Abstract
Background
Respiratory infections are the most common infections in humans. The prevalence of respiratory viruses in adults is largely underestimated, and relevant data mostly concern infants and children.
Objectives
To evaluate the prevalence of respiratory viruses in adults hospitalized in Italy.
Study design
During April 2004–May 2005, 510 consecutive lower respiratory tract samples were prospectively collected. These were evaluated with a molecular panel that detected 12 respiratory viruses.
Results
Two hundred and fifteen samples were positive for at least one viral pathogen, with an overall sample prevalence of 42.2%. Human rhinoviruses (HRVs) were the most commonly detected viruses (32.9%), followed by influenza virus (FLU)-A (9.0%); the other viruses were 2% or less. Multiple agents were detected in 30 samples from 29 patients, resulting in a co-infection rate of 6.7%.
Conclusions
This study shows a high prevalence of viruses in the lower respiratory tract samples of hospitalized adults, mostly HRV and FLU-A. It is not possible to establish the role of viruses detected at low frequency, but our findings suggest the necessity to consider them as potential causes or precursors of lower respiratory tract infections (LRTIs).
Abbreviations: HRV, human rhinovirus; FLU, influenzavirus; hCoV, human coronavirus; AdV, adenovirus; PIV, parainfluenzavirus; RSV, respiratory syncytial virus; hMPV, human metapneumovirus; LRTIs, lower respiratory tract infections; SP, sputum; EA, endotracheal aspirate; BAL, bronchoalveolar lavage; COPD, chronic obstructive pulmonary disease; ARI, acute respiratory illness; hBoV, human bocavirus
Keywords: Lower respiratory tract infection, Respiratory viruses, Adults, Hospitalized
1. Introduction
The etiology of lower respiratory tract infections (LRTIs) in adults, which are an important cause of morbidity, remains undetermined in more than 50% of cases (Fine et al., 1999, Garbino et al., 2002, File, 2003), mostly due to limited diagnostic procedures. The prevalence of respiratory viruses in adults is largely unexplored, as most relevant data concern infants and children (Choi et al., 2006, Costa et al., 2006, Kusel et al., 2006, Ma et al., 2006, Ordas et al., 2006, Pierangeli et al., 2007). Respiratory syncytial virus (RSV), parainfluenzaviruses (PIVs), influenzaviruses (FLUs) and adenoviruses (AdVs) are considered the leading causes of acute viral LRTIs (Henderson et al., 1979, Hall et al., 1990, Yun et al., 1995, Hong et al., 2001). However, many other respiratory viruses have been recently identified as possible causative agents of LRTIs. Human rhinoviruses (HRVs), which previously were believed to cause only mild upper respiratory illnesses, have been found in association with acute and chronic LRTIs, including asthma exacerbations and chronic obstructive pulmonary disease (COPD), although their role as causative agents has only been established for asthma (Singh and Busse, 2007, Papadopoulos et al., 2002, Friedlander and Busse, 2005, Xatzipsalti et al., 2005). Newly discovered viruses, such as human metapneumovirus (hMPV) and human coronaviruses (hCoVs) have been associated with LRTIs in children (van den Hoogen et al., 2001, Fouchier et al., 2004, van der Hoek et al., 2004, Choi et al., 2006).
The aim of this study was to assess the point prevalence of a full spectrum of respiratory viruses in the lower respiratory tract specimens of hospitalized adults by using a molecular diagnostic panel.
