In a large study of children hospitalized with community-acquired pneumonia, virus-bacterium coinfections resulted in worse outcomes than virus-only infections. Patterns of coinfections varied with the pathogen.
Keywords: Coinfection, community-acquired pneumonia, children, etiology, severity
Abstract
Background
Recognition that coinfections are common in children with community-acquired pneumonia (CAP) is increasing, but gaps remain in our understanding of their frequency and importance.
Methods
We analyzed data from 2219 children hospitalized with CAP and compared demographic and clinical characteristics and outcomes between groups with viruses alone, bacteria alone, or coinfections. We also assessed the frequency of selected pairings of codetected pathogens and their clinical characteristics.
Results
A total of 576 children (26%) had a coinfection. Children with only virus detected were younger, more likely to be black, and more likely to have comorbidities such as asthma, compared with children infected with typical bacteria alone. Children with virus-bacterium coinfections had a higher frequency of leukocytosis, consolidation on chest radiography, parapneumonic effusions, intensive care unit admission, and need for mechanical ventilation and an increased length of stay, compared with children infected with viruses alone. Virus-virus coinfections were generally comparable to single-virus infections, with the exception of the need for oxygen supplementation, which was higher during the first 24 hours of hospitalization in some virus-virus pairings.
Conclusions
Coinfections occurred in 26% of children hospitalized for CAP. Children with typical bacterial infections, alone or complicated by a viral infection, have worse outcomes than children infected with a virus alone.
(See the Editorial commentary by Kaplan, on pages 173–5.)
Community-acquired pneumonia (CAP) is an important cause of morbidity and mortality in children [1]. The advent of molecular testing has markedly enhanced the detection of respiratory viruses and has demonstrated that virus-bacterium and virus-virus coinfections are common among children with CAP and contribute to pathogenesis [1–3]. For example, during the 2009 influenza A(H1N1) pandemic, a coinfecting bacterial pathogen was identified in about one third of all severe or fatal infections due to 2009 pandemic influenza A(H1N1) virus [4–6], and identification of Streptococcus pneumoniae was an independent risk factor for severe disease [7]. Other bacteria, such as Staphylococcus aureus, have been detected in patients with serious complications of influenza [8]. Although the impact of influenza virus–bacterium coinfection has been well described, fewer data exist for other virus-bacterium or virus-virus coinfections.
A number of barriers limit our ability to understand important aspects of coinfections. Most of the burden of CAP occurs in outpatient settings, where comprehensive etiologic molecular testing is not performed. Studies of hospitalized children from a single site are generally too small to conduct meaningful analyses of pathogen-pathogen interactions. Common testing algorithms for determining the etiology of CAP also suffer from poor sensitivity and specificity, particularly for bacterial infections, since blood culture is predominantly used to determine the bacterial etiology of pneumonia in children [9]. The use of polymerase chain reaction (PCR) analysis aids in identifying viral causes of pneumonia, but pathogens detected in the upper respiratory tract may not correlate with pathogens obtained directly from the lung. In addition, sequential infections in which one pathogen alters the host’s defenses facilitating a secondary infection are particularly difficult to accurately diagnose, as the preceding pathogen may no longer be detectable when the patient comes to medical attention.
The Etiology of Pneumonia in the Community (EPIC) study presented an opportunity to comprehensively study coinfections [1]. This large, multisite, prospective study included a comprehensive assessment of etiology, using multiple state-of-the-art diagnostic tests linked to clinical outcomes. The size of the study and the breadth of demographic, clinical, and diagnostic data available presented an opportunity to overcome many of the barriers cited above to understanding causes of pneumonia, as well as analyzing the impact of coinfection. Coinfections with multiple pathogens were identified in 26% of the children with radiographically defined CAP [1]. Here we explore the interactions of various viral and bacterial organisms in pneumonia and the impact of coinfections on clinical characteristics and outcomes.
METHODS
Study Population and Pathogen Detection
Children (<18 years old) were enrolled in the EPIC study at 3 children’s hospitals in Memphis, Tennessee, Nashville, Tennessee, and Salt Lake City, Utah [1]. Between January 2010 and June of 2012, 2638 children were enrolled, of whom 2358 had final radiographic evidence of pneumonia. Of this cohort, 2219 had specimens tested for both viruses and bacteria and were included in the current analyses (Figure 1). Children with recent hospitalization, tracheostomy, cystic fibrosis, or severe immunosuppressive conditions were excluded [1]. Demographic and clinical data were obtained using previously described methods. Typical bacteria were detected by cultures of blood, pleural fluid, and/or bronchoalveolar lavage specimens and by PCR testing of blood or pleural fluid specimens as described elsewhere [1]. Atypical bacteria, Chlamydophila pneumoniae and Mycoplasma pneumoniae, were detected by PCR analysis of combined nasopharyngeal and oropharyngeal swab specimens. Human adenovirus (hAdV), human coronavirus (hCoV), human metapneumovirus (hMPV), human rhinovirus (hRV), influenza A and B virus, parainfluenza virus (PIV), and respiratory syncytial virus (RSV) were also detected by PCR analysis of combined nasopharyngeal and oropharyngeal swab specimens. Serologic testing for hAdV, hMPV, influenza A and B viruses, PIV, and RSV was also performed on acute and convalescent specimens in 44% of children [1]. The parent study protocol was approved by the University of Tennessee Health Science Center Institutional Review Board, informed consent was obtained from parents or guardians in all cases, and assent was obtained from participants when appropriate.
