Abstract
Background: The detection of multiple viruses is important for pathogenic diagnosis and disease control. Microarray detection is a good method, but requires complex procedures for multiple virus detection.
Methods: We developed a novel PCR assay, the microarray-in-a-tube system, which integrates multiple PCR processes and DNA microarrays for multiple virus detection. A 5 × 5 oligonucleotide microarray for detecting 4 respiratory tract viruses (severe acute respiratory syndrome–associated coronavirus, influenza A virus, influenza B virus, and enterovirus) with inner controls was arranged on the inner surface of a specially designed Eppendorf cap with a flat, optically transparent window.
Results: We were able to perform all detection processes in the encapsulated system without opening the cap. The 4 viruses were successfully amplified by one-step reverse transcription–PCR in the encapsulated tube. After the PCR process, the microarray-in-a-tube was inverted, and the fluorescence-labeled PCR products were directly hybridized on the microarray. Hybridization signals were obtained with an ordinary fluorescent microscope. The sensitivity of the system for virus detection reached 102 copies/μL. With the help of inner controls, the system provided reliable results without false negatives and false positives.
Conclusions: The microarray-in-a-tube system is a rapid, labor-saving tool for multiple virus detection with several advantages, such as convenience, prevention of cross-contamination of the PCR products, and potential for multiple-gene detection.
Miniaturization of DNA diagnostic assays and integration for multipathogen detection are advantageous features for use in epidemiology, food safety, and antiterrorism. Two different microfluidic systems, continuous flow and fixed hole, were recently developed for use with PCR or reverse transcription (RT)1 –PCR, with fluorescence and capillary electrophoresis as the major detection systems for the integrated devices. Although these devices can perform many functions, all of them include high-cost microfabrication techniques and complicated processing steps (1)(2)(3)(4)(5)(6).
Microarray techniques have great potential in high-throughput analysis for genomic screening. Current DNA microarray technology, however, involves the complex and strict execution of multiple experimental processes, and cross-contamination can lead to false results. The combination of microfluidics with a microarray in a single device is one reported method for solving these problems (7)(8)(9).
Severe acute respiratory syndrome (SARS) is a serious infectious disease with global impact (10). A virus, human coronavirus, strain SARS (HCoV-SARS) has been isolated from tissues of patients with SARS (10)(11)(12). The early stage of SARS infection is characterized by fever, dyspnea, lymphopenia, and rapid lung changes visible on x-ray (10)(13), and thus it can be misdiagnosed as influenza. There are 2 prevailing serologic diagnostic methods for SARS, ELISA (14)(15)(16) and indirect immunofluorescence assays (12), which are very suitable for the SARS diagnosis during the convalescent phase, when the virus titer is relatively high (17). For early diagnosis of SARS, which is critical for control of the spread of the disease, nucleic acid testing is more sensitive than the above assays (18). Many qualitative PCR assays have been reported (19)(20)(21), and electrochemical (22), electrospray ionization mass spectrometry (23), and microarray techniques (24) have been used for sequence-specific detection.
We aimed to develop a microarray-in-a-tube that integrates RT-PCR and a DNA microarray for detecting and distinguishing 4 viruses causing human acute respiratory tract infection, SARS coronavirus, influenza A and B viruses, and enterovirus.
Materials and Methods
configuration of the microarray-in-a-tube
The microarray-in-a-tube system integrates the microarray and multiple PCR processes in an Eppendorf tube. The system (Fig. 1 ) has 3 parts, which include an optically transparent plastic cap with an oligonucleotide microarray on the inner surface, a black inner vessel that contains hybridization solution, and the body of the Eppendorf tube. The cap, designed for sealing the commercial 200-μL Eppendorf tube, was a flat, optically transparent window made of polycarbonate. The inner vessel, which covered half of the circumference within the horizontal section and contained the hybridization solution, was installed in the Eppendorf tube. The microarray-in-a-tube was adapted to a commercial thermocycler. The cap and the vessel were manufactured by plastic injection molding.
