Abstract
Acute lower respiratory tract infections (LRTIs) are a major worldwide health problem, particularly in childhood. About 30–50% of acute LRTIs are viral in origin with influenza A infection a key cause of explosive community outbreaks. Many different influenza A viruses occur naturally in animal reservoirs and present a constant threat of zoonotic infections and global pandemics. Since 2009, when pandemic (H1N1) influenza A emerged from a swine origin, there have been a number of different zoonotic influenza A transmissions into the human population, including H1N1 and H3N2 variant viruses in North America and H7N9 viruses in China. The segmented nature of the influenza A virus genome and the circulation of these viruses in wild bird, domestic poultry and mammalian reservoirs presents a continuous opportunity for reassortment of viral genes and the emergence of a novel pandemic virus. Constant vigilance is required.
The emergence of severe acute respiratory syndrome in 2003 and Middle East respiratory syndrome coronavirus in 2012, highlights the fact that other serious respiratory viral infections in humans may originate in animals. Enhanced awareness of the potential for serious human respiratory disease in association with travel, or animal exposure, should form part of clinical assessment. Rapid developments in genomic technology improve the ability to diagnose previously undetected pathogens.
Preventative measures for influenza include annual vaccination and treatment with antiviral drugs such as neuraminidase inhibitors, oseltamivir and zanamivir. Subtype-dependent resistance to antivirals can develop and should be closely monitored.
Keywords: coronavirus, emerging, influenza, pandemic, respiratory, severe, virus, zoonosis
What's new?
-
•
Emergence of Middle East respiratory syndrome coronavirus as a cause of sporadic, severe acute respiratory illness with a high fatality rate linked to unknown animal exposure in the Arabian peninsula
-
•
Emergence of H7N9 influenza virus in humans exposed to poultry in China
-
•
Recombination of avian, pig and human influenza viruses giving rise to new variants of H3N2 and H1N1 in swine in North America
-
•
Introduction of live attenuated influenza vaccines in childhood in the UK
Respiratory viruses
Transmission of infections between humans through a respiratory route is well established and dependent on pathogens being carried in secretions that are transmitted between individuals as aerosols, droplets, secretions or via direct mucosal contact. Viruses have adapted to the human population over many millennia,1 and many have now evolved to exclusive transmission in humans through the respiratory route, yet the disease they cause is not restricted to the respiratory system. Examples of viruses causing systemic disease transmitted through the respiratory route include measles and erythrovirus B19. Other human viral infections cause respiratory system disease and transmit through respiratory secretions; for example, respiratory syncytial virus (RSV), rhinoviruses and human adapted strains of influenza A, which cause seasonal epidemics, and influenza B and C, which are human pathogens. The outcome of infection and clinical disease presentation is partly dependent on virus factors, which restrict viral replication to certain cell types (tissue tropism).
Viruses emerging into humans from animal hosts
Sporadic transmission of viruses from their natural animal host to humans can cause zoonotic infections with disease of varying severity and epidemic scale, depending on the virus. For many of these infections, humans are dead-end hosts. Such infections are usually well recognized, with effective control measures to limit human disease; examples include rabies. However, new infectious agents of man are described at regular intervals. For the last decade at least one significant new human pathogen has been identified almost every year, involving a diversity of organisms. The rate of identification of new viruses has increased in recent years with the application of genomics, which has facilitated the detection of organisms that could not previously be cultivated (Table 1 ).2
Table 1.
Viruses in humans
| Co-evolution of viruses with humans | Acquisition from animals, now permanently established | Temporary residents, occasional zoonotic transmission |
|---|---|---|
| α β γ herpesviruses Papilloma + polyoma Hepatitis B |
Measles HIV Smallpox Human Influenza A H3N2 Influenza B RSV |
Rabies Ebola Lassa Hanta Nipah SARS H5N1 |
RSV, respiratory syncytial virus; SARS, severe acute respiratory syndrome.