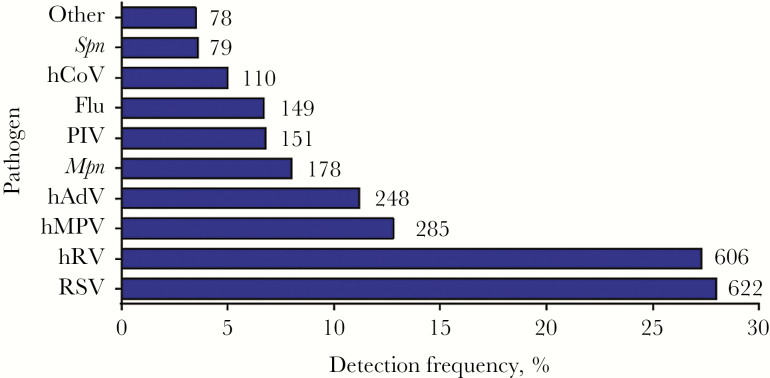
Figure 1.
Etiology of pneumonia in hospitalized children. In 2219 children hospitalized in the Etiology of Pneumonia in the Community Study, 2506 pathogens were detected. Shown is the percentage of patients with a positive result for each pathogen (on the x-axis) and the total numbers of each pathogen that were detected (numbers beside the bars). Pathogens other than those highlighted here were detected in 78 children, including Staphylococcus aureus (in 22 children), Streptococcus pyogenes (in 16), viridans streptococci (in 13), Chlamydophila pneumoniae (in 12), Haemophilus influenzae (in 9), other gram-negative bacteria (in 9), and other Streptococcus species (in 4). Flu, influenza virus; hADV, human adenovirus; hCoV, human coronavirus; hMPV, human metapneumovirus; hRV, human rhinovirus; Mpn, Mycoplasma pneumoniae; PIV, parainfluenza virus; RSV, respiratory syncytial virus; Spn, Streptococcus pneumoniae.
Statistical Analyses
Demographic and clinical characteristics of all cases of CAP were described and stratified in the following groups: viruses, bacteria, or atypical bacteria; viruses-bacteria or viruses–atypical bacteria; and no detection of viruses or bacteria. Children with single-virus infections were similar demographically and clinically to children with virus-virus coinfections and, for clarity, were therefore combined in the viruses group in the tables. The same is true for those with single and multiple typical bacterial pathogens detected, who were combined in the typical bacteria group. Comparisons between types of infection, as defined by these groups, were made using χ2 tests or the Fisher exact test as appropriate for categorical variables and t tests or the Wilcoxon rank sum test as appropriate for continuous variables.
We used 2-by-2 tables to estimate whether certain codetections occurred more frequently than would be expected by chance. For each pair of potential coinfections, we tabulated the individual pathogen detections against each other. The frequencies of observed codetected pairs were compared with how often they would be expected to occur by chance. Deviations from expected frequencies were quantified using a χ2 test and then by calculating the odds ratio (OR) and 95% confidence interval (CI) for each pair of pathogens. Age and infection season were not included in the a priori analyses but were assessed post hoc to develop hypotheses that might explain the findings.
To assess differences in severity among types of infections, we compared the categorical outcomes of supplemental oxygen use in the first 24 hours, intensive care unit admission, invasive mechanical ventilation use, and hospital length of stay.
RESULTS
Of the 2219 children studied, 1801 (81%) had ≥1 respiratory pathogen detected (Figure 1). Of these, 576 (26%) had a coinfection (Supplementary Table 1), including 417 children with ≥ 2 viruses detected, 99 with ≥ 1 virus and ≥ 1 typical bacterial pathogen, 56 with ≥ 1 virus and ≥ 1 atypical bacterial pathogen, and four with two bacterial pathogens. Children with only viruses detected were younger than children with only typical bacteria detected, and children with only atypical bacteria detected were older than either of these groups (Table 1). Children with only viruses detected were more likely to be black than children with only typical bacterial or atypical bacterial pathogens. The age distribution and race of children with virus-bacterium coinfection most closely resembled those with viral infection alone (Table 1). Children with typical bacterial infection alone, atypical bacterial infection alone, or virus-typical bacterium coinfection were more likely to have no previous underlying medical conditions and less likely to have asthma than children with viral infection alone. No striking differences in body mass index or vaccination history were observed among types of infection. Children with a diagnosis of atypical bacterial infection alone were more likely to have received antibiotics prior to admission than the other groups (Table 1).
Table 1.
Demographic Characteristics of 2219 Participating Children, by Infection Group
| Characteristic | Viruses (n = 1472) | Typical Bacteria (n = 41) | Atypical Bacteria (n = 133) | Viruses-Typical Bacteria (n = 99) | Viruses–Atypical Bacteria (n = 56) | No Detection (n = 418) |
|---|---|---|---|---|---|---|
| Age, y, mean ± SD | 3.2 ± 3.5 | 5.7 ± 4.9a | 8.8 ± 4.3b,c | 3.1 ± 3.7 | 6.2 ± 4.4 | 5.9 ± 4.9 |