2. Methods
A prospective collection of 510 lower respiratory samples from 433 adult patients with pulmonary diseases, admitted to three Italian hospitals was performed. Clinical-epidemiological data were collected through a standardized questionnaire. Patients with suspect Mycobacterium tuberculosis infection were not included in the analysis. The study was approved by the Institutional Ethics Committees, and informed consent signed by participants. The selection criterion for patients’ enrolment was to have any one of the following samples sent to the laboratory for routine microbiological evaluation: sputum (SP), bronchoalveolar lavages (BALs), or endotracheal aspirate (EA). To minimize the possibility that positive results reflected contamination from the upper respiratory tract, only samples containing more than 25 polymorphonuclear leucocytes per field (40×) were processed (Murray and Washington, 1975, Geckler et al., 1977). Less than 4% of the samples did not meet this criterion and were discarded. Half of each sample was submitted to standard microbiological analysis (common microbiological culture and, in case of positive result, identification and antibody susceptibility test), while the remainder was stored at −80 °C for the molecular analysis. During April 2004–May 2005, a total of 510 consecutive lower tract respiratory samples (329 SP, 165 EA and 16 BAL), which were considered appropriate on the basis of microscope inspection, were analyzed.
The samples were thawed and liquefied by using 1:1 Sputasol (Oxoid Ltd., Basingstoke, England), and then underwent DNA/RNA extraction (Boom et al., 1990). RNA was reverse transcribed with random primers with a Reverse Transcriptase Kit (Invitrogen, Milan, Italy). Control DNA ((-globin DNA) and RNA (MS2 phage genome) preparations were spiked into the samples before the extraction, to control their suitability for PCR/RT-PCR.
The molecular diagnostic panel included: FLU-A and -B, hMPV, AdV, PIV-1, -2 and -3, RSV, HRV, hCoV-OC43, -229E and -NL63. Specific primers and PCR or RT-PCR conditions were as previously described (Allard et al., 1994, Pitkaranta et al., 1997, Echevarria et al., 1998, Gröndahl et al., 1999, Allard et al., 2001, Steininger et al., 2001, Maggi et al., 2003, Fouchier et al., 2004, Bastien et al., 2005, Esper et al., 2005, Woo et al., 2005, Minosse et al., 2007). The sensitivity of the assays ranged between 1 and 10 copies for FLU-A, AdV, PIV-1, -2 and -3, RSV, HRV and hCoV-NL63; between 10 and 50 copies for FLU-B and hMPV A; and about 300 copies for hCoV-OC43. These sensitivity values, established by probit analysis on plasmids containing the target region as the insert, were confirmed by participation in the Quality Control for Molecular Diagnostics External Quality Assurance (QCMD EQA) programmes (Glasgow, UK). From the hMPV QCMD EQA programme MPV.RSV06, we determined that the sensitivity for hMPV B of our test was much lower (>1000 times) than for other respiratory viruses.
The amplified products were separated by agarose gel electrophoresis and UV-visualized after ethidium bromide staining.
3. Results
A molecular diagnostic panel for detection of 12 viruses was applied to 510 lower respiratory samples from 433 adult patients hospitalized with pulmonary diseases.
The most common respiratory illnesses in the patients were: COPD, 45.3%, pneumonia (30.5%) and acute respiratory illness (ARI, 17.1%); 19.6% of patients were in the intensive care unit, and 17.5% were receiving assisted ventilation (Table 1 ). Two hundred and fifteen specimens (42.2%) were positive for a respiratory virus (Table 2 ). The most common viruses were HRV (32.9%), FLU-A (9.0%) and hCoV-NL63 (2.0%). The frequency of the other viruses ranged from 0.2 to 1.4%. In 30 out of 510 samples (5.9%), corresponding to 29 out of 433 patients (6.7%), more than one virus was detected. Multiple detection included HRV + FLU-A (2.9% of total samples), HRV + FLU-B and HRV + hCoV-NL63 (0.6% each combination), HRV + hCoV-OC43 (0.4%), HRV + AdV, HRV + PIV-1, HRV + RSV, FLU-A + hCoV-OC43, AdV + hCoV-NL63, HRV + FLU-A + PIV-1 and HRV + FLU-A + AdV (0.2% each combination). The association with the sample type (either EA, or SP, or BAL) could be evaluated only for the most frequent viruses, i.e. FLU-A and HRV, showing that HRV, and not FLU-A, was unevenly distributed (more frequent in SP and BAL than in EA, Table 2). Data on the smoking status was available for 403/433 patients: the frequency of patients positive to any virus was similar in smokers and non-smokers (49.4% vs. 50.6%, p = 0.742), and the frequency of individual viruses did not significantly differ according to the smoker status (for instance, HRV positive patients were 42.2% in smokers and 34.5% in non-smokers, p = 0.137).
