Abstract
Comparative genomics has begun to elucidate the genomic basis of social life in insects, but insight into the genomic basis of spider sociality has lagged behind. To begin, to characterize genomic signatures associated with the evolution of social life in spiders, we performed one of the first spider comparative genomics studies including five solitary species and two social species, representing two independent origins of sociality in the genus Stegodyphus. We found that the two social spider species had a large expansion of gene families associated with transport and metabolic processes and an elevated genome-wide rate of molecular evolution compared with the five solitary spider species. Genes that were rapidly evolving in the two social species relative to the five solitary species were enriched for transport, behavior, and immune functions, whereas genes that were rapidly evolving in the solitary species were enriched for energy metabolism processes. Most rapidly evolving genes in the social species Stegodyphus dumicola were broadly expressed across four tissues and enriched for transport functions, but 12 rapidly evolving genes showed brain-specific expression and were enriched for social behavioral processes. Altogether, our study identifies putative genomic signatures and potential candidate genes associated with spider sociality. These results indicate that future spider comparative genomic studies, including broader sampling and additional independent origins of sociality, can further clarify the genomic causes and consequences of social life.
Keywords: comparative genomics, molecular evolution, sociality, social spider
Introduction
A major goal of evolutionary biology is to elucidate the genomic underpinnings of phenotypic innovations across the tree of life. Over the past decade, comparative genomics has been used as a powerful tool to begin to elucidate the genetic basis of phenotypic innovations and adaptation in a wide range of organisms (Goodman et al. 2009; Gou et al. 2014; Wu et al. 2014; Zhang et al. 2014; Xu et al. 2017; Exposito-Alonso et al. 2018; Gaither et al. 2018; Chen et al. 2019; Wang et al. 2019). These studies have often identified changes in gene repertoire (e.g., expansions of certain gene families) or genomic signatures of molecular evolution at orthologous genes, by comparing the genomes of species with and without a phenotype of interest (Goodman et al. 2009; Gou et al. 2014; Wu et al. 2014; Xu et al. 2017; Gaither et al. 2018; Wang et al. 2019).
The evolution of group living is a conspicuous phenotypic innovation found across diverse groups of animals, including many vertebrates, insects, and spiders (Rubenstein and Abbot 2017). A series of comparative genomic and transcriptomic studies in eusocial insects has begun to identify putative genomic signatures of the evolution of complex societies (Simola et al. 2013; Kapheim et al. 2015). These putative signatures include the expansion of certain gene families associated with functions such as chemical communication (McKenzie et al. 2014, 2016; Harrison et al. 2018) and signatures of elevated molecular evolution at certain genes (Woodard et al. 2011; Kulmuni et al. 2013; Roux et al. 2014; Jia et al. 2018; Privman et al. 2018). Studies in social insects and other social organisms have also commonly emphasized the importance of genes associated with metabolism (Woodard et al. 2011; Rittschof et al. 2014) and reproduction (Warner et al. 2019) as potentially making up a conserved social toolkit or groundplan (Linksvayer and Wade 2005; Amdam et al. 2006; Toth and Robinson 2007; Johnson and Linksvayer 2010; Rittschof and Robinson 2016).
Sociality has also evolved multiple times independently in spiders (Johannesen et al. 2007). Specifically, while the vast majority of the more than 40,000 known species of spiders are solitary (Lubin and Bilde 2007), approximately 25 species, across nine genera in six families, are social (Agnarsson et al. 2006; Avilés; Lubin and Bilde 2007). The high complexity and large size of spider genomes have constrained the development of genomic resources for spiders (Sanggaard et al. 2014; Babb et al. 2017; Schwager et al. 2017; Garb et al. 2018; Liu et al. 2019), so that comparative genomic studies of spider sociality have lagged behind other groups such as social insects. Previous comparative genomic studies including spiders have compared newly sequenced spider genomes to available insect genomes (Sanggaard et al. 2014; Babb et al. 2017), but not across spiders or among other arachnids. These previous studies have mainly focused on identifying sets of venom and silk genes in spiders (Garb et al. 2018, 2019), but have also provided evidence for whole-genome duplication and gene family expansion during arachnid evolution (Sanggaard et al. 2014; Babb et al. 2017; Schwager et al. 2017).
