Abstract
The calcium-sensing receptor (CaR) regulates transepithelial calcium transport into milk by mammary epithelial cells. Using a genome-wide screening strategy, we identified the plasma membrane calcium ATPase isoform 2 (PMCA2) as a potential downstream target of the CaR. We show that PMCA2 expression in the mouse mammary gland increases during lactation and that PMCA2 is localized solely to the apical plasma membrane of mammary epithelial cells. In milk from deafwaddler mice, which have mutations in the gene encoding PMCA2, calcium concentrations were reduced, confirming its importance in calcium transport into milk. Furthermore, in cultured primary and EpH4 mouse mammary epithelial cells, CaR stimulation up-regulated calcium-dependent ATPase activity in plasma membrane preparations. By small interfering RNA-mediated gene knockdown of PMCA2, we show that PMCA2 accounts for the preponderance of calcium-ATPase activity. We also show that reduction of CaR expression with small interfering RNA eliminates the ability of extracellular calcium to elicit an increase in calcium-dependent ATPase activity in EpH4 cell membranes. These results demonstrate that activation of the CaR increases PMCA2 activity in mouse mammary epithelial cells, providing a mechanism for the regulation of transepithelial calcium transport by calcium in the lactating mouse mammary gland.
DURING LACTATION, THE mammary gland produces milk, which contains all of the nutrients needed for neonatal growth, including calcium. To supply enough calcium in milk, mammary epithelial cells (MECs) must transport large amounts against a steep concentration gradient over an extended time period. Furthermore, despite the large throughput of calcium, MECs must maintain low cytoplasmic free calcium concentrations to avoid toxicity. There is no paracellular passage of calcium into milk (1, 2). Rather, it is secreted from MECs through a transepithelial process. Little is known about the entry of calcium into MECs, although it has been suggested that a stretch-activated channel may be involved (2, 3). Once in the epithelial cell, calcium has been thought to be pumped into the Golgi in an ATP-dependent fashion, where it becomes associated with caseins, citrate, phosphate, and other calcium-binding molecules (1, 2). The majority of milk calcium is bound to caseins (1), phosphoproteins that form large, highly organized macromolecular micelles in the presence of calcium (4). Therefore, the widely held view has been that calcium enters milk through the secretory pathway primarily contained within casein micelles and that little or no free calcium is transported directly across the apical membrane (1, 5).
There is relatively little information regarding the identity of specific molecules responsible for calcium transport in MECs. Several studies have described Ca-ATPase activity in the Golgi fraction of the lactating mammary gland (5–7), which may be due to the expression of two secretory pathway Ca-ATPases (SPCA1 and SPCA2) (1, 8–10). In addition, mammary gland expression of plasma membrane calcium ATPase isoform 2 (PMCA2) has been shown to increase during lactation in several species (10–13). Reinhardt and colleagues (13) recently reported that mice deficient for PMCA2 have a substantial reduction in milk calcium concentration, suggesting that this transporter is important to calcium transport into milk.
PMCAs are P-type primary ion transport ATPases. There are four PMCA genes, and alternative splicing produces up to 30 different individual PMCA isoforms (14–16). In addition to the lactating mammary gland, PMCA2 mRNA is highly expressed only in certain regions of the brain (17). Recent work has shown that specific splice variants of the PMCA2 gene are differentially sorted. Inclusion of three alternative exons at site A, within the first intracellular loop between membrane spanning domains 2 and 3, defines a variant, PMCA2w, that traffics to the apical membrane instead of the basolateral membrane (18–20). Interestingly, PMCA2w isoforms have been reported to be expressed in the lactating mammary gland (10). Therefore, despite the traditional model, calcium, in fact, may be transported across the apical surface of MECs.
We have previously shown that the calcium-sensing receptor (CaR) participates in regulating calcium transport in the mouse mammary gland (21, 22). The CaR is a G protein-coupled receptor that enables the parathyroid glands to regulate PTH secretion in response to changes in the extracellular calcium concentration (23, 24). The CaR has subsequently been found to be expressed in many other tissues, where it participates in a wide variety of physiological functions (25). In the mammary gland, CaR expression is greatly up-regulated during lactation, and activation of the CaR on the basolateral surface of MECs stimulates calcium transport into milk in vivo and in vitro (21, 22). As a result, increased calcium delivery to the mammary gland augments calcium transport into milk (26).
In this study, we examined PMCA2 as a potential target of CaR signaling that might mediate the effects of calcium-stimulated calcium transport in the mammary gland. Given the paucity of knowledge about the specific molecules employed by MECs to transport calcium, we used a genomic approach to verify PMCA2 as a likely target of CaR signaling. We found PMCA2 to be the predominant calcium transporter in the mammary gland, where it is expressed on the apical surface of MECs. Two separate genetic models of loss of PMCA2 function confirmed that this pump is responsible for the majority of calcium transport into milk. Finally, we found that CaR signaling regulates PMCA2 function, explaining the ability of calcium to stimulate its own transport across MECs. These data suggest a new model for calcium transport into milk.
Materials and Methods
Animals
All experiments were carried out with the approval of the Yale University Animal Care and Use Committee. Deafwaddler mice (dfw, strain C3H/HeJ-Atp2b2dfw/J), deafwaddler-2J mice (dfw-2J, strain CByJ.A-Atp2b2dfw-2J/J), C3H mice (strain C3H/HeJ), and BALB/c mice (strain BALB/cByJ) were purchased from the Jackson Laboratory (Bar Harbor, ME). CD1 mice were purchased from Charles River Laboratories (Wilmington, MA). The deafwaddler mice have a G to A transition mutation in Atp2b2, the gene encoding PMCA2 (27, 28). The dfw-2J mice have a two-nucleotide deletion that results in a frameshift mutation with a premature stop codon at position 471 (28). Wild-type C3H mice served as controls for the dfw mice, whereas wild-type BALB/c mice were controls for the dfw-2J mice. Female mice of each genotype were mated at 8 wk of age and were milked on d 12 of lactation as previously described (22). The no. 4 inguinal mammary glands were harvested for RNA isolation.
Microarray analysis
RNA was purified from mammary glands of BALB/c (22) and FVB (29) mice at various stages of postnatal mammary gland development and used for microarray analysis with Affymetrix Mu74Av2 microarray chips. Animals, tissue collection, RNA isolation, and microarray analysis have been described previously (22, 29).
RT-PCR and quantitative RT-PCR (QRT-PCR)
RNA was isolated from tissues and cells using TRIzol (Invitrogen, Carlsbad, CA) and further purified with on-column DNase treatment using RNeasy Mini columns (QIAGEN, Valencia, CA). The High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) was used for cDNA synthesis from 10 μg RNA in a 100-μl reaction volume. For RT-PCR and QRT-PCR, 1 μl cDNA was used in each 25-μl reaction. QRT-PCR was performed in the Opticon II DNA Engine (Bio-Rad, Hercules, CA) using the Full Velocity SYBR Green QPCR system (Stratagene, La Jolla, CA). Each reaction was run in duplicate, and the comparative cycle threshold (CT) method was used to determine relative expression levels. QRT-PCR primers are shown in Table 1.
TABLE 1.
Primers used for quantitative RT-PCR
| Common name | Primer sequences (5′–3′) | Gene symbol |
|---|---|---|
| HPRT | ||
| Forward | CAGTACAGCCCCAAAATGGT | Hprt1 |
| Reverse | CAGTACAGCCCCAAAATGGT | |
| GAPDH | ||
| Forward | CGTCCCGTAGACAAAATGGT | Gapdh |
| Reverse | TCAATGAAGGGGTCGTTGAT | |
| β-Actin | ||
| Forward | GTCCACCTTCCAGCAGATGT | Actb |
| Reverse | AAGGGTGTAAAACGCAGCTC | |
| PMCA2 | ||
| Forward | AGAGAAGTCGGTGCTTCAGG | Atp2b2 |
| Reverse | GAAGGTGTCCACGGTGAAGT | |
| SERCA2 | ||
| Forward | CCCGAAACTACCTGGAACAA | Atp2a2 |
| Reverse | AGGGCTGGTAGATGTGTTGC | |
| SPCA1 | ||
| Forward | TCTCTGTGGGGGACAGAGTT | Atp2c1 |
| Reverse | GCTGAGGGGCTGTCACTTTA | |
| SPCA2 | ||
| Forward | GAGCGTCCTCACCAAAGAAT | Atp2c2 |
| Reverse | CCAGAGACTTCTCCGACCTG | |
| K18 | ||
| Forward | CTTGCGAATTCTGTGGACAA | Krt1–18 |
| Reverse | AGTCCATGGATGTCGCTCTC |
We used the AccuPrime Taq DNA Polymerase High Fidelity PCR system (Invitrogen) to determine splice variants of PMCA2. The following primers spanning splice site A of the mouse Atp2b2 gene were modified from those used previously in rats (10): 5′-TGAAAGCTCACTCACAGGG-3′ and 5′-TCAGAGGCTGCATCTCCAT-3′. Products were analyzed by 1.5% agarose gel electrophoresis and confirmed by sequencing.