Characteristics of emerging virus infections
Approximately 75% of all newly described human pathogens (emerging diseases) are RNA viruses, many of which are transmitted through a mucosal or respiratory route. Not all of these cause serious human infections. Some may be viruses that have been in the human population for decades or even centuries without an animal reservoir, but through the application of improved diagnostic techniques are now recognized, such as human metapneumovirus (hMPV) (Table 2 ). Other newly described infections may have emerged very recently into the human population as a result of a species jump, such as human immunodeficiency virus (HIV), pandemic influenza, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome coronavirus (MERS-CoV). Such infections may first come to attention as a result of a severe undiagnosed respiratory illness, or an unusual severe illness requiring critical care, without obvious animal exposures, and may be detected as a result of an astute clinician requesting detailed diagnostic testing to rule out known infections. Evaluating the importance of a newly emerging virus is dependent on understanding the host range, disease and pathogenesis in man, transmissibility and likelihood of sustained transmission in human population.
Table 2.
Zoonotic origin of emerging viral pathogens in human disease
| Virus (genus or type species) | Human disease | Animal source | Probability L = Likely P = Possible C = Confirmed |
|---|---|---|---|
| Calicivirus | Diarrhoea | Swine, cattle | L |
| Rotavirus | Diarrhoea | Swine, cattle | L |
| Ebola virus | Haemorrhagic fever | Monkeys | P |
| Sin nombre virus | HFRS, HPS | Rodents | C |
| HIV | AIDS | Monkeys | L/C |
| HEV | Hepatitis | Swine | P |
| Influenza virus | Influenza | Pigs Horses Birds |
Reassortants L Reassortants L C |
| Hendravirus | Meningo-encephalitis | Fruit bat Tree shrew |
L L |
| Nipahvirus | Encephalitis | Pigs Dogs |
L P |
| Metapneumovirus | Respiratory tract infection | Birds | P |
| West Nile virus | Encephalitis | Birds | C |
| Cantalagovirus | Vesicular rash | Cattle | C |
| Borna disease virus | Encephalomyelitis | Horses | P |
| BSE agent | nvCJD | Cattle | P |
BSE, bovine spongiform encephalopathy; HEV, hepatitis E virus; HFRS, haemorrhagic fever with renal syndrome; HPS, hantavirus pulmonary syndrome; nvCJD, new variant Creutzfeldt-Jacob disease.
Viruses with pandemic potential
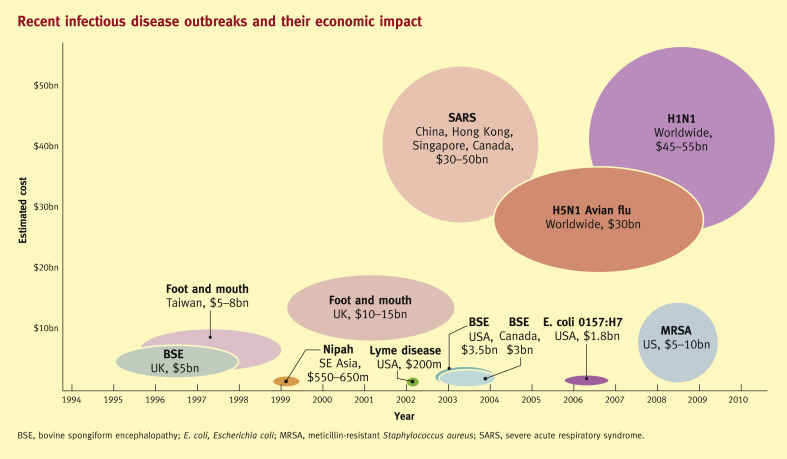
The term pandemic is usually taken to indicate a very widespread epidemic with an infection that has the capacity to cause disease on a global scale. A new influenza A virus in the human population, known as pandemic influenza, has the greatest impact on human populations as a result of morbidity and mortality in all age groups,2 but several other viruses, such as SARS coronavirus are also capable of causing significant clinical, economic and financial impact on a global level (Figures 1 and 2 ). In 2013, there is an unprecedented pandemic threat involving severe virus infections new to human populations. Two different viruses, influenza A H7N93 and MERS-CoV,4 circulating in different animal reservoirs in China and the Arabian peninsula respectively, have caused fatalities in humans and demonstrated limited human-to-human transmission through the respiratory route, with potential to cause much more widespread disease. The genetic characteristics of viruses that emerge to cause serious human respiratory disease, with pandemic potential, may be diverse.
Figure 1.
(Copyright of Bio-era LLC. Reproduced with kind permission.)
