Graphical abstract
RNA silencing suppression with combination of three gene slinging suppressors as well as 450 mM NaCl increased the production of Griffithsin.
Keywords: GRFT, HIV-1, Nicotiana benthamiana, E. coli, Agroinfiltration, Silencing suppressors
Highlights
-
•
HIV inhibitor expressed via three gene silencing suppressors.
-
•
The salt treatment increased the amount of Griffithsin production.
-
•
Anti-Hiv activity was confirmed via in vitro.
Abstract
The exploration of emerging host organisms for the economic and efficient production of protein microbicides against HIV is urgently needed in resource-poor areas worldwide. In this study, the production of the novel HIV entry inhibitor candidate, griffithsin (GRFT), was investigated using Nicotiana benthamiana as the expression platform based on a non-viral vector. To increase the yield of recombinant GRFT, the RNA silencing defense mechanism of N. benthamiana was abolished by using three gene silencing suppressors. A transient expression system was used by transferring the GRFT gene, which encodes 122 amino acids, under the control of the enhanced CaMV 35S promoter. The presence of correctly assembled GRFT in transgenic leaves was confirmed using immunoglobulin-specific sandwich ELISA. The data demonstrated that the use of three gene silencing suppressors allowed the highest accumulation of GRFT, with a yield of 400 μg g−1 fresh weight, and this amount was reduced to 287 μg g−1 after purification, representing a recovery of 71.75%. The analysis also showed that the ability of GRFT expressed in N. benthamiana to bind to glycoprotein 120 is close to that of the GRFT protein purified from E. coli. Whole-cell assays using purified GRFT showed that our purified GRFT was potently active against HIV. This study provides the first high-level production of the HIV-1 entry inhibitor griffithsin with a non-viral expression system and illustrates the robustness of the co-agroinfiltration expression system improved through the use of three gene silencing suppressors.
1. Introduction
Human immunodeficiency virus (HIV) infection has been considered a major threat to global public health since it was first deciphered in 1981 in the United States [1]. According to the World Health Organization, approximately 36.7 million people were living with HIV at the end of 2015 (WHO) (http://www.who.int/mediacentre/factsheets/fs360/en/), and HIV remains the leading cause of death in sub-Saharan Africa and was responsible for the death of 1.1 million people throughout the world in 2105 [World Health Organization; Global Health Observatory (GHO) data, The Top 10 Causes of Death; http://www.who.int/mediacentre/factsheets/fs310/en/ (accessed March 31, 2017)]. The current predictions suggest that 5753 people will become infected with HIV every day, and this corresponds to approximately 240 people every hour. Macrophages, T cells, and dendritic cells are the preferred destination of HIV. The viral glycoprotein 120 (gp120) binds to the CD4 receptor to initiate a series of conformational changes in gp120 that induce fusion of the virus with the host cell membrane. The viral infection progresses by transformation of the viral RNA genome into a proviral DNA, and the integrated provirus is eventually transcribed by cellular RNA polymerases into a set of mRNAs that generate progeny genomic mRNA and spliced mRNA encoding viral proteins [2].
The production and manufacturing of HIV entry inhibitors via recombinant DNA technology has been reported [3], but despite the discovery of potent HIV prophylactics, their high production costs have prevented the development of corresponding manufacturing processes in the resource-poor areas of the world. In this context, application of microbicides that would efficiently prohibit the initiation transmission of HIV virus is promising tool. As microbicides show significant potential for control and prevention of HIV transmission, they are alternative approach to prevent the heterosexual transmission of HIV [4]. The mechanism of protection against spread of sexually transmitted pathogens included in prohibition of viral replication, creation of physical barriers between HIV virus and cell membrane, boosting defense mechanism of vagina and cervix and etc. [5]. Griffithsin (GRFT) is a lectin HIV entry inhibitor that targets the terminal mannose residues on HIV N-linked glycan as long as the HIV is in contact with GRFT. GRFT was structurally characterized [[6], [7]]. The anti-HIV activity of GRFT derived from red algae has been estimated to have an EC50 value of 40 pM [[8], [9]]. The antiviral activity of GRFT, the fact that it lacks in vitro and in vivo toxicity, and its environmental stability, such as its stability in media with a broad range of pH values and temperatures, even temperatures close to the boiling point, indicate that GRFT is a good anti-HIV microbicide candidate [[10], [11]].
The large-scale production of high-quality GRFT is required for the development of this recombinant protein as an anti-HIV microbicide. As a result, plant and bacterial platforms have been used for the expression and accumulation of GRFT [[6], [12]]. The formation of insoluble inclusion bodies (33%), the high manufacturing cost and the presence of bacterial endotoxin have been considered factors that limit the production of GRFT using an E. coli system [13]. Moreover, the production of recombinant GRFT has been reported in Nicotiana benthamiana using a vector based on tobacco mosaic virus (TMV) [3] and transgenic seeds of Oryza sativa [14]. Regardless of these findings, the high cost of in vitro RNA transcription is a marked problem with virus-based system and also the purification of GRFT based on tobacco mosaic virus (TMV) is considered to show some contamination with TMV coat protein and protein degradation [6]. In addition, seed-based expression systems have been shown to exhibit a high degree of versatility regarding the production of recombinant proteins due to protein stability at ambient temperature [15], but the long-term process of seed production and space requirements might be preclusive [16]. In this study, we used a transient expression system based on a non-viral vector to improve the large-scale production of GRFT.
In comparison to stable expression, transient gene expression display considerable advantages in term of overall protein accumulation as well as an improved time of protein production, as protein will be extracted approximately 1–2 weeks after agroinfiltration [17]. The most efficient method for the high-level expression of heterologous protein is a transient expression system based on vectors derived from RNA plant virus due to the ability of RNA viruses to replicate to high titers within infected cells. As research and development of plant virus expression vectors progress, the means of introducing them to the host plant has also advanced [18]. However, this method cannot be used for the insertion of foreign genes without affecting replication and compromising the fidelity of the transcripts due to the lack of RNA-dependent RNA polymerases for proofreading and the movement of viral replicons throughout the plant, resulting in biocontamination problems and yielding undesirable features [[19], [20], [21]]. In this context, a recent significant development in the area of agroinfiltration is using “deconstructed” viral vectors. In this new type of vector, unnecessary viral genome components for the function of plasmid expression are removed, which leads to the assembly of larger transgenes while keeping viral replication and transcription. The MagnICON system represents an efficient and robust gene transferring technology for the transient expression of biopharmaceuticals in plant platforms. Using this system, the need for plasmid delivery based on complicated methods of generating RNA is eliminated. The MagnICON system provides an efficient system without functional infectious proteins because of the deletion of the CP gene, whereas the yield and speed of viral system are maintained [22].
Non-viral gene delivery systems were developed as an alternative to viral-based systems. One of the most important advantages of these systems is that they develop transfection [23]. Moreover, lower induction of the immune system and no limitation in terms of the size of transgenic DNA add versatility to this system. In addition to its speed and high yield, the transient expression platform presents versatility in terms of the expression and accumulation of personalized recombinant proteins, including therapeutics for patient-specific cancers and vaccines for viruses that show rapid antigenic drift and/or multiple strains with unpredictable outbreaks [[24], [25]].
However, restricted protein production based on transient expression system remains a significant drawback in term of economic production. In this context, the gene silencing is major limitation for expression of genes by Agrobacterium-mediated transient system as the foreign nucleic acids recognized and subsequently degraded through mRNA stability process or translational levels [26]. RNA silencing (also known as post-transcriptional gene silencing) is considered an adaptive defense mechanism by eukaryotic organisms whereby foreign transgenes and/or viruses eliminate through highly conserved mechanism. This mechanism starts with formation of the double stranded (ds)RNA and subsequently processing of dsRNA to small (s) 20–26-nt dsRNAs with staggered ends and finally terminate with inhibitions of selected sRNA strand within effector complexe RISC (RNA Induced Silencing Complex) acting on partially or fully complementary RNA or DNA [27]. However, this mechanism can be inhibited by gene silencing suppressors derived from viruses. In this case viruses are developed as beneficial tool to counteract gene silencing by viral protein [28]. RNA silencing suppression activity of viral protein of virus stems from its ability to bind siRNA as well as proteins involved in antiviral silencing ([[42], [43]]. The P19 suppressor from the Tomato bushy stunt virus (TBSV) has been widely used to boost transient or constitutive expression level of recombinant proteins in the plant as it selectively inhibits the 21-nt and 22-nt classes of siRNA and also reduce the availability of free siRNA for RISC loading [[58], [61]].
In the present study, a high production level of recombinant GRFT was obtained from transgenic plants seven days after gene delivery. Our transient expression significantly shortened the timeline of GRFT production, i.e., 7 days compared with 12 days, as was previously reported. Additionally, in comparison with a stable transgenic seed of O. sativa, our transient assays with agroinfiltration showed high levels of transgene expression. Therefore, the short timeline and higher production of GRFT based on our transient expression could be considered advantageous for GRFT development and commercialization.