Immunofluorescence and immunoelectron microscopy
PMCA2 staining was performed with the PA1-915 rabbit polyclonal, affinity-purified antibody (Affinity Bioreagents, Golden, CO). Tissue was fixed with Nakane’s fixative and Epon-embedded, after which 1- to 2-μm sections were stained using standard techniques. Immunoperoxidase staining of the lactating d-12 CD1 mouse mammary tissue and electron microscopy were also performed using standard techniques (30). Immunofluorescent staining for PMCA2 in EpH4 cells was carried out after fixing the cells in 4% paraformaldehyde for 15 min at room temperature and permeabilizing them in PBS with 0.05% Triton X-100. The secondary antibody was goat antirabbit Alexa594 (Invitrogen).
Western blotting
Plasma membrane protein fractions were prepared from mammary gland tissue from lactating mice (d 12) (essentially as in Ref. 10) and from cultured cells, as described below for the Ca-ATPase assay. Western blotting was performed using standard techniques, and the anti-PMCA2 antibody PA1-915 (Affinity Bioreagents) at 1:1000 followed by anti-β-actin (clone AC-74; Sigma-Aldrich, St. Louis, MO) at 1:50,000. Densitometric quantitation of protein band intensity was done on a Kodak Image Station 440CF (Kodak, Rochester, NY).
Milk calcium and milk protein analysis
Milk was diluted 1:100 in distilled water, and total milk calcium was measured using the colorimetric QuantiChrom calcium assay kit (BioAssay Systems, Hayward, CA). Protein concentrations were determined using the Bradford reagent (Bio-Rad) with BSA standards.
Cells and transfection
Primary mouse MECs were prepared as previously described (22). MECs were cultured in DMEM/F12 with 5% FBS (Invitrogen) plus 5 μg/ml insulin, 1 μg/ml hydrocortisone, 3 μg/ml prolactin (Cambrex Corp., Walkersville, MD), 50 μg/ml gentamycin (Invitrogen), and 2.5 μg/ml amphotericin-B (Sigma-Aldrich). EpH4 mouse MECs were grown in DMEM with 10% FBS, 50 μg/ml gentamycin, and 2.5 μg/ml amphotericin-B.
Transfection of small interfering RNA (siRNA) was performed with Lipofectamine 2000 (Invitrogen). All transfections were done without antibiotics, and all analyses were done 48 h after transfections. To optimize transfection, 12.5, 25, or 50 pmol of the BLOCK-iT Fluorescent Oligo (Invitrogen) was transfected into EpH4 cells using 0.5, 1, or 1.5 μl Lipofectamine 2000 per well of four-well glass chamber slides. The best transfection efficiency was obtained with 50 pmol RNA oligo per 1.5 μl Lipofectamine 2000 (see Fig. 7C), which also maximally reduced GAPDH mRNA levels using Silencer GAPDH siRNA (mouse, human, and rat; Ambion, Austin, TX). All subsequent transfections used these conditions. Atp2b2 Stealth Select RNAis (MSS202252 or MSS202250) were used to knock down PMCA2, and Casr Stealth Select RNAis (MSS202652 or MSS202654) (Invitrogen) were used to knock down CaR. The Stealth RNAi Negative Control Med GC (Invitrogen) was transfected in parallel as a control in all experiments.
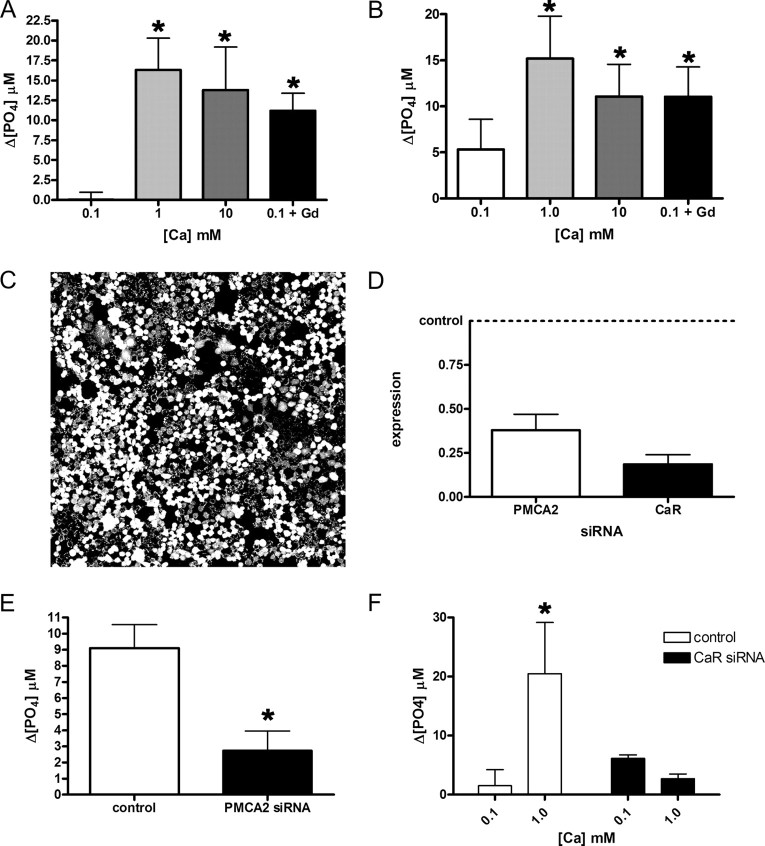
Fig. 7.
Activation of the CaR augments PMCA2 activity. Ca-ATPase activity in the plasma membrane fraction of mouse MECs (A) and EpH4 cells (B) increases when the concentration of calcium in the media is raised from 0.1 to 1 or 10 mm. Bars in A represent the mean of three experiments, whereas bars in B represent the mean of five experiments. In A, P < 0.05 for 0.1 mm calcium vs. 1 and 10 mm calcium; in B, P < 0.01 for 0.1 vs. 1 mm calcium and P < 0.05 for 0.1 calcium vs. 10 mm calcium. In addition, if 100 μm GdCl3, a CaR agonist, is added to these cells in media with 0.1 mm calcium, an increase in Ca-ATPase activity is also seen, compared with 0.1 mm calcium alone (P < 0.05 in A; P < 0.05 in B). C, We achieved high transfection efficiency as demonstrated with fluorescent oligonucleotides transfected into EpH4 cells. D, Transfection of siRNA for mouse PMCA2 reduced mRNA levels by almost 70%, whereas transfection of CaR siRNA reduced CaR mRNA by approximately 80%. E, Ca-ATPase activity in the plasma membrane fraction of EpH4 cells grown at 2 mm calcium was reduced by 75% after transfection with PMCA2 siRNA compared with transfection with the control siRNA (P < 0.05, three experiments). These data suggest that PMCA2 accounts for most of the Ca-dependent ATPase activity present in the plasma membrane of MECs. F, Ca-ATPase activity increased significantly when the extracellular calcium concentration was raised from 0.1 to 1 mm calcium in EpH4 cells transfected with control siRNA (P < 0.05, three experiments). However, there was no increase in Ca-ATPase activity in the plasma membrane fractions from cells transfected with CaR siRNA. These data suggest that the CaR mediates the response of PMCA2 to extracellular calcium. Statistically significant differences are denoted by asterisks.
Calcium-dependent ATPase activity assay
Cells were incubated in calcium-free DMEM with hydrocortisone, insulin, prolactin, and 0.1 mm CaCl2 for 15 min at 37 C. The medium was then adjusted to 0.1 mm, 1 mm, or 10 mm CaCl2, or 0.1 mm CaCl2 plus 100 μm GdCl3, and cells were incubated at 37 C for an additional 3 h. The cells were washed with 5 ml 10 mm Tris-maleate (pH 7.4) (Sigma-Aldrich), and cell lysates were prepared by scraping cells in 10 mm Tris-maleate (pH 7.4) with Complete Protease Inhibitor Cocktail (EDTA-free; Roche Molecular Biochemicals, Indianapolis, IN) and passing them through a 22-gauge needle 12 times. The lysate was centrifuged at 5000 × g for 10 min, and the supernatant was removed to a fresh tube. The plasma membrane fraction was obtained by centrifuging the supernatant for 30 min at 100,000 × g. The pellet was resuspended in 10 mm Tris-maleate with protease inhibitors. Protein concentrations were determined using the Bio-Rad assay as described for milk protein analysis. Ca-ATPase activity was measured according to the method of Kosk-Kosicka (31). Assay buffer consisted of 50 mm Tris-maleate (pH 7.4), 8 mm MgCl2, 120 mm KCl, 1 mm EGTA, and 10 μm thapsigargin (Sigma-Aldrich) with or without 1.008 mm CaCl2. Two micrograms of membrane preparation in 10 μl 10 μm Tris-maleate were added to 90 μl assay buffer with and without calcium. Reactions were started by the addition of 5 μl 60 mm Tris-ATP and incubated for 15 min at 37 C, after which 300 μl Lin-Morales reagent, diluted 1:3 in distilled water, was added, and phosphate concentrations were determined by reading the absorbance at 350 nm after 30 min. The concentration of phosphate was determined from the linear calibration curve, prepared by adding varying concentrations of K2HPO4 to the assay buffer without membrane preparations. The Ca-ATPase activity was calculated as the difference between the phosphate generated in the presence of calcium and the phosphate generated in the absence of calcium.