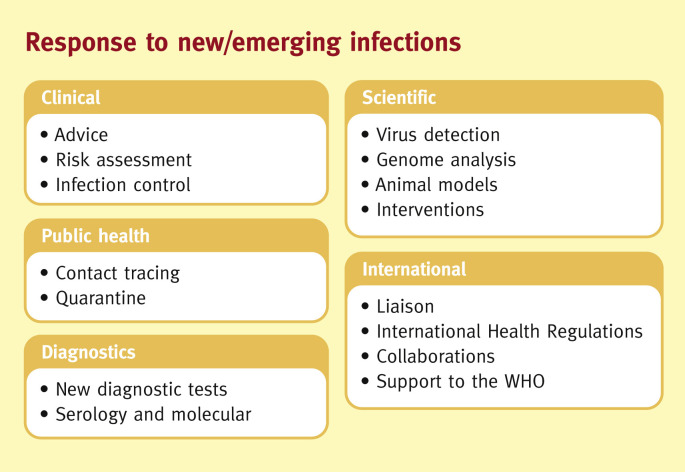
Figure 2.
Response to new/emerging infections.
Influenza viruses
Influenza viruses have a segmented negative-sense genome of approximately 15 kb, and exist as three different types: A, B and C. Influenza B and C, which are well-recognized human respiratory pathogens, have limited genetic diversity and are adapted to transmission exclusively in humans. Disease caused by influenza C is markedly milder than that caused by influenza A or B. Occasional transmission of influenza B to mammalian species such as seals and influenza C to pigs has been described. In contrast, there are many different subtypes of influenza A viruses, all of which naturally infect water-based wild birds, usually with very little disease.2
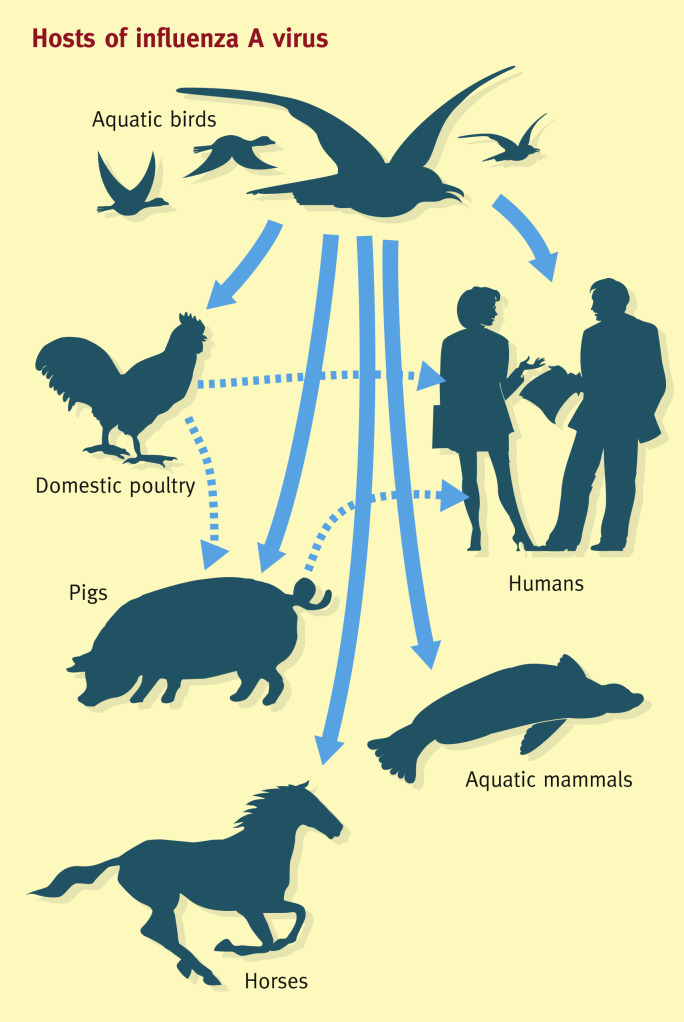
Influenza A viruses have a broad host range with the potential to infect a wide range of animals (Figure 3 ). Influenza A viral subtypes are distinguished according to their surface proteins, haemagglutinin (HA) and neuraminidase (NA). Seventeen HA subtypes and nine NA subtypes are found circulating in the wild, giving rise to over 150 different subtype combinations. There is great genetic diversity within influenza viruses in wild bird populations as a result of the ability of viruses to recombine segments. Not all different influenza A subtypes are established in man or mammalian hosts. Only a limited number of subtypes have adapted to circulation in mammalian species, including humans, horses and swine. Currently seasonal influenza A epidemics are caused by influenza A H1N1 or H3N2, which have adapted from the original avian host to transmit through respiratory secretions and cause respiratory tract infections in humans.2
Figure 3.