2. Material and methods
2.1. Expression vector construction
A 369-bp sequence encoding GRFT (accession number FJ594069) was optimized using the N. benthamiana codon usage table, synthesized by Epoch Life Science, Inc. (Missouri City, TX, USA) and then cloned into the pBin61 plant expression vector under the control of the double-enhanced Cauliflower mosaic virus (CaMV) 35S promoter and the nopaline synthase (NOS) terminator sequence. A hexahistidine tag (His) was included to facilitate protein detection and purification from transgenic N. benthamiana leaves. The p35:GRFT construct was introduced into Agrobacterium tumefaciens strain GV3101 via electroporation [29].
2.2. Plant material and agroinfiltration
Plants of N. benthamiana were grown in controlled greenhouse conditions consisting of 25 °C/16-h light and 20 °C/8-h dark and 40–65% relative humidity for four to six weeks. At this stage, the plant has at least six fully developed true leaves and no visible flower buds. A. tumefaciens strains were grown in LB medium supplemented with 50 mg/L kanamycin, 50 mg/L gentamycin and 50 mg/L rifampicin overnight at 28 °C with shaking at 200–250 rpm. The following day, the A. tumefaciens strains were centrifuged at 5000 × g and 4 °C for 15 min, resuspended in induction medium containing 10 mM MgCl2, pH 5.7, and 100–200 μM acetosyringone to an OD600 of 0.5 and maintained at room temperature for 2–4 h before infiltration into the intercellular space of N. benthamiana leaves. Agrobacterium GV3101 cultures carrying the pBin61:GRFT gene and the silencing suppressors pBin61: P19, pBin61:P0 and pCambia1300:P1 were mixed at a ratio of 3:1 and infiltrated into plants.
2.3. Protein extraction
Agroinoculeted leaves were homogenized with Tissue Lyser Adapter Set 2 × 24 (QIAGEN, Germany) according to the protocol recommended with the kit. We modified the protocol for the preparation of protein extraction buffer that was previously described [30]. Briefly, 400 mg of grounded material was extracted with 2 ml of ice-cold 20% TCA/acetone supplemented with 20 mM DTT. The samples were maintained at −20 °C for 30 min and subsequently centrifuged at 16,000 rpm and 0 °C for 20 min. The supernatant was discarded, and the pellet was gently resuspended on 2 ml of ice-cold acetone supplemented with 20 mM DTT. The washing process with acetone was performed twice at −20 °C for 30 min, with centrifugation at 16,000 rpm and 0 °C for 20 min. Finally, the collected pellet that was deprived of chlorophyll was air-dried, and the total protein were extracted with 700 μl of protein extraction buffer (50 mM sodium phosphate (pH 7), 50 mM ascorbic acid, 10 mM di-sodium EDTA, and 1 mM PMSF). The protein concentration was assayed based on the method developed [31] using a SpectraMax® 190 Absorbance Plate Reader (Molecular Devices, USA).
2.4. SDS-PAGE and Western blot analysis
To identify the 121-amino-acid sequence of GRFT, 50 μg of soluble protein was resuspended in loading buffer containing 200 mM Tris (pH 6.8), 10% glycerol, 10% SDS, 10 mM dithiothreitol, and 0.05% bromophenol blue, and the samples were then heated at 95 °C for 10 min and separated by 16% glycine-SDS-PAGE at 0.02 mA and 80 V. The nitrocellulose membrane (Amersham Protran 0.45 NC 300 mm × 4 m, GE Healthcare Life Sciences) and filter papers were incubated in 5 ml of transfer buffer (125 mM Tris base, pH 8.3, and 960 mM tricin), 35 ml of H20 and 10 ml of methanol for 15. The proteins were subsequently transferred electrophoretically using a Trans-Blot® SD Semi-Dry Transfer Cell (Bio-Rad, USA) at a constant 2 mA/cm2 for 25 min. The proteins transferred to a nitrocellulose membrane were then blocked overnight at room temperature in 3% non-fat dry milk and Tris-buffered saline (TBS) (50 mM Tris and 150 mM NaCl, pH 7.4) containing 0.05% Tween (TBST). The following day, after three washes with TBST, the membrane was probed with primary anti-GRFT rabbit polyclonal antibody (National Cancer Institute-Frederick Cancer Research and Development Center, USA) at a dilution of 1:1000 for 4 h at room temperature and secondary horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody at a dilution of 1:5000 for 1 h at room temperature. Finally, the membrane was developed with developing buffer (CAS No. Nr77861, Bio-Rad, USA), buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM Kcl, 0.5 mM DTT and 0.05% NP-40, pH 7.9) and buffer B (5 mM HEPES, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 26% glycerol) prior to signal detection based on the manufacturer’s instructions.
2.5. Enzyme-linked immunosorbent assay (ELISA)
The accumulation of GRFT in transgenic plants was determined by ELISA using a previously described protocol [32]. Briefly, a 96-well plate was coated with 100 ng of the protein extracted from transgenic plants and incubated in carbonate/bicarbonate buffer (15 mM Na2CO3 and 35 mM NaHCO3, pH 9.6) overnight at 4 °C. The wells were rinsed with phosphate buffered saline PBS containing 0.1% Tween-20 (PBST) and then blocked with 1% bovine serum albumin (BSA) for 2 h at 37 °C. The plates were then incubated with the primary anti-GRFT rabbit polyclonal antibody at a dilution of 1:1000 for 2 h at 37 °C and then with the secondary horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody at dilution of 1:5000 for 1 h in 1% PBST. Finally, the substrate 3,30,5,50-tetramethylbenzidine (TMB) was added to the reaction, and the reaction was stopped with 5 M H2SO4. The absorbance was read at 450 nm.
2.6. Purification by affinity chromatography
Transgenic leaves of N. benthamiana were ground in liquid nitrogen and extracted at 4 °C into lysis buffer (50 mM sodium phosphate, 50 mM ascorbic acid, 10 mM disodium EDTA, and 1 mM PMSF, pH 7.4). The insoluble material was removed by centrifugation at 14,000 rpm and 4 °C for 30 min, and the extract was passed through a 0.45-μm filter and loaded onto a Profinity IMAC column at the rate of 1 ml/min. The column was washed with washing buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 7.4), and the GRFT protein was eluted from the column twice with elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 500 mM imidazole, pH 7.4). Fractions containing GRFT were mixed and concentrated by ultrafiltration using spin columns with a 10-kDa molecular weight cut-off (Amicon Ultra, EMD Millipore, Darmstadt, Germany). The purity of the concentrated GRFT was confirmed by SDS-PAGE and Western blot analysis. Additionally, to further assessment of NT-GRFT activity the protein sample was exchange into PBS buffer by ultrafiltration using spin columns with a 10-kDa molecular weight cut-off (Amicon Ultra, EMD Millipore, Darmstadt, Germany) based on manufacturing instruction.
2.7. In vitro gp120-binding assay
To detect the specific antigen-binding activity of GRFT, 100 ng of HIV-1 gp120 protein (group M, subtype CRF07_BC) (Sino Biological Inc., China) was coated onto 96-well ELISA plates, and the plate was incubated overnight at 4 °C. The plates were washed three times with PBST-0.05% and blocked with 1% bovine serum albumin (BSA). After washing with PBST-0.05%, serial dilutions of plant-produced GRFT were added to the plates, and the wells were subsequently incubated with GRFT anti-rabbit primary antibody (1:1000 in PBST) and HRP anti-rabbit secondary antibody (1:2000 in PBST). The plates were washed with PBST, and the TMB substrate was added to each well. The reaction was stopped with 5 M H2SO4, and the absorbance was read at 450 nm.
2.8. Whole-cell HIV neutralization assays
A whole-cell cytopathicity assay was conducted to evaluate the protective effect of the GRFT preparations as described previously, but modified for a 384-well assay plate format [33]. Briefly, a quantity of 2000 exponentially growing CEM-SS cells, maintained in RPM1 1640 medium (Lonza) without phenol red and supplemented with 5% fetal bovine serum (FBS) (Hyclone), 2 mM t-glutamine and 50 μg/ml gentamicin (Gibco), were combined with serial dilutions of GRFT in assay medium and incubated with or without HIV-1RF in a final volume of 50 μl. After six days incubation, cellular viability was assessed spectrophotometrically by measuring the reduction of XTT to the chromogenic formazan product at 450 nm. Experiments were carried out in quadruplicate. The tetrazolium reagent XTT was kindly supplied by the Developmental Therapeutics Program at the NCI-Frederick. The CEM-SS cell line was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: CEM-SS Cells from Dr. Peter L. Nara [[34], [35], [36]]. To evaluate functional activity of NT-GRFT-450 mM NaCl at ambient temperature, purified NT-GRFT-450 mM NaCl in PBS solution was subject to lyophilisation process by Alpha 1-4 LD plus freeze-dryer (Martin Christ, Germany) based on manufacturing instruction and whole -cell cytophathicity assay was conducted for lyophilized GRFT (Lyo-GRFT-450 mM NaCl).