Figure 1.
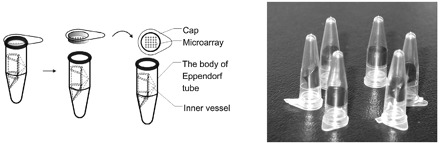
Schematic structures and image of microarray-in-a-tube.
The new cap with a flat, optically transparent window was designed and fabricated to fit the body of the 200-μL Eppendorf tube. The microarray of the DNA probes has been immobilized on the inner side of the window. An inner vessel has been installed inside the Eppendorf tube to store the hybridization solution.
modification of the inner surface of the new type cap
Microwave-plasma was used to generate a hydrophilic surface on the (hydrophobic) polycarbonate cap so that the agarose film would adhere to its inner surface. The microwave-plasma chamber used here was a sealed quartz glass cylinder (25). The stable ammonia plasma discharge remained for 15 min at a chamber pressure <60–70 Pa and with the microwave output power at 100 W. After plasma exposure, the cap board was transferred and immersed in 1 mL/L glutaraldehyde solution for 2 h at room temperature.
preparation of activated agarose film
To immobilize the oligonucleotide probes on the inner surface of the Eppendorf cap, an activated agarose film was prepared on the polycarbonate surface. The method for agarose film fabrication was a modification of a previously reported protocol (26)(27). The 0.5% agarose (Sigma) solution was prepared by mixing and boiling for 5 min. To prepare the agarose film-coated microarray-in-a-tube, 10 μL of the agarose solution, prewarmed in a 60 °C water bath, was poured over each of the specially designed caps. After gelation of the agarose, the cap was dried overnight at 37 °C in an oven. The dried cap could be stored at 4 °C for future use. Before immobilization of the probes, the agarose films were activated by immersion in 20 mmol/L NaIO4 (Sigma) in 0.1 mol/L PBS buffer (pH 7.2) for 30 min at room temperature, then thoroughly rinsed with deionized distilled water and dried. The microarray-in-a-tube was stored under nitrogen at 4 °C for future use.
design and synthesis of dna probes and pcr primers
The system was designed to detect and distinguish the SARS coronavirus, influenza A and B viruses, and enterovirus. We designed the primer and the probes based on the GenBank data at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/Genbank/GenbankSearch.html). Three additional probes were designed as controls, one probe for the negative control, one for the inner positive control, which was designed according to the measles virus, and one for the position marker, which was designed for identification of the probe position, as described in Table 1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol53/issue1). The position marker was designed for detection of the immobilization of the probes and detection of the position of the probes. The negative control probe was designed for the hybridization background. The most important control was the inner positive control that was used for evaluating the validity of the molecular biology operating processes such as RNA/DNA purification, RT-PCR, microarray hybridization, and hybridization image capture. The primers are shown in Table 2 in the online Data Supplement. The probes and primers were synthesized by TaKaRa Biotechnology (Dalian) Co., Ltd.
setup of the microarray
Spotting solutions were obtained by dissolving DNA probes in sodium carbonate buffer (0.1 mol/L, pH 9.0) at the concentration of 80 μmol/L The special cap holder was designed to fix the caps on the platform of the spotting robot. Approximately 500 pL of spotting solution was delivered onto the activated agarose film coating the cap inner surface with a 120-μm spot diameter and 300-μm spacer. The probes were spotted in a pattern as shown in Fig. 2 . After spotting, the agarose film–coated caps were incubated overnight in a humid chamber at room temperature, washed with Tween, 1 mL/L, in deionized distilled water, and dried. The microarray-in-a-tube could be used for immediate detection or stored under nitrogen at 4 °C for future use.
Figure 2.