| Male sex | 779 (53) | 27 (66) | 82 (62) | 67 (68)d | 29 (52) | 240 (57) |
| Race/ethnicity | ||||||
| Non-Hispanic white | 500 (34) | 24 (59) | 87 (65)b | 37 (37) | 28 (51) | 196 (47) |
| Non-Hispanic black | 546 (37) | 7 (17)a | 21 (16)b | 30 (30) | 14 (25) | 144 (35) |
| Hispanic | 300 (20) | 7 (17) | 19 (14) | 22 (22) | 11 (20) | 55 (13) |
| Other | 122 (9) | 3 (7) | 6 (5)b | 10 (10) | 2 (4) | 18 (4) |
| Age group | ||||||
| <2 y | 773 (53) | 11 (27)a | 10 (8)b,c | 55 (56) | 12 (21) | 118 (28) |
| 2–4 y | 397 (27) | 12 (29) | 19 (14)b,c | 25 (25) | 14 (25) | 92 (22) |
| 5–9 y | 202 (14) | 12 (29) | 52 (39)b | 11 (11) | 17 (30) | 114 (27) |
| 10–17 y | 100 (7) | 6 (15) | 52 (39)c | 8 (8) | 13 (23) | 94 (22) |
| Comorbidities | ||||||
| None | 687 (47) | 29 (71)a | 71 (54)c | 65 (66)d | 25 (45) | 194 (46) |
| Asthma/reactive airway disease | 538 (37) | 8 (20)a | 37 (28)b | 19 (19)d | 21 (38) | 119 (28) |
| Preterm birth (if age <2 y) | 155 (20) | 1 (9) | 5 (50)b | 10 (18) | 3 (25) | 31 (26) |
| Neurologic disorder | 106 (7) | 1 (2) | 8 (6) | 4 (4) | 1 (2) | 60 (14) |
| Congenital heart disease | 101 (7) | 2 (5) | 13 (10) | 6 (6) | 3 (5) | 34 (8) |
| Chromosomal disorder | 64 (4) | 2 (5) | 11 (8)b | 4 (4) | 6 (11) | 39 (9) |
| BMI statuse | ||||||
| Underweight | 130 (20) | 2 (7) | 13 (11) | 5 (12) | 5 (12) | 40 (15) |
| Normal | 353 (54) | 15 (54) | 70 (58) | 23 (55) | 23 (56) | 136 (52) |
| Overweight | 61 (9) | 2 (7) | 18 (15) | 7 (17) | 4 (10) | 28 (11) |
| Obese | 105 (16) | 9 (32) | 20 (17) | 7 (17) | 9 (22) | 57 (22) |
| Up-to-date vaccination statusf | ||||||
| H. influenzae type B | 1282 (90) | 35 (82) | 13 (90) | 85 (92) | 53 (95) | 359 (90) |
| Influenza | 394 (28) | 8 (22) | 30 (23) | 27 (30) | 13 (24) | 107 (27) |
| Pneumococcal | 1246 (88) | 31 (82) | 90 (68)b | 79 (86) | 46 (82) | 315 (79) |
| Preadmission medications | ||||||
| Antibioticg | 238 (16) | 11 (26) | 52 (39)b,c | 19 (20) | 13 (25) | 78 (19) |
| Corticosteroid | 126 (9) | 1 (2) | 8 (6) | 4 (4) | 5 (9) | 31 (7) |
| Influenza antiviral | 8 (<1) | 3 (7)a | 1 (1)c | 6 (6)d | 2 (4) | 7 (2) |
Data are no. (%) of children, unless otherwise indicated. Percentages were calculated using the number of children with available data, rather than the number in the column headings, as the denominator. “Viruses” and “Bacteria” encompass single-pathogen detections and multiple detections.
Abbreviations: H. influenzae, Haemophilus influenzae.
a P < .05, for comparison of viral infections to typical bacterial infections.
b P < .05, for comparison of viral infections to atypical infections.
c P < .05, for comparison of typical bacterial infections to atypical infections.
d P < .05, for comparison of viral infections to combined viral and typical bacterial infections.
eBody mass index (BMI) was calculated as the weight in kilograms divided by the height in meters squared and was determined for children ≥2 years of age.
fDefined as having received age-appropriate dosing for H. influenzae type B and pneumococcal vaccine and current seasonal influenza vaccine.
gWithin 5 days of admission.
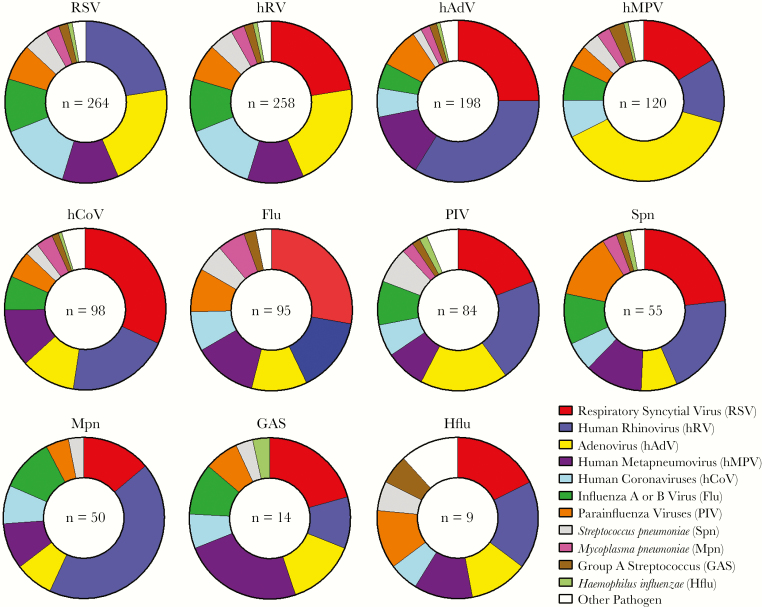
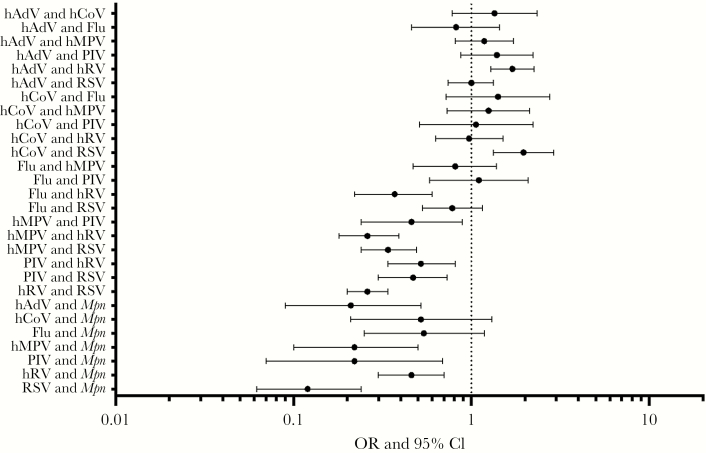
The most common pathogens among codetection pairings are shown in Figure 2. The viruses most commonly identified in CAP, RSV and hRV, were also the viruses most frequently found in combination with other viruses and/or bacteria. Among commonly identified pathogens, influenza virus was not detected with Haemophilus influenzae, and M. pneumoniae was not detected with S. pyogenes or H. influenzae. S. pneumoniae was detected in association with the major viruses detected here in roughly equal proportions (Figure 2). For many other pathogen pairings, however, the distribution of pathogen pairings was different than expected by chance (Figures 2 and 3). For example, hAdV and hRV were detected together more commonly as a pair than expected, based on their relative frequency of detection in the EPIC study; a similar pattern was observed for hCoV and RSV. Although the fifth most common pathogen detected overall, M. pneumoniae was the ninth most common pathogen codetected with other organisms; the relative infrequency of coinfections with M. pneumoniae was noted, particularly when pairings with respiratory viruses were examined (Figure 3). Several virus pairs were also observed less frequently than expected, including hMPV with hRV, PIV, or RSV; PIV with hRV or RSV; and hRV with RSV.