Statistical analysis
GraphPad Prism, version 4.02 for Windows, was used for all statistical analysis. Comparisons of three or more groups used one-way ANOVA with the Newman-Keuls multiple comparison test; comparisons of two groups used an unpaired, two-tailed Student’s t test.
Results
PMCA2 is the principal mammary calcium transporter expressed during lactation
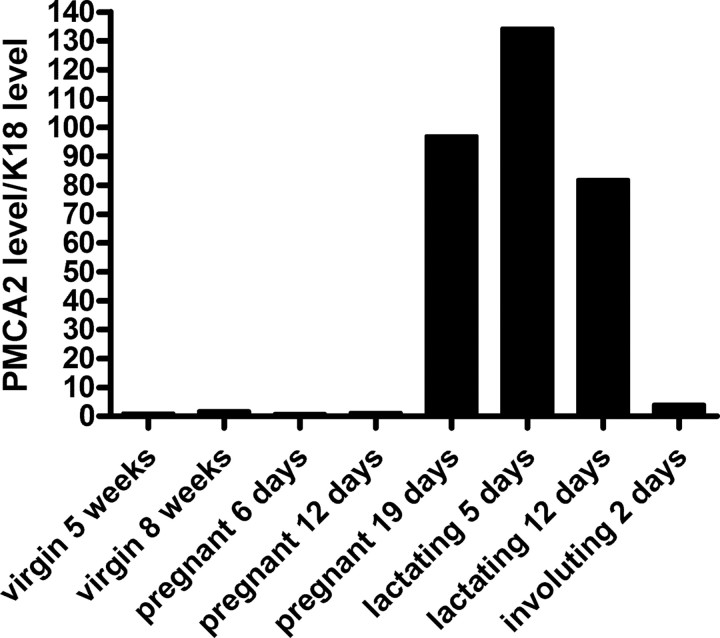
To elucidate the molecular pathway by which the CaR regulates calcium transport in the mammary gland, we first needed to know what calcium channels or transporters were expressed during lactation. We therefore performed a genome-wide screen using Affymetrix microarray analysis of RNA from nulliparous, pregnant, lactating, and involuting mouse mammary glands to identify genes involved in calcium handling and/or transport whose expression was specifically up-regulated during lactation. Table 2 lists the genes on the array identified as calcium channels, calcium transporters, or calcium-binding proteins that were expressed at any level in the mouse mammary gland. Of these genes, Atp2b2, the gene encoding PMCA2, was most dramatically up-regulated specifically during lactation. Based on the array data, expression of Atp2b2 increased over 60-fold on d 9 of lactation compared with nulliparous controls. Two days after weaning, expression of Atp2b2 was greatly reduced. The profile of Atp2b2 (PMCA2) expression in the mouse mammary gland was confirmed by real-time QRT-PCR (Fig. 1). When normalized to the epithelial cell marker keratin 18 (K18), Atp2b2 mRNA levels were low in the mammary glands of virgin CD1 mice, began to increase in late pregnancy, rose to a peak during lactation of about 134 times higher than virgin controls, and dropped precipitously upon weaning. Therefore, PMCA2 is the predominant calcium transporter expressed within the mammary gland, and its profile of expression is typical of genes involved in milk production (32).
TABLE 2.
Genes on the array identified as calcium channels, calcium transporters, or calcium-binding proteins that were expressed at any level in the mouse mammary gland
| Gene | Protein | Virgin | Pregnant | Lactating | Involuting |
|---|---|---|---|---|---|
| Ca-ATPases | |||||
| PMCAs | |||||
| Atp2b2 | PMCA2 | 1 | 1.56 ± 0.17 | 62.95 ± 6.58 | 1.40 ± 0.17 |
| SERCAs | |||||
| Atp2a1 | SERCA1 | 1 | 2.60 ± 0.95 | 0.73 ± 0.10 | 0.89 ± 0.21 |
| Atp2a2 | SERCA2 | 1 | 0.46 ± 0.03 | 0.68 ± 0.10 | 0.38 ± 0.02 |
| Atp2a3 | SERCA3 | 1 | 1.20 ± 0.03 | 1.51 ± 0.03 | 1.27 ± 0.06 |
| Calcium channels | |||||
| Voltage-dependent channels | |||||
| Cacna1a | α1-a subunit | 1 | 1.11 ± 0.02 | 1.39 ± 0.09 | 1.14 ± 0.03 |
| Cacna1b | α1-b subunit | 1 | 1.16 ± 0.02 | 1.40 ± 0.03 | 1.19 ± 0.03 |
| Cacna1c | α1-c subunit | 1 | 0.81 ± 0.04 | 1.26 ± 0.09 | 0.96 ± 0.10 |
| Cacna1e | α1-e subunit | 1 | 1.17 ± 0.04 | 1.53 ± 0.05 | 1.27 ± 0.05 |
| Cacna1g | α1-g subunit | 1 | 1.07 ± 0.04 | 1.49 ± 0.07 | 1.13 ± 0.08 |
| Cacna1 h | α1-h subunit | 1 | 1.23 ± 0.05 | 1.64 ± 0.13 | 1.23 ± 0.05 |
| Cacna1s | α1-s subunit | 1 | 1.23 ± 0.02 | 1.65 ± 0.08 | 1.32 ± 0.05 |
| Cacna2d1 | α2-δ subunit 1 | 1 | 1.24 ± 0.12 | 1.58 ± 0.05 | 1.23 ± 0.08 |
| Cacna2d3 | α2-δ subunit 3 | 1 | 1.50 ± 0.07 | 2.44 ± 0.13 | 1.65 ± 0.09 |
| Cacnb2 | β2-subunit | 1 | 1.12 ± 0.03 | 1.43 ± 0.03 | 1.20 ± 0.07 |
| Cacng2 | γ2 subunit | 1 | 1.44 ± 0.07 | 2.17 ± 0.46 | 1.47 ± 0.11 |
| Transient receptor potential channels | |||||
| Trpc1 | 1 | 1.28 ± 0.03 | 1.71 ± 0.09 | 1.36 ± 0.07 | |
| Trpc4 | 1 | 1.37 ± 0.05 | 1.84 ± 0.07 | 1.54 ± 0.17 | |
| Trpc5 | 1 | 1.31 ± 0.04 | 1.87 ± 0.09 | 1.42 ± 0.10 | |
| Trpc6 | 1 | 1.57 ± 0.08 | 2.80 ± 0.20 | 1.81 ± 0.22 | |
| Trpv2 | 1 | 0.27 ± 0.02 | 0.33 ± 0.02 | 0.28 ± 0.02 | |
| Intracellular calcium release | |||||
| Inositol 1,4,5-triphosphate receptors | |||||
| Itpr1 | 1 | 0.77 ± 0.05 | 0.90 ± 0.05 | 1.01 ± 0.20 | |
| Itpr2 | 1 | 0.29 ± 0.04 | 0.06 ± 0.00 | 0.71 ± 0.07 | |
| Itpr5 | 1 | 1.11 ± 0.03 | 1.51 ± 0.06 | 1.21 ± 0.07 | |
| Ryanodine receptors | |||||
| Ryr1 | 1 | 0.20 ± 0.00 | 0.26 ± 0.01 | 0.22 ± 0.01 | |
| Ryr2 | 1 | 1.10 ± 0.02 | 1.45 ± 0.06 | 1.15 ± 0.04 | |
| Ryr3 | 1 | 1.13 ± 0.05 | 1.75 ± 0.08 | 1.30 ± 0.13 | |
| Calcium-binding proteins | |||||
| Calmodulins | |||||
| Calm1 | Calmodulin 1 | 1 | 0.66 ± 0.02 | 0.38 ± 0.04 | 0.55 ± 0.05 |
| Calm2 | Calmodulin 2 | 1 | 1.28 ± 0.02 | 0.50 ± 0.04 | 1.20 ± 0.02 |
| Calm3 | Calmodulin 3 | 1 | 1.13 ± 0.10 | 0.53 ± 0.13 | 0.87 ± 0.01 |
| Calm4 | Calmodulin 4 | 1 | 1.06 ± 0.03 | 1.56 ± 0.13 | 1.10 ± 0.04 |
| Calml4 | Calmodulin-like 4 | 1 | 0.28 ± 0.01 | 0.37 ± 0.01 | 0.32 ± 0.02 |
| Calbindins | |||||
| Calb1 | Calbindin 1 | 1 | 1.11 ± 0.04 | 1.59 ± 0.07 | 1.19 ± 0.07 |
| Calb2 | Calbindin 2 | 1 | 1.10 ± 0.02 | 1.35 ± 0.02 | 1.17 ± 0.04 |
| Nucleobindins | |||||
| Nucb1 | Nucleobindin 1 | 1 | 0.50 ± 0.06 | 0.10 ± 0.01 | 0.13 ± 0.01 |
| Nucb2 | Nucleobindin 2 | 1 | 3.05 ± 0.36 | 8.14 ± 1.67 | 14.42 ± 1.48 |
| Calreticulins | |||||
| Calr | Calreticulin | 1 | 1.13 ± 0.04 | 1.52 ± 0.06 | 1.18 ± 0.05 |
| Calr4 | Calreticulin 4 | 1 | 1.10 ± 0.03 | 1.86 ± 0.11 | 1.22 ± 0.07 |
| S100 proteins | |||||
| S100a1 | 1 | 0.63 ± 0.08 | 0.19 ± 0.02 | 0.76 ± 0.06 | |
| S100a3 | 1 | 1.68 ± 0.12 | 3.72 ± 0.32 | 2.00 ± 0.23 | |
| S100a4 | 1 | 0.39 ± 0.02 | 0.26 ± 0.04 | 0.32 ± 0.02 | |
| S100a6 | 1 | 0.23 ± 0.04 | 0.08 ± 0.01 | 0.30 ± 0.07 | |
| S100a8 | 1 | 0.71 ± 0.07 | 0.71 ± 0.03 | 0.80 ± 0.18 | |
| S100a9 | 1 | 1.12 ± 0.07 | 1.40 ± 0.05 | 1.20 ± 0.09 | |
| S100a10 | 1 | 0.45 ± 0.05 | 0.06 ± 0.01 | 0.44 ± 0.06 | |
| S100a11 | 1 | 0.44 ± 0.06 | 0.06 ± 0.01 | 1.07 ± 0.05 | |
| S100a13 | 1 | 0.67 ± 0.07 | 0.20 ± 0.03 | 0.71 ± 0.05 | |
| S100pbp | 1 | 0.86 ± 0.06 | 1.01 ± 0.06 | 0.95 ± 0.07 | |
| Other | |||||
| Rcvrn | Recoverin | 1 | 1.12 ± 0.03 | 1.50 ± 0.04 | 1.21 ± 0.07 |
| Anxa6 | Annexin A6 | 1 | 0.74 ± 0.07 | 0.68 ± 0.03 | 0.84 ± 0.07 |
Expression levels are given relative to the virgin mammary gland samples.