3. Results
3.1. A combination of P19, P0 and P1 suppressors highly improves the expression efficiency
In this study, we investigated the effect of the use of three gene-silencing viral suppressors (P19, P0 and P1) with the syringe co-agroinfiltration method on the expression of GRFT protein. To demonstrate that, the widely and classically used P19 protein from TBSV was selected to boost gene expression level [[37], [38]]. We further selected the P1 suppressor from the Rice yellow mottlevirus (RYMV) [39] and the P0 suppressor from Beet western yellows virus (BWYMV) to inhibit the accumulation of 24 nt siRNA [[39], [40], [41]] and RISC activity on RNA silencing pathways [[42], [43]], respectively in combination of P19. To figure out that our transient expression system based on three gene-silencing suppressors could be successfully used to boost the expression level of GRFT, six-week-old N. benthamiana plants were infiltrated with A. tumefaciens strain GV3101 containing the pBIN61 vector harbouring the sequence encoding GRFT under the control of the double-enhanced Cauliflower mosaic virus (CaMV) 35S promoter (Fig. 1 ). The plants were also co-infiltrated with either three A. tumefaciens clones carrying the pBIN61:P19, pBIN61:P0 and pCambia1300:P1 gene silencing suppressors together and also separately. We also studied the expression efficiency obtained using the syringe co-agroinfiltration method with the addition of Tween-20 at concentration of 0.015% and 0.03% without any suppressor application. Leaves from the infiltrated plants were harvested at 7 days’ post-infiltration (d.p.i.), and the protein production level was analysed.
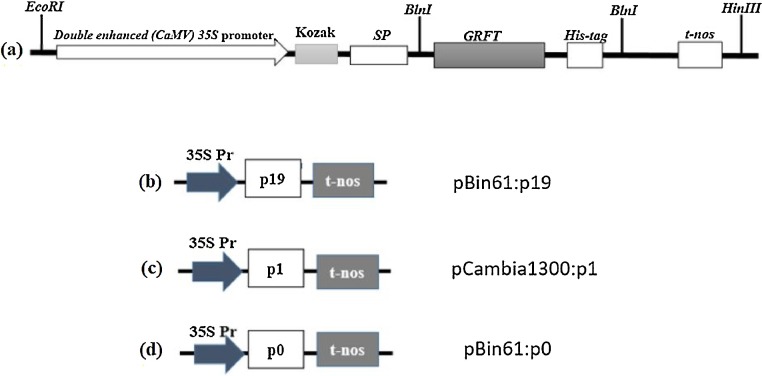
Fig. 1.
Schematic representation of the expression cassette and viral silencing suppressors used for the agroinfiltration of N. benthamiana leaves. (a) The expression cassette contained the double-enhanced Cauliflower mosaic virus (CaMV) 35S promoter (35S Pr), the Kozak consensus sequence to influence the initiation of the translation process, (SP) the signal peptide of the tobacco PR1a protein, the GRFT coding sequence, His-tag6, and the NOS terminator (t-NOS). (b) 35S Pr, (P19) Tomato bushy stunt virus P19 gene and (t-nos) NOS terminator (c) 35S Pr, (P1) Sweet potato feathery mottle virus P1 gene (t-nos) NOS terminator and (d) 35S Pr, (P0) the beet-infecting poleroviruses beet chlorosis virus (BChV) P0 gene and (t-nos) NOS terminator. All plasmids were based on the vector pBin61 backbone except P0, which was based on pCambia1300.EcoRI, BlnI and HindIII are restriction enzymes.
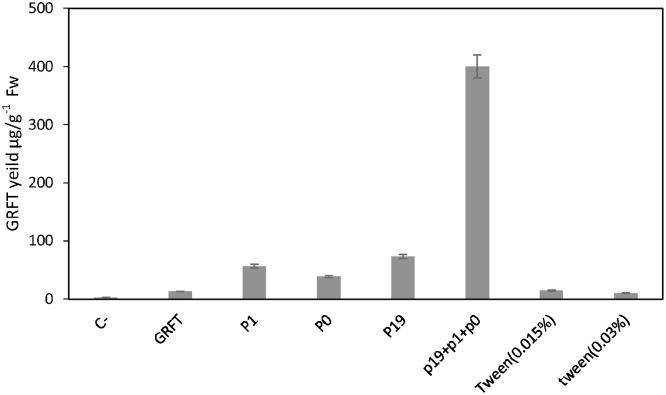
To detect the expression level of soluble GRFT, agroinfiltrated leaves (200 mg) were ground in liquid nitrogen for quantitative ELISA. The well-grounded leaves were then homogenized in PBS buffer supplemented with 1 mM PMSF as a protease inhibitor. The proteins extracted from the leaves were then centrifuged at 14,000 rpm and then quantified using the Bradford colourimetric assay [31]. The ELISAs included three biological replicas. Primary anti-GRFT rabbit polyclonal antibody (at a dilution of 1:1000) and the secondary horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody were used for detection. The ELISA results showed that the maximal expression level was obtained with the combination of three gene-silencing suppressors and demonstrated that the application of Tween-20 at different concentrations without any suppressors did not increase the expression efficiency, suggesting the importance of suppressor application in agroinfiltration (Fig. 2 ). We then excluded the plant extracts that expressed GRFT at a low level from the subsequent experiments.
Fig. 2.
Analysis of the effects of 0.015% and 0.03% Tween-20 separately and three gene silencing suppressors (p19 + p1 + p0) together and separately on syringe agroinfiltration efficiency by GRFT expression. C− = NT leaves without expression construct as negative control. GRFT = NT-GRFT without co-agroinfiltration with suppressors and Tween-20. Values are the average of three experiments ± SD.
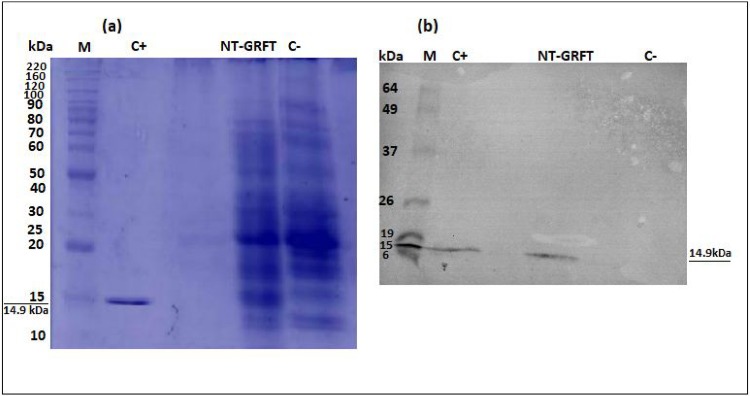
To confirm the presence of a band of the expected size (14.9 kDa) of GRFT, the plant extracts were analysed by SDS-PAGE and Western blotting analysis. The presence of GRFT with the size corresponding to that of His6‐tagged GRFT (14.9 kDa) was confirmed by Western blot analysis, showing that the soluble form of the protein was expressed by our transient expression system (Fig. 3 ).
Fig. 3.
Production and detection of anti-HIV inhibitor GRFT in N. benthamiana. The presence of expected recombinant GRFT was detected by western blot analysis. a) SDS page analysis of expressed GRFT in leaf of N. benthamiana harvested at 7 dpi. Lane M, 4 μl of the benchmark protein ladder (Thermo Scientific); lane C+ = GRFT expressed from E. coli as the positive control; Lane NT-GRFT = 30 μg of total soluble protein extracted from N. benthamiana plants agroinfiltrated with the pBIN61:GRFT construct and three gene-silencing suppressor proteins (P19, P0 and P1); and lane C− = 30 μg of total soluble protein extracted from N. benthamiana plants agroinfiltrated with pBIN61 construction and three gene-silencing viral suppressor proteins (P19, P0 and P1) as a control. b) Western blot analysis of expressed GRFT in N. benthamiana plants co-agroinfiltrated with three gene-silencing viral suppressor proteins. Lane M, BenchMark Pre-stained Protein Ladder (Thermo Scientific); lane C+ = GRFT expressed from E. coli as the positive control; Lane NT-GRFT = 30 μg of total soluble protein extracted from N. benthamiana plants agroinfiltrated with the pBIN61:GRFT construct and three gene-silencing viral suppressor proteins (P19, P0 and P1); and lane C−, 30 μg of total soluble protein extracted from N. benthamiana plants agroinfiltrated with pBIN61 construction and three gene-silencing viral suppressor proteins (P19, P0 and P1) as a control.