Figure 2.
Frequency of pair-wise associations between pathogens in coinfections. Pair-wise associations, by pathogen, are shown for all coinfections involving the 11 most common pathogens detected in the study; other pathogens are grouped together under “Other Pathogen” (see Supplementary Table 1 for a complete listing of all pathogens detected). A single coinfection with ≥3 pathogens present may be represented multiple times to show all associations.
Figure 3.
Frequency of actual as compared to expected coinfections. The frequencies of commonly observed coinfection pairs are shown relative to how often they would be expected to occur by chance, using the frequencies of detection of the individual pathogens as the baseline. Deviations from expected frequencies were quantified using a χ2 test and then by calculating the odds ratio (OR) and 95% confidence interval (CI) for each pair of pathogens. Flu, influenza virus; hADV, human adenovirus; hCoV, human coronavirus; hMPV, human metapneumovirus; hRV, human rhinovirus; Mpn, Mycoplasma pneumoniae; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Clinical symptoms associated with CAP were generally similar among different infection groupings, and length of illness prior to admission did not differ statistically (Table 2). The total white blood cell count and the percentage of white blood cells that were band forms were significantly higher in the typical bacteria and viruses-typical bacteria groups compared to the viruses group. Chest radiography findings, including consolidation and pleural effusion, were also more common in these 2 groups as compared to the viruses group, whereas infiltrates and a pattern consistent with complicated bronchiolitis were less common (Table 2). Serious outcomes or complications, including parapneumonic effusion, admission to an intensive care unit, and use of mechanical ventilation, were significantly more common in the typical bacteria and viruses-typical bacteria groups as compared to the viruses group; median length of stay was also about twice as long in the typical bacteria and viruses-typical bacteria groups (Table 2). Outcome measures, including intensive care admission, invasive mechanical ventilation, and length of stay, were similar for the typical bacteria group as compared to the viruses-typical bacteria group.
Table 2.
Clinical Characteristics of 2219 Participating Children, by Infection Group
| Characteristic | Viruses (n = 1472) | Typical Bacteria (n = 41) | Atypical Bacteria (n = 133) | Viruses-Typical Bacteria (n = 99) | Viruses–Atypical Bacteria (n = 56) | No detection (n = 418) |
|---|---|---|---|---|---|---|
| Time from illness onset to admission, d | 3 (2–5) | 4 (2–6) | 7 (5–9) | 4 (2–6) | 5 (3–7) | 3 (1–6) |
| Mean ± SD | 3.6 ± 2.8 | 4.6 ± 3.7 | 6.9 ± 3.1 | 4.1 ± 2.8 | 5.1 ± 3.4 | 4.1 ± 3.6 |
| Median (IQR) | 3 (2–5) | 4 (2–6) | 7 (5–9) | 4 (2–6) | 5 (3–7) | 3 (1–6) |
| Symptoms | ||||||
| Cough | 1417 (96) | 37 (90)a | 127 (95) | 89 (90)b | 54 (96) | 373 (89) |
| Fever / feverishness | 1340 (91) | 39 (95) | 124 (93) | 87 (88) | 52 (93) | 377 (89) |
| Anorexia | 1105 (75) | 36 (88) | 102 (77) | 80 (81) | 41 (73) | 293 (70) |
| Dyspnea | 1061 (72) | 31 (76) | 82 (62) | 71 (72) | 42 (75) | 273 (65) |
| Fatigue | 997 (68) | 33 (81) | 104 (78) | 69 (70) | 41 (73) | 298 (71) |
| Excessive crying | 984 (67) | 25 (61) | 47 (35) | 59 (60) | 28 (50) | 236 (56) |
| Wheezing | 993 (67) | 17 (41)a | 57 (43) | 50 (51)b | 30 (54) | 206 (49) |
| Nausea | 809 (55) | 30 (73)a | 79 (59) | 47 (47)b | 33 (59) | 212 (51) |
| Chest indrawing/retractions | 693 (47) | 18 (44) | 32 (24) | 45 (45) | 17 (30) | 164 (39) |
| Chills | 468 (32) | 21 (51)a | 80 (60) | 39 (39) | 24 (43) | 189 (45) |
| WBC count, cells × 1000/mL, mean ± SD | 13.1 ± 6.9 | 17.6 ± 8.2a | 10.8 ± 5.2 | 17.6 ± 11.5b | 11.9 ± 6.1 | 16.5 ± 8.9 |
| Neutrophils, %, mean ± SD | 57.8 ± 22.4 | 64.4 ± 23.2 | 63.9 ± 15.8 | 54.6 ± 23.6b | 60.2 ± 18.9 | 65.4 ± 39.4 |
| Bands, %, mean ± SD | 12.9 ± 13.6 | 16.6 ± 15.6a | 13.1 ± 11.9 | 18.6 ± 15.1b | 15.4 ± 16.3 | 12.4 ± 13.4 |
| Chest radiography pattern at admissionc | ||||||
| Consolidation | 796 (54) | 33 (80)a | 75 (56) | 71 (72)a | 35 (63) | 289 (69) |
| Infiltrate | 818 (56) | 11 (27)a | 70 (53) | 39 (39)a | 22 (39) | 161 (39) |
| Complicated bronchiolitis | 388 (26) | 4 (10)a | 15 (11) | 14 (14)a | 3 (5) | 51 (12) |
| Pleural effusion | 102 (7) | 22 (54)a | 33 (25) | 43 (43)a | 15 (27) | 87 (21) |
| O2 use in first 24 hours | 856 (58) | 20 (49) | 81 (61) | 55 (56) | 30 (54) | 183 (44) |
| Antibiotics during admission | ||||||
| No antibiotic | 224 (15) | 0 (0) | 3 (2) | 4 (4) | 4 (7) | 31 (7) |
| One antibiotic | 338 (23) | 1 (2) | 17 (13) | 6 (6) | 7 (13) | 58 (14) |
| Multiple antibiotics | 910 (62) | 40 (98) | 113 (85) | 89 (90) | 45 (80) | 329 (79) |