Table 1.
Demographic and clinical characteristics of 433 adult hospitalized patients
| Age | |
| Mean (±S.D.) | 56.3 (18.2) |
| Median (IQR) | 57.0 (41–72) |
| Sex | |
| Male (%) | 291 (67.2) |
| Female (%) | 136 (31.4) |
| Not known (%) | 6 (1.4) |
| Smoking status | |
| Smokers (%) | 168 (38.8) |
| Non-smokers (%) | 235 (54.3) |
| Not known (%) | 30 (6.9) |
| Clinical findings | |
| COPD (%) | 196 (45.3) |
| Pneumonia (%) | 132 (30.5) |
| Acute respiratory failure (%) | 76 (17.5) |
| Chronic respiratory failure (%) | 15 (3.5) |
| Assisted ventilation (%) | 76 (17.5) |
| Asthma (%) | 9 (2.1) |
COPD, chronic obstructive pulmonary disease.
Table 2.
Detection of respiratory agents in 510 samples from 433 adult hospitalized patients
| Respiratory agent | Distribution of positive specimens, n (%) |
|||
|---|---|---|---|---|
| Overall positivity | SP | EA | BAL | |
| FLU-A* | 46 (9.0) | 24 (7.3) | 20 (12.1) | 2 (12.5) |
| FLU-B | 5 (1.0) | 5 (1.5) | 0 | 0 |
| PIV-1 | 6 (1.2) | 6 (1.8) | 0 | 0 |
| PIV-2 | 0 | 0 | 0 | 0 |
| PIV-3 | 1 (0.2) | 0 | 1 (0.6) | 0 |
| RSV | 1 (0.2) | 1 (0.3) | 0 | 0 |
| hMPV | 0 | 0 | 0 | 0 |
| HRV** | 168 (32.9) | 124 (37.7) | 39 (23.6) | 5 (31.2) |
| hCoV-OC43 | 7 (1.4) | 4 (1.2) | 3 (1.8) | 0 |
| hCoV-229E | 0 | 0 | 0 | 0 |
| hCoV-NL63 | 10 (2.0) | 7 (2.1) | 3 (1.8) | 0 |
| AdV | 6 (1.2) | 6 (1.8) | 0 | 0 |
| Samples positive to at least 1 agent*** | 215 (42.2) | 156 (47.4) | 58 (35.1) | 6 (37.5) |
| Samples positive to >1 agent | 30 (5.9) | 22 (6.7) | 8 (4.8) | 0 |
| Total analyzed samples | 510 (100) | 329 (100) | 165 (100) | 16 (100) |
SP, sputum; EA, endotracheal aspirate; BAL, bronchoalveolar lavage; FLU, influenzavirus; PIV, parainfluenzavirus; RSV, respiratory syncytial virus; hMPV, human metapneumovirus; HRV, human rhinovirus; hCoV, human coronavirus; AdV, adenovirus.
p = 0.186.
p = 0.007.
p = 0.031 for frequency in SP, EA and BAL in χ2 test.
In addition, similar frequencies of virus-positive samples were obtained when comparing patients with COPD vs. patients without COPD (57.1% vs. 56.7%); patients with pneumonia vs. subjects with no record of pneumonia (54.4% in both cases); or patients with ARI vs. subjects without ARI (44.7% vs. 51.5%, p = 0.342). In addition, the frequency of virus-positive samples did not significantly differ among patients with pneumonia, COPD and ARI (p = 0.207).