3.2. One step-protocol for purifying GRFT from crude extract
The recombinant GRFT protein was purified from the total soluble protein (TSP) of N. benthamiana using an immobilized metal affinity chromatography (IMAC) and the AKTA™ Prime Plus system (GE Healthcare Bio-Sciences, Uppsala, Sweden). The protein was captured with a 1-ml His Trap FF column (GE Healthcare Bio-Sciences, Uppsala, Sweden) and eluted with buffer containing 500 mm imidazole. The final yield of purified GRFT was 238 μg/g FW, which represent a recovery of 59.5%. The presence of purified GRFT was confirmed by SDS-PAGE and immunoblotting analysis (Fig. 4 ). The immunoblotting analysis results for purified GRFT in comparison with His6‐tagged GRFT revealed that the protein was correctly folded and of the correct size. No degradation was observed, indicating that the protein remained stable during upstream and downstream processes.
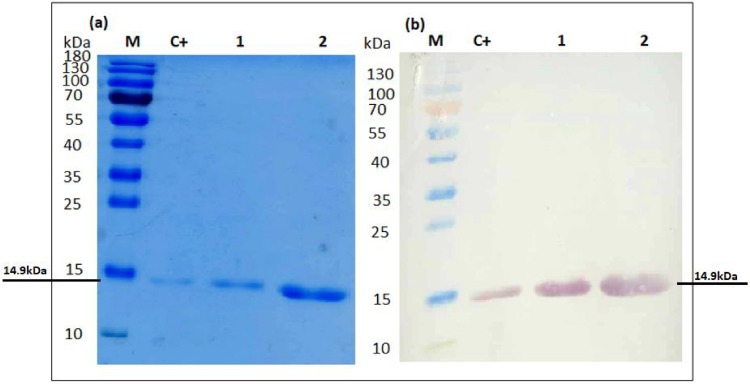
Fig. 4.
(a) Separation of purified NT-GRFT under denaturing condition by SDS-page showing a 14.9-kDa band representing GRFT. Lane M = 5 μl of the PageRuler Prestained Protein Ladder (Thermo Scientific); lane c+ = 500 ng of GRFT purified from E. coli, lane 1 = 1.5 μg of NT-GRFT; lane 2 = 3.5 μg of NT-GRFT; (b) Western blot analysis of expressed GRFT in N. benthamiana plants co-agroinfiltrated with three gene-silencing viral suppressor proteins. lane c+ = 500 ng of GRFT purified from E. coli; lane 1 = 1.5 μg of NT-GRFT; lane 2 = 3.5 μg of NT-GRFT.
3.3. Effect of salt concentration on GRFT extraction
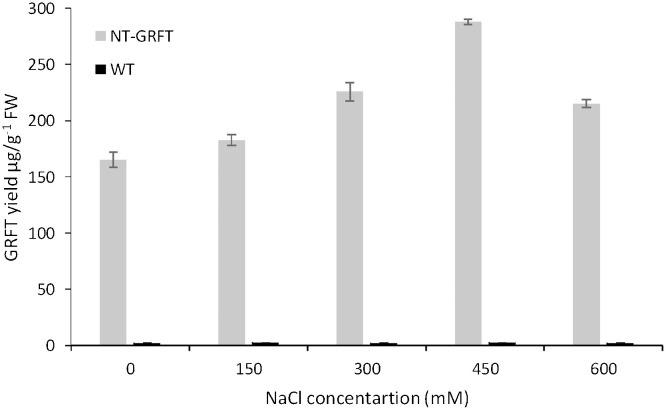
To study the effect of various salt concentrations on the recovery of GRFT, NaCl was added at different concentrations to the extraction buffer (50 mM sodium phosphate (pH 7), 50 mM ascorbic acid, 10 mM di-sodium EDTA, and 1 mM PMSF). As shown in Fig. 5 , increasing the concentration of NaCl to 450 mM improved the GRFT content from 238 μg/g FW to 287 μg/g FW, representing a recovery of 71.75%. However, the GRFT yield was decreased by the addition of salt at a concentration above 450 mM, suggesting that the optimal concentration of salt for GRFT recovery was 450 mM.
Fig. 5.
Effect of salt concentration in extraction buffer on GRFT recovery from TSP of N. benthamiana extracts. Wild type (WT) was used as negative control. The extraction buffer (pH 7.4) contained 50 mM sodium phosphate, 50 mM ascorbic acid, 10 mM di-sodium EDTA, 1 mM PMSF and different concentrations of NaCl. Values are the average of three experiments ± SD.
3.4. The purified GRFT showed gp120 binding activity comparable with that of GRFT produced in E. coli
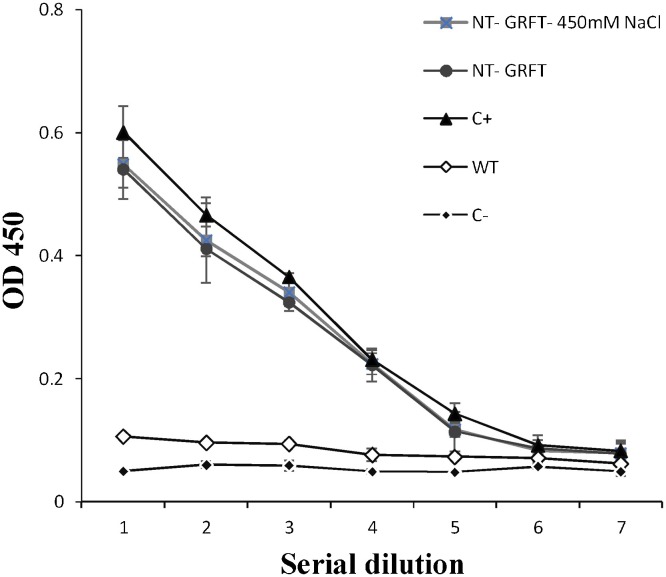
The in vitro binding activity of the purified GRFT from leaves of N. benthamiana harvested at 7 d.p.i. was analysed by ELISA against HIV gp120. In this experiment, purified GRFT from E. coli (EC-GRFT) and PBS were used as the positive and negative controls, respectively. The ELISA results showed that purified NT-GRFT had higher gp120-binding activity than the wild type as negative control, and the gp120-binding activity of NT-GRFT was consistent with that obtained with the same concentration of purified EC-GRFT, suggesting that its activity was nearly identical to that of GRFT produced in E. coli (Fig. 6 ). Our results demonstrate that the assembled GRFT protein that accumulated in leaves of N. benthamiana was functional.
Fig. 6.
Antigen-binding activity of crude extract from leaves of N. benthamiana harvested at 7 d.p.i. and purified GRFT from E. coli. To detect the specific antigen-binding activity of plant GRFT, plates were coated with 100 ng of HIV-1 gp120 protein, and bound of purified GRFT was detected with GRFT anti-rabbit primary antibody (1:1000 in PBST) and HRP anti-rabbit secondary antibody (1:2000 in PBST). NT-GRFT-450 mM NaCl = GRFT expressed by extraction buffer contained 450 mM NaCl; NT-GRFT = GRFT expressed by the pBIN6:GRFT cassette in N. benthamiana; C+ = GRFT expressed from E. coli as the positive control; WT = wild-type plants; C− = negative control (PBS). OD, optical density at 450 nm. Both purified NT-GRFT and NT- GRFT 450 mM NaCl showed higher binding activity compared with PBS as the negative control and WT. Values are the average of three experiments ± SD.
3.5. Purified GRFT showed potent activity against HIV in whole-cell HIV neutralization assays
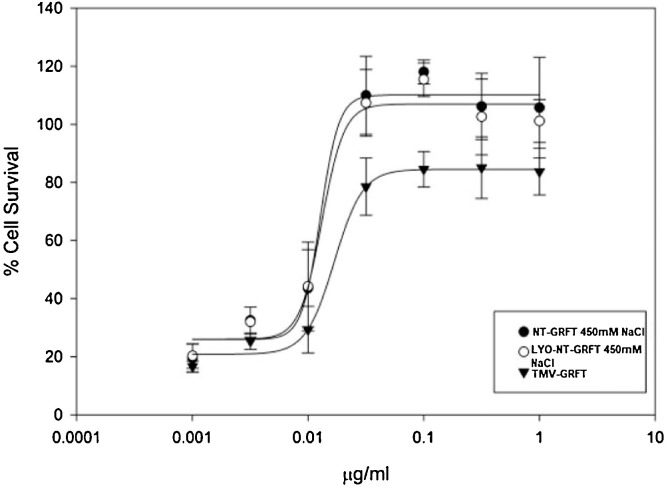
A whole-cell cytopathicity assay was conducted to evaluate the protective effect of the purified GRFT in aqueous solution and lyophilized form against HIV-1 in CEM-SS (Fig. 7 ). The result showed potent and similar concentration-dependent protection against HIV-induced cell death. The activity of purified NT-GRFT-450 mM NaCl (EC50 value of 0.0128 μg/ml or 0.9 nM) was comparable to that of GRFT protein produced using a TMV-based vector (TMV-GRFT; EC50 value of 0.0169 μg/ml or 1.3 nM) [49]). In addition, the activity of lyophilized NT-GRFT-450 mM NaCl (LYO-GRFT-450 mM NaCl; EC50 of 0.0167 μg/ml or 1.2 nM) against HIV-1 was comparable to both non-lyophilized NT-GRFT-450 mM NaCl and to TMV-based vector GRFT, suggesting the functional activity of GRFT can withstand the lyophilization process.