Building on previous comparative genomic work in social insects, we performed one of the first spider comparative genomic studies and focused in particular on identifying putative genomic signatures associated with the evolution of social life in spiders. Specifically, we used recently available genomes from seven spider species (Sanggaard et al. 2014; Babb et al. 2017; Schwager et al. 2017; Liu et al. 2019), including two social spiders with independent origins of sociality (Johannesen et al. 2007; Settepani et al. 2016) from the genus Stegodyphus (Sanggaard et al. 2014; Liu et al. 2019). We aimed to identify genome content and genome-wide patterns of molecular evolution that differ between social and solitary spiders. In addition, using new tissue-specific expression data, we generated from the social spider Stegodyphus dumicola, we also characterized the expression pattern of genes identified as having elevated rates of molecular evolution in the two Stegodyphus social spider species.
Materials and Methods
Data Retrieval and Sequence Analysis
We searched all available sequences, annotations, and predicted proteins of spider genomes and downloaded from public online databases (fig. 1A and supplementary table S1, Supplementary Material online), including NCBI (https://www.ncbi.nlm.nih.gov/) and HGSC (https://www.hgsc.bcm.edu/). Specifically, we used the genomic data of the social spiders Stegodyphus mimosarum (GCA_000611955.2, NCBI) (Sanggaard et al. 2014) and S. dumicola (Liu et al. 2019), and the solitary spiders Parasteatoda tepidariorum (GCA_000365465.3, NCBI) (Schwager et al. 2017), Acanthoscurria geniculata (GCA_000661875.1, NCBI) (Sanggaard et al. 2014), Nephila clavipes (GCA_002102615.1, NCBI) (Babb et al. 2017), Loxosceles reclusa (GCA_001188405.1, NCBI and HGSC), and Latrodectus hesperus (GCA_000697925.2, NCBI and HGSC).
Fig. 1.
—Genomic content and evolution in social spider relative to solitary species. (A) Maximum likelihood phylogenetic trees of seven spider species reconstructed by 2,824 of core single-copy orthologous genes with 100% ML bootstrap values for all nodes. Divergence time of seven spiders estimated by Timetree (http://www.timetree.org/). The node bars indicate 95% posterior probability intervals. (B) Comparison of number of orthologous gene family in seven spider genomes. Gene family expansion and contraction indicated by plus (+) and minus (−), respectively. (C) Average dN/dS ratios of concatenated all orthologs in seven spider species estimated by branch model in PAML. (D) Violin plot showed the dN/dS ratios of each orthologous genes in seven spider species estimated by branch-site mode in PAML.
We downloaded the curated orthology map of Arachnida from OrthoDB (Kriventseva et al. 2015) which contains 8,805 orthologous gene groups (OGGs) shared by nine Arachnid species with sequenced genomes. Of these 8,805 seed orthologous groups in HaMStR (Ebersberger et al. 2009), we identified the orthologs in each spider species with E values of <10−20. We aligned and trimmed the nucleotide sequences of the 8,805 orthologous groups using PRANK (Löytynoja and Goldman 2005) with the parameter “-codon” and MATFF (https://mafft.cbrc.jp/alignment/software/), and trimmed using trimAl (Capella-Gutiérrez et al. 2009) with the parameter “-automated1.”
We identified 1:1, one-to-many, and many-to-many orthologs among all seven spider genomes, and strict 1:1 orthologs (i.e., genes for which only one gene from each species matches the given OrthoDB8 ortholog group). For each 1:1 ortholog pair, we only selected the longest transcript associated with the gene for each pair of species. We ran CAFE (De Bie et al. 2006) to analyze the gene family expansion and contraction based on the above identified 1:1 ortholog pairs, and annotated by gene ontology (GO) with the R software, package topGO (https://bioconductor.org/packages/release/bioc/html/topGO.html).
We also identified strict 1:1:1:1:1:1:1 orthologs among all seven spider genomes as single-copy orthologs. We defined orthologs that had more than two homologs as multiple-copy orthologs, and orthologs that had only two homologs as unique paralogs. We also identified the core single-copy orthologs that were shared by all seven spider genomes.