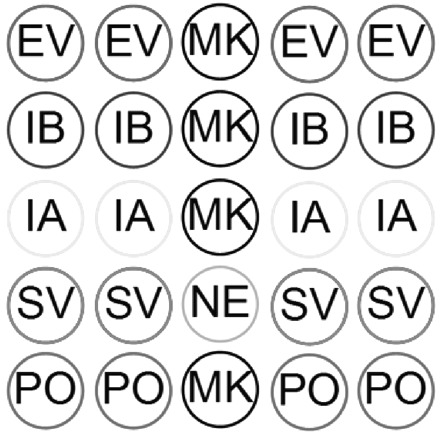
The pattern of the capturing probe arrangement.
From top to bottom rows, the dots represent enterovirus virus, influenza B virus, influenza A virus, SARS-CoV, and positive control. Abbreviations: EV, enterovirus; MK, position marker; IB, influenza B virus; IA, influenza A virus; SC, SARS-associated coronavirus; NE, negative control; PO, inner positive control using measles rubeola virus RNAs.
sample preparation
All standard samples, treated with Trizol, were provided by the National Institute for the Control of Pharmaceutical and Biological Products. HCoV-SARS (n = 20), influenza A virus (n = 20), influenza B virus (n = 20), and enterovirus (n = 20) were treated with Trizol and stored at −20 °C. The 6 clinical serum samples of HCoV-SARS, treated with Trizol, were provided by the Jiangsu Center for Disease Prevention and Control. The inner control virus was the measles virus, which was the further-attenuated measles vaccine.
We added 50 μL of measles virus to Trizol-treated samples and used the chloroform method for RNA purification.
hybridization solution
The hybridization buffer should be previously sealed in the system before gene fragment amplification. The hybridization buffer contained 50.0 mmol/L MgCl2, 6× SSC buffer, pH 7.2, which was put into the inner vessel. The volume of the hybridization buffer was 25 μL.
one-step rt-pcr
We placed 12.5 μL of sample RNA solution into the bottom of the Eppendorf tube and mounted the microarray-in-a-tube on the top of the tube to seal the Eppendorf tube. The specified sequences were then amplified. Amplification was performed with a thermocycler (MJ Research) programmed with 1 cycle at 45 °C for 60 min and 95 °C for 5 min, followed by 45 cycles of 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 30 s, and then 72 °C for 10 min.
We dispensed the one-step RT-PCR solution (Takara) into the bottom of the Eppendorf tube (at the outside of inner vessel). The ∼12.5-μL volume of the RT-PCR solution contained 5 units of AMV Reverse Transcriptase XL (Takara), 40 units of RNase Inhibitor, 5 units of AMV-Optimized Taq (Takara), 400 μmol/L each of 4 dNTPs, and 1 μmol/L of each antisense primer, 0.2 μmol/L of each sense primer, 4.0 mmol/L MgCl2, and 2× One-Step RNA PCR Buffer.
hybridization and detection
When the amplification was complete, the solution temperature was maintained at 95 °C for 5 min so that the DNA was denatured. The microarray-in-a-tube was then transferred into ice water. We then turned over the tube and let the 2 solutions mix on the cap of the microarray-in-a-tube with centrifugation (Eppendorf 5804) at 27g for 30 s at room temperature, so that the labeled target DNA would hybridize to the probe immobilized on the cap inner surface. The microarray-in-a-tube should be kept at 37 °C during hybridization. After hybridization, the tube was turned over again to remove the solution from the cap. The microarray-in-a-tube was recentrifuged at 500 rpm for 30 s. Then the hybridization image was collected by a fluorescence microscope (Nikon E200) and a CCD camera. The operation steps are shown in File 2 in the online Data Supplement. The data were analyzed with in-house developed software (28) to develop a hybridization intensity plot.
Results and Discussion
detection results
A typical negative result of the 4 positional marker dots and 4 inner control dots, which was obtained from a sample that contained only measles virus as the inner positive control, is shown in Fig. 3A . The negative image should contain a low fluorescence intensity for the negative probe (NE) and high fluorescence intensity for the position probe (MK) and inner positive probes (PO), as shown in Fig. 3B .