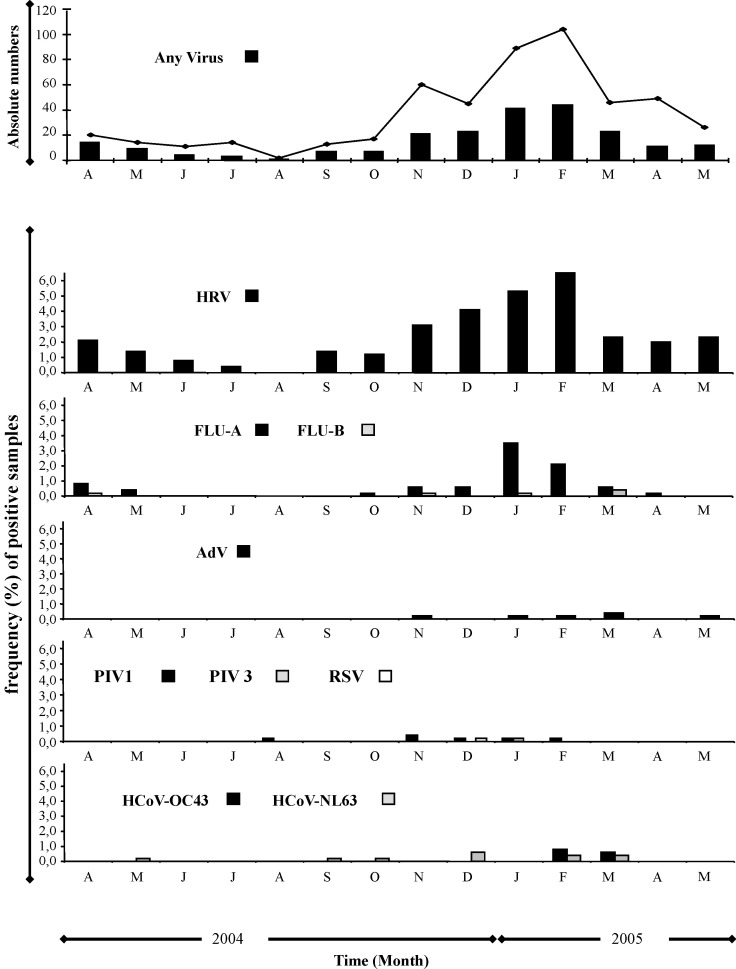
The monthly distribution of respiratory agents over the study period (Fig. 1 ) highlights a peak in January–February for HRV and FLU-A.
Fig. 1.
Monthly distribution of lower respiratory tract samples positive for respiratory viruses, during April 2004–May 2005. The line represents the total collected samples, while the bars represent the samples positive for the indicated viruses. First panel: absolute numbers; lower panels: frequency (%) of samples positive for the indicated viruses.
4. Discussion
Many reports have highlighted the possible role of viruses as causative agents, or as possible co-factors, in LRTIs. We applied a large molecular diagnostic panel which included 12 respiratory viruses to evaluate the prevalence of respiratory pathogens in adults hospitalized in Italy, during April 2004–May 2005. Although we would be unable to detect some viral respiratory pathogens, such as bocavirus, coronavirus HKU1 and parainfluenzavirus 4, this is the first extensive investigation of lower respiratory tract secretions from adults that was capable of detecting such a wide selection of respiratory pathogens. We have made the assumption that the samples represent the lower respiratory tract, as a recognized microscope inspection criterion was used to validate their quality. However, as expectorated sputum is necessarily contaminated with oral contents, it is not possible to rule out that the virus detected in the samples (mainly SP) could actually derive from contamination by upper respiratory tract secretions. In fact, the rate of positivity in SP is higher than in EA and BAL (47.4% vs. 35.1% and 37.5%, respectively); however, this is not the case for all viruses (i.e. FLU-A). In addition, the presence of several viruses (FLU-A, PIV-3, HRV, hCoV-OC43 and hCoV-NL63) on both EA and BAL, suggests that contamination with oral secretions is not a major source of the virus present in the analyzed samples.