Genome-Wide Phylogeny Construction and Divergence Time Estimation
We aligned each core single-copy ortholog using MUSCLE v3.8.31 (https://www.ebi.ac.uk/Tools/msa/muscle/) with default parameters and trimmed using trimAl (Capella-Gutiérrez et al. 2009) with parameter “-automated1.” To maximize the information content of the sequences and to minimize the impact of missing data, we filtered the core single-copy orthologs with strict constraints, including length (minimum 200 aa) and sequence alignment (maximum missing data 50% in CDS alignments). Next, we prepared two types of gene data sets after filtering. First, we concatenated all core single-copy genes of each species into one-line sequence as a supergene using a custom python script (genome-scale concatenation-based, supergene). Second, we conducted a genome-scale coalescent-based data set including 2,824 of core single-copy genes. We used ModelTest2 (Posada and Crandall 1998) to detect the best model for phylogeny construction and then used RAxML 8 (Stamatakis 2014) to build the maximum likelihood (ML) tree. We built the ML trees using the two types of data sets (concatenation- and coalescent-based) described above in RAXML, respectively. Finally, we reconstructed the species tree using ASTRAL 4.4.4 (Mirarab et al. 2014).
We generated two data sets from the CDS alignments to estimate the divergence time of each species. One data set contained the first two partitions, including the first and second codon positions of the sequences. The other data set contained all three partitions corresponding to all three codon positions in the sequences. We estimated the divergence times under a relaxed clock model using the MCMCTree package in PAML4.7a (Yang 2007), with the “independent rates model (clock = 2)” and the “JC69 model.” We used 4,000,000 iterations after a burn-in of 2,000,000 iterations, with other parameters as the default settings of MCMCTree. We ran this analysis twice for each data set to confirm that the results were consistent between runs. We used the time calibrations from TIMETree (http://www.timetree.org/), a public knowledge-base of divergence times among organisms, demonstrating the high reliability of this molecular clock dating strategy.
Nucleotide Substitution Rate Estimation
To estimate lineage-specific evolutionary rates for each branch of the phylogeny of the seven spiders, we aligned orthologous protein sequences using MUSCLE v3.8.31 (https://www.ebi.ac.uk/Tools/msa/muscle/), derived nucleotide alignments from protein alignments using PAL2NAL (Suyama et al. 2006), and estimated pairwise dN/dS of nucleotide alignments using the CodeML package in PAML 4.7a (Yang 2007). Specifically, we used the free-ratio model to calculate the ratio of nonsynonymous (dN) to synonymous (dS) nucleotide changes separately for each ortholog and a concatenation of all alignments of 2,824 single-copy orthologs from the seven species. Parameters, including dN, dS, dN/dS, N*dN, and S*dS values, were estimated for each branch, and genes were discarded if N*dN or S*dS < 1, or dS >2, following a previous study (Goodman et al. 2009).
Rapidly Evolving Gene Identification
To identify the rapidly evolving genes (REGs) in social and solitary spiders separately, we built multiple single-copy orthologs data sets for social-solitary comparisons, including one social spider versus the five solitary spiders (fig. 2A) and one solitary spider versus the two solitary spiders (fig. 2B). Each clipped species tree including target spider species were prepared, respectively (fig. 2). We ran the CodeML package to identify REGs of each spider species with corresponding gene sets and appointed species tree separately, with the null model assuming that all branches have been evolving at the same rate and the alternative model allowing the focal foreground branch to evolve under a different evolutionary rate. We used a likelihood ratio test in R software (MASS package) with df = 1 to discriminate between the alternative model and the null model for each single-copy ortholog in the gene set. We only considered the genes as evolving with a significantly faster rate in the foreground branch if the adjusted P value < 0.05 and if the dN/dS in the focal foreground branch was higher than that in the focal background branches (i.e., other six spiders). Finally, we identified the core REGs shared by two social spiders or five solitary spiders, and annotated by GO using the R software, package topGO (https://bioconductor.org/packages/release/bioc/html/topGO.html).
Fig. 2.
—Illustration of comparisons made between social and solitary species to identify rapidly evolving genes (REGs) in social and solitary species. (A) Each of the two social spider species was separately compared with the five solitary spiders and (B) each of the five solitary spider species was separately compared with the two social spiders. Each phylogenetic tree was clipped from the previously reconstructed species tree.
