Vpu protein: new concepts in virus release and CD4 down-modulation. Curr. HIV Res. 2010;8:240–252. doi: 10.2174/157016210791111124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh D.K. The presence of the casein kinase II phosphorylation sites of Vpu enhances the CD4+ T cell loss caused by the simian-human immunodeficiency virus SHIVKU-lbMC33 in pig-tailed macaques. Virology. 2003;313:435–451. doi: 10.1016/s0042-6822(03)00339-8. [DOI] [PubMed] [Google Scholar]
- 9.Sato K. Vpu augments the initial burst phase of HIV-1 propagation and downregulates BST2 and CD4 in humanized mice. J. Virol. 2012 doi: 10.1128/JVI.07062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewart G.D. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol. 1996;70:7108–7115. doi: 10.1128/jvi.70.10.7108-7115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubert U. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 12.Ewart G.D. Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur. Biophys. J. 2002;31:26–35. doi: 10.1007/s002490100177. [DOI] [PubMed] [Google Scholar]
- 13.Ewart G.D. Potential new anti-human immunodeficiency virus type 1 compounds depress virus replication in cultured human macrophages. Antimicrob. Agents Chemother. 2004;48:2325–2330. doi: 10.1128/AAC.48.6.2325-2330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khoury G. Antiviral efficacy of the novel compound BIT225 against HIV-1 release from human macrophages. Antimicrob. Agents Chemother. 2010;54:835–845. doi: 10.1128/AAC.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert U. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 1996;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolduan S. Ion channel activity of HIV-1 Vpu is dispensable for counteraction of CD317. Virology. 2011;416:75–85. doi: 10.1016/j.virol.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Kuhl B.D. The HIV-1 Vpu viroporin inhibitor BIT225 does not affect Vpu-mediated tetherin antagonism. PLoS One. 2011;6:E27660. doi: 10.1371/journal.pone.0027660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah A.H. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe. 2010;8:397–409. doi: 10.1016/j.chom.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moll M. Inhibition of lipid antigen presentation in dendritic cells by HIV-1 Vpu interference with CD1d recycling from endosomal compartments. Blood. 2010;116:1876–1884. doi: 10.1182/blood-2009-09-243667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 2011;17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 21.Vermehren J., Sarrazin C. New hepatitis C therapies in clinical development. Eur. J. Med. Res. 2011;16:303–314. doi: 10.1186/2047-783X-16-7-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashfaq U.A. An overview of HCV molecular biology, replication and immune responses. Virol. J. 2011;8:161. doi: 10.1186/1743-422X-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrere-Kremer S. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke D. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 2006;281:37057–37068. doi: 10.1074/jbc.M602434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffin S.D. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 26.Pavlovic D. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Premkumar A. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 28.Jones C.T. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinmann E. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:E103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai A. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gastaminza P. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouille Y. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 2006;80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boson B. A concerted action of hepatitis C virus p7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Pathog. 2011;7:E1002144. doi: 10.1371/journal.ppat.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tedbury P. The subcellular localization of the hepatitis C virus non-structural protein NS2 is regulated by an ion channel-independent function of the p7 protein. J. Gen. Virol. 2011;92:819–830. doi: 10.1099/vir.0.027441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wozniak A.L. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 2010;6:E1001087. doi: 10.1371/journal.ppat.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luscombe C.A. A novel Hepatitis C virus p7 ion channel inhibitor, BIT225, inhibits bovine viral diarrhea virus in vitro and shows synergism with recombinant interferon-alpha-2b and nucleoside analogues. Antiviral Res. 2010;86:144–153. doi: 10.1016/j.antiviral.2010.02.312. [DOI] [PubMed] [Google Scholar]
- 37.Guzman M.G. Dengue: a continuing global threat. Nat. Rev. Microbiol. 2010;8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urcuqui-Inchima S. Recent developments in understanding dengue virus replication. Adv. Virus Res. 2010;77:1–39. doi: 10.1016/B978-0-12-385034-8.00001-6. [DOI] [PubMed] [Google Scholar]
- 39.Yu I.M. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 40.Premkumar A. Dengue virus M protein C-terminal peptide (DVM-C) forms ion channels. J. Membr. Biol. 2005;204:33–38. doi: 10.1007/s00232-005-0744-9. [DOI] [PubMed] [Google Scholar]
- 41.Wong S.S. Dengue virus PrM/M proteins fail to show pH-dependent ion channel activity in Xenopus oocytes. Virology. 2011;412:83–90. doi: 10.1016/j.virol.2010.12.050. [DOI] [PubMed] [Google Scholar]
- 42.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeDiego M.L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeDiego M.L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieto-Torres J.L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres J. Model of a putative pore: the pentameric alpha-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 2006;91:938–947. doi: 10.1529/biophysj.105.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson L. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330:322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres J. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16:2065–2071. doi: 10.1110/ps.062730007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pervushin K. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:E1000511. doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson L. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353:294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreoletti L. Viral causes of human myocarditis. Arch Cardiovasc. Dis. 2009;102:559–568. doi: 10.1016/j.acvd.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Cameron C.E. Expanding knowledge of P3 proteins in the poliovirus lifecycle. Future Microbiol. 2010;5:867–881. doi: 10.2217/fmb.10.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferrer-Orta C. Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase. Curr. Opin. Struct. Biol. 2009;19:752–758. doi: 10.1016/j.sbi.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 54.Steil B.P., Barton D.J. Cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res. 2009;139:240–252. doi: 10.1016/j.virusres.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrison D.N. Amiloride derivatives inhibit coxsackievirus B3 RNA replication. J. Virol. 2008;82:1465–1473. doi: 10.1128/JVI.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gazina E.V. Amiloride is a competitive inhibitor of coxsackievirus B3 RNA polymerase. J. Virol. 2011;85:10364–10374. doi: 10.1128/JVI.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Jong A.S. Determinants for membrane association and permeabilization of the coxsackievirus 2B protein and the identification of the Golgi complex as the target organelle. J. Biol. Chem. 2003;278:1012–1021. doi: 10.1074/jbc.M207745200. [DOI] [PubMed] [Google Scholar]
- 58.van Kuppeveld F.J. Homomultimerization of the coxsackievirus 2B protein in living cells visualized by fluorescence resonance energy transfer microscopy. J. Virol. 2002;76:9446–9456. doi: 10.1128/JVI.76.18.9446-9456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zemach L. The interaction of amiloride with acetylcholinesterase and butyrylcholinesterase. FEBS Lett. 1990;263:166–168. doi: 10.1016/0014-5793(90)80730-7. [DOI] [PubMed] [Google Scholar]
- 60.Zeslawska E. Crystals of the urokinase type plasminogen activator variant beta(c)-uPAin complex with small molecule inhibitors open the way towards structure-based drug design. J. Mol. Biol. 2000;301:465–475. doi: 10.1006/jmbi.2000.3966. [DOI] [PubMed] [Google Scholar]