Our results confirm previous data that HRV are the most frequent viruses in lower respiratory tract samples (Garbino et al., 2004, Puro et al., 2005), and strongly support the association of HRV with LRTIs, already hypothesized in previous studies based on upper respiratory samples (Tsolia et al., 2004). However, based on the present data it is not possible to establish the precise role of HRV as a leading cause, as well as its possible involvement as co-factor in LRTIs.
Among the other viruses, accounting for 7.2% of positive samples, hCoV-NL63 was detected in 2.0% of samples. The involvement of hCoV (namely NL63) in lower respiratory diseases has been largely hypothesized, but available data are mostly based on upper respiratory samples, mostly on the basis of studies in pediatric patients (Arden et al., 2005, Bastien et al., 2005, Boivin et al., 2005, Chiu et al., 2005, Ebihara et al., 2005, Moes et al., 2005, Pierangeli et al., 2007). The circulation of hCoV-NL63 in Italy is probably underreported. There is only one other report concerning this virus and lower respiratory samples from adults (Gerna et al., 2006).
We did not detect hMPV in our patients, although this is a relatively common respiratory pathogen (van den Hoogen et al., 2001, Choi et al., 2006). The absence of hMPV is probably due to the low sensitivity of the RT-PCR assay used in this study (particularly for hMPV B). Alternatively, it is possible that sputasol, used to liquefy the samples, could have affected the sensitivity of the RT-PCR used. This issue could also apply to other viruses investigated in this study, such as RSV, and needs further investigation.
In conclusion, our study shows a high prevalence of viruses, especially HRV and influenza virus, in the lower respiratory tract samples of adult hospitalized patients with pulmonary diseases, without any significant difference in the frequency of virus-positivity among patients with pneumonia, COPD and ARI. Additional viruses were detected at low frequency. It is not possible to establish the role of viruses detected at low frequency as causative agents or as co-factors in LRTIs, but these results highlight the necessity to consider them as potential causes of LRTIs.
Acknowledgments
This work was supported in part by grants from the Italian Ministry of Health (“Fondi Ricerca Corrente”, “Ricerca Finalizzata”, and “Fondi per la creazione di un polo centralizzato per la crioconservazione”) to INMI L. Spallanzani. Neither funding source influenced the design, conduct or reporting of this study.
Anna Prygodzicz and Carla Nisii are gratefully acknowledged for assistance in writing the manuscript. We acknowledge the enthusiastic contribution of M. Visca, M. Cava, G. Gualano, who assisted in clinical investigation and collaborated with sample and data collection.
References
- Allard A., Kajon A., Wadell G. Simple procedure for discrimination and typing of enteric adenoviruses after detection by polymerase chain reaction. J Med Virol. 1994;44(3):250–257. doi: 10.1002/jmv.1890440307. [DOI] [PubMed] [Google Scholar]
- Allard A., Albinsson B., Wadell G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J Clin Microbiol. 2001;39(2):498–505. doi: 10.1128/JCM.39.2.498-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden K.E., Nissen M.D., Sloots T.P., Mackay I.M. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J Med Virol. 2005;75(3):455–462. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N., Anderson K., Hart L., Van Caeseele P., Brandt K., Milley D. Human coronavirus NL63 infection in Canada. J Infect Dis. 2005;191(4):503–506. doi: 10.1086/426869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin G., Baz M., Cote S., Gilca R., Deffrasnes C., Leblanc E. Infections by human coronavirus-NL in hospitalized children. Pediatr Infect Dis J. 2005;24(12):1045–1048. doi: 10.1097/01.inf.0000183743.68569.c7. [DOI] [PubMed] [Google Scholar]