| Parapneumonic effusion during admission | 78 (5) | 27 (66)a | 28 (21) | 52 (53)a | 6 (11) | 78 (19) |
| ICU admission | 293 (20) | 18 (44)a | 13 (10) | 43 (43)a | 8 (14) | 88 (21) |
| Invasive mechanical ventilation, % | 78 (5) | 12 (29)a | 1 (1) | 26 (26)a | 2 (4) | 30 (7) |
| Length of stay, d | ||||||
| Mean ± SD | 4.0 ± 7.5 | 7.8 ± 5.3a | 3.2 ± 3.0 | 8.7 ± 7.1d | 3.5 ± 3.7 | 4.1 ± 5.1 |
| Median (IQR) | 3 (2–4) | 7 (4–10) | 2 (2–4) | 7 (3–12) | 2 (2–4.5) | 3 (1–5) |
Data are no. (%) of children, unless otherwise indicated.
Abbreviations: ICU, intensive care unit; IQR, interquartile range; WBC, white blood cell.
a P < .05, for comparison of viral infections to typical bacterial infections.
b P < .05, for comparison of typical bacterial infections to combined viral and typical bacterial infections.
cMore than 1 pattern may be present, so numbers do not add up to 100%.
Extending these analyses beyond the broad groupings in Tables 1 and 2, the differences in outcomes remained significant in comparisons of single viruses or multiple viruses to coinfections between viruses and single typical bacterial pathogens or multiple typical bacterial pathogens (Table 3). Intensive care unit admission, use of invasive mechanical ventilation, and length of stay were all significantly greater when single or multiple viruses were complicated by ≥1 typical bacterial pathogen as compared to infections due to single or multiple viruses alone. Coinfections by viruses and atypical bacteria did not lead to worse outcomes. Two virus-virus coinfections (RSV and hAdV vs hAdV alone; influenza virus and RSV vs influenza virus alone) demonstrated higher rates of supplemental oxygen use in the first 24 hours of hospitalization, compared with single infections alone. Other virus-virus coinfections did not lead to similar findings, and no differences in intensive care unit admission or need for mechanical ventilation were identified (Table 3). Interestingly, children with influenza virus and RSV coinfection had a shorter length of stay than those infected with either influenza virus or RSV alone. Typical bacterial coinfection with RSV was associated with statistically significant increases in the length of hospital admission and the need for invasive mechanical ventilation as compared to infection with RSV alone (Table 3). Intensive care unit admission was also twice as frequent among children with RSV-typical bacterium coinfections, but this difference did not reach statistical significance.
Table 3.
Outcomes Comparing Single-Pathogen Infections to Coinfections
| Etiology | O2 Use in the First 24 h | ICU Admission | Invasive Mechanical Ventilation | Length of Stay, d | ||||
|---|---|---|---|---|---|---|---|---|
| Infections, % | P a | Infections, % | P a | Infections, % | P a | Mean (IQR) | P a | |
| By group | ||||||||
| Any viral pathogen | ||||||||
| Overall (n = 1462) | 58 | 20 | 5 | 4.0 (2–4) | ||||
| Plus any typical bacterial pathogen (n = 97) | 55 | .501 | 42 | <.001 | 26 | <.001 | 8.6 (3–11) | <.001 |
| Plus any single typical bacterial pathogen (n = 78) | 53 | .333 | 33 | .004 | 19 | <.001 | 7.7 (3–10) | <.001 |
| Any single viral pathogen | ||||||||
| Overall (n = 1054) | 57 | 21 | 6 | 3.8 (2–4) | ||||
| Plus any single typical bacterial pathogen (n = 56) | 48 | .202 | 36 | .007 | 21 | <.001 | 8.0 (3–9.5) | <.001 |
| Plus any single atypical-bacterial pathogen (n = 40) | 53 | .582 | 18 | .699 | 5 | 1.00c | 3.3 (2.0–4.5) | .233 |
| Any single atypical bacterial pathogen | ||||||||
| Overall(n = 133) | 61 | 10 | 1 | 3.2 (2–4) | ||||
| Plus any viral pathogen (n = 55) | 53 | .300 | 15 | .323 | 4 | .205c | 3.5 (2–5) | .525 |
| Plus any single viral pathogen (n = 40) | 53 | .344 | 18 | .165c | 5 | .134c | 3.3 (2–4.5) | .874 |
| Any single typical bacterial pathogen (n = 31) | 52 | 35 | 23 | 6.8 (3–9) | ||||
| Plus any viral pathogen (n = 61) | 51 | .943 | 31 | .675 | 13 | .245 | 7.1 (3–10) | .829 |
| Plus any single viral pathogen (n = 56) | 48 | .761 | 36 | .983 | 21 | .901 | 8.0 (3–9.5) | .386 |
| Plus RSV (n = 17) | 59 | .632 | 41 | .697 | 41 | .201 | 10.1 (3–11) | .143 |
| By pathogenb | ||||||||
| hRV | ||||||||
| Overall (n = 348) | 47 | 22 | 6 | 3.6 (2–4) | ||||
| Plus hAdV (n = 61) | 61 | .054 | 20 | .668 | 10 | .252c | 7.4 (1–3) | .304 |
| Plus RSV (n = 54) | 56 | .257 | 22 | .987 | 2 | .334c | 3.7 (2–4) | .742 |
| Plus influenza A/B virus (n = 12) | 50 | .852 | 8 | .475c | 8 | .520c | 3.2 (1.5–3.5) | .767 |
| RSV | ||||||||
| Overall (n = 358) | 66 | 20 | 6 | 3.9 (2–5) | ||||
| Plus any single typical bacterial pathogen (n = 17) | 59 | .536 | 41 | .061c | 41 | <.001c | 10.1 (3–11) | .006 |
| Plus hRV (n = 54) | 56 | .131 | 22 | .722 | 2 | .336c | 3.8 (2–4) | .797 |
| Plus hAdV (n = 43) | 70 | .631 | 14 | .335 | 2 | .714c | 3.6 (2–4) | .650 |