Fig. 7.
Anti-HIV activity of NT-GRFT-450 mM NaCl, Lyo-GRFTand TMV based vector GRFT. The activity of purified GRFT was analysed based on CEM-SS cellular viability infected with HIV-1RF. The activity of NT-GRFT was comparable to that of GRFT protein produced using a TMV-based vector [49]. In addition, the activity of lyophilized NT-GRFT-450 mM NaCl against HIV-1 was comparable to both non-lyophilized NT-GRFT-450 mM NaCl and to TMV-based vector GRFT. The cell viability was assessed using the XTT assay as described in the method section. All data points are averages (±SE) of quadruplicate (N = 4) measurements.
4. Discussion
The lectin griffithsin is one of the potent HIV inhibitory candidates isolated from red algae that effectively binds to the HIV envelope protein gp120 and prevents virus infection [[44], [45]]. A cellular analysis revealed that GRFT exerts inhibitory effects against HIV infection at picomolar concentrations (EC50 of ∼50 pM) [32]. With the aim of assessing the inhibitory activity of GRFT against HIV viral entry, the antiviral activity of GRFT against several glycosylated viruses, such as the murine herpes simplex virus type 2 [46], the Japanese encephalitis virus [47], the hepatitis C virus [48], and the coronavirus responsible for SARS [49], were demonstrated. GRFT shows no toxicity and no immunogenicity, is thermostable under a wide range of conditions, and has a low cost, and these features make GRFT an interesting candidate for the development of antiviral therapies [[3], [11], [50]]. The production and purification of native GRFT from red algae have been reported, but only a low concentration has been obtained [32]. Therefore, the large-scale production of GRFT using a plant platform might open up a new avenue for the commercialization of this protein as a potent HIV inhibitory candidate. A previous study demonstrated the production and purification of GRFT in the cytoplasm of an E. coli system [13]. The production of GRFT into inclusion bodies of E. coli is the main challenge in the development of an E. coli system for the large-scale production of GRFT because the use of various detergents and highly polymerized cycloamylose as a refolding agent or the co-expression of His-GRFT with chaperone components, such as DnaK-DnaJ-GrpE and/or GroEL-GroES, have not yielded sufficiently high recovery of biologically active GRFT [13]. The production of GRFT using N. benthamiana based on a viral vector and the O. sativa endosperm was previously investigated by [3] and [14], respectively. Even so, the purification of GRFT based on a viral vector showed contamination with the TMV coat protein [6], protein purification and protein degradation [14] because the protein produced was decreased from 1 mg/g FW to 0.3 mg/g after purification. Although a seed-based expression system has been shown to exhibit a high degree of versatility for the production of recombinant proteins due to protein stability at ambient temperature [15], potential cross-contamination, the long-term process of seed production, space requirements, unintentional mixing of transgenic and non-transgenic seeds, and public and food industry concerns regarding the production of biopharmaceuticals using food crops might preclude the implementation of this system [[16], [51]].
We investigated the production of GRFT in a non-viral-vector-based transient expression system using N. benthamiana as a convenient platform for the accumulation of biopharmaceuticals. This study provides the first demonstration of the expression and production of GRFT using a non-viral-vector-based transient expression system with N. benthamiana and three gene-silencing suppressors. A high level of recombinant GRFT was recovered from transgenic plants seven days after gene delivery. Thus, our transient expression significantly shortened the time for GRFT production compared with the previously reported timeline of 12 days. Additionally, in comparison to that of stable transgenic seeds of O. sativa, our transient expression with agroinfiltration yielded high levels of transgene expression. Therefore, the short timeline and higher production of GRFT obtained with our transient expression system could be considered advantageous for GRFT development and commercialization.
Transient gene expression based on agroinfiltration is an efficient, cost-effective and time-saving strategy for obtaining high amounts of recombinant proteins, as stable genetic transformation is a slow process and requires months or years to generate transgenic plants due to regeneration protocols. In addition, transient expression is genome integration-independent strategy, which will not be affected by position effects existing in stable transformation, once that the expression vector remains an episomal DNA molecule [22]. Our result is in accordance with previous works, which indicated that agroinfiltration could efficiently resulted in transferring of Agrobacterium into the plant leaves and robust expression of recombinant proteins [[24], [52], [53], [54]]. In our study, the infiltration of agrobacterium into the intercellular space of the leaf was performed with syringe method (Fig. 2b). Syringe infiltration has demonstrated as favorable tool that provides remarkable advantages such as simple procedure without requiring any specialized equipment, possibility of either infiltrating the whole leaf with one transgene construct or transferring multiple constructs into different areas of one leaf and multiple assessment in one leaf [[53], [55]]. However, syringe infiltration just can be used for production, purification and preclinical functional investigations of recombinant protein in laboratory scale and this method is not affordable in large scale production of recombinant protein. In this context, agroinfiltration with vacuum method was established to produce pharmaceutical protein in terms of scalability, economically and safety.
GRFT production was previously shown with a yield 819 mg/L, but the formation of insoluble inclusion bodies (33%), the low recovery of 66% soluble protein, the high manufacturing cost and the presence of bacterial endotoxins are considered limitation factors for the production of GRFT using an E. coli system [13]. Moreover, the production of recombinant GRFT has been reported in N. benthamiana [3] and O. sativa [14]. Even so, a recovery of 30% GRFT protein from N. benthamiana after purification, contamination with TMV coat protein [6], and degradation are considered some drawbacks. [14] achieved a yield of 301 μg/g dry seed weight, and this amount was reduced to 223 μg/g dry seed weight after purification, representing a recovery of 74%. In this case, as described above, the long-term process of seed production and space requirements might be preclusive [16].
Transient expression systems have proven to obtain several similar yields. For the transient expression of recombinant protein by agroinfiltration, bacterial suspension could be infiltrated into fresh tissue via vacuum or syringe infiltration, which allows the production of different biopharmaceutical proteins in a large scale and within a short time. However, the expression of recombinant protein could be notably decreased by post-transcriptional gene silencing (PTGS) in the plant host. In this work, we studied the addition of three gene-silencing viral suppressor proteins (P19, P0 and P1) combined with the syringe co-agroinfiltration method for high GRFT protein expression. We also assayed the effect of syringe co-agroinfiltration method with Tween-20 at concentrations of 0.015% and 0.03% and without any suppressor application on expression efficiency. The results illustrated herein demonstrated that leaves co-agroinfiltrated with the three gene-silencing suppressors exhibited the highest level of GRFT at 7 d.p.i. compared with the application of P19, P1 and P0 individually as well as in the absence of suppressors. Our results suggest that suppressing the post-transcriptional gene silencing mechanism with three gene silencing suppressors in N. benthamiana increased the number of GRFT gene transcripts and subsequently elevated the transient expression of recombinant GRFT.
RNA silencing is critical process involved in decreasing of foreign genes expression in plant system. In this context, using of plant virus encoding suppressors of RNA silencing is considered as an applicable strategy to counteract RNA silencing mechanism and subsequently boost protein expression content in plants [[37], [56], [57]]. Use of RNA silencing suppressors has been widely reported for increasing of recombinant proteins content in plant. In this context P19 from TBSV involved in siRNA sequestration has been gained more attention. The P19 protein is a strong silencing suppressor protein and can selectively inhibits the 21-nt and 22-nt classes of siRNA [58]. The P1 protein from Rice Yellow Mottle Virus has own mechanism to inhibit the accumulation of the 22-nt and 24-nt siRNA [59]. P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation [60]. Our result is similar to that of Lacombe et al. [61], who recently showed the effect of the combination of P19, P1 and P0 to enhance expression levels of anti-leishmaniasis vaccine candidate by transient expression system in N. benthamiana. The synergetic effect of P0 and P1 on RNA silencing suppression was shown in combination with P19. It is hypothesized that the P1suppressor could affect system RNA silencing suppression via inhibition of 24 nts siRNA production. As 24 nts siRNA might induce the formation of systemic RNA silencing signal and the combination of the P1 suppressor with P19 and P0 would strongly affect the RNA gene silencing, and consequently increase recombinant protein production in plant not only locally but also on systemic level through viral based tool [61]. Based on our data, we can speculate that combination of P19, P0, and P1 suppressors could be efficiently applied to boost expression level of other recombinant protein by transient expression in N. benthamiana. However, the use of the three P19, P0, and P1 gene suppressors together might not be recommended for stable transgenic plants since the combination of gene silencing suppressors could trigger harmful developmental damage on plant cell [[61], [62]].