Expression Profiles of REGs in Four Tissues of the Social Spider S. dumicola
The social spider S. dumicola is native to central and southern Africa (Johannesen et al. 2007) and has been widely used to study the evolutionary ecology of group living (Grinsted et al. 2013; Berger-Tal et al. 2014; Pruitt et al. 2018, 2019). This species builds a complex silken retreat permeated by an elaborate series of tunnels that can house hundreds of spiders. Often radiating outward from the retreat are one or more 2D capture webs. Spiders are recruited to the capture web from the retreat in response to vibratory cues indicative of struggling prey. Notably, these spiders also exhibit consistent individual variation in their boldness–shyness personality (Wright et al. 2015). Boldness and aggressiveness have been shown to be linked with participation in foraging tasks in S. dumicola (Keiser et al. 2014), intriguingly, extremely bold individuals appear to have the ability to catalyze otherwise sedentary nestmates into aggressive foraging behavior (Pruitt and Keiser 2014). In this study, we collected three colonies of ∼80 individuals each from Kalkrand, Namibia, in February 2017 and housed them in the lab, feeding them with crickets ad libitum until RNA extraction. By July 2017, when we extracted the RNA, there were ∼30–40 individuals in each colony. These individuals were assayed for boldness, by measuring the latency to respond to a simulated predator attack (Keiser et al. 2014; Pruitt and Keiser 2014). We failed to collect sufficient bold individuals for RNA extraction before sequencing library preparation. Therefore, only shy individuals, that is, those that took 400 s or longer to recover from the simulated predator attack, were used in this study. All individuals were adult females of the same age, that is, in their final molt and over a year old. We haphazardly selected ten shy individuals from each colony for the analysis, resulting in a total of 30 individuals. Individuals were decapitated immediately before tissue extraction. We dissected four tissues: brain, venom gland, legs (excluding pedipalps), and abdomen from each individual and immediately stored in RNAlater (Sigma). Samples were stored individually at 4 °C overnight for the RNAlater to penetrate the tissues before being finally stored at −80 °C. Subsequently, samples of each tissue were separately pooled according to colony of origin, resulting in three biological replicate pools (i.e., originating from the three replicate colonies), for each of the four tissues.
The total RNA of each pool was extracted using RNeasy kits (Qiagen, CA), and the quality and quantity of RNAs were detected with Nanodrop 1000 (NanoDrop Technologies, DE) and Agilent Bioanalyzer 2100 (Agilent Technologies, CA). Only RNA with high quality (RNA integrity number >7) were used for cDNA synthesis and amplification. Libraries were prepared with the Nextera XT DNA Library Prep Kit (Illumina, CA) using ∼350-bp inserted fragments for transcriptome sequencing as previously described (Tong, Fei, et al. 2017; Tong, Tian, et al. 2017). Libraries were individually barcoded and run on a single lane of an Illumina NovaSeq (Novogene, CA) yielding 150-bp paired-end reads.
Sequencing reads were checked for quality using Bioconda software, package FastQC. Adapters and reads with a quality score <20 were trimmed with Trimmomatic (Bolger et al. 2014). We mapped all the clean reads to the S. dumicola genome (Liu et al. 2019) using RSEM (Li and Dewey 2011) to obtain expected counts and transcripts per million (TPM). We removed genes with low expression that did not meet one of two criteria: 1) transcripts per million (TPM) >1 in at least half the samples or 2) TPM >1 in all samples of a given tissue. To classify genes by their tissue specificity, we calculated τ, a commonly used metric of expression specificity (Yanai et al. 2005). τ ranges from 0 to 1, where 0 indicates that genes are ubiquitously expressed and 1 indicates that genes are exclusively expressed in one tissue (Warner et al. 2019). Finally, we compared the expression profiles of REGs across the four tissues and identified the broadly expressed and tissues-specific REGs.
Results
Genome Content and Evolution
We identified 8,805 OGGs belonging to Arachnida according to the curated orthology map from OrthoDB (https://www.orthodb.org/). The social spiders S. mimosarum (N = 27,135) and S. dumicola (N = 37,601) had the most protein coding genes and the largest number of OGGs (N = 7,780, 8,516) relative to the other solitary spider species (table 1). In addition, even though the two social spiders had fewer orthologs than the solitary species, they had more single-copy orthologs but fewer multiple-copy orthologs (table 1). The two solitary spider species P. tepidariorum and N. clavipes had the most orthologs (N = 12,763, 12,784) and multiple-copy orthologs (N = 2,654, 2,579).
Table 1.
