Abstract
Background
Currently, the role of the novel human polyomaviruses, KI (KIV) and WU (WUV) as agents of human disease remains uncertain.
Objectives
We sought to determine the prevalence of these viruses and their rate of co-detection with other viral respiratory pathogens, in an Australian population.
Study design
Polymerase chain reaction assays previously described were used to examine the presence of KIV and WUV in 2866 respiratory specimens collected from January to December 2003 from Australian patients with acute respiratory infections.
Results
KIV and WUV were present in our population with an annual prevalence of 2.6% and 4.5%, respectively. There was no apparent seasonal variation for KIV, but a predominance of infection was detected during late winter to early summer for WUV. The level of co-infection of KIV or WUV with other respiratory viruses was 74.7% and 79.7%, respectively. Both viruses were absent from urine and blood specimens collected from a variety of patient sources.
Conclusions
KIV and WUV circulate annually in the Australian population. Although there is a strong association with the respiratory tract, more comprehensive studies are required to prove these viruses are agents causing respiratory disease.
Keywords: Human polyomavirus, Acute respiratory infection, Respiratory viruses, Epidemiology, Molecular, Clinical, Emerging virus
1. Introduction
Intensive investigations over the last 6 years have led to the discovery of a number of new or emerging putative respiratory viruses, including human metapneumovirus (HMPV) (van den Hoogen et al., 2001), human coronaviruses, SARS (Ksiazek et al., 2003), NL63 (van der Hoek et al., 2004) and HKU1 (Woo et al., 2005), human bocavirus (HBoV) (Allander et al., 2005) and very recently, two new human polyomaviruses, KI virus (KIV) (Allander et al., 2007) and WU virus (WUV) (Gaynor et al., 2007). Their role as causative agents of respiratory disease has been conclusively demonstrated for some of these (HMPV, SARS, NL63), but for others their classification as a respiratory pathogen remains highly speculative in the absence of fulfilling Koch's postulates. This is particularly so for the newly discovered human KIV and WUV.
KIV and WUV were discovered independently in Sweden and the USA within months of each other. Allander et al. (2007) showed that KIV is phylogenetically related to other primate polyomaviruses in the early region of the genome, but had little homology in the late region. Similarly, by multiple sequence alignments of the predicted STAg, LTAg, VP1 and VP2 open reading frames, Gaynor et al. (2007) confirmed that WUV was clearly another novel polyomavirus that is most closely related to KIV. They also showed that WUV differed significantly from the other human polyomaviruses BK (BKV) and JC (JCV) in genome sequence and suggested that it is likely to have unique biological properties.
As a first step in elucidating the distribution and a possible role in human disease of KIV and WUV, we sought to determine the presence of these viruses in the population and their association with the respiratory tract. For this purpose we had at our disposal respiratory samples collected during 2003 from patients with acute respiratory tract infection (ARTI) that had been extensively tested for other known respiratory viruses. This sample cohort was investigated for the presence of KIV and WUV, seasonality, and rate of co-infection with each other or other common respiratory viruses.
2. Materials and methods
2.1. Respiratory specimens and nucleic acid extraction
We examined 2866 respiratory samples consisting of 2733 nasopharyngeal aspirates (NPAs), 91 bronchoalveolar lavage (BAL), 33 bronchial washings (BW) and 9 endotracheal aspirates (ETA) for the presence of KIV and WUV. Also, in consideration of the fact that BKV and JCV may be frequently found in urine and blood, we examined 215 urine samples, and 102 blood samples.
NPAs were collected from January 2003 until January 2004, from hospitalized patients or patients presenting for assessment at hospital emergency departments in Queensland, Australia, with ARTI. Patients ranged in age from 3 days to 95 years (mean = 9.2 years; median age = 1.38 years), and 76.5% of specimens were from children 5 years of age or younger. Nucleic acids were extracted from 0.2 mL of each NPA specimen using the High Pure Viral Nucleic Acid kit (Roche Diagnostics, Australia), according to the manufacturer's instructions.
To ensure that a diverse urine sample population was tested in this study, we included specimens that were obtained from a general hospital population and submitted for routine investigation of micro-organisms. Urine samples were collected between October 1999 and August 2005 from pediatric and adult patients ranging in age from 1 day to 76 years (mean = 16.9 years; median age = 24.6 years). Fifty-five samples were included from adult immunocompromised (bone marrow transplant) patients. Nucleic acids were extracted from 0.2 mL of each urine specimen with a QIAamp DNA blood mini kit (Qiagen, Clifton Hill, Australia). This procedure has been shown to be suitable for the extraction of viral DNA from urine specimens because of the high DNA yield and the removal of PCR inhibitors (Echavarria et al., 1998). Similarly, 102 blood samples collected between October 2004 and June 2005 for the monitoring of aspergillosis in 27 children with leukemia ranging in age from 2 to 16 years (mean = 7.6 years; median age = 6.0 years) were tested for KIV and WUV. A 200 μL volume of each blood sample was extracted as described for urine samples above.