Hosts of influenza A virus.
Emergence of pandemic influenza: role of poultry and swine
Avian influenza A subtypes are transmitted through a faeco-oral route and virus can be shed in high quantity in the environment following replication in the gastrointestinal tract in wild birds. There is frequent spillover of avian influenza A subtypes into domestic poultry reservoirs, as a result of shared habitats with wild birds. Replication in poultry may be associated with disease in poultry of varying severity or may be asymptomatic, depending on the viral subtype and the poultry species. Domestic poultry may therefore act as a source of transmission of zoonotic influenza A subtypes to humans. This has been demonstrated with H5N1, which is highly pathogenic in poultry and more recently with H7N9, which is less highly pathogenic in poultry.2
H7N9 in China
Since March 2013, approximately 150 cases of human infection with influenza A (H7N9) have been reported from eastern China. The pathogenicity of the virus is high in humans, and higher age appears to be a risk factor; the median age of cases is 64 years, with a range between 4 and 89 years. Approximately one-third of cases have been fatal to date. There is currently no evidence of sustained human-to-human transmission. The latest epidemiological data are available from the WHO: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/en/index.html.
Genetic analysis of A (H7N9) isolates has identified mutations, which suggests that the virus may have a greater ability to infect mammalian species, including humans, than most other avian influenza viruses. Small family clusters have been reported. There is limited evidence for human-to-human transmission of this emerging influenza virus. The source and mode of transmission of influenza A (H7N9) have not yet been confirmed, but most cases have reported contact with poultry or live animal markets.5, 6, 7
Swine influenza viruses
Pigs are also considered a mixing vessel for the reassortment of avian, swine, and human influenza viruses. Until the 1990s, classic swine influenza A (H1N1), the most commonly circulating swine influenza virus among pigs, remained fairly constant. However, by the late 1990s, different subtypes (H1N1, H3N2, and H1N2) had emerged and became predominant among North American pig herds. These swine influenza A viruses acquired avian, human and swine virus gene segments through reassortment, and various genetic lineages can be distinguished within each subtype. In Europe, swine influenza is caused primarily by the same subtypes. However, the antigenic and genetic characteristics differ significantly from those found in North America and Asia. Genetic diversity has been expanded through multiple introductions of influenza viruses from other animal hosts into pig herds including from humans, most recently demonstrated with H1N1 pdm 2009 virus in Europe, Asia, and the Americas.2
Recent events confirm the important role of swine in the emergence of novel influenza viruses capable of causing a human pandemic, with the emergence of H1N1 pdm 2009 from a swine reservoir in North America. This H1N1 was sufficiently different from seasonal H1N1 previously circulating since 1977 to show sustained transmission in humans and eventually to replace the previously circulating H1N1 variant. Since 2009, other variant influenza H1N1 and H3N2 viruses circulating in the swine population of North America have also emerged to cause sporadic infections.8
Spectrum of clinical presentation of influenza
Seasonal influenza
Seasonal influenza A and B illness in humans ranges from subclinical or mild upper respiratory tract symptoms to more severe illness, including laryngotracheitis and pneumonia, or less commonly, death from respiratory system failure. The most common presenting symptoms are cough, high temperature, joint pain and general malaise. The rapid onset and short incubation period are characteristic, though incubation can last up to 4 days. Individuals at greatest risk of complications are those with pre-existing cardiac and respiratory disease, the elderly, and those with impaired immunity. The severity of illness reflects pre-existing host immunity and the prevailing virus strain.2
Pandemic influenza 2009
Disease caused by pandemic influenza A in 2009 was considered to be of moderate severity, with a similar range of presentation to that of seasonal influenza A, including typical symptoms of fever, cough and sore throat. Younger people were more commonly affected, particularly for fatal or severe disease. This is in stark contrast to seasonal influenza, which generally has a more severe impact on the elderly (over 65 years) population. Severe respiratory failure is seen in a small proportion of cases, with certain underlying medical conditions posing an increased risk of severe or fatal disease, with an increased risk of complications in pregnancy.2