The effect of Tween-20 on GRFT production through syringe agroinfiltration was investigated. In the present study, the application of Tween-20 did not improve the expression efficiency. Our result contrasts those reported by Zhao et al. [63], who found that Tween-20 increased GUS expression by influencing the agroinfiltration efficiency. Co-agroinfiltration with Tween-20 caused fast tissue necrosis and cell death, potentially due to the induction of damage by Tween 20 to plant cells in the agroinfiltration system.
To confirm the expression of GRFT by our transient expression system, SDS-PAGE and immunoblotting analyses were performed. The results demonstrated the specific band of His-tagged GRFT, representing the expression of GRFT under the control of the strong CaMV 35S promoter and in the presence of gene silencing suppressors. Our results are in agreement with those reported previously [[3], [14]]. Vamvaka et al. [14] recently showed the results of an immunoblotting analysis for GRFT detection, which showed a pair of intense bands at 14.6 kDa and 16–17 kDa, and concluded that incomplete removal of the rice α‐amylase (RAmy3D) signal peptide might have resulted in the presence of additional band at 16–17 kDa. The result of our immunoblotting using primary anti-GRFT rabbit polyclonal antibody did not show this additional band under denaturing conditions, suggesting complete removal of the signal peptide used in our transient expression system.
Purification of the His-tagged GRFT was performed by one-step purification through affinity chromatography. The purification of GRFT was performed under native conditions and without ammonium sulphate precipitation. We first obtained a calculated yield of 238 μg/g FW, which represent a recovery of 59.5%. The number and nature of a purification step are considered significant factors affecting the viability of recombinant proteins produced in plants [14]. The GRFT produced based on a viral vector in N. benthamiana decreased from 1 mg/g to 300 μg/g after purification, representing 30% recovery. The presence of the TMV vector coat protein required further purification to remove the coat protein, resulting in a low recovery of GRFT [3]. Vamvaka et al. [14] demonstrated that protein extraction using a phosphate buffer combined with IMAC achieved 74% recovery of GRFT expressed in the rice endosperm based on a stable expression system, and this value is slightly higher than the recovery obtained using our system. To study the effect of the salt concentration on GRFT recovery, we added different concentrations of NaCl to the extraction buffer, and the results clearly illustrated that the yield of GRFT was significantly increased by the addition of salt at a concentration of 450 mM. After salt treatment, the yield of GRFT increased from 238 μg/g to 287 μg/g, representing a recovery of 71.75%. This result is in agreement with Mayani et al. [64], who showed that the addition of salt at a concentration of 450 mM to the extraction buffer can improve the recovery of recombinant protein in N. benthamiana. The protein extraction process is as essential step of the recovery process because it tailors the total extract volume, concentrations and purity of the recombinant protein as well as the isolation of the desired protein from impurities or/and contaminations that should be removed during the purification process [[65], [66]]. Our data demonstrate that protein recovery increases after salt treatment due to the combination of a reduction in electrostatic interactions between the recombinant protein and plant protein components, such as cellulose that carry a negative charge, and/or increases in osmotic pressure in the extraction buffer, which would result in the dehydration of tissue components [64]. However, increasing the salt concentration in the extraction buffer to a concentration above 450 mM decreased the GRFT yield. It is possible that the solubility of GRFT was reduced at a high salt concentration, which would subsequently increase the hydrophobic interactions with plant tissue components, and this mechanism can feasibly occur in the presence of a higher concentration of salt. Our results demonstrate that optimization of the early steps in the recovery and purification processes are crucial for obtaining viable plant-based recombinant protein production. We used a non-viral-vector-based transient expression system to eliminate the step for purifying the expressed GRFT. The expression of GRFT in the non-viral vector was prepared under non-denaturing conditions to maintain the native form of GRFT and thus avoid an additional refolding step. The GRFT protein expressed by our system was obtained at a yield of 287 μg/g FW, which is higher than that obtained in transgenic seed rice (223 μg/g dry seed weight). Our transient expression system significantly shortened the timeline of GRFT production compared with the previously reported timeline of 12 days. Therefore, the short timeline and the higher production of GRFT obtained with our transient expression could be considered advantageous toward GRFT development and commercialization.
GRFT is a potent HIV inhibitor candidate that tightly binds to high mannose-saccharides on the surface of the glycoproteins gp120, gp41 and gp160 and efficiently inhibits virus infection [[45], [48]]. GRFT has been shown to bind the HIV glycoprotein by blocking CD4 binding as well as by binding other anti-HIV antibodies [67]. GRFT has a smaller recognition epitope and lower binding stoichiometry compared to other HIV lectin. The suitable content of GRFT for binding to a single gp120 glycoprotein has been reported approximately to equal 10 GRFT units [49]. In this study, we therefore analysed the biological activity NT-GRFT- 450 mM NaCl protein exhibits a comparable EC50 value of 0.9 nM to tobacco-produced GRFT transduced with a viral vector (TMV-GRFTEC50 1.3 nM). Our analysis showed that the ability of GRFT expressed in N. benthamiana to bind to gp120 was close to that of the protein purified from E. coli. Similarly, O’Keefe et al. and Vamvaka et al. [[3], [14]] showed that the binding characteristics of plant-based GRFT exhibited similar or even better gp120-binding activity than GRFT expressed in E. coli.
We also estimated the anti-HIV activity of GRFT through a whole-cell HIV cytopathicity assay. We found that GRFT produced in E. coli presented whole-cell HIV cytopathicity with an EC50 value of 0.089 nM as well as EC50 values of 0.054 nM for native GRFT. Additionally, O’Keefe et al. [3] reported an EC50 value of 0.156 nM for GRFT. But in our experiments, we report the anti-HIV activity of TMV-GRFT at an EC50 value of 1.3 nM. In this case, the discrepancy in EC50 values is the result of reformatting the whole-cell neutralization assay from a 96-well to a 384-well higher throughput format, as described in the material and methods section. Recently, an EC50 value of 0.27 was obtained for GRFT expressed in the rice endosperm. The differences among the EC50 values obtained in these studies reflect the variability of the syncytium inhibition assay, which is used as a comparison tool in an experiment. In vivo safety and efficacy are two critical issues related to potential anti-HIV microbicide activity. The HIV neutralization activity observed in this study confirmed that GRFT is correctly folded in plants and maintains its biological activity.
Physico-chemical modifications such as modification of amino acid side chain in aqueous solution can extremely affect stability and/or functional activity of protein, and subsequently influence the risk of adverse side effect in term of protein drug. Therefore, the inherent instability protein-based pharmaceuticals as well as product storage and shipping condition preclude the preparation of protein as shipping and storing product at controlled condition are not technical and economically practicable [68]. Lyophilisation process was used to prepare dehydrated GRFT. In this context lyophilized GRFT showed functional activity in room temperature. This characteristic allow the product to be handled and stored conveniently to wider market.
5. Conclusion
In conclusion, here series of experiments were carried out to boost GRFT transient production in N. benthamiana leaves, and to demonstrate that this protein maintained its immunogenic properties. In this study, RNA silencing suppression with combination of three gene silencing suppressors as well as the effect of the salt concentration on GRFT recovery were performed to establish the most effective situation to effectively accumulate the recombinant immunogenic GRFT protein in a rapid, efficient, and low-cost way for further its development and commercialization.
Author contribution statement
Peyman habibi conceived and designed research, analyzed data, acquisition of data, conducted experiments, and wrote the manuscript. Carlos Ricardo Soccol contributed to manuscript. Barry R. O’Keefe directed anti-HIV and analytical experimentation, analyzed data and contributed to the manuscript. Lauren R.H. Krumpe analyzed data and contributed to figure preparation and the manuscript. Jennifer Wilson conducted anti-HIV assay and contributed to data analysis. Leonardo Lima Pepino de Macedo, Muhammad Faheem, Vanessa Olinto Dos Santos and Guilherme Souza Prado contributed to analytical expriments. Marco Antonio Botelho contributed to manuscript. Severine Lacombe supported the project with vectors harboring gene-silencing suppressors, Maria Fatima Grossi-de- Sa directed experiments, analyzed data and contributed to the manuscript.
Acknowledgments
The authors would like to acknowledge EMBRAPA, CNPq, CAPES, and FAP-DF for the funding and support. The authors report no conflicts of interest. This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research.
Contributor Information
Peyman Habibi, Email: peymanhabibi@ufpr.br.
Maria Fatima Grossi-de-Sa, Email: fatima.grossi@embrapa.br.