Figure 3.
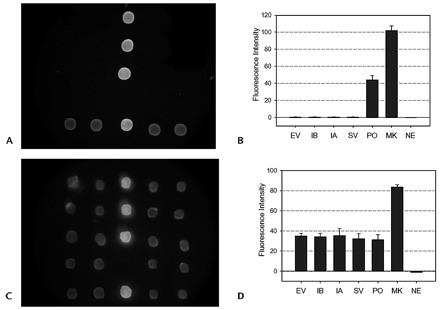
Images and fluorescent intensities of the microarray-in-a-tube with complete negative samples (A, B) and complete positive samples (C, D).
From top to bottom rows, fluorescence dots represent enterovirus virus, influenza B virus, influenza A virus, and SARS-CoV positive result, respectively. From the fluorescent intensities of the probes of PO, MK, and NE, the reliability of the detection results can be checked. See legend of Fig. 2 for abbreviations.
The 4 detected viruses and the inner control virus were mixed to demonstrate multivirus detection in a single run (Fig. 3C ). The 4 viruses and the inner control showed similar fluorescence intensities (Fig. 3D ).
The respiratory tract virus detection system, based on the microarray-in-a-tube, was tested with standard samples from the National Institute for the Control of Pharmaceutical and Biological Products (NICPBP). In the NICPBP, the standard samples were BJ04 strain for HCoV-SARS, Puerto Rico/8/34/Mount Sinai(H1N1) segment 8 for influenza A, Memphis/12/97-MA for influenza B, and echovirus type 12 (prototype Travis wild-type genome) for enterovirus. A total of 86 samples, which included 20 negative, 60 standard, and 6 clinical serum samples, were detected with our microarray-in-a-tube. Examples of hybridization results and plots of the fluorescent intensities of the samples with single detected virus are shown in Fig. 4 . All results with the microarray-in-a-tube were consistent those with the methods based on RT-PCR and electrophoresis. Neither negative nor false-positive results were found with the microarray-in-a-tube system. The virus could be confirmed from the hybridization image. The detection sensitivity was 102 copies/μL, and the results were confirmed by the NICPBP (Table 1 ). Meanwhile, the Jiangsu CDC provided us with a quantified HCoV-SARS RNA sample (the RealArt HPA coronavirus LC Kit, Artus). The RNA sample contained 7500 copies/μL RNA of HCoV-SARS. With a 1/10 to 1/1000 dilution series of the sample, sample concentrations >75 copies/μL were detected.
Figure 4.
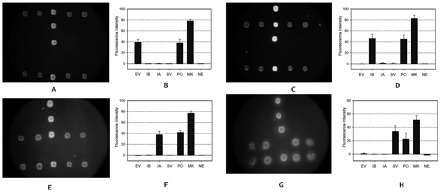
Images and fluorescent intensities of the microarray-in-a-tube with single virus samples.
The nucleotide probes on the inner surface of the cap. All the probe positions are as shown in Fig. 2 . (A, C, E, G), images of fluorescence images; (B, D, F, H), plots of the fluorescence intensities of the fluorescence images. When detection was performed, the measles rubeola virus was added into the sample as the inner positive control. If the sample contained one of the viruses, the positive and the virus probe could be detected with the same fluorescence level. The images shown in A, C, E, and G are the fluorescence images of the samples of enterovirus virus, influenza B virus, influenza A virus, and SARS-CoV, respectively. See legend of Fig. 2 for abbreviations.
Table 1.