| Plus hCoV (n = 31) | 55 | .206 | 16 | .593 | 6 | .691 | 3.7 (2–4) | .742 |
| Plus influenza A/B virus (n = 24) | 79 | .188 | 21 | 1.00c | 4 | 1.00c | 2.8 (2–3) | .015 |
| Plus hMPV (n = 18) | 72 | .592 | 22 | .768c | 6 | 1.00c | 5.7 (3–5) | .363 |
| hAdV | ||||||||
| Overall (n = 50) | 45 | 16 | 8 | 3.4 (2–4) | ||||
| Plus hRV (n = 61) | 61 | .099 | 20 | .617 | 10 | 1.00c | 7.4 (1–3) | .284 |
| Plus RSV (n = 43) | 70 | .016 | 14 | .783 | 2 | .368c | 3.6 (2–4) | .722 |
| Influenza A/B virus | ||||||||
| Overall (n = 54) | 56 | 22 | 7 | 4.9 (2–5) | ||||
| Plus RSV (n = 24) | 79 | .046 | 21 | .891 | 4 | 1.00c | 2.8 (2–3) | .044 |
| Plus hRV (n = 12) | 50 | .727 | 8 | .433c | 8 | 1.00c | 3.2 (1.5–3.5) | .220d |
| hMPV | ||||||||
| Overall (n = 165) | 65 | 24 | 5 | 4.0 (2–5) | ||||
| Plus RSV (n = 18) | 72 | .532 | 22 | 1.00c | 6 | 1.00c | 5.7 (3–5) | .376 |
Abbreviations: hADV, human adenovirus; hCoV, human coronavirus; hMPV, human metapneumovirus; hRV, human rhinovirus; ICU, intensive care unit; IQR, interquartile range; RSV, respiratory syncytial virus.
aCompared to the overall group or pathogen.
bInfections due to 1 or 2 pathogens only.
cBy the Fisher exact test.
dBy the Wilcoxon rank sum test.
DISCUSSION
Because of the difficulties attributing a specific etiology to a pathogen or to multiple pathogens, the current state of knowledge of the role of coinfections in CAP is limited in several critical areas. Specific potential risk factors for coinfections are not known. Clearly, severe immunosuppression from cancer chemotherapy or untreated human immunodeficiency virus infections predispose to coinfections, but what other factors might be in play? The morbidity and mortality associated with influenza and bacterial superinfections is well described [6, 8, 10–12], but does pairing of other pathogens engender similar modifications to the disease course relative to an infection from a single agent? Are some coinfections milder than single infections because of earlier or stronger engagement of innate immune responses? Are there particular pairs (or other multiples) of pathogens that cooperate or compete, leading to different outcomes?
In our large multicenter study, children with viral pneumonia were younger and more likely to have comorbidities such as asthma than other children with pneumonia, but no specific risk factors or distinguishing demographic characteristics were identified for children with virus-bacterium coinfections. When viral and typical bacterial pathogens were codetected, virus-bacterium coinfection clinically more closely resembled typical bacterial pneumonia than viral pneumonia, with these children having a higher frequency of leukocytosis, consolidation, parapneumonic effusion, intensive care unit admission, and need for mechanical ventilation and an increased length of stay, compared with the virus only group. Differences in the outcomes measured in this study did not differ between the typical bacteria group and the viruses-typical bacteria group. RSV infection alone was less severe than RSV-typical bacterium coinfection. These are important observations, as testing algorithms in current use often restrict testing for bacterial pathogens if an initial screen for viral pathogens is positive, to reduce costs and antibiotic use. This might result in some typical bacterial coinfections going undiagnosed, leading to worse outcomes. Virus-virus infections were generally comparable to single-virus infections, with the exception of supplemental oxygen use, which was higher during the first 24 hours of hospitalization in select virus-virus pairings.
Children with typical bacterial infections were less likely to have a comorbidity such as asthma as compared to children with viruses alone. Asthma is a well-established risk factor for hospitalization with viruses [13]. The finding that asthma and reactive airway disease were more frequent as comorbidities in the viruses group as compared to the typical bacteria group is therefore not surprising (Table 1). Interestingly, the viruses-typical bacteria group had a low prevalence rate of asthma, which, when coupled with worse outcomes, could lead to the speculation that asthmatics with viral pneumonia were more likely to be admitted for reasons related to their underlying chronic diagnosis, rather than for the severity of their CAP. Alternatively, the immune status of the allergic lung could provide protection from the consequences of these infections. As has been suggested from work in animal models, this could mitigate disease severity [14]. Obesity was identified as a risk factor for severe influenza during the 2009 influenza A(H1N1) pandemic [15]. Although our analyses were limited because of the preponderance of young children in this study, obesity did not appear to be a risk factor for coinfections (Table 1).