References
- 1.Barre-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vezinet-Brun F., Rouzioux C., Rozenbaum W., Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science (New York, N.Y.) 1983;220(4599):868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Shors T. second ed. Jones and Bartlett Learning; Wisconsin: 2011. Understanding Viruses. [Google Scholar]
- 3.O'Keefe B.R., Vojdani F., Buffa V., Shattock R.J., Montefiori D.C., Bakke J., Mirsalis J., d'Andrea A.L., Hume S.D., Bratcher B., Saucedo C.J., McMahon J.B., Pogue G.P., Palmer K.E. Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc. Natl. Acad. Sci. U. S. A. 2009;106(15):6099–6104. doi: 10.1073/pnas.0901506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shattock R.J., Rosenberg Z. Microbicides: topical prevention against HIV. Cold Spring Harb. Perspect. Med. 2012;2(2):a007385. doi: 10.1101/cshperspect.a007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez-Sanchez M.P., Martin-Illana A., Ruiz-Caro R., Bermejo P., Abad M.J., Carro R., Bedoya L.M., Tamayo A., Rubio J., Fernandez-Ferreiro A., Otero-Espinar F., Veiga M.D. Chitosan and kappa-carrageenan vaginal acyclovir formulations for prevention of genital herpes. In Vitro Ex Vivo Eval. Mar. Drugs. 2015;13(9):5976–5992. doi: 10.3390/md13095976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuqua J.L., Hamorsky K., Khalsa G., Matoba N., Palmer K.E. Bulk production of the antiviral lectin griffithsin. Plant Biotechnol. J. 2015;13(8):1160–1168. doi: 10.1111/pbi.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moulaei T., Shenoy S.R., Giomarelli B., Thomas C., McMahon J.B., Dauter Z., O'Keefe B.R., Wlodawer A. Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure (London, England: 1993) 2010;18(9):1104–1115. doi: 10.1016/j.str.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veazey R.S., Ketas T.A., Klasse P.J., Davison D.K., Singletary M., Green L.C., Greenberg M.L., Moore J.P. Tropism-independent protection of macaques against vaginal transmission of three SHIVs by the HIV-1 fusion inhibitor T-1249. Proc. Natl. Acad. Sci. U. S. A. 2008;105(30):10531–10536. doi: 10.1073/pnas.0802666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veazey R.S., Shattock R.J., Pope M., Kirijan J.C., Jones J., Hu Q., Ketas T., Marx P.A., Klasse P.J., Burton D.R., Moore J.P. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 2003;9(3):343–346. doi: 10.1038/nm833. [DOI] [PubMed] [Google Scholar]
- 10.Emau P., Tian B., O'Keefe R.B., Mori T., McMahon J.B., Palmer K.E., Jiang Y., Bekele G., Tsai C.C. Griffithsin, a potent HIV entry inhibitor, is an excellent candidate for anti-HIV microbicide. J. Med. Primatol. 2007;36(4–5):244–253. doi: 10.1111/j.1600-0684.2007.00242.x. [DOI] [PubMed] [Google Scholar]
- 11.Kouokam J.C., Huskens D., Schols D., Johannemann A., Riedell S.K., Walter W., Walker J.M., Matoba N., O'Keefe B.R., Palmer K.E. Investigation of griffithsin’s interactions with human cells confirms its outstanding safety and efficacy profile as a microbicide candidate. PLoS One. 2011;6(8):e22635. doi: 10.1371/journal.pone.0022635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vafaee Y., Staniek A., Mancheno-Solano M., Warzecha H. A modular cloning toolbox for the generation of chloroplast transformation vectors. PLoS One. 2014;9(10):e110222. doi: 10.1371/journal.pone.0110222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giomarelli B., Schumacher K.M., Taylor T.E., Sowder R.C., 2nd, Hartley J.L., McMahon J.B., Mori T. Recombinant production of anti-HIV protein, griffithsin, by auto-induction in a fermentor culture. Protein Expr. Purif. 2006;47(1):194–202. doi: 10.1016/j.pep.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Vamvaka E., Arcalis E., Ramessar K., Evans A., O’Keefe B.R., Shattock R.J., Medina V., Stoger E., Christou P., Capell T. Rice endosperm is cost-effective for the production of recombinant griffithsin with potent activity against HIV. Plant Biotechnol. J. 2016;14(6):1427–1437. doi: 10.1111/pbi.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obembe O.O., Popoola J.O., Leelavathi S., Reddy S.V. Advances in plant molecular farming. Biotechnol. Adv. 2011;29(2):210–222. doi: 10.1016/j.biotechadv.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Boothe J., Nykiforuk C., Shen Y., Zaplachinski S., Szarka S., Kuhlman P., Murray E., Morck D., Moloney M.M. Seed-based expression systems for plant molecular farming. Plant Biotechnol. J. 2010;8(5):588–606. doi: 10.1111/j.1467-7652.2010.00511.x. [DOI] [PubMed] [Google Scholar]
- 17.Leuzinger K., Dent M., Hurtado J., Stahnke J., Lai H., Zhou X., Chen Q. Efficient agroinfiltration of plants for high-level transient expression of recombinant proteins. J. Visualized Exp. JoVE. 2013;(77) doi: 10.3791/50521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hefferon K. Plant virus expression vector development: new perspectives. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/785382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahlquist P., Schwartz M., Chen J., Kushner D., Hao L., Dye B.T. Viral and host determinants of RNA virus vector replication and expression. Vaccine. 2005;23(15):1784–1787. doi: 10.1016/j.vaccine.2004.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro C., Arnold J.J., Cameron C.E. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 2005;107(2):141–149. doi: 10.1016/j.virusres.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sainsbury F., Lomonossoff G.P. Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 2008;148(3):1212. doi: 10.1104/pp.108.126284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habibi P., Prado G.S., Pelegrini P.B., Hefferon K.L., Soccol C.R., Grossi-de-Sa M.F. Optimization of inside and outside factors to improve recombinant protein yield in plant. Plant Cell Tissue Organ Cult. (PCTOC) 2017;130(3):449–467. [Google Scholar]
- 23.Cevher E., Sezer A.D., Çağlar E.S.E. Recent Advances in Novel Drug Carrier Systems. InTech; 2012. Gene delivery systems: recent progress in viral and non-viral therapy. [Google Scholar]
- 24.Chen Q., Lai H. Gene delivery into plant cells for recombinant protein production. BioMed Res. Int. 2015;2015:10. doi: 10.1155/2015/932161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nayerossadat N., Maedeh T., Ali P.A. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012;1:27. doi: 10.4103/2277-9175.98152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansen L.K., Carrington J.C. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 2001;126 doi: 10.1104/pp.126.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brodersen P., Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22(5):268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Incarbone M., Dunoyer P. RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 2013;18(7):382–392. doi: 10.1016/j.tplants.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Mersereau M., Pazour G.J., Das A. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene. 1990;90(1):149–151. doi: 10.1016/0378-1119(90)90452-w. [DOI] [PubMed] [Google Scholar]
- 30.Sarnighausen E., Wurtz V., Heintz D., Van Dorsselaer A., Reski R. Mapping of the Physcomitrella patens proteome. Phytochemistry. 2004;65(11):1589–1607. doi: 10.1016/j.phytochem.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 31.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Mori T., O’Keefe B.R., Sowder R.C., 2nd, Bringans S., Gardella R., Berg S., Cochran P., Turpin J.A., Buckheit R.W., Jr., McMahon J.B., Boyd M.R. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia spp. J. Biol. Chem. 2005;280(10):9345–9353. doi: 10.1074/jbc.M411122200. [DOI] [PubMed] [Google Scholar]
- 33.Gulakowski R.J., McMahon J.B., Staley P.G., Moran R.A., Boyd M.R. A semiautomated multiparameter approach for anti-HIV drug screening. J. Virol. Methods. 1991;33(1–2):87–100. doi: 10.1016/0166-0934(91)90010-w. [DOI] [PubMed] [Google Scholar]
- 34.Foley G.E., Lazarus H., Farber S., Uzman B.G., Boone B.A., McCarthy R.E. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer. 1965;18(4):522–529. doi: 10.1002/1097-0142(196504)18:4<522::aid-cncr2820180418>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 35.Nara P., Fischinger P. Quantitative infectivity assay for HIV-1 and-2. Nature. 1988;332(6163):469–470. doi: 10.1038/332469a0. [DOI] [PubMed] [Google Scholar]
- 36.Nara P., Hatch W., Dunlop N., Robey W., Arthur L., Gonda M., Fischinger P. Simple, rapid, quantitative, syncytium-forming microassay for the detection of human immunodeficiency virus neutralizing antibody. AIDS Res. Hum. Retroviruses. 1987;3(3):283–302. doi: 10.1089/aid.1987.3.283. [DOI] [PubMed] [Google Scholar]