The result of the sample detection.
| Sample source | Sample type | Detection method | Sample numbers | Microarray-in-a-tube | Coincidence rate, % | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||
| Jiangsu CDC | SARS positive | Clinical diagnosis (US CDC standard) | 6 | 6 | 0 | 100 | |
| NICPBP | SARS positive | NICPBP | 20 | 20 | 0 | 100 | |
| NICPBP | Enterovirus positive | NICPBP | 20 | 20 | 0 | 100 | |
| NICPBP | Influenza B virus | NICPBP | 20 | 20 | 0 | 100 | |
| NICPBP | Influenza A virus positive | NICPBP | 20 | 20 | 0 | 100 | |
| NICPBP | Normal serum | NICPBP | 20 | 0 | 20 | 100 | |
simplification of features in the microarray-in-a-tube
The practical application of most reported PCR microfluidic devices integrated with microarrays is limited because they must be equipped with complicated pumps and valves to drive the reagents, delicate temperature-controllers to perform on-chip PCR processes, and optical instruments to read the fluorescent signals. Some devices need high air pressure, special pumps, or special design implementation for solution position control and signal capture (7)(8).
The microarray-in-a-tube described here is a simple system that allows the assay to be performed with available instruments, such as a hot-cover thermocycler, centrifuge, constant-temperature oven, and conventional fluorescence microscope. The design is based on the conventional Eppendorf tube and features 2 additional parts, the specially designed cap and the inner vessel, which were manufactured by plastic injection and made with polycarbonate plastic. Plastic injection molding is easy to adapt to larger scale fabrication than are conventional photolithographic production techniques (6) or laser machining techniques (9).
prevention of cross-contamination
The gene amplification method is hypersensitive. A sealed tube for nested-PCR and a 2nd PCR could be performed with a protocol including 2 separated gene amplifications to avoid cross-contamination (29). The disadvantage, however, is that during the assays the amplicon could leak when detected by gel electrophoresis or hybridization analysis. With microelectromechanical system techniques, many other devices are able to prevent the vapor exchange during PCR amplification and DNA hybridization within a closed device (6)(7)(8)(30). These methods are inconvenient to perform, however, whereas the microarray-in-a-tube system was developed to enhance structural simplicity, labor savings, and low cost. In this system, all the steps from gene amplification to image capture could be performed in the sealed device. The system prevents cross-contamination because there is no gene amplicon leakage during any detection process.
determination of the reliability from the hybridization image
After capturing the hybridization image, we calculated all the fluorescence intensities with in-house developed software. Three kinds of signal groups could be detected on the microarray for analyzing the reliability of the hybridization results. From the position signals we obtained the immobilized efficiency and the relative position of the probes. In general, because of the effect of hybridization efficiency, the fluorescence intensity of the position probes should be higher than that of hybridization probes. Therefore the fluorescence signals of the hybridization image were classified into 3 groups: the position signal, the negative signal, and the hybridization signals, which include inner control signals. All reliable results of fluorescent hybridization images should contain the above 3 signal groups. In the in-house developed software for the microarray-in-a-tube system, the fluorescence intensities were <5 for the negative result and >50 for the position marker. The fluorescence intensity of the inner positive control should be >20 and less than that of the position marker, otherwise the result should be discarded and the detection should be performed again. If the fluorescence intensity of the detection probes was >50% of the inner positive control it was considered a positive result. If <40%, it was considered a negative result.
comparison of the oligonucleotide probes with the molecular beacon probes
To develop the no-washing microarray in the microarray-in-a-tube, we originally designed molecular beacon probes that differed from the present oligonucleotide probes. Two molecular beacons, modified at the 5′-terminal with NH2 groups and immobilized on activated agarose film, were used for HCoV-SARS detection, as shown in Table 3 in the online Data Supplement). The hybridization buffer contained 50 mmol/L MgCl2 in 20 mmol/L Tris-HCl (pH 8.0) buffer. Before hybridization, the microarray-in-a-tube system was incubated in the hybridization buffer for 30 min at room temperature. The asymmetrical RT-PCR and hybridized were described as above. Fluorescence images before and after hybridization are shown in Fig. 5 . We were able to use these molecular beacons for HCoV-SARS detection in our microarray-in-a-tube system, but there was relatively high fluorescence background attributable to the unstable stem structure in the molecular beacon probes immobilized on the surface, and the molecular beacon probes are much more expensive than ordinary oligonucleotide probes, which we used successfully for virus detection in our system.