2.2. Detection of KIV and WUV by PCR
Extracts were analyzed for the presence of KIV and WUV sequences by conventional PCR. A common PCR reaction mix was used for all conventional assays consisting of 1.25 units of QIAGEN HotStart Taq (Qiagen), 2.5 pmol of each primer, 0.625 μL of 10 mM dNTPs, 0.5 μL of 25 mM MgCl2, 2.5 μL of 10× QIAGEN PCR buffer (Qiagen) and 2.5 μL of nucleotide extract made up to a 25 μL volume. Amplification was performed on an ABI GeneAmp 2700 instrument (Applied Biosystems Pty Ltd., Scoresby, Australia). For KIV, specimens were screened using primers POLVP1-39F and POLVP1-363R and positive results were confirmed with a second set of primers (POLVP1-118F and POLVP1-324R) as originally described by Allander et al. (2007). The KIV PCR assays utilized cycling conditions of 15 min incubation at 95 °C, followed by 40 cycles of 95 °C for 30 s, 54 °C for 30 s and 72 °C for 1 min, followed by a final extension of 10 min at 72 °C. WUV was initially detected by conventional PCR using primers AG0044 and AG0045, as described by Gaynor et al. (2007), utilizing 15 min incubation at 95 °C, followed by 40 cycles of 95 °C for 30 s, 56 °C for 30 s and 72 °C for 1 min, and a final extension of 10 min at 72 °C. PCR amplification products were realized by electrophoresis on 2% agarose gels and visualized with ethidium bromide staining. Positive results were confirmed using a WUV-specific real-time PCR assay, which was recently developed in our laboratory, incorporating a hydrolysis (TaqMan) probe (Bialasiewicz et al., 2007).
2.3. PCR analysis for other respiratory viruses
Samples had been previously tested for the presence of other significant respiratory viruses (respiratory syncytial virus (RSV), influenza virus A and B (INF A & B), parainfluenza viruses 1,2,3 (PIV 1,2,3), adenoviruses (ADV), HMPV) as previously described (Syrmis et al., 2004, Maertzdorf et al., 2004). All KIV and WUV positive specimens were further tested to determine the simultaneous presence of human rhinoviruses (HRV), human coronaviruses (HCoV) OC43, 229E, NL63 and HKU1 (Arden et al., 2006, Sloots et al., 2006) and HBoV. The presence of HBoV was determined by real-time PCR using primers STBoVP-1f (GGCAGAATTCAGCCATACTCAAA) and STBoVP-1r (TCTGGGTTAGTGCAAACCATGA) and a hydrolysis probe STBoVP-1probe (AGAGTAGGACCACAGTCATCAGACACTGCTCC-Yakima) targeting the HBoV VP-1 gene. All HBoV-positive results were confirmed by a second real-time PCR assay developed in our laboratory targeting the NP-1 gene (unpublished data).
2.4. Sequencing of KIV and WUV genomes
We sequenced the complete genomes of KIV detected in three patients in this study, and compared those sequences to KIV sequences available for the Swedish isolates (Allander et al., 2007). Similarly we sequenced six complete WUV genomes using previously described protocols (Gaynor et al., 2007) and sought to determine the genetic relationship of WUV with each other and compared to the other human polyomaviruses BKV and JCV. Raw KIV and WUV sequence data were aligned together with JCV, BKV and SV40 sequences using BioEdit version 7.0.5.3. Fractional nucleotide sequence identity matrices were used to examine the relationship between KIV, WUV, JCV, and BKV, with SV40 being utilized as the rooting out-group.