Fig. 1.
Levels of mammary PMCA2 mRNA as determined by QRT-PCR. PMCA2 mRNA levels were assayed at the different developmental stages noted and were first normalized to HPRT and then to K18 expression to adjust for the changing epithelial content of the gland at the different stages of development. Data are expressed as the relative expression compared with 5-wk-old virgin mice. At the end of pregnancy on d 19, there is a large increase in epithelial PMCA2 expression. During lactation, PMCA2 expression peaks at up to 134 times its level in the mammary gland of a 5-wk-old nulliparous mouse. Within 2 d after forced weaning (done on d 12 of lactation), expression of PMCA2 has fallen almost to baseline.
PMCA2 is expressed on the apical plasma membrane
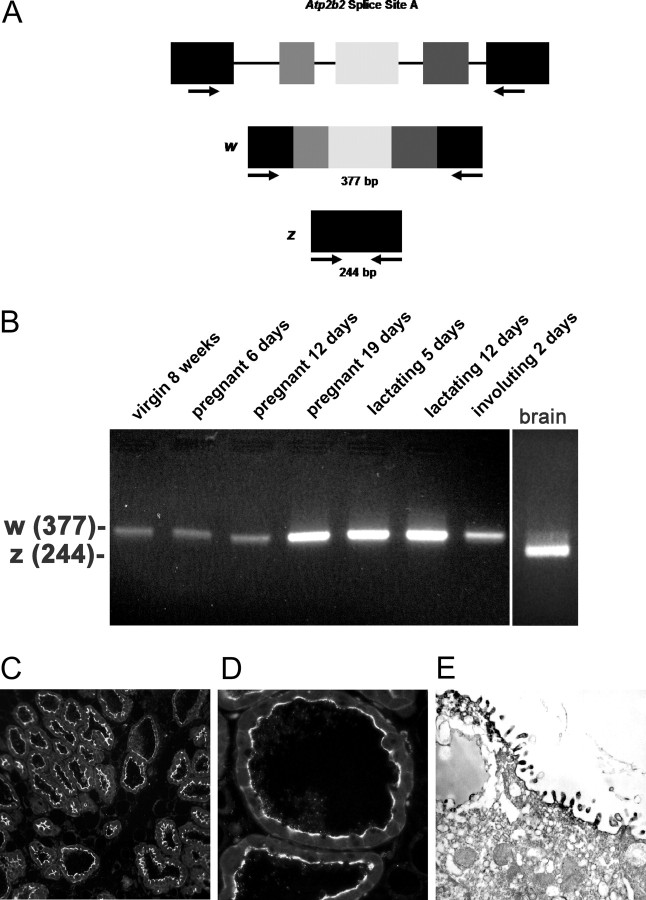
Because specific splice variants of PMCA2 are differentially sorted, we determined which site A variant was expressed in the mouse mammary gland. There are four possible splicing options at this site (20), which depend on the inclusion of different combinations of three possible exons. The 2w isoform includes all three alternative exons, the 2x variant includes only the third exon, the 2y includes only the first two exons, and the 2z variant excludes all three of these exons (20). We modified previously published PCR primers (10) to perform RT-PCR across splice site A of the mouse Atp2b2 transcript (Fig. 2A). As shown in Fig. 2B, we amplified a 377-bp product, corresponding to the PMCA2w splice variant from RNA harvested from the mouse mammary gland throughout its postnatal development. In mouse brain, where the predominant variant is PMCA2z, the smaller 244-bp amplicon was obtained. The identity of the PCR products as genuine PMCA2 splice site A variants was confirmed by sequencing (data not shown). The w variant of splice site A is sufficient to target PMCA2 to the apical plasma membrane. Accordingly, in semi-thin sections of lactating mammary glands from CD1 mice, immunofluorescent staining revealed a distinct apical localization (Fig. 2, C and D). Immunoperoxidase electron microscopy confirmed the apical membrane localization of PMCA2 in the lactating mouse mammary gland (Fig. 2E). We observed very little intracellular staining, and PMCA2 immunoreactivity was not associated with either the Golgi apparatus or with secretory vesicles.
Fig. 2.
A, The genomic organization of the Atp2b2 (PMCA2) gene at splice site A is illustrated with the alternatively spliced variants PMCA2w and PMCA2z shown below. RT-PCR was performed with primers (represented by arrows) flanking the splice site. B, RT-PCR was performed on RNA from mammary glands of mice at the developmental stages noted in the figure. Mouse brain RNA served as a control for the PMCA2z splice variant (the brain sample was run on a different gel). PCR products were of the predicted sizes for either the w splice variant (377 bp) or the PMCA2z splice variant (244 bp), but PCR products were sequenced to confirm their identity. Throughout postnatal development, the mouse mammary gland expresses only the PMCA2w variant of splice site A. C and D, Immunofluorescent staining for PMCA2 in mammary tissue from a lactating mouse (d 12) demonstrated that it was located at the apical plasma membrane. E, An electron micrograph of lactating mammary tissue stained for PMCA2 using immunoperoxidase techniques confirmed the apical membrane localization. There was no PMCA2 staining in secretory vesicles or other intracellular compartments.
PMCA2 deficiency lowers calcium transport into milk
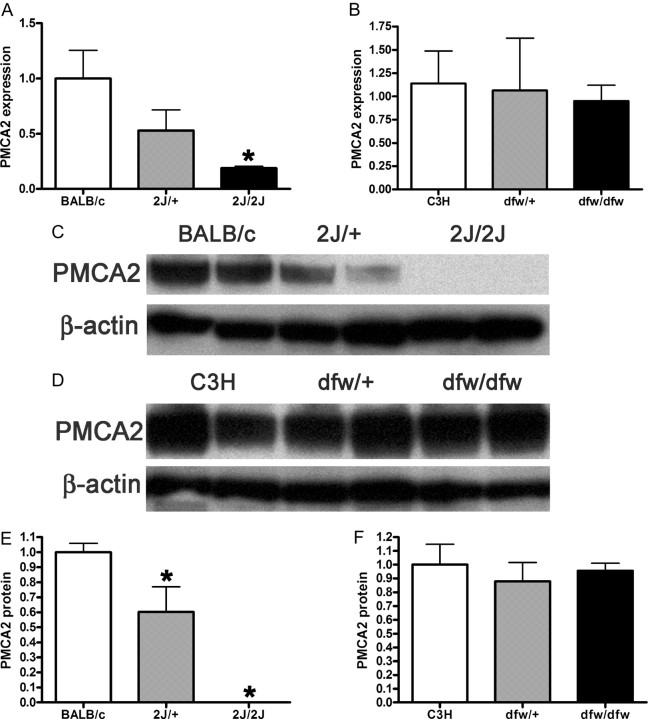
To confirm the importance of PMCA2 in the transport of calcium into milk, we analyzed two strains of deafwaddler mice with spontaneously arising, loss-of-function mutations in the Atp2b2 gene. The dfw strain harbors a point mutation leading to a glycine to serine substitution at residue 283 that reduces the enzymatic activity of the PMCA2 protein by approximately 70% (27). The dfw-2J mutation is essentially a null mutation resulting from a 2-bp deletion that causes a frame-shift and premature stop codon (28). Homozygous mice from both strains are deaf and ataxic, hence the name deafwaddler (28). As seen in Fig. 3A, by QRT-PCR, mammary gland PMCA2 RNA expression was reduced (over 80%) in dfw-2J/dfw-2J mice compared with BALB/c controls. Dfw-2J/+ mice have approximately half of the PMCA2 expression seen in BALB/c controls. In contrast, expression of PMCA2 mRNA in dfw/dfw mice was similar to its expression in C3H control mice (Fig. 3B). In parallel to RNA levels, PMCA2 protein was readily detectable in the plasma membrane fraction of BALB/c mammary glands, was 60% reduced in heterozygous mammary glands, and was undetectable in dfw-2J/dfw-2J mice by Western blotting (Fig. 3, C and E). As with mRNA levels, PMCA2 protein levels in mammary gland membrane preparations from dfw/dfw and dfw/+ mice were similar to those from C3H control mice (Fig. 3, D and F).