Figure 5.
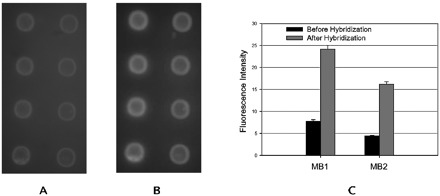
Images and fluorescent intensities of the molecular beacon array on agarose film on the inner surface of microarray-in-a-tube.
The molecular beacons were spotted on the film in quaternion format. From left to right, the probes are MB1 and MB2. Hybridization buffer: 40 mmol/L Tris-HCl (pH 8.0) containing 100 mmol/L MgCl2. Asymmetrical RT-PCR production was added in an equal volume hybridization buffer for the 30-min hybridization. Images in A and B are fluorescence images before and after hybridization, respectively. The plots in C are fluorescence intensity before and after hybridization.
In conclusion, the microarray-in-a-tube system is a rapid, labor-saving tool for multiple virus detection with several advantages, such as convenience, prevention of cross-contamination of the PCR products, and potential for multiple-gene detection.
Supplementary Material
Acknowledgments
The clinical samples of HCoV-SARS were provided by the Jiangsu CDC. Other virus culture samples, influenza A and B viruses, enterovirus, and measles rubeola viruses were kindly donated by Nanjing Medical University and Beijing Genomics Institute, Chinese Academy of Science. This study was supported by Projects 60121101 and 60671019 of the National Natural Science Foundation of China and by Grant 2003AA2Z2007 from the National High Tech Program of China.
Footnotes
Nonstandard abbreviations: RT, reverse transcription; SARS, severe acute respiratory syndrome; HCoV-SARS, human coronavirus, strain SARS; NICPBP, National Institute for the Control of Pharmaceutical and Biological Products.
References
- 1.Kopp MU, de Mello AJ, Manz A. Chemical amplification: continuous-flow PCR on a chip. Science 1998;280:1046-1048. [DOI] [PubMed] [Google Scholar]
- 2.Curcio M, Roeraade J. Continuous segmented-flow polymerase chain reaction for high-throughput miniaturized DNA amplification. Anal Chem 2003;75:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Obeid PJ, Christopoulos TK, Crabtree HJ, Backhouse CJ. Microfabricated device for DNA and RNA amplification by continuous-flow polymerase chain reaction and reverse transcription-polymerase chain reaction with cycle number selection. Anal Chem 2003;75:288-295. [DOI] [PubMed] [Google Scholar]
- 4.Khandurina J, McKnight TE, Jacobson SC, Waters LC, Foote RS, Ramsey JM. Integrated system for rapid PCR-based DNA analysis in microfluidic devices. Anal Chem 2000;72:2995-3000. [DOI] [PubMed] [Google Scholar]
- 5.Woolley AT, Hadley D, Landre P, deMello AJ, Mathies RA, Northrup MA. Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device. Anal Chem 1996;68:4081-4086. [DOI] [PubMed] [Google Scholar]
- 6.Burns MA, Johnson BN, Brahmasandra SN, Handique K, Webster JR, Krishnan M, et al. An integrated nanoliter DNA analysis device. Science 1998;282:484-487. [DOI] [PubMed] [Google Scholar]
- 7.Liu YJ, Rauch CB, Stevens RL, Lenigk R, Yang JN, Rhine DB, et al. DNA amplification and hybridization assays in integrated plastic monolithic devices. Anal Chem 2002;74:3063-3070. [DOI] [PubMed] [Google Scholar]
- 8.Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Anal Chem 2004;76:1824-1831. [DOI] [PubMed] [Google Scholar]
- 9.Anderson RC, Su X, Bogdan GJ, Fenton J. A miniature integrated device for automated multistep genetic assays. Nucleic Acids Res 2000;28:E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003;361:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enserink M, Vogel G. Infectious diseases: deferring competition, global net closes in on SARS. Science 2003;300:224-225. [DOI] [PubMed] [Google Scholar]
- 12.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 13.Tsang KW, Ho PL, Ooi GC, Yee WK, Wang T, Chan-Yeung M, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003;348:1977-1985. [DOI] [PubMed] [Google Scholar]