Swine influenza variants in North America (H1N1, H1N2 and H3N2)
Disease caused by novel swine variants of influenza arising from circulating viruses in swine populations is largely indistinguishable clinically from seasonal influenza, but the mean age of presentation occurs in the young population, under 16 years, with an exposure history to swine.9
Disease due to H5N1 or H7
Disease presentation following zoonotic infection with these avian subtypes of influenza tends to be more severe, with a longer prodrome. Often the presentation may be that of a viral pneumonia, without the prominent features of cough and upper respiratory tract syndromes with rapid progression to acute respiratory distress syndrome (ARDS). Following zoonotic transmission of non-human subtypes of influenza A to humans, viral receptors and replication competence will affect the tropism of the virus, and the tissues and body fluids in which the virus may be detected, as has been demonstrated for H5N1 and H7 viruses. In this situation, a broad range of tissues may yield infectious virus recovery and a different spectrum of illness, in contrast to seasonal influenza virus infection. This would include evidence of leucopenia at presentation, which is not a typical feature of seasonal influenza. A history of exposure to poultry, with recent travel to parts of the world where avian influenza is endemic, may be an important diagnostic clue; also, conjunctivitis has been a clinical presenting feature associated with H7 infections involving H7N7, H7N2 and H7N3, but does not seem to have featured in H7N9 infections.9
Control of influenza
An influenza pandemic poses a significant risk to human health, and spreads rapidly around the globe, as was seen in 2009. Every attempt is made to ensure that influenza viruses with pandemic potential are controlled in their animal reservoirs, but once a new variant has emerged into the population several countermeasures are undertaken. In order to pose a pandemic threat, a novel influenza virus (or any other virus with pandemic potential) must satisfy two separate criteria. First, the virus itself must be capable of efficient transmission between humans; and second, the virus must be antigenically novel, such that a large proportion of the population are serologically naïve and fully susceptible to infection, and so will support its onward transmission.
The presence of a large mobile animal reservoir of influenza A virus suggests that eradication of this agent will be impossible. Control strategies focus on limiting the opportunities for cross-species transmission of novel subtypes; for example:
-
•
housing domestic poultry in shelters to avoid contact with migratory/wild birds (improving biosecurity)
-
•
eliminating/reducing live bird markets
-
•
housing aquatic birds and domestic poultry separately
-
•
slaughtering flocks infected with avian influenza H5 and H7
-
•
raising awareness of potential for acquisition of influenza from swine.
These measures may achieve some success in prevention of zoonotic transmission of influenza A to humans, but have little impact on its annual cycle in humans. Unprecedented levels of H5 and H7 circulating in South East Asia and elsewhere present a high risk for emergence of a novel pandemic influenza A strain, particularly as it is recognized that relatively few mutations are required to alter the transmission characteristics of avian viruses to those more adapted to respiratory transmission in humans.
Clinical countermeasures for influenza
The mainstay of control of influenza is vaccination. The basis of vaccination against influenza is the observation that infection with influenza generates neutralizing antibodies to the viral surface proteins, predominantly HA. These antibodies will protect against infection with the same strain of influenza, and may give limited protection against closely related strains. Vaccines used in humans are designed to elicit high levels of antibody to viral HA protein. The more distant an influenza virus, the less protective the antibodies generated by natural immunity or vaccination will be. As human influenza drift variants arise frequently, it is necessary to update vaccines on a regular basis. Influenza vaccines are given annually, before the influenza season, so that those with the highest risk factor may benefit from boosted immunity. In developed countries, there has been an expansion of vaccination policies to ensure that individuals aged over 65 are vaccinated, irrespective of pre-existing illness, and every effort is made to target those with risk factors in younger age groups. Vaccination rates vary considerably between developed countries, and between the developed and the developing world.