In support of a body of literature examining influenza-associated coinfections [(6)], virus-typical bacterium coinfections were more severe than virus infections alone, even when multiple viruses were codetected (Tables 2 and 3). In influenza studies, bacterial coinfection is common in severe and fatal cases [7, 11, 16]. In particular, the presence of S. pneumoniae and S. aureus is associated with high morbidity and mortality [7, 8, 17, 18]. Similarly, while very few deaths were observed during this study, serious outcomes occurred more frequently in children with virus-bacterium coinfections as compared to those virus infections, and their length of hospital stay was longer. This finding did not extend to coinfection with atypical bacteria, which may be related to a greater frequency of antibiotic use before admission (Table 1). Intensive care unit admission, use of mechanical ventilation and length of stay were also all increased when RSV was complicated by a typical bacterial pathogen. Unfortunately, despite the extremely large size of the study, the numbers of individual pathogen pairings were too low to examine other individual combinations. Of note, severe outcomes were seen in single-virus infections with all viruses studied but were more common when typical bacteria were involved.
Whether some viruses such as hRV could cause severe acute respiratory illness in the absence of a coinfection has been questioned [19]. We observed severe disease at a low frequency with both single hRV infections and coinfections where hRV was detected, but we did not observe a difference between these groups, as others have reported [18]. In general, virus-virus coinfections were not more severe than single-virus infections and did not have worse outcomes (Tables 2 and 3), in agreement with a smaller study [20]. Two combinations of virus-virus pairs yielded increased supplemental oxygen use, compared with infections with a single virus (Table 3). Interestingly, RSV coinfections with influenza virus and hAdV were associated with increased need for supplemental oxygen, suggesting that coinfection with these relatively virulent viruses led to increased severity.
One of the most interesting observations from this study was the distribution of frequencies of specific codetections. A wealth of literature from animal models and epidemiologic studies [6] suggests that viral infections enhance acquisition of bacterial pathogens by hampering immune defenses through a variety of mechanisms. Similarly, studies of the common cold in crowded living conditions suggest similar changes to susceptibility in humans to acquisition of colonization with pathogenic bacteria [21, 22]. However, prior studies have not been large enough to adequately compare the actual frequency of specific pathogen-pathogen pairs in a coinfection to the frequency expected by chance. We report that the observed prevalence of several specific codetections was lower than expected, particularly when examining virus pairings with M. pneumoniae (Figure 3). A smaller study of M. pneumoniae infections noted that codetection with viruses was rare [23]. One hypothesis to explain the relative paucity of M. pneumoniae and virus codetections is that infections with these pathogens exhibit different age and seasonal distributions. Indeed, the mean age of children presenting with M. pneumoniae in the study was 98 months, while the mean age for children hospitalized with respiratory viruses ranged from 25 months (for hAdV and RSV) to 63 months (for influenza virus) (Supplementary Figure 1). Stratification of the analyses by age (<5 years and ≥5 years) demonstrated a similar magnitude of effect as compared to the overall study population, although the CIs were wider and crossed 0 in some cases, suggesting decreased power to examine the interactions (Supplementary Figures 2 and 3). In addition, M. pneumoniae was not detected with H. influenzae or S. pyogenes, common bacterial pathogens of the upper respiratory tract. This may be due to the low numbers of bacterial infections in the study, or it may be related to competition within the upper respiratory tract between bacteria, as has been observed in bacterial coinfection models [24].
Several virus-virus pairings were also uncommonly observed. Interestingly, an additional virus-virus interaction, hAdV and RSV, was also significantly less common than expected when analyzing only children aged <5 years (Supplementary Figure 2). The simplest explanation would be differences in seasonality that limited temporal circulation together and thus minimized the chance of a codetection (Supplementary Figure 4). Because we were studying children hospitalized with pneumonia and not a broader population, further analyses regarding the impact of seasonality were not possible. Another hypothesis is that induction of interferon by a first virus protects against the second virus. At least 2 studies have suggested that circulation of rhinoviruses delays or reduces the circulation of other viruses, as was seen in this study with hRV pairings with RSV, PIV, and hMPV [25, 26]. This and other hypotheses should be studied in model systems. Unfortunately, the prevalence of specific bacterial infections was too low for a similar analysis to be performed for specific virus and bacterium pairings. From existing observational studies and animal models (eg, mouse and ferret models of influenza virus and S. pneumoniae coinfection [27, 28]) one would have expected an increased frequency of specific pathogen pairings relative to chance. Why this was seen for hAdV and RSV and for hCoV and RSV but not other pairings is unclear; further study is indicated to replicate and potentially explain these findings.
This study has several limitations and challenges. Detection of bacteria in patients with CAP is difficult and may have caused misclassification of some virus-bacterium coinfections into the viruses-only group. The assays to determine etiology and define classes of infections did not include all possible viral and bacterial respiratory pathogens, which may also have caused misclassification, and only 44% of children had acute and convalescent serum specimens available for serologic testing. Some results may be confounded by factors such as age, comorbidities, or strong seasonal circulation of certain viruses; the analyses were unadjusted for these potential confounders. We also could not adjust for the likelihood of hospitalization with each pathogen, which will bias the determination of the frequency of codetection in the hospital downward if patients infected with the pathogen in the community are more likely to be admitted than patients not infected with the pathogen. Finally, the contribution of any specific pathogen to the severity of infection cannot be determined with certainty; on average, 3% of asymptomatic controls (and up to 17% with hRV) in the EPIC study had a virus detected [1].