3. Results
3.1. Prevalence and temporal distribution of KIV and WUV
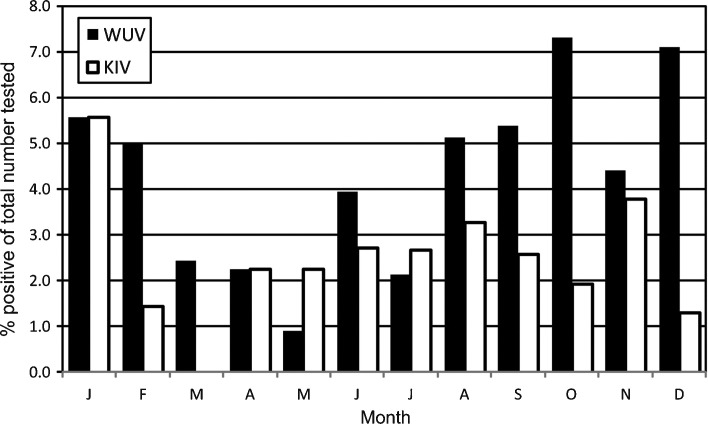
Of 2866 specimens tested, 75 (2.6%) NPA were positive for KIV and 128 (4.5%) NPA were positive for WUV. The youngest KIV positive subject was 4 months of age, and the oldest 69 years, and for WUV, 19 days and 52 years, respectively. 78.6% of KIV-positive results were from children 5 years of age or younger and 93.0% of WUV-positive results fell in this age group. Although both viruses had a consistent presence during 2003, there appeared to be no seasonal pattern of peak infection associated with KIV in our sample population. However, there appeared to be a predominance of infection with WUV in Queensland patients during the period from late winter to early summer, peaking in December (Fig. 1 ). The prevalence of WUV from August to December was 5.5% compared to an average incidence of 2.9% for the rest of the year. In contrast, the prevalence of KIV for this period was 2.9% compared to 2.5% for the remaining year. However, this distribution across different seasons proved not to be statistically significant for either virus. In this specimen set for 2003, complete annual prevalence data were only available for those respiratory viruses considered to be of most clinical significance. Of these the predominant viral pathogen detected was RSV (9.2%), followed by HMPV (7.1%); INF A (3.5%); PIV 3 (2.3%); and ADV (1.3%). Examination of the urine and blood samples failed to detect the presence of either KIV or WUV in any samples.
Fig. 1.
Seasonal distribution of KIV and WUV positive results shown as a percentage of total number of specimens tested per month. Respiratory secretions tested were collected from an Australian population with acute respiratory tract infection during 2003.
3.2. Co-detection of KIV and WUV with other respiratory viruses
KIV and WUV were associated with the co-detection of another respiratory virus in 56 of 75 (74.7%) and 102 of 128 (79.7%) cases, respectively. Both were most commonly co-detected with HRV, followed by HBoV as the second most frequently detected virus (Table 1 ). Interestingly, in 14 patients, KIV was co-detected with WUV and in only one of these no other respiratory virus was present.
Table 1.
Co-detection rates of other respiratory viruses with KIV and WUV
| Number positive | HRV | HBoV | RSV | INF | HMPV | ADV | PIV3 | OC43 | 229E | KIV | WUV | Any virus | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KIV | 75 | 39 (52.0) | 14 (18.7) | 6 (8.0) | 6 (8.0) | 2 (2.7) | 2 (2.7) | 1 (1.3) | 4 (5.3) | 1 (1.3) | – | 14 (18.7) | 56 (74.7) |
| WUV | 128 | 58 (45.3) | 26 (20.3) | 10 (7.8) | 14 (10.9) | 7 (5.5) | 5 (3.9) | 3 (2.3) | 12 (9.4) | 0 | 14 (10.9) | – | 102 (79.7) |
Figures shown are numbers of samples tested positive, with the percent positive indicated in brackets. Some samples tested positive for more than two viruses. HRV, human rhinoviruses; HBoV, human bocavirus; RSV, respiratory syncytial virus; INF, influenza virus; HMPV, human metapneumovirus; ADV, adenoviruses; PIV3, parainfluenza virus type 3; OC43, human coronavirus OC43; 229E, human coronavirus 229E; KIV, KI polyomavirus; WUV, WU polyomavirus.
3.3. Clinical presentation of KIV and WUV infected patients
The clinical signs at presentation to hospital from 51 of 75 KIV-positive and 98 of 128 WUV-positive patients were scrutinized, and presenting clinical features were recorded. There was no observable difference in presenting signs between patients infected with KIV and those infected with WUV, and these were consistent with ARTI. These signs also did not differ from those observed in patients with ARTI attributed to other respiratory viruses, or in fact those patients for whom a cause of infection could not be identified. Clinical presentation for these patients was comprised predominantly of cough, fever and malaise (Table 2 ).
Table 2.