New vaccines
In 2013, for the first time in the UK, live attenuated vaccines (LAIV) will be used to vaccinate children over 2 years and under 16 years. LAIV are particularly suitable for children as they can be given intranasally without the use of needles. The vaccines undergo limited replication in the nasal cavity and induce both humoral and cellular immunity, which is more broadly protective. Vaccines used for seasonal vaccination of older adults and elderly are subunit-inactivated vaccines that do not replicate. They are intended to boost pre-existing immunity, to reduce the likelihood of severe infection.
There are no widely available vaccines for H7 or H5 influenza, and although there have been several clinical trials of vaccines against H5, these viral targets are not included in seasonal vaccines.
Antiviral drugs
Despite the protective efficacy of vaccination, the need for treatment of severe influenza remains, particularly in the case of zoonotic infections. NA inhibitors (NIs) are the most widely used class of anti-influenza compounds. They act on the viral surface protein, NA, prevent release of viral particles from infected cells and are most effective when given early in illness. Use in the pandemic of 2009 has confirmed that, when given early during illness, drugs can prevent severe outcomes (death and critical care hospitalization) and may also have benefit in severe infection, even if given late.10 In the UK in 2000, the National Institute for Health and Care Excellence (NICE) has recommended that NI drugs may be used for prophylaxis and treatment, with certain restrictions.
Other anti-influenza drugs that target other viral proteins such as polymerase are being progressed through drug development pipelines. When such drugs are licensed they will enhance the ability to treat infections and avoid the emergence of drug resistance that occurs in the severely immunocompromised host, and with higher frequency in treated children. In immunocompromised individuals, influenza A replicates to high viral load in the absence of immune responses, and infection may become persistent, giving prolonged shedding of unusual drug-resistant variants that are difficult to treat. Intravenous immunoglobulin usually contains high levels of neutralizing antibody to influenza, and may be useful in treating severely immunocompromised individuals who are unable to clear seasonal influenza infections. Children may be more likely to generate resistant viruses following treatment, as a result of higher viral shedding in the absence of pre existing immunity.
Drug-resistant influenza
The emergence of drug-resistant influenza is dependent on drug, viral and host factors. Viral factors include the NA subtype of influenza, with N1-containing viruses more likely to sustain the mutations in viral NA to inhibit drug binding (and thereby generate resistance) compared with N2-containing viruses. The most commonly described mutation associated with NA drug resistance is the histidine-to-tyrosine shift (H275Y) seen in H1N1 or H5N1 viruses, which reduces oseltamivir susceptibility by over 600-fold. This is associated with clinical resistance to drug. However, even when resistant virus emerges in an individual during the course of treatment as a species detectable towards the end of a treatment course, it may not have any clinical consequence for the individual in whom resistance is detected if that individual is immunologically normal, or the infection is with seasonal influenza. The ability of drug-resistant influenza to transmit occurred during 2007/2008 with seasonal H1N1, although this virus has now been replaced with pandemic H1N1, which remains sensitive to oseltamivir.11
Individuals with a compromised immune system are more likely to generate drug-resistant variants, and require additional therapy with drugs directed at alternative targets or the use of immunotherapy. There is increasing interest in combination therapies with some of the unlicensed alternative antiviral drugs (polymerase inhibitors) or diverse neuraminidase inhibitors that are given intravenously, which may have a slightly different resistance profile.
Coronaviruses
Coronaviruses have a large, positive-sense segmented RNA genome of approximately 30 kb and infect a diverse range of animals, including domestic and companion animals of man. It is considered most likely that coronaviruses in these animals originated from a bat reservoir, and became established in many secondary hosts. Coronaviruses show great adaptability in host range, and as a result can establish and sustain, in a wide range of species, infections that may then spillover into man. This was seen in 2003 with SARS being transmitted from civet cats, which were a permissive secondary host.
Since the identification of SARS in 2003,12 several other new respiratory coronaviruses of humans have been identified, including NL63 and HKU1 virus. These are in addition to known human coronaviruses OC43 and 229E, which have been recognized as a source of milder respiratory infections in humans for several decades. As a result of SARS, much greater attention has been given to this group of viruses generally, with the result that many new viruses in animals have also been identified. Coronaviruses group genetically into four major groups, with human viruses found in three of the four major groups, although the greatest diversity of coronaviruses is found in bats.13
SARS-CoV
The first indication of a new viral infection in humans was the detection of clusters of illness in South East Asia. This virus spread worldwide in early 2003, with over 8000 cases and approximately 10% fatalities before the disease was brought under control through application of public health control measures, such as quarantine of infected individuals and strict infection control in hospital settings. Risk factors for SARS were:
-
•
close contact with civet cats
-
•
eating/preparing civet meat
-
•
laboratory work with SARS
-
•
contact with a known case.