- Boom R., Sol C.J., Salimans M.M., Jansen C.L., Wertheim-van Dillen P.M., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.S., Chan K.H., Chu K.W., Kwan S.W., Guan Y., Poon L.L. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40(12):1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.H., Lee H.J., Kim S.J., Eun B.W., Kim N.H., Lee J.A. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children 2000–2005. Clin Infect Dis. 2006;43(5):585–592. doi: 10.1086/506350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L.F., Yokosawa J., Mantese O.C., Oliveira T.F., Silveira H.L., Nepomuceno L.L. Respiratory viruses in children younger than five years old with acute respiratory disease from 2001 to 2004 in Uberlandia, MG, Brazil. Mem Inst Oswaldo Cruz. 2006;101(3):301–306. doi: 10.1590/s0074-02762006000300014. [DOI] [PubMed] [Google Scholar]
- Ebihara T., Endo R., Ma X., Ishiguro N., Kikuta H. Detection of human coronavirus NL63 in young children with bronchiolitis. J Med Virol. 2005;75(3):463–465. doi: 10.1002/jmv.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarria J.E., Erdman D.D., Swierkosz E.M., Holloway B.P., Anderson L.J. Simultaneous detection and identification of human parainfluenza viruses 1, 2 and 3 from clinical samples by multiplex PCR. J Clin Microbiol. 1998;36(5):1388–1391. doi: 10.1128/jcm.36.5.1388-1391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J Infect Dis. 2005;191(4):492–498. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- File T.M. Community-acquired pneumonia. Lancet. 2003;362(9400):1991–2001. doi: 10.1016/S0140-6736(03)15021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine M.J., Stone R.A., Singer D.E., Coley C.M., Marrie T.J., Lave J.R. Processes and outcomes of care for patients with community-acquired pneumonia: results from the Pneumonia Patient Outcomes Research Team (PORT) cohort study. Arch Intern Med. 1999;159(9):970–980. doi: 10.1001/archinte.159.9.970. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA. 2004;101(16):6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander S.L., Busse W.W. The role of rhinovirus in asthma exacerbations. J Allergy Clin Immunol. 2005;116(2):267–273. doi: 10.1016/j.jaci.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Garbino J., Sommer R., Gerber A., Regamey C., Vernazza P., Genne D. Prospective epidemiologic survey of patients with community-acquired pneumonia requiring hospitalization in Switzerland. Int J Infect Dis. 2002;6(4):288–293. doi: 10.1016/s1201-9712(02)90163-3. [DOI] [PubMed] [Google Scholar]
- Garbino J., Gerbase M.W., Wunderli W., Deffernez C., Thomas Y., Rochat T. Lower respiratory viral illnesses: improved diagnosis by molecular methods and clinical impact. Am J Respir Crit Care Med. 2004;170(11):1197–1203. doi: 10.1164/rccm.200406-781OC. [DOI] [PubMed] [Google Scholar]
- Geckler R.W., Gremillion D.H., McAllister C.K., Ellenbogen C. Microscopic and bacteriological comparison of paired sputa and transtracheal aspirates. J Clin Microbiol. 1977;6:396–399. doi: 10.1128/jcm.6.4.396-399.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G., Campanini G., Rovida F., Percivalle E., Sarasini A., Marchi A. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol. 2006;78(7):938–949. doi: 10.1002/jmv.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröndahl B., Puppe W., Hoppe A., Kuhne I., Weigl J.A., Schmitt H.J. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol. 1999;37(1):1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C.B., Walsh E.E., Schnabel K.C., Long C.E., McConnochie K.M., Hildreth S.W. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis. 1990;162(6):1283–1290. doi: 10.1093/infdis/162.6.1283. [DOI] [PubMed] [Google Scholar]
- Henderson F.W., Clyde W.A., Jr, Collier A.M., Denny F.W., Senior R.J., Sheaffer C.I. The etiologic and epidemiologic spectrum of bronchiolitis in pediatric practice. J Pediatr. 1979;95(2):183–190. doi: 10.1016/s0022-3476(79)80647-2. [DOI] [PubMed] [Google Scholar]
- Hong J.Y., Lee H.J., Piedra P.A., Choi E.H., Park K.H., Koh Y.Y. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin Infect Dis. 2001;32(10):1423–1429. doi: 10.1086/320146. [DOI] [PubMed] [Google Scholar]