Fig. 3.
A, QRT-PCR demonstrates a progressive reduction in the expression of the PMCA2 gene (Atp2b2) in the mammary glands of dfw-2J/+ mice and dfw-2J/dfw-2J mice compared with wild-type BALB/c mice. PMCA2 mRNA levels were reduced by 50% in Dfw-2J/+ glands and by 85% in dfw-2J/dfw-2J glands. B, In contrast, PMCA2 mRNA levels in mice with the dfw mutation (dfw/+ and dfw/dfw) were not different from those in control C3H mice. C, Western blotting for PMCA2 in the plasma membrane fraction of lactating mouse mammary glands revealed abundant PMCA2 in BALB/c mice (lanes 1 and 2), reduced PMCA2 in dfw-2J/+ mice (lanes 3 and 4), and no PMCA2 in dfw-2J/dfw-2J mice (lanes 5 and 6). D, PMCA2 protein levels were similar in C3H (lanes 1 and 2), dfw/+ (lanes 3 and 4), and dfw/dfw (lanes 5 and 6) mice. E and F, Quantitation of PMCA2 protein normalized to β-actin protein levels in blots from C and D. Protein levels generally paralleled mRNA levels, with the exception that PMCA2 mRNA was detectable in dfw-2J/dfw-2J mice, but PMCA2 protein was not. An asterisk denotes statistical significance.
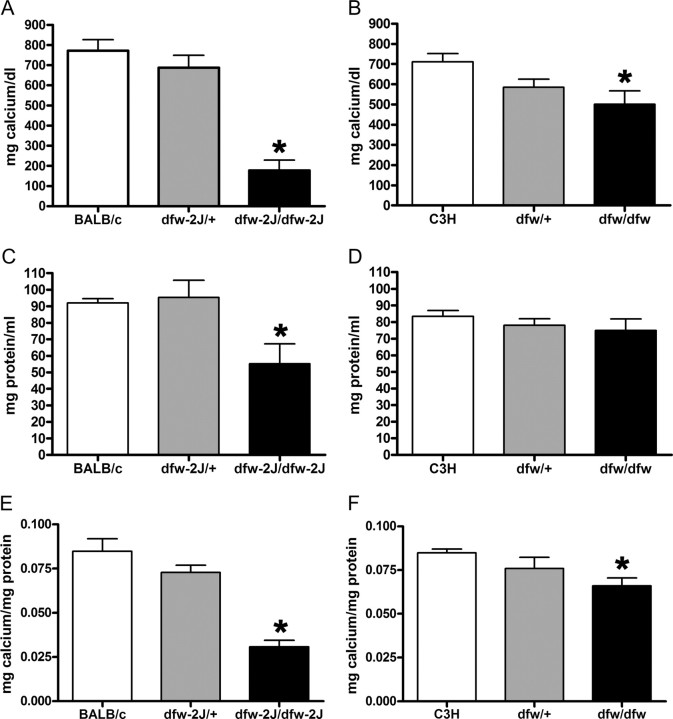
The absence of PMCA2 in dfw-2J/dfw-2J mice caused a 70% reduction in milk calcium concentration compared with BALB/c controls (Fig. 4A). Dfw-2J/+ mice had only a slight reduction in milk calcium. Calcium was reduced by 30% in milk from dfw/dfw mice compared with C3H controls (Fig. 4B). Heterozygous dfw/+ mice had an intermediate milk calcium concentration. The protein concentration of milk from dfw-2J/dfw-2J mice (Fig. 4C) was significantly lower than controls, but even when corrected for protein concentration, the calcium concentration of milk from dfw-2J homozygotes was reduced by 64% (Fig. 4E). Milk protein concentrations were similar in controls compared with dfw heterozygotes and homozygotes (Fig. 4D).
Fig. 4.
Milk calcium concentrations are reduced in PMCA2 mutant mice. A, On d 12 of lactation, milk calcium concentrations were approximately 70% lower (P < 0.01) in dfw-2J/dfw-2J mice (n = 4) than in BALB/c controls (n = 7). B, Milk from dfw/dfw mice (n = 4) had 30% less calcium (P < 0.05) than did milk from C3H controls (n = 5). In both cases, the heterozygous mice (dfw/+, n = 6; dfw-2J/+, n = 4) had milk calcium concentrations between the wild-type and homozygous mutant mice (A and B) but were not significantly different from wild-type mice. Milk protein levels (C and D) were also reduced in the dfw-2J/dfw-2J but not the dfw/dfw mutant mice (P < 0.01). Because most milk calcium is bound to proteins (mainly caseins), the milk calcium concentration was corrected for differences in protein content (E and F). Even when corrected for protein content, milk calcium levels were significantly reduced in dfw-2J/dfw-2J (P < 0.01) and dfw/dfw (P < 0.05) mutant mice. Statistical significance is represented with an asterisk.
Increased expression of other Ca-ATPase genes does not compensate for deficiency in PMCA2
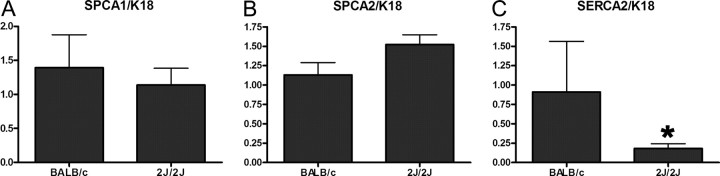
Reinhardt et al. (13) reported increases in SPCA1 and sarco/endoplasmic reticulum Ca-ATPase (SERCA2) levels in the mammary glands of lactating PMCA2−/− mice, suggesting that these pumps might partially compensate for the loss of PMCA2. Therefore, we examined SPCA1, SPCA2, and SERCA2 mRNA levels, normalized to K18, in the mammary glands of lactating BALB/c controls and dfw-2J/dfw-2J mice. The expression of SPCA1 was similar in controls and dfw-2J homozygotes (Fig. 5A). SPCA2 expression was slightly higher (20–30%) in dfw-2J homozygous mice (Fig. 5B), whereas SERCA2 expression was actually reduced by almost 80% in the dfw-2J/dfw-2J mice (Fig. 5C). These data suggest that there is no significant up-regulation of secretory pathway calcium pump gene transcription to compensate for the loss of PMCA2.
Fig. 5.
Expression of other Ca-ATPase genes in the mammary glands of dfw-2J/dfw-2J mice. To determine whether the loss of PMCA2 in dfw-2J/dfw-2J mice causes a compensatory up-regulation of the calcium-handling capacity in the secretory pathway, we measured the levels of SPCA1, SPCA2, and SERCA2 mRNA in mammary glands from d-12 lactating BALB/c and dfw-2J homozygous mice. Expression of these Ca-ATPase genes was normalized to expression of the epithelial cell marker K18. Expression of SPCA1 and SPCA2 do not change, but the expression of SERCA2 is significantly decreased in the mutant mammary glands. Asterisks indicate statistically significant differences.