- 37.Garabagi F., Gilbert E., Loos A., McLean M.D., Hall J.C. Utility of the P19 suppressor of gene-silencing protein for production of therapeutic antibodies in Nicotiana expression hosts. Plant Biotechnol. J. 2012;10(9):1118–1128. doi: 10.1111/j.1467-7652.2012.00742.x. [DOI] [PubMed] [Google Scholar]
- 38.Lombardi R., Circelli P., Villani M.E., Buriani G., Nardi L., Coppola V., Bianco L., Benvenuto E., Donini M., Marusic C. High-level HIV-1 Nef transient expression in Nicotiana benthamiana using the P19 gene silencing suppressor protein of Artichoke Mottled Crinckle Virus. BMC Biotechnol. 2009;9(1):96. doi: 10.1186/1472-6750-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siré C., Bangratz-Reyser M., Fargette D., Brugidou C. Genetic diversity and silencing suppression effects of Rice yellow mottle virus and the P1 protein. Virol. J. 2008;5(1):55. doi: 10.1186/1743-422X-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamilton A., Voinnet O., Chappell L., Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21(17):4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacombe S., Bangratz M., Vignols F., Brugidou C. The rice yellow mottle virus P1 protein exhibits dual functions to suppress and activate gene silencing. Plant J. 2010;61(3):371–382. doi: 10.1111/j.1365-313X.2009.04062.x. [DOI] [PubMed] [Google Scholar]
- 42.Baumberger N., Tsai C.-H., Lie M., Havecker E., Baulcombe D.C. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 2007;17(18):1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 43.Bortolamiol D., Pazhouhandeh M., Marrocco K., Genschik P., Ziegler-Graff V. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 2007;17(18):1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 44.Moulaei T., Alexandre K.B., Shenoy S.R., Meyerson J.R., Krumpe L.R., Constantine B., Wilson J., Buckheit R.W., Jr., McMahon J.B., Subramaniam S., Wlodawer A., O'Keefe B.R. Griffithsin tandemers: flexible and potent lectin inhibitors of the human immunodeficiency virus. Retrovirology. 2015;12:6. doi: 10.1186/s12977-014-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue J., Hoorelbeke B., Kagiampakis I., Demeler B., Balzarini J., Liwang P.J. The griffithsin dimer is required for high-potency inhibition of HIV-1: evidence for manipulation of the structure of gp120 as part of the griffithsin dimer mechanism. Antimicrob. Agents Chemother. 2013;57(8):3976–3989. doi: 10.1128/AAC.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nixon B., Fakioglu E., Stefanidou M., Wang Y., Dutta M., Goldstein H., Herold B.C. Genital herpes simplex virus type 2 infection in humanized HIV-transgenic mice triggers HIV shedding and is associated with greater neurological disease. J. Infect. Dis. 2014;209(4):510–522. doi: 10.1093/infdis/jit472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishag H.Z., Li C., Huang L., Sun M.X., Wang F., Ni B., Malik T., Chen P.Y., Mao X. Griffithsin inhibits Japanese encephalitis virus infection in vitro and in vivo. Arch. Virol. 2013;158(2):349–358. doi: 10.1007/s00705-012-1489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meuleman P., Albecka A., Belouzard S., Vercauteren K., Verhoye L., Wychowski C., Leroux-Roels G., Palmer K.E., Dubuisson J. Griffithsin has antiviral activity against hepatitis C virus. Antimicrob. Agents Chemother. 2011;55(11):5159–5167. doi: 10.1128/AAC.00633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Keefe B.R., Giomarelli B., Barnard D.L., Shenoy S.R., Chan P.K., McMahon J.B., Palmer K.E., Barnett B.W., Meyerholz D.K., Wohlford-Lenane C.L., McCray P.B., Jr. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 2010;84(5):2511–2521. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barton C., Kouokam J.C., Lasnik A.B., Foreman O., Cambon A., Brock G., Montefiori D.C., Vojdani F., McCormick A.A., O’Keefe B.R., Palmer K.E. Activity of and effect of subcutaneous treatment with the broad-spectrum antiviral lectin griffithsin in two laboratory rodent models. Antimicrob. Agents Chemother. 2014;58(1):120–127. doi: 10.1128/AAC.01407-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilken L.R., Nikolov Z.L. Recovery and purification of plant-made recombinant proteins. Biotechnol. Adv. 2012;30(2):419–433. doi: 10.1016/j.biotechadv.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 52.Abdelghani M., El-Heba G.A., Abdelhadi A.A., Abdallah N.A. Expression of synthetic human tropoelastin (hTE) protein in Nicotiana tabacum. GM Crops Food. 2015;6(1):54–62. doi: 10.1080/21645698.2015.1026524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Q., Lai H., Hurtado J., Stahnke J., Leuzinger K., Dent M. Agroinfiltration as an effective and scalable strategy of gene delivery for production of pharmaceutical proteins. Adv. Tech. Biol. Med. 2013;1(1):103. doi: 10.4172/atbm.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leuzinger K., Dent M., Hurtado J., Stahnke J., Lai H., Zhou X., Chen Q. Efficient agroinfiltration of plants for high-level transient expression of recombinant proteins. J. Visualized Exp. JoVE. 2013;(77) doi: 10.3791/50521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaghchhipawala Z., Rojas C.M., Senthil-Kumar M., Mysore K.S. Agroinoculation and agroinfiltration: simple tools for complex gene function analyses. Methods Mol. Biol. (Clifton, N.J.) 2011;678:65–76. doi: 10.1007/978-1-60761-682-5_6. [DOI] [PubMed] [Google Scholar]
- 56.Lombardi R., Circelli P., Villani M.E., Buriani G., Nardi L., Coppola V., Bianco L., Benvenuto E., Donini M., Marusic C. High-level HIV-1 Nef transient expression in Nicotiana benthamiana using the P19 gene silencing suppressor protein of Artichoke Mottled Crinckle Virus. BMC Biotechnol. 2009;9 doi: 10.1186/1472-6750-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vézina L.P., Faye L., Lerouge P., D’Aoust M.A., Marquet-Blouin E., Burel C., Lavoie P.O., Bardor M., Gomord V. Transient co-expression for fast and high-yield production of antibodies with human-like N-glycans in plants. Plant Biotechnol. J. 2009;7(5):442–455. doi: 10.1111/j.1467-7652.2009.00414.x. [DOI] [PubMed] [Google Scholar]
- 58.Arzola L., Chen J., Rattanaporn K., Maclean J.M., McDonald K.A. Transient co-expression of post-transcriptional gene silencing suppressors for increased in planta expression of a recombinant anthrax receptor fusion protein. Int. J. Mol. Sci. 2011;12(8):4975–4990. doi: 10.3390/ijms12084975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li F., Ding S.W. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fusaro A.F., Correa R.L., Nakasugi K., Jackson C., Kawchuk L., Vaslin M.F., Waterhouse P.M. The Enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology. 2012;426(2):178–187. doi: 10.1016/j.virol.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 61.Lacombe S., Bangratz M., Brizard J.P., Petitdidier E., Pagniez J., Sereme D., Lemesre J.L., Brugidou C. Optimized transitory ectopic expression of promastigote surface antigen protein in Nicotiana benthamiana, a potential anti-leishmaniasis vaccine candidate. J. Biosci. Bioeng. 2017;125(1):116–123. doi: 10.1016/j.jbiosc.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Saxena P., Hsieh Y.C., Alvarado V.Y., Sainsbury F., Saunders K., Lomonossoff G.P., Scholthof H.B. Improved foreign gene expression in plants using a virus-encoded suppressor of RNA silencing modified to be developmentally harmless. Plant Biotechnol. J. 2011;9(6):703–712. doi: 10.1111/j.1467-7652.2010.00574.x. [DOI] [PubMed] [Google Scholar]
- 63.Zhao H., Tan Z., Wen X., Wang Y. An improved syringe agroinfiltration protocol to enhance transformation efficiency by combinative use of 5-azacytidine, ascorbate acid and Tween-20. Plants. 2017;6(1):9. doi: 10.3390/plants6010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mayani M., McLean M.D., Christopher Hall J., Filipe C.D.M., Ghosh R. Recovery and isolation of recombinant human monoclonal antibody from transgenic tobacco plants. Biochem. Eng. J. 2011;54(2):103–108. [Google Scholar]
- 65.Hassan S., Van Dolleweerd C.J., Ioakeimidis F., Keshavarz-Moore E., Ma J.K.C. Considerations for extraction of monoclonal antibodies targeted to different subcellular compartments in transgenic tobacco plants. Plant Biotechnol. J. 2008;6(7):733–748. doi: 10.1111/j.1467-7652.2008.00354.x. [DOI] [PubMed] [Google Scholar]
- 66.Nikolov Z.L., Regan J.T., Dickey L.F., Woodard S.L. John Wiley & Sons, Inc.; 2008. Purification of Antibodies From Transgenic Plants, Process Scale Purification of Antibodies; pp. 387–406. [Google Scholar]
- 67.Lusvarghi S., Bewley C.A. Griffithsin an antiviral lectin with outstanding therapeutic potential. Viruses. 2016;8(10):296. doi: 10.3390/v8100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Avis K.E., Wu V.L. CRC Press; 1996. Biotechnology and Biopharmaceutical Manufacturing, Processing, and Preservation. [Google Scholar]