- 14.He Q, Du Q, Lau S, Manopo I, Lu L, Chan SW, et al. Characterization of monoclonal antibody against SARS coronavirus nucleocapsid antigen and development of an antigen capture ELISA. J Virol Methods 2005;127:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Che XY, Hao W, Wang Y, Di B, Yin K, Xu YC, et al. Nucleocapsid protein as early diagnostic marker for SARS. Emerg Infect Dis 2004;10:1947-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau SK, Woo PC, Wong BH, Tsoi HW, Woo GK, Poon RW, et al. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in SARS patients by enzyme-linked immunosorbent assay. J Clin Microbiol 2004;42:2884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yam WC, Chan KH, Poon LL, Guan Y, Yuen KY, Seto WH, et al. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J Clin Microbiol 2003;41:4521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu AC, Lau LT, Fung YW. Boosting the sensitivity of real-time polymerase-chain-reaction testing for SARS. N Engl J Med 2004;350:1577-1579. [DOI] [PubMed] [Google Scholar]
- 19.Poon LL, Wong OK, Chan KH, Luk W, Yuen KY, Peiris JS, et al. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS). Clin Chem 2003;49:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poon LL, Wong BW, Chan KH, Ng SS, Yuen KY, Guan Y, et al. Evaluation of real-time reverse transcriptase PCR and real-time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J Clin Microbiol 2005;43:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houng HS, Norwood D, Ludwig GV, Sun W, Lin M, Vaughn DW. Development and evaluation of an efficient 3′-noncoding region based SARS coronavirus (SARS-CoV) RT-PCR assay for detection of SARS-CoV infections. J Virol Methods 2004;120:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abad-Valle P, Fernandez-Abedul MT, Costa-Garcia A. Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. Biosens Bioelectron 2005;20:2251-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampath R, Hofstadler SA, Blyn LB, Eshoo MW, Hall TA, Massire C, et al. Rapid identification of emerging pathogens: coronavirus. Emerg Infect Dis 2005;11:373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juang JL, Chen TC, Jiang SS, Hsiung CA, Chen WC, Chen GW, et al. Coupling multiplex RT-PCR to a gene chip assay for sensitive and semiquantitative detection of severe acute respiratory syndrome-coronavirus. Lab Invest 2004;84:1085-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Q, Liu Z, Xiao P, Liang R, He N, Lu Z. Preparation of hydrophilic poly (dimethylsiloxane) stamps by plasma-induced grafting. Langmuir 2003;19:6982-6986. [Google Scholar]
- 26.Wang H, Li J, Liu H, Liu Q, Mei Q, Wang Y, et al. Label-free hybridization detection of a single nucleotide mismatch by immobilization of molecular beacons on an agarose film. Nucleic Acids Res 2002;30:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afanassiev V, Hanemann V, Wolfl S. Preparation of DNA and protein micro arrays on glass slides coated with an agarose film. Nucleic Acids Res 2000;28:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Zhou Q, Bai Y, Ge Q, Lu Z. Detection and analysis system for hybridization images of lab-in-a-tube microarray. Chinese Sci Bull 2005;50:2896-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao SC, Jiang D, Lu HL, Xing WL, Zhou YX, Cheng J. One-tube nested RT-PCR enabled by using a plastic film and its application for the rapid detection of SARS-virus. Biotechnol Lett 2004;26:179-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenigk R, Liu RH, Athavale M, Chen Z, Ganser D, Yang J, et al. Plastic biochannel hybridization devices: a new concept for microfluidic DNA arrays. Anal Biochem 2002;311:40-49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