- Kusel M.M., de Klerk N.H., Holt P.G., Kebadze T., Johnston S.L., Sly P.D. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr Infect Dis J. 2006;25(8):680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- Ma X., Endo R., Ishiguro N., Ebihara T., Ishiko H., Ariga T. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J Clin Microbiol. 2006;44(3):1132–1134. doi: 10.1128/JCM.44.3.1132-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi F., Pifferi M., Vatteroni M., Fornai C., Tempestini E., Anzilotti S. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J Clin Microbiol. 2003;41(7):2987–2991. doi: 10.1128/JCM.41.7.2987-2991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minosse C., Selleri M., Zaniratti M.S., Lauria F.N., Puro V., Carletti F. Improved detection of human influenza A and B viruses in respiratory tract specimens by hemi-nested PCR. J Virol Methods. 2007;141(2):225–228. doi: 10.1016/j.jviromet.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes E., Vijgen L., Keyaerts E., Zlateva K., Li S., Maes P. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis. 2005;5(1):6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.R., Washington J.A., II Microscopic and bacteriologic analysis od expectored sputum. Mayo Clin Proc. 1975;50:339–344. [PubMed] [Google Scholar]
- Ordas J., Boga J.A., Alvarez-Arguelles M., Villa L., Rodriguez-Dehli C., de Ona M. Role of metapneumovirus in viral respiratory infections in young children. J Clin Microbiol. 2006;44(8):2739–2742. doi: 10.1128/JCM.00164-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos N.G., Moustaki M., Tsolia M., Bossios A., Astra E., Prezerakou A. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med. 2002;165(9):1285–1289. doi: 10.1164/rccm.200112-118BC. [DOI] [PubMed] [Google Scholar]
- Pierangeli A., Gentile M., Di Marco P., Pagnotti P., Scagnolari C., Trombetti S. Detection and typing by molecular techniques of respiratory viruses in children hospitalized for acute respiratory infection in Rome, Italy. J Med Virol. 2007;79(4):463–468. doi: 10.1002/jmv.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkaranta A., Arruda E., Malmberg H., Hayden F.G. Detection of rhinovirus in sinus brushings of patients with acute community-acquired sinusitis by reverse transcription-PCR. J Clin Microbiol. 1997;35(7):1791–1793. doi: 10.1128/jcm.35.7.1791-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puro V., Minosse C., Cappiello G., Lauria F.N., Capobianchi M.R. Rhinovirus and lower respiratory tract infection in adults. Clin Infect Dis. 2005;40(7):1068–1069. doi: 10.1086/428359. [DOI] [PubMed] [Google Scholar]
- Singh A.M., Busse W.W. Human rhinovirus models in asthma. Contrib Microbiol. 2007;14:12–20. doi: 10.1159/000107051. [DOI] [PubMed] [Google Scholar]
- Steininger C., Aberle S.W., Popow-Kraupp T. Early detection of acute rhinovirus infections by a rapid reverse transcription-PCR assay. J Clin Microbiol. 2001;39(1):129–133. doi: 10.1128/JCM.39.1.129-133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolia M.N., Psarras S., Bossios A., Audi H., Paldanius M., Gourgiotis D. Etiology of community-acquired pneumonia in hospitalized school-age children: evidence for high prevalence of viral infections. Clin Infect Dis. 2004;39(5):681–686. doi: 10.1086/422996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7(6):719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xatzipsalti M., Kyrana S., Tsolia M., Psarras S., Bossios A., Laza-Stanca V. Rhinovirus viremia in children with respiratory infections. Am J Respir Crit Care Med. 2005;172(8):1037–1040. doi: 10.1164/rccm.200502-315OC. [DOI] [PubMed] [Google Scholar]
- Yun B.Y., Kim M.R., Park J.Y., Choi E.H., Lee H.J., Yun C.K. Viral etiology and epidemiology of acute lower respiratory tract infections in Korean children. Pediatr Infect Dis J. 1995;14(12):1054–1059. doi: 10.1097/00006454-199512000-00005. [DOI] [PubMed] [Google Scholar]