MERS-CoV
In 2012, a new coronavirus, MERS-CoV, was identified in cases of severe respiratory illness as a result of clinical vigilance.14, 15 Since original notification, this virus has affected over 100 people in nine countries with a high (50%) case fatality rate,16 as a result of acute lung injury following viral replication in lower respiratory tract. All cases so far have a link to the Arabian peninsula. The virus has a zoonotic origin, probably from an animal species indigenous to this region. Cases occur in a sporadic pattern, with human-to-human transmission in small family or healthcare clusters.16, 17, 18
Risk factors for MERS-CoV include:
-
•
contact with known case
-
•
travel to the Arabian peninsula.
Exposures or behaviours giving rise to this infection are currently unknown. Once the condition has been diagnosed, supportive care and lung protective ventilation are required, if the case requires hospitalization, with strict attention to infection control. No specific interventions are available. Update on cases is available from the WHO website.
References
- 1.Reperant L.A., Cornaglia G., Osterhaus A.D. The importance of understanding the human-animal interface: from early hominins to global citizens. Curr Top Microbiol Immunol. 2012 Oct 7 doi: 10.1007/82_2012_269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster, Monto, editors. Textbook of influenza. 2nd edn. Wiley; 2013. [Google Scholar]
- 3.World Health Organization. Number of confirmed human cases of avian influenza A(H7N9) reported to WHO. Available at: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/en/index.html (accessed 13 August 2013).
- 4.WHO. Global Alert and Response (GAR): novel coronavirus infection http://www.who.int/csr/don/2013_08_01/en/index.html (accessed 01 August).
- 5.Qi X., Qian Y.-H., Bao C.-J. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. Br Med J. 2013;347:f5452. doi: 10.1136/bmjj.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan M.C.W., Chan R.W.Y., Chan L.Y.L. Tropism and innate host responses of a novel avian influenza AH7N9 virus: an analysis of ex vivo and in vitro cultures of the human respiratory tract. Lancet Resp Med. 2013 doi: 10.1016/S2213-2600(13)70138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong X., Martin S.R., Haire L.F. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013;499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 8.Wong K.K., Gambhir M., Finelli L., Swerdlow D.L., Ostroff S., Reed C. Transmissibility of variant influenza from swine to humans: a modeling approach. Clin Infect Dis. 2013;57(suppl 1):S16–S22. doi: 10.1093/cid/cit303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belser J.A., Bridges C.B., Katz J.M., Tumpey T.M. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009;15:859–865. doi: 10.3201/eid1506.090072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muthuri S., Myles P., Venkatesan S. Impact of neuraminidase inhibitor treatment on outcomes of public health importance during the 2009–2010 influenza A(H1N1) pandemic: a systematic review and meta-analysis in hospitalized patients. J Infect Dis. 2013;207:553–563. doi: 10.1093/infdis/jis726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurt A.C., Chotpitayasunondh T., Cox N.J. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis. 2012;12:240–248. doi: 10.1016/S1473-3099(11)70318-8. [DOI] [PubMed] [Google Scholar]
- 12.Lee N., Hui D., Wu A. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 13.Van Boheeman S., de Graaf M., Lauber C. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3:e00473–12. doi: 10.1128/mBio.00473-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaki A.M., van Boheemen S., Bestebroer T.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 15.Bermingham A., Chand M.A., Brown C.S. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20290 pii=20290. Available online: [PubMed] [Google Scholar]
- 16.WHO website GOARN MERS CoV.
- 17.Assiri A., McGeer A., Perl T.M. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:P407–P416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breban R., Riou J., Fontanet A. Interhuman transmissibility of middle east respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013 doi: 10.1016/S0140-6736(13)61492-0. Early Online Publication. [DOI] [PMC free article] [PubMed] [Google Scholar]