PMCA2 expression is not regulated by the CaR
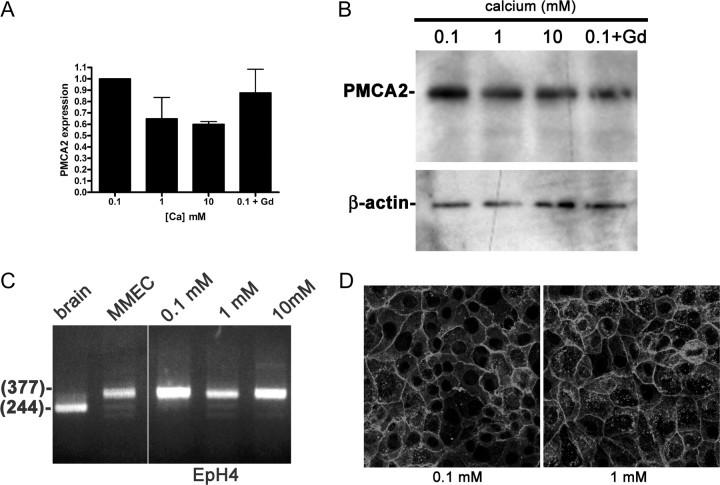
Because PMCA2 accounts for most of the calcium transported into milk, we next sought to determine whether it might be a target of CaR regulation during lactation. We first asked whether CaR signaling altered the expression of PMCA2 mRNA. We cultured primary mouse MECs or EpH4 cells overnight in media containing 0.1, 1, or 10 mm calcium or 0.1 mm calcium with 100 μm gadolinium and analyzed expression of PMCA2 by QRT-PCR and Western blotting. EpH4 cells are immortalized, but nontransformed mouse MECs that have been used as a model of normal MECs (33). We first documented that EpH4 cells expressed the CaR (data not shown). As shown in Fig. 6A, these cells also express the Atp2b2 gene. Levels of PMCA2 mRNA were highest at 0.1 mm calcium and were actually reduced at 1, 10, and 0.1 mm calcium with gadolinium. Western blotting showed that PMCA2 protein levels were unchanged by increasing the extracellular calcium concentration or by treating the cells with gadolinium (Fig. 6B). Therefore, calcium receptor signaling does not augment calcium transport by increasing PMCA2 expression.
Fig. 6.
A, PMCA2 mRNA expression measured by QRT-PCR in EpH4 mouse MECs exposed to varying levels of extracellular calcium concentrations. Increasing doses of calcium or gadolinium do not increase PMCA2 mRNA expression. B, Western analysis of PMCA2 protein in EpH4 cells incubated overnight in 0.1, 1, or 10 mm calcium or 0.1 mm calcium with 100 mm gadolinium. The calcium concentration and gadolinium treatment had no effect on the amount of PMCA2 protein or its electrophoretic mobility. C, RT-PCR analysis of PMCA2 splice site A in brain (lane 1), primary mouse MECs (MMEC, lane 2), and EpH4 cells incubated in media containing 0.1 (lane 3), 1 (lane 4), or 10 (lane 5) mm calcium reveals that the PMCA2w variant is expressed in MECs, regardless of the calcium concentration. The samples were all run on the same gel, and the space between lanes 2 and 3 is where intervening lanes were removed from the image. D, Immunofluorescent staining for PMCA2 in EpH4 cells incubated for 3 h in media containing 0.1 or 1 mm calcium. The pattern of PMCA2 staining is consistent with plasma membrane localization and is unchanged at 0.1 vs. 1 mm extracellular calcium.
CaR signaling does not alter localization or splicing of PMCA2
In kidney collecting duct cells, activation of the CaR antagonizes the ability of vasopressin to cause translocation of aquaporin 2 from intracellular stores to the apical plasma membrane (34, 35). Therefore, we asked whether the CaR could potentially regulate calcium transport in MECs by similarly regulating the insertion of PMCA2 into the plasma membrane. To determine whether the CaR regulates the localization of PMCA2, we stained EpH4 cells and primary mouse MECs incubated in media with 0.1 or 1 mm calcium for PMCA2. Figure 6D shows the results for EpH4 cells; identical results were seen with MECs. At both low and high calcium concentrations, PMCA2 was located at the plasma membrane. As noted before, because exon usage at splice site A is important in the localization of PMCA2 (20), we also analyzed whether extracellular calcium exerts any effect on the splice variants of PMCA2 expressed by MECs. As seen previously with whole mammary glands (Fig. 2B), the PMCA2w splice variant was the only one expressed in mouse MECs and EpH4 cells incubated at either 0.1, 1, or 10 mm calcium (Fig. 6C). Therefore, CaR signaling does not appear to alter trafficking of PMCA2 in MECs.
The CaR regulates PMCA2 activity
To determine whether the CaR regulates PMCA2 enzymatic activity during lactation, we examined the effect of varying extracellular calcium on the Ca-ATPase activity present in the plasma membrane fraction of primary MECs and EpH4 cells (Fig. 7, A and B). The dose of thapsigargin used in the assay has been shown to inhibit both SERCA and SPCA activity (36–39). Therefore, the Ca-dependent ATPase activity we observed in the plasma membrane preparations reflects PMCA function. As shown in Fig. 7, A and B, increasing the extracellular calcium concentration from 0.1 to 1 or 10 mm significantly increased Ca-ATPase activity in both primary MECs and EpH4 cells. Ca-ATPase activity also increased significantly in response to gadolinium, a CaR agonist (25), suggesting that the CaR is responsible for the extracellular calcium-induced increase in Ca-ATPase activity. We next sought to confirm that the Ca-ATPase activity that we measured in the plasma membranes of mammary cells was due to PMCA2 by knocking down its expression with oligonucleotide siRNAs. We were able to transfect EpH4 cells with fluorescently labeled siRNAs at a high efficiency (Fig. 7C). However, we could not efficiently transfect primary cultures of mouse MECs with siRNAs. Therefore, subsequent knockdown experiments were performed only in EpH4 cells. Figure 7D shows that in cells transfected with siRNAs specific for PMCA2, we were able to reduce the level of its mRNA expression by 70% relative to the expression of PMCA2 mRNA in EpH4 cells transfected with nonspecific control siRNAs. In those EpH4 cells with reduced PMCA2 expression, Ca-ATPase activity was reduced by 75% compared with the control-transfected cells with normal levels of PMCA2 mRNA (Fig. 7E). These results indicate that most of the Ca-ATPase activity that we measured in EpH4 cells was, indeed, due to PMCA2. To confirm that the CaR mediated the regulation of PMCA2 activity by extracellular calcium in these cells, we also employed siRNAs specific to the CaR to knock down its expression by 80% (Fig. 7D). This reduction in CaR expression did not affect basal PMCA activity, but it completely abrogated the expected increase in Ca-ATPase activity in response to a shift from 0.1 to 1.0 mm extracellular calcium (Fig. 7F). In contrast, cells transfected with nonspecific control siRNAs demonstrated a significant increase in PMCA activity in response to raising extracellular calcium to 1.0 mm. For the siRNA experiments, similar results were obtained with more than one siRNA oligo, making off-target effects unlikely. In the aggregate, these results strongly suggest that activation of the CaR increases PMCA2 activity in MECs.
Discussion
In this study, we performed a genomic screen to identify potential targets through which CaR signaling might regulate calcium transport into milk by MECs. We found PMCA2 to be the most prominently expressed calcium transporter in the lactating mammary gland. Little PMCA2 is expressed in the nulliparous mammary gland, but mRNA levels increase by over 100-fold in the lactating gland, matching the timing for the greatly increased demand for calcium transport capacity. The mammary gland expresses only PMCA2w splice variants, which are predicted to traffic to the apical rather than the basolateral membranes of MECs. We confirmed the apical location of PMCA2 by immunofluorescence and immunoelectron microscopy, suggesting that calcium is actively pumped across the apical membrane. PMCA2 is clearly necessary for calcium transport into milk, because loss-of-function mutations in the Atp2b2 gene in the two deaf-waddler strains deplete milk calcium. Complete loss of function of PMCA2 in the Dfw-2J strain causes a 70% reduction in calcium content. Finally, we show that activation of the CaR on MECs activates PMCA2 activity, providing a mechanistic framework to explain our previous observations that increased extracellular calcium concentrations stimulate transepithelial calcium transport by MECs.
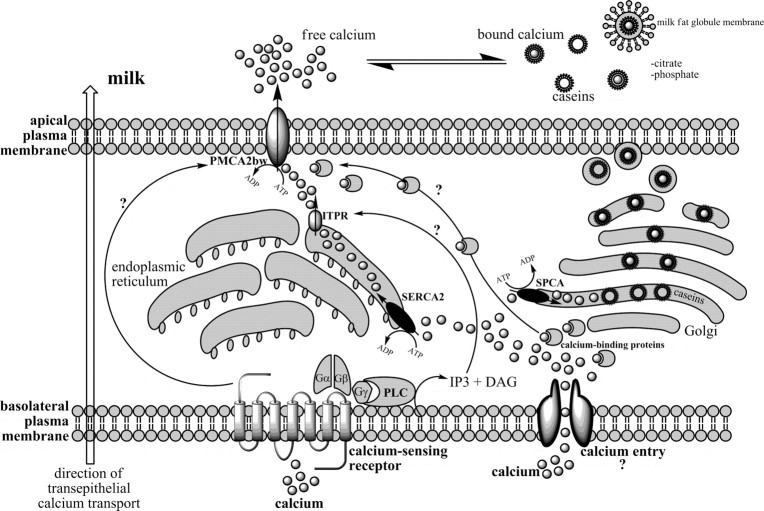
Our data regarding PMCA2 confirm and extend prior results from Reinhardt, Horst, and colleagues (10–13). In a survey of the calcium-ATPases expressed in the lactating mammary gland, they also documented up-regulation of PMCA2 (10, 12), and in a study of a PMCA2−/− mouse strain created by homologous recombination, they observed a 60% reduction in milk calcium concentration (13). Given their results, the results of our genomic survey and our finding of a reduction in milk calcium in two additional, independent genetic models of loss of PMCA2 function, it can be concluded with some certainty that PMCA2 is responsible for transporting the majority of calcium found in milk. Furthermore, our immunofluorescence and immunoelectron microscopy studies clearly establish that PMCA2 is found in the apical membrane of mammary cells. Importantly, we did not find evidence for significant amounts of PMCA2 concentrated within the Golgi or secretory vesicle membranes. Therefore, most of the calcium entering milk must do so directly from the cytoplasm across the apical membrane and not through the secretory pathway as previously believed (1, 40). Some calcium does enter the Golgi, endoplasmic reticulum, and secretory vesicles to participate in casein micelle formation, because SERCAs and SPCAs are expressed within the lactating gland (10, 12), and deletion of PMCA2 does not eliminate all calcium transport in either the PMCA2−/− (13) or Dfw-2J mice. However, transit of calcium through the secretory pathway is quantitatively less important than apical transport, and we suggest a new working model of calcium transport into milk as illustrated in Fig. 8. The nature of the calcium entry thorough the basolateral membrane remains unclear, although as mentioned before, prior evidence implicates a stretch-activated calcium channel (2, 3). Once within the cytoplasm, some calcium enters the Golgi and/or secretory vesicles to permit casein micelle formation, but the majority of calcium is shuttled through the cytoplasm to the apical surface where it is extruded into milk by PMCA2 and becomes associated with casein already secreted into the alveolar lumen.