Summary of the Gene Content of Seven Spider Genomes
| Species | Sociality | Genes | Orthologous Gene Groups | Orthologs | Average Ortholog Number Per Group | Single-Copy Orthologs | Multiple-Copy Orthologs |
|---|---|---|---|---|---|---|---|
| Acanthoscurria geniculata | Solitary | 23,277 | 5,933 | 9,940 | 1.6754 | 3,682 | 2,251 |
| Latrodectus hesperus | Solitary | 17,364 | 5,504 | 6,429 | 1.1681 | 4,994 | 508 |
| Loxosceles reclusa | Solitary | 20,617 | 5,064 | 6,755 | 1.3340 | 4,387 | 6,77 |
| Nephila clavipes | Solitary | 20,241 | 5,895 | 12,763 | 2.1651 | 3,241 | 2,654 |
| Parasteatoda tepidariorum | Solitary | 27,515 | 7,002 | 12,784 | 1.8258 | 4,423 | 2,579 |
| Stegodyphus mimosarum | Social | 27,135 | 7,590 | 7,780 | 1.0250 | 7,540 | 50 |
| Stegodyphus dumicola | Social | 37,601 | 7,118 | 8,516 | 1.1964 | 6,366 | 752 |
In the two social spider genomes, we detected gene family expansions for 214 families in S. dumicola and 686 families in S. mimosarum, whereas the five solitary spider species tended to have many fewer gene family expansions and more gene family contractions relative to their most recent common ancestor (fig. 1B). In the S. dumicola genome, these expanded gene families were significantly enriched for 128 related GO categories, mainly consisting of two large groups (supplementary table S2, Supplementary Material online). The first group was related to transport function, such as transmembrane transport (GO:0055085, P = 0.00725), water transport (GO:0006833, P = 0.00456), and calcium ion transmembrane transport (GO:0070588, P = 0.0072). The second largest group was associated with metabolic processes, including carbohydrate metabolic process (GO:0005975, P = 0.00456) and UDP-glucose metabolic process (GO:0006011, P = 0.0000088). We also found similar enrichment for gene families expanded for the second social species, S. mimosarum (supplementary table S2, Supplementary Material online).
Genome-Wide Phylogeny
We identified 2,824 core single-copy orthologs that were shared by the seven spider species. We constructed four ML phylogenetic trees of the seven spiders based on the concatenated (supergene) and coalesced single-copy orthologs. Each phylogenetic reconstruction method (coalescent-based, concatenation-based) and sequence type (nucleotide or amino acid) generated the same topology with 100% ML bootstrap values for all nodes (fig. 1A and supplementary fig. S1, Supplementary Material online). This strongly supported phylogeny also had similar topology to recent phylogenetic studies on Stegodyphus species using multiple nuDNA loci (Garrison et al. 2016; Settepani et al. 2016). In addition, we estimated the divergence times for all nodes on the phylogenetic tree (fig. 1A). The two social Stegodyphus spiders were estimated to have diverged ∼14.5921 Ma with confidence intervals 13.5859–16.5332 Ma, and diverged from the remaining solitary spider species ∼59 Ma (fig. 1A).
Genome-Wide Pattern of Nucleotide Substitution Rate
We identified the nucleotide substitution rates in each spider species based on the 2,824 core single-copy genes using PAML (Yang 2007). We found that the two social spiders S. dumicola (dN/dS = 0.138194) and S. mimosarum (dN/dS = 0.148917) had elevated terminal genome-wide dN/dS compared with their most recent ancestral branch (dN/dS = 0.031015) (supplementary fig. S2, Supplementary Material online). In contrast, the five solitary spider species did not show elevated terminal genome-wide dN/dS relative to their most recent ancestral branch (supplementary fig. S2, Supplementary Material online). Furthermore, the two social Stegodyphus spider species had elevated dN/dS compared with the five solitary spider species based on both concatenation-based (fig. 1C) and coalescent-based gene sets (fig. 1D).