Clinical signs on presentation to hospital of patients with ARTI, who tested positive for KIV or WUV
| Presenting signs | KIV (N = 51) | WUV (N = 98) |
|---|---|---|
| Cough | 56% | 63% |
| Fever | 41% | 45% |
| Malaise | 39% | 32% |
| Bronchiolitis | 15% | 23% |
| Diarrhoea | 8% | 7% |
| Pneumonia | 0% | 2% |
3.4. Phylogenetic analysis of KIV and WUV
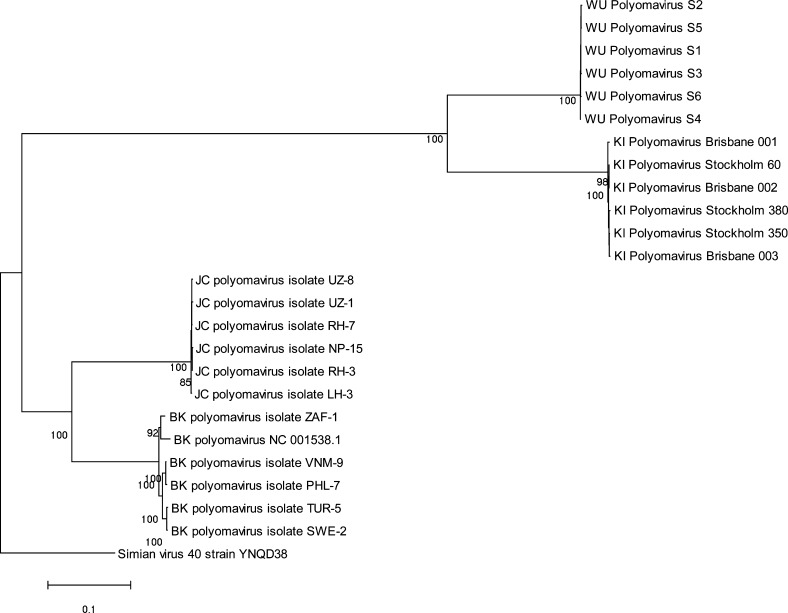
The results of phylogenetic analysis showed a high level of genome conservation for KIV, with 99.4–100% similarity at a nucleotide level between the strains detected in Australia (GenBank accession numbers EF520287–EF520289) compared to KIV sequences reported by the Swedish group. Similarly little sequence diversity was found amongst local WUV strains. Detailed analysis of the viral DNA sequence confirmed that KIV and WUV were closely related genetically, but were clearly divergent from BKV and JCV (Fig. 2 ).
Fig. 2.
Strain variation of KIV and WUV compared with JCV, BKV and SV40. Whole genome sequence was aligned using ClustalX. WUV strains were obtained from patients in Brisbane, Australia, and St. Louis, USA.
4. Discussion
This is the first study examining the association of two newly described human polyomaviruses, KIV and WUV, with illness in the human respiratory tract. In our sample of respiratory specimens collected from individuals presenting to a hospital emergency department for assessment or admission, KIV and WUV were found at a prevalence of 2.6% and 4.5%, respectively. It is not possible to prove a causal relationship between the detection of KIV and WUV and respiratory disease from these findings; this will require further more comprehensive studies, ideally involving those that satisfy Koch's postulates.
However, a number of features in the detection of these newly discovered viruses mean further exploration of a causal role in respiratory disease is warranted. KIV and WUV were present in respiratory secretions from patients with ARTI at rates similar to those reported for the recently described respiratory viruses, human coronavirus HKU1, NL63 (Lau et al., 2006), and HBoV (Kesebir et al., 2006). Neither of the viruses were detected in other sample sets consisting of urine from a variety of sources, and blood from immunocompromised children. This was contrary to observations for the other human polyomaviruses BKV and JCV, which are commonly present in clinical urine samples and blood specimens, particularly from immunocompromised patients (Rossi et al., 2007, Polo et al., 2004, Azzi et al., 1996). These results would suggest that the biology of KIV and WUV may be similar to each other, but quite different from BKV and JCV.
Determining the role of KIV and WUV as human respiratory pathogens is confounded by the high level of co-detection of these viruses with other respiratory agents, particularly HRV. This feature is very similar to observations reported for HBoV where the rates of co-detection have ranged from 33.3% (Naghipour et al., 2007) to 83.7% (Fry et al., 2007), and, although epidemiologically the virus is associated with pneumonia, it is postulated that infection and illness of HBoV may be dependent on co-infection with other viruses (Fry et al., 2007). Similarly, this may be true for infection and disease involving KIV and WUV.
The clinical features associated with KIV and WUV infection in our study population were not sufficiently distinctive to differentiate them from other respiratory viral infections in children. However, the majority of these patients, including those in whom KIV (25.3%) or WUV (20.3%) were the only pathogen detected, were sufficiently sick to require medical attention, thereby representing a significant burden on the health care system.