Nevertheless, this study was very large and comprehensive in its assessment of the etiology of coinfections and the demographic characteristics and outcomes of these children hospitalized for CAP. This allowed pathogen-specific analyses that could not have been attempted with smaller studies. The findings of differences in clinical characteristics and severity of outcomes with virus-typical bacterium coinfections as compared to viruses alone and the few observed differences in comparisons of single-virus infections to multiple-virus infections or of single-typical bacterium infections to virus-typical bacterium infections have important implications for the diagnosis and management of CAP in children. In particular, better detection methods are needed for a broad array of pathogens, and studies to update clinical testing patterns should take into account these findings.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Financial support. This work was supported by the Centers for Disease Control and Prevention (grant U18IP000489 to J. A. M.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Jain S, Williams DJ, Arnold SR et al. ; CDC EPIC Study Team. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med 2015; 372:835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Honkinen M, Lahti E, Österback R, Ruuskanen O, Waris M. Viruses and bacteria in sputum samples of children with community-acquired pneumonia. Clin Microbiol Infect 2012; 18:300–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wei L, Liu W, Zhang XA et al. . Detection of viral and bacterial pathogens in hospitalized children with acute respiratory illnesses, Chongqing, 2009–2013. Medicine 2015; 94:e742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Estenssoro E, Rios FG, Apezteguia C et al. . Pandemic 2009 influenza A in Argentina: a study of 337 patients on mechanical ventilation. Am J Respir Crit Care Med 2010; 182:41–8. [DOI] [PubMed] [Google Scholar]
- 5. Domínguez-Cherit G, Lapinsky SE, Macias AE et al. . Critically Ill patients with 2009 influenza A(H1N1) in Mexico. JAMA 2009; 302:1880–7. [DOI] [PubMed] [Google Scholar]
- 6. McCullers JA. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 2014; 12:252–62. [DOI] [PubMed] [Google Scholar]
- 7. Palacios G, Hornig M, Cisterna D et al. . Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS One 2009; 4:e8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finelli L, Fiore A, Dhara R et al. . Influenza-associated pediatric mortality in the United States: increase of Staphylococcus aureus coinfection. Pediatrics 2008; 122:805–11. [DOI] [PubMed] [Google Scholar]
- 9. Nolte FS. Molecular diagnostics for detection of bacterial and viral pathogens in community-acquired pneumonia. Clin Infect Dis 2008; 47(Suppl 3):S123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 2008; 198:962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gill JR, Sheng ZM, Ely SF et al. . Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med 2010; 134:235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dawood FS, Chaves SS, Perez A et al. . Complications and associated bacterial coinfections among children hospitalized with seasonal or pandemic influenza, United States, 2003–2010. J Infect Dis 2014; 209:686–94. [DOI] [PubMed] [Google Scholar]
- 13. American Academy of Pediatrics Committee on Infectious Diseases. Recommendations for Prevention and Control of Influenza in Children, 2015–2016. Pediatrics 2015; 136:792–808. [DOI] [PubMed] [Google Scholar]
- 14. Samarasinghe AE, Woolard SN, Boyd KL, Hoselton SA, Schuh JM, McCullers JA. The immune profile associated with acute allergic asthma accelerates clearance of influenza virus. Immunol Cell Biol 2014; 92:449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguyen-Van-Tam JS, Openshaw PJ, Hashim A et al. . Risk factors for hospitalisation and poor outcome with pandemic A/H1N1 influenza: United Kingdom first wave (May-September 2009). Thorax 2010; 65:645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Centers for Disease Control and Prevention. Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1) - United States, May-August 2009. MMWR Morb Mortal Wkly Rep 2009; 58:1071–4. [PubMed] [Google Scholar]
- 17. Weinberger DM, Simonsen L, Jordan R, Steiner C, Miller M, Viboud C. Impact of the 2009 influenza pandemic on pneumococcal pneumonia hospitalizations in the United States. J Infect Dis 2012; 205:458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nascimento-Carvalho AC, Ruuskanen O, Nascimento-Carvalho CM. Comparison of the frequency of bacterial and viral infections among children with community-acquired pneumonia hospitalized across distinct severity categories: a prospective cross-sectional study. BMC Pediatr 2016; 16:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moesker FM, van Kampen JJ, van Rossum AM et al. . Viruses as Sole Causative Agents of Severe Acute Respiratory Tract Infections in Children. PLoS One 2016; 11:e0150776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Asner SA, Rose W, Petrich A, Richardson S, Tran DJ. Is virus coinfection a predictor of severity in children with viral respiratory infections?Clin Microbiol Infect 2015; 21:264.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gwaltney JM Jr, Sande MA, Austrian R, Hendley JO. Spread of Streptococcus pneumoniae in families. II. Relation of transfer of S. pneumoniae to incidence of colds and serum antibody. J Infect Dis 1975; 132:62–8. [DOI] [PubMed] [Google Scholar]
- 22. Regev-Yochay G, Raz M, Dagan R et al. . Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis 2004; 38:632–9. [DOI] [PubMed] [Google Scholar]
- 23. Song Q, Xu BP, Shen KL. Effects of bacterial and viral co-infections of Mycoplasma pneumoniae pneumonia in children: analysis report from Beijing Children’s Hospital between 2010 and 2014. Int J Clin Exp Med 2015; 8:15666–74. [PMC free article] [PubMed] [Google Scholar]
- 24. Lysenko ES, Clarke TB, Shchepetov M et al. . Nod1 signaling overcomes resistance of S. pneumoniae to opsonophagocytic killing. PLoS Pathog 2007; 3:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Casalegno JS, Ottmann M, Duchamp MB et al. . Rhinoviruses delayed the circulation of the pandemic influenza A (H1N1) 2009 virus in France. Clin Microbiol Infect 2010; 16:326–9. [DOI] [PubMed] [Google Scholar]
- 26. Greer RM, McErlean P, Arden KE et al. . Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections?J Clin Virol 2009; 45:10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis 2002; 186:341–50. [DOI] [PubMed] [Google Scholar]
- 28. Peltola VT, Boyd KL, McAuley JL, Rehg JE, McCullers JA. Bacterial sinusitis and otitis media following influenza virus infection in ferrets. Infect Immun 2006; 74:2562–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.