REG Repertoire
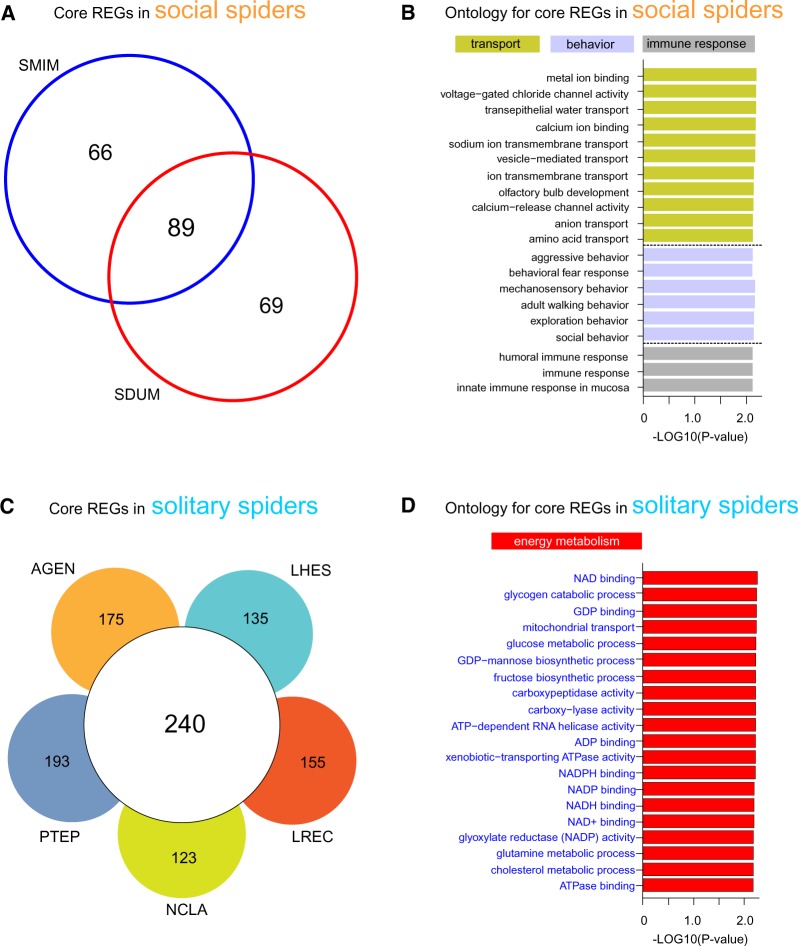
We identified 89 REGs (P < 0.05) that had elevated rates of molecular evolution (dN/dS) in both social spider species relative to the five solitary spider species (fig. 3A). Specifically, we identified a total of 158 REGs in the S. mimosarum genome, 155 REGs in S. dumicola genome, and 89 of these overlapped (supplementary table S3, Supplementary Material online). Significantly enriched GO functional categories for these core REGs in social spiders mainly included three groups: transport, behavior, and immune response processes (fig. 3B). REGs associated with behavioral processes included the steroid receptor seven-up involved in social behavior (GO:0035176) and aggressive behavior (GO:0002118), sodium channel protein involved in mechanosensory behavior (GO:0007638), and oxidation resistance protein 1 related to adult walking behavior (GO:0007628). Transport-related REGs in the two social spiders included genes functioning in metal ion binding, calcium ion binding, and sodium ion transmembrane transport, such as secretory carrier-associated membrane protein 1 and RING finger protein 170 (supplementary table S4, Supplementary Material online). REGs associated with immune response function included toll, CCAAT enhancer binding protein gamma, and transcription factor E3.
Fig. 3.
—Identification and annotation of shared REGs in social and solitary spider species. (A) Venn diagram showing the number of species-specific and shared REGs in the two social spiders. (B) Bar plot depicting the top 20 gene ontologies significantly enriched in REGs shared between the two social spiders. Ontology for these shared REGs in social spiders was associated with transport, behavior, and immune response functions. (C) Venn diagram showing the number of species-specific and shared REGs in the five solitary spider species. (D) Bar plot depicting the top 20 gene ontologies significantly enriched in REGs shared between the five solitary spiders. Ontology for these shared REGs in solitary spiders mainly involved energy metabolism processes.
We identified a total of 240 core REGs shared by the five solitary spider species (fig. 3C and supplementary table S5, Supplementary Material online). More specifically, we identified the largest number of REGs (N = 433) in the P. tepidariorum genome and the smallest number of REGs (N = 323) in N. clavipes genome. Interestingly, we found that the significantly enriched GO categories of core REGs in the solitary spiders were mainly involved in energy metabolism processes (fig. 3D and supplementary table S6, Supplementary Material online), such as fatty acid synthase, mitochondrial processing peptidase beta subunit, and acyl carrier protein involved into glucose metabolic process (supplementary table S6, Supplementary Material online).