Phylogenetic analysis of KIV and WUV sequences confirmed that these viruses are genetically distinct. No evidence for the existence of viral genetic subtypes was found, but these observations are limited by the small number of specimens examined in this study. Given this, it would be prudent to perform phylogenetic analysis on a larger population of KIV and WUV before firm conclusions on circulating viral (sub) types may be proposed. Such studies are currently proceeding in our laboratory.
This preliminary study examines the presence of two newly described human polyomaviruses, KIV and WUV, in respiratory samples from patients with ARTI. Through more comprehensive studies, further data will need to be obtained before the role of these viruses as respiratory pathogens can be firmly established. It remains possible that KIV and WUV are not involved in respiratory disease and their presence in the respiratory tract simply reflects their mode of transmission. Establishing the association of these viruses with a particular disease will be challenging, but considering the oncogenic potential of polyomaviruses in mammals, the results of such studies may have important medical implications.
Acknowledgements
This study was supported by the Royal Children's Hospital Foundation Grant I 922-034, sponsored by the “Woolworths Fresh Futures” Appeal. All the necessary ethics approval for this study was obtained from the Institute's (Royal Children's Hospital) Ethics Committee. We wish to thank the staff of the Microbiology Division of the Pathology Queensland Central Laboratory.
References
- Allander T., Andreasson K., Gupta S., Bjerkner A., Bogdanovic G., Persson M.A. Identification of a third human polyomavirus. J Virol. 2007;81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi A., De Santis R., Ciappi S., Leoncini F., Sterrantino G., Marino N. Human polyomaviruses DNA detection in peripheral blood leukocytes from immunocompetent and immunocompromised individuals. J Neurovirol. 1996;2:411–416. doi: 10.3109/13550289609146907. [DOI] [PubMed] [Google Scholar]
- Bialasiewicz S., Whiley D.M., Lambert S.B., Gould A., Nissen M.D., Sloots T.P. Development and evaluation of real-time PCR assays for the detection of the newly identified KI and WU polyomaviruses. J Clin Virol. 2007;40:9–14. doi: 10.1016/j.jcv.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echavarria M., Forman M., Ticehurst J., Dumler J.S., Charache P. PCR method for detection of adenovirus in urine of healthy and human immunodeficiency virus-infected individuals. J Clin Microbiol. 1998;36:3323–3326. doi: 10.1128/jcm.36.11.3323-3326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Lu X., Chittaganpitch M., Peret T., Fischer J., Dowell S.F. Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J Infect Dis. 2007;195:1038–1045. doi: 10.1086/512163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor A.M., Nissen M.D., Whiley D.M., Mackay I.M., Lambert S.B., Wu G. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;4:e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesebir D., Vazquez M., Weibel C., Shapiro E.D., Ferguson D., Landry M.L. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis. 2006;194:1276–1282. doi: 10.1086/508213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Yip C.C., Tse H., Tsoi H.W., Cheng V.C. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Wang C.K., Brown J.B., Quinto J.D., Chu M., de Graaf M. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol. 2004;42:981–986. doi: 10.1128/JCM.42.3.981-986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghipour M., Cuevas L.E., Bakhshinejad T., Dove W., Hart C.A. Human bocavirus in Iranian children with acute respiratory infections. J Med Virol. 2007;79:539–543. doi: 10.1002/jmv.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo C., Perez J.L., Mielnichuck A., Fedele C.G., Niubo J., Tenorio A. Prevalence and patterns of polyomavirus urinary excretion in immunocompetent adults and children. Clin Microbiol Infect. 2004;10:640–644. doi: 10.1111/j.1469-0691.2004.00882.x. [DOI] [PubMed] [Google Scholar]
- Rossi A., Delbue S., Mazziotti R., Valli M., Borghi E., Mancuso R. Presence, quantitation and characterization of JC virus in the urine of Italian immunocompetent subjects. J Med Virol. 2007;79:408–412. doi: 10.1002/jmv.20829. [DOI] [PubMed] [Google Scholar]
- Sloots T.P., McErlean P., Speicher D.J., Arden K.E., Nissen M.D., Mackay I.M. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrmis M.W., Whiley D.M., Thomas M., Mackay I.M., Siebert D.J., Nissen M.D. A sensitive, specific and cost-effective multiplex reverse-transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J Mol Diagn. 2004;6:125–131. doi: 10.1016/S1525-1578(10)60500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]