Fig. 8.
A model of transcellular calcium transport into milk. Mechanisms leading to calcium entry through the basolateral membrane are unknown but may involve the action of a stretch-activated calcium channel. Once inside the cell, calcium is pumped into the ER by SERCAs or the Golgi by SPCAs or is bound by cytosolic calcium-binding proteins. Some calcium transits through the secretory pathway, takes part in casein micelle formation, and is secreted into milk by exocytosis. However, the majority of calcium moves to the apical plasma membrane where it is pumped directly into milk from the cytoplasm by PMCA2bw. Calcium may reach the apical surface of the cell by traveling through the cytoplasm bound to buffering proteins, or it may transit the endoplasmic reticulum and be released back into the cytoplasm in proximity to PMCA2bw. Once it crosses the apical membrane, calcium then equilibrates with caseins, phosphate, and citrate, so that ionized calcium represents only 10–30% of total milk calcium. Finally, the CaR on the basolateral membrane allows interstitial calcium levels to regulate calcium transport into milk by altering the activity of the apical PMCA2bw calcium pump. DAG, Diacylglycerol; IP3, inositol 1,4,5-trisphosphate; ITPR, inositol 1,4,5-trisphosphate receptor; PLC, phospholipase C.
The presence of apical transport in MECs suggests that calcium handling by the mammary gland is similar to calcium transport in other sites such as salivary glands, the duodenum, and the distal nephron (41–43). In all three, calcium is thought to enter the cells through epithelial calcium channels (TRPV5 or TRPV6), to transit through the cytoplasm bound to calbindins, and to exit on the opposite side through the actions of PMCAs and a Na/Ca exchanger (NCX1) (41). Although calcium extrusion in MECs relies on PMCA2 activity, it appears to deviate from these other sites in several important ways. First, expression of the epithelial calcium entry transporters (TRPV5 and TRPV6) is low during lactation (VanHouten, J. N., and J. J. Wysolmerski, unpublished data) excluding them as candidates for mediating calcium entry. Second, it has previously been reported that Na/Ca exchange is not important to transepithelial calcium transport in MECs (5), and expression of NCX1 (Slc8a1) did not increase during lactation in our microarray data. Finally, in the intestine, kidney, and salivary gland, calbindins serve to limit the free intracellular calcium in transit through the cytoplasm (41). However, calbindin expression in MECs does not increase during lactation (Table 2), raising the question as to how free calcium is sequestered within MECs and how it reaches PMCA2 on the apical surface. Calcium is likely removed from the cytoplasm into the endoplasmic reticulum and Golgi through the actions of SERCA2, SPCA1, and SPCA2 (10, 12). In pancreatic acinar cells, it has been suggested that after being pumped from the cytoplasm by SERCAs, calcium can transit or tunnel though the endoplasmic reticulum network from the basal surface of the cell to the apical surface. In mammary cells, a similar mechanism might allow for the bulk movement of calcium from its site of entry to the apical surface in close proximity to PMCA2, without necessitating a dramatic increase in the production of calbindins or other calcium shuttle proteins (Fig. 8).
We previously documented that the CaR is expressed on the basolateral surface of MECs and that transepithelial calcium transport is stimulated by activation of this receptor both in vitro and in vivo (21, 22). Given the importance of PMCA2 in transporting calcium into milk, we sought to determine whether CaR signaling regulated PMCA2 expression, localization, and/or activity. Our data suggest that the CaR does not increase PMCA2 expression, nor does it alter the splicing of PMCA2 mRNA or the intracellular localization of PMCA2 protein. Rather, CaR activation achieved either with calcium or the type I calcimimetic, gadolinium, was able to simulate Ca-dependent ATPase activity in the plasma membrane of mammary cells. This activity was not caused by contaminating SERCAs or SPCAs, because it was insensitive to high concentrations of thapsigargin (36–39). Furthermore, knockdown of PMCA2 expression using siRNAs greatly reduced the membrane Ca-ATPase activity, confirming that it was generated by PMCA2. Therefore, PMCA2 activity is activated by increases in extracellular calcium, a response mediated by the CaR as shown by its replication with gadolinium and abolition by CaR-specific siRNAs.
Up-regulation of CaR expression during lactation enables the mammary gland to become a calcium-sensing organ, which gauges its supply of calcium and adjusts its usage accordingly (22). The ability of the CaR to regulate PMCA2 function may have implications for pathophysiology as well. Breast tumors have a propensity to metastasize to bone, where they are particularly difficult to eradicate. Breast cancer cells have been shown to express both the CaR and PMCA2, and data from Lee and colleagues (40, 44) suggest that PMCA function may enhance proliferation of breast cancer cell lines in culture. Thus, it is interesting to postulate that the high local calcium concentrations of the bone microenvironment may activate the CaR and stimulate PMCA2 function, which, in turn, might contribute to the growth of bone metastases. We hope a greater understanding of calcium handling by the lactating mammary gland will shed light on both the control of milk production as well as the pathophysiology of breast cancer.
Acknowledgments
We thank Dr. Michael Caplan for helpful consultations regarding this study.
This work was supported by the National Institutes of Health (NIH) (DK064206 to J.V.H., CA094175 and DK069542 to J.W., and NIH P01 HD38129 to M.C.N.) and the American Cancer Society (IRG 58-012-48 to J.V.H.).
Disclosure Statement: The authors have nothing to declare.
Abbreviations
- CaR
Calcium-sensing receptor;
- K18
keratin 18;
- MEC
mammary epithelial cell;
- PMCA2
plasma membrane Ca-ATPase isoform 2;
- QRT-PCR
quantitative RT-PCR;
- SERCA2
sarco/endoplasmic reticulum Ca-ATPase;
- siRNA
small interfering RNA;
- SPCA
secretory pathway Ca-ATPase.