Expression Pattern of REGs in the Social Spider S. dumicola
Transcriptome sequencing of four tissues (brain, venom gland, legs, and abdomen) from the social spider S. dumicola generated ∼86.5-Gb raw reads. After trimming, we obtained nearly 28.6 million PE 150-bp clean reads (supplementary table S7, Supplementary Material online). We focused on the expression patterns between the four tissues of the shared 89 REGs identified in S. dumicola and S. mimosarum compared with the five solitary spiders (supplementary table S8, Supplementary Material online). Most REGs (N = 81) were expressed in the brain, whereas fewer REGs (N = 57) were expressed in the venom gland (fig. 4A). We found 53 REGs (59.55%) were broadly expressed in all tissues (fig. 4A). Only a few REGs showed a tissue-specific expressed pattern, including 12 brain-specific genes, 3 abdomen-specific genes, 2 leg-specific genes, and 0 venom gland-specific genes. Among the broadly expressed REGs, many of them had higher expression levels in the brain compared with the other three tissues (fig. 4B), such as glycoprotein 3-alpha-l-fucosyltransferase A, AP-2 complex subunit sigma, and AP-1 complex subunit beta-1. GO enrichment analysis showed that this set of broadly expressed REGs was enriched in transport processes (fig. 4C and supplementary table S9, Supplementary Material online), such as protein transport (GO:0015031, P = 0.00283), ion transmembrane transport (GO:0034220, P = 0.00323), and ion channel activity (GO:0005216, P = 0.00983). Among the tissue-specific REGs, we found brain-specific REGs had higher expression levels than abdomen-specific REGs (fig. 4D). Notably, significantly enriched GO functional categories of brain-specific REGs included behavioral processes (fig. 4E and supplementary table S10, Supplementary Material online), such as aggressive behavior (GO:0002121, P = 0.00137) and social behavior (GO:0035176, P = 0.00263).
Fig. 4.
—Expression patterns of REGs in Stegodyphus dumicola. (A) Distribution of broadly expressed, tissue-specific, and other REGs in each tissue, including brain, legs, abdomen, and venom gland. (B) Violin plot depicting the distribution of log10(TPM) values of broadly expressed genes labeled by REGs in four tissues. (C) Bar plot depicting the top six gene ontology for broadly expressed REGs. Ontology for broadly expressed REGs involved in transport processes (signal transduction). (D) Violin plot depicting the expression ranges of brain- and abdomen-specific genes labeled by REGs. (E) Bar plot depicting the top six gene ontology for broadly expressed REGs. Ontology for brain-specific REGs involved into social behavior processes.
Discussion
Building on previous comparative genomic research into the genomic underpinnings of social life in social insects (Simola et al. 2013; Kapheim et al. 2015), we compared the genomes of two social and five solitary spider species. We identified putative genomic signatures of spider social evolution, including expansions of gene families associated with transport and metabolism, and genome-wide elevated rates of molecular evolution in the two social species relative to the five solitary species. Furthermore, we found specific changes in the rate of molecular evolution between the social and solitary species for genes with transport, behavior, immune, and metabolism function. Finally, we found that most REGs in the social spider S. dumicola were broadly expressed across four tissues and enriched for transport (e.g., signal transduction) function, but 12 of the REGs in S. dumicola showed brain-specific expression and were enriched for genes annotated for behavioral processes.
Extensive Gene Family Evolution in Stegodyphus Social Spiders Targets Transport and Metabolism Functions
Gene families are sets of paralogs that often display similar gene functions. Expansions (gene gain) or contractions (gene lose) in gene families may correspond to adaptive events coupled to life-history transitions (Ranson et al. 2002). We found that the social spiders S. mimosarum and S. dumicola had large expansions of gene families compared with the five solitary spider species with available genomes (fig. 1A). These extensive gene family expansions of the two social spider species in particular involved gene families associated with metabolism (e.g., carbohydrate metabolic process) and transport (e.g., transmembrane transport) functions. This finding suggests that large-scale transport and metabolism function-associated gene duplication (or perhaps whole-genome duplication) could be associated with social evolution in spiders.
Previous studies in social insects (ants and bees) identified expansions of gene families associated with metabolism and chemical communication functions (Wurm et al. 2011; Simola et al. 2013; Kapheim et al. 2015; McKenzie et al. 2016). Similarly, previous comparative studies across animals have emphasized genes associated with metabolism as being important in the response to social challenges (Rittschof et al. 2014; Saul et al. 2019), suggesting that changes in genes with metabolic functions might often be involved in social evolution across animals. Indeed, previous research indicates that social spiders may have lower metabolic rates than their solitary relatives (Zimmerman 2007), further implicating changes in metabolic-related genes to the evolution of social life in spiders. Unlike for social insects, we did not detect expansions of chemosensory gene families in the two social spiders. Although previous studies have identified large changes in chemosensory gene families across chelicerates (Vizueta et al. 2017, 2018), including spiders, pheromonal communication may be relatively less important for social life in social spiders than in social insects (Vander Meer et al. 1998; Zhou et al. 2015; Leonhardt et al. 2016).