References
- 1. Neville MC. 2005. Calcium secretion into milk. J Mammary Gland Biol Neoplasia 10:119–128 [DOI] [PubMed] [Google Scholar]
- 2. Shennan DB, Peaker M. 2000. Transport of milk constituents by the mammary gland. Physiol Rev 80:925–951 [DOI] [PubMed] [Google Scholar]
- 3. Shennan DB. 1998. Mammary gland membrane transport systems. J Mammary Gland Biol Neoplasia 3:247–258 [DOI] [PubMed] [Google Scholar]
- 4. Farrell Jr HM, Kumosinski TF, Malin EL, Brown EM. 2002. The caseins of milk as calcium-binding proteins. Methods Mol Biol 172:97–140 [DOI] [PubMed] [Google Scholar]
- 5. Neville MC, Watters CD. 1983. Secretion of calcium into milk: review. J Dairy Sci 66:371–380 [DOI] [PubMed] [Google Scholar]
- 6. Bingham EW, McGranaghan MB, Wickham ED, Leung CT, Farrell Jr HM. 1992. Ca2+- and Mg2+-ATPases in the Golgi apparatus and microsomes of the lactating mammary glands of cows. Ann NY Acad Sci 671:418–420 [DOI] [PubMed] [Google Scholar]
- 7. Watters CD. 1984. A Ca2+-stimulated adenosine triphosphatase in Golgi-enriched membranes of lactating murine mammary tissue. Biochem J 224:39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vanoevelen J, Dode L, Van Baelen K, Fairclough RJ, Missiaen L, Raeymaekers L, Wuytack F. 2005. The secretory pathway Ca2+/Mn2+-ATPase 2 is a Golgi-localized pump with high affinity for Ca2+ ions. J Biol Chem 280:22800–22808 [DOI] [PubMed] [Google Scholar]
- 9. Wuytack F, Raeymaekers L, Missiaen L. 2003. PMR1/SPCA Ca2+ pumps and the role of the Golgi apparatus as a Ca2+ store. Pflugers Arch 446:148–153 [DOI] [PubMed] [Google Scholar]
- 10. Reinhardt TA, Filoteo AG, Penniston JT, Horst RL. 2000. Ca2+-ATPase protein expression in mammary tissue. Am J Physiol Cell Physiol 279:C1595–C1602 [DOI] [PubMed] [Google Scholar]
- 11. Prapong S, Reinhardt TA, Goff JP, Horst RL. 2005. Short communication: Ca2+-adenosine triphosphatase protein expression in the mammary gland of periparturient cows. J Dairy Sci 88:1741–1744 [DOI] [PubMed] [Google Scholar]
- 12. Reinhardt TA, Horst RL. 1999. Ca2+-ATPases and their expression in the mammary gland of pregnant and lactating rats. Am J Physiol 276:C796–C802 [DOI] [PubMed] [Google Scholar]
- 13. Reinhardt TA, Lippolis JD, Shull GE, Horst RL. 2004. Null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2 impairs calcium transport into milk. J Biol Chem 279:42369–42373 [DOI] [PubMed] [Google Scholar]
- 14. Grover AK, Khan I. 1992. Calcium pump isoforms: diversity, selectivity and plasticity. Review article. Cell Calcium 13:9–17 [DOI] [PubMed] [Google Scholar]
- 15. Zylinska L, Kawecka I, Lachowicz L, Szemraj J. 2002. The isoform- and location-dependence of the functioning of the plasma membrane calcium pump. Cell Mol Biol Lett 7:1037–1045 [PubMed] [Google Scholar]
- 16. Strehler EE, Zacharias DA. 2001. Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev 81:21–50 [DOI] [PubMed] [Google Scholar]
- 17. Strehler EE, Treiman M. 2004. Calcium pumps of plasma membrane and cell interior. Curr Mol Med 4:323–335 [DOI] [PubMed] [Google Scholar]
- 18. Hill JK, Williams DE, LeMasurier M, Dumont RA, Strehler EE, Gillespie PG. 2006. Splice-site A choice targets plasma-membrane Ca2+-ATPase isoform 2 to hair bundles. J Neurosci 26:6172–6180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grati M, Aggarwal N, Strehler EE, Wenthold RJ. 2006. Molecular determinants for differential membrane trafficking of PMCA1 and PMCA2 in mammalian hair cells. J Cell Sci 119:2995–3007 [DOI] [PubMed] [Google Scholar]
- 20. Chicka MC, Strehler EE. 2003. Alternative splicing of the first intracellular loop of plasma membrane Ca2+-ATPase isoform 2 alters its membrane targeting. J Biol Chem 278:18464–18470 [DOI] [PubMed] [Google Scholar]
- 21. Ardeshirpour L, Dann P, Pollak M, Wysolmerski J, VanHouten J. 2006. The calcium-sensing receptor regulates PTHrP production and calcium transport in the lactating mammary gland. Bone 38:787–793 [DOI] [PubMed] [Google Scholar]
- 22. VanHouten J, Dann P, McGeoch G, Brown EM, Krapcho K, Neville M, Wysolmerski JJ. 2004. The calcium-sensing receptor regulates mammary gland parathyroid hormone-related protein production and calcium transport. J Clin Invest 113:598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chattopadhyay N, Brown EM. 2006. Role of calcium-sensing receptor in mineral ion metabolism and inherited disorders of calcium-sensing. Mol Genet Metab 89:189–202 [DOI] [PubMed] [Google Scholar]
- 24. Tfelt-Hansen J, Brown EM. 2005. The calcium-sensing receptor in normal physiology and pathophysiology: a review. Crit Rev Clin Lab Sci 42:35–70 [DOI] [PubMed] [Google Scholar]
- 25. Brown EM, MacLeod RJ. 2001. Extracellular calcium sensing and extracellular calcium signaling. Physiol Rev 81:239–297 [DOI] [PubMed] [Google Scholar]
- 26. VanHouten JN. 2005. Calcium sensing by the mammary gland. J Mammary Gland Biol Neoplasia 10:129–139 [DOI] [PubMed] [Google Scholar]
- 27. Penheiter AR, Filoteo AG, Croy CL, Penniston JT. 2001. Characterization of the deafwaddler mutant of the rat plasma membrane calcium-ATPase 2. Hear Res 162:19–28 [DOI] [PubMed] [Google Scholar]
- 28. Street VA, McKee-Johnson JW, Fonseca RC, Tempel BL, Noben-Trauth K. 1998. Mutations in a plasma membrane Ca2+-ATPase gene cause deafness in deafwaddler mice. Nat Genet 19:390–394 [DOI] [PubMed] [Google Scholar]
- 29. Rudolph MC, McManaman JL, Phang T, Russell T, Kominsky DJ, Serkova NJ, Stein T, Anderson SM, Neville MC. 2007. Metabolic regulation in the lactating mammary gland: a lipid synthesizing machine. Physiol Genom 28:323–336 [DOI] [PubMed] [Google Scholar]
- 30. Machamer CE, Mentone SA, Rose JK, Farquhar MG. 1990. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc Natl Acad Sci USA 87:6944–6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kosk-Kosicka D. 2006. Measurement of Ca2+-ATPase activity (in PMCA and SERCA1). Methods Mol Biol 312:343–354 [PubMed] [Google Scholar]
- 32. Hennighausen L, Westphal C, Sankaran L, Pittius CW. 1991. Regulation of expression of genes for milk proteins. Biotechnology 16:65–74 [PubMed] [Google Scholar]
- 33. Montesano R, Soriano JV, Fialka I, Orci L. 1998. Isolation of EpH4 mammary epithelial cell subpopulations which differ in their morphogenetic properties. In Vitro Cell Dev Biol Anim 34:468–477 [DOI] [PubMed] [Google Scholar]
- 34. Valenti G, Procino G, Tamma G, Carmosino M, Svelto M. 2005. Minireview: aquaporin 2 trafficking. Endocrinology 146:5063–5070 [DOI] [PubMed] [Google Scholar]
- 35. Procino G, Carmosino M, Tamma G, Gouraud S, Laera A, Riccardi D, Svelto M, Valenti G. 2004. Extracellular calcium antagonizes forskolin-induced aquaporin 2 trafficking in collecting duct cells. Kidney Int 66:2245–2255 [DOI] [PubMed] [Google Scholar]
- 36. Dode L, Andersen JP, Vanoevelen J, Raeymaekers L, Missiaen L, Vilsen B, Wuytack F. 2006. Dissection of the functional differences between human secretory pathway Ca2+/Mn2+-ATPase (SPCA) 1 and 2 isoenzymes by steady-state and transient kinetic analyses. J Biol Chem 281:3182–3189 [DOI] [PubMed] [Google Scholar]
- 37. Reinhardt TA, Horst RL, Waters WR. 2004. Characterization of Cos-7 cells overexpressing the rat secretory pathway Ca2+-ATPase. Am J Physiol Cell Physiol 286:C164–C169 [DOI] [PubMed] [Google Scholar]
- 38. Jimenez-Gonzalez C, Michelangeli F, Harper CV, Barratt CL, Publicover SJ. 2006. Calcium signalling in human spermatozoa: a specialized ‘toolkit’ of channels, transporters and stores. Hum Reprod Update 12:253–267 [DOI] [PubMed] [Google Scholar]
- 39. Harper C, Wootton L, Michelangeli F, Lefievre L, Barratt C, Publicover S. 2005. Secretory pathway Ca2+-ATPase (SPCA1) Ca2+ pumps, not SERCAs, regulate complex [Ca2+]i signals in human spermatozoa. J Cell Sci 118:1673–1685 [DOI] [PubMed] [Google Scholar]
- 40. Lee WJ, Monteith GR, Roberts-Thomson SJ. 2006. Calcium transport and signaling in the mammary gland: targets for breast cancer. Biochim Biophys Acta 1765:235–255 [DOI] [PubMed] [Google Scholar]
- 41. Ashby MC, Tepikin AV. 2002. Polarized calcium and calmodulin signaling in secretory epithelia. Physiol Rev 82:701–734 [DOI] [PubMed] [Google Scholar]
- 42. Belan P, Gardner J, Gerasimenko O, Gerasimenko J, Mills CL, Petersen OH, Tepikin AV. 1998. Isoproterenol evokes extracellular Ca2+ spikes due to secretory events in salivary gland cells. J Biol Chem 273:4106–4111 [PubMed] [Google Scholar]
- 43. Homann V, Kinne-Saffran E, Arnold WH, Gaengler P, Kinne RK. 2006. Calcium transport in human salivary glands: a proposed model of calcium secretion into saliva. Histochem Cell Biol 125:583–591 [DOI] [PubMed] [Google Scholar]
- 44. Lee WJ, Robinson JA, Holman NA, McCall MN, Roberts-Thomson SJ, Monteith GR. 2005. Antisense-mediated Inhibition of the plasma membrane calcium-ATPase suppresses proliferation of MCF-7 cells. J Biol Chem 280:27076–27084 [DOI] [PubMed] [Google Scholar]