Social Spider Genomes Exhibit the Distinct Genome-Wide Signature of Accelerated Molecular Evolution
We found an elevated genome-wide rate of molecular evolution (dN/dS) in the two social spider species relative to the five solitary species. Our results are consistent with a previous spider study that compared dN/dS ratios for 13 randomly chosen nuclear loci for three social and seven subsocial Stegodyphus species (Settepani et al. 2016). Similarly, four eusocial insects were also found to have higher rates of molecular evolution (dN/dS) compared with solitary insects (Romiguier et al. 2014). Studies very often interpret high dN/dS as a putative sign of positive selection (Privman et al. 2018). However, relaxed purifying selection instead of elevated positive selection can also lead to higher average gene-specific or genome-wide dN/dS. Relaxed purifying selection is expected to be especially common in species with low effective population size (Ne), and social species such as social spiders are expected to experience low Ne as a result of reproductive skew, female-biased sex ratios, and inbreeding (Romiguier et al. 2014; Settepani et al. 2016; Galtier et al. 2018). Indeed, previous studies in social spiders (Settepani et al. 2014, 2016, 2017; Bechsgaard et al. 2019) and also social insects have found evidence for low Ne and genome-wide relaxed purifying selection when compared with solitary species (Romiguier et al. 2014; Kapheim et al. 2015; Galtier et al. 2018). Future studies using both polymorphism data and divergence data will be necessary to further tease apart the contribution of elevated positive selection and relaxed purifying selection (Yang and Bielawski 2000; Nielsen 2005) to spider genome evolution.
Divergent Signatures of REGs between Social and Solitary Spiders
Genes that were rapidly evolving (REGs; i.e., genes with elevated dN/dS) in the two social spider species relative to the five solitary species were significantly enriched in the functions categories associated with transport, social behavior, and immune response. These include genes such as sodium channel protein, which has previously been implicated in neuronal function and behavior (Ren 2011; Miller 2013). Genes involved in immune response have also been emphasized in social insect studies and may be generally important for the evolution of group living (Sadd et al. 2005; Harpur and Zayed 2013; Vojvodic et al. 2015). In addition, a set of genes associated with social behavior have been highlighted in social insects and vertebrates (Robinson et al. 2008).
Genes that were rapidly evolving in each of the five solitary spider species relative to the two social species were enriched for energy metabolism function, which together with our gene family expansion results described above, indicate that changes in metabolism may be involved in the evolution of spider sociality. Previous comparative studies have also identified elevated rates of molecular evolution for metabolism-associated genes in eusocial relative to solitary bees (Woodard et al. 2011), and in ants (Roux et al. 2014), although these studies find evidence for higher dN/dS for metabolic genes in more highly social species, whereas we found the opposite pattern. Altogether, these previous results together with our results suggest that the evolution of social life may often involve changes in genes associated with metabolic function.
Social Spider Brain-Specific REGs Exhibit Distinct Signature of Social Behavior Bias
To gain insight into the expression pattern of REGs shared in the two social spiders, we characterized the expression profiles of four tissue types (brain, venom gland, abdomen, legs) in S. dumicola. Most REGs were broadly expressed across all four tissues, suggesting that genes experiencing rapid molecular evolution in the two social spiders may have general functions. These broadly expressed REGs were enriched for transport-associated functions (e.g., signal transduction) and tended to be more highly expressed in the brain. Most REGs that showed tissue-specific expression were found in the brain. Interestingly, these brain-specific REGs were enriched for behavior annotations (e.g., aggressive behavior, social behavior, mechanosensory behavior) (fig. 4 and supplementary table S10, Supplementary Material online), and could be candidates for genes influencing social behavior that were involved in the evolution of spider sociality.
Conclusions
The differences we observed between the genomes of the two Stegodyphus social spiders and five solitary spiders are putatively causes or consequences of spider sociality, including both adaptive and nonadaptive evolutionary processes. Alternatively, these differences between the genomes of the two Stegodyphus social spiders and the genomes of the other five solitary spiders could also be associated with lineage-specific adaptation or nonadaptive evolutionary processes. In order to further clarify the genomic underpinnings of social life in the spiders, and to disentangle adaptive and nonadaptive genomic signatures associated with spider social evolution, future studies will need to combine more genomic and transcriptomic data from species representing more independent origins of sociality using formal phylogenetic comparative methods (Garamszegi 2014; Cornwell and Nakagawa 2017; Linksvayer and Johnson 2019). Such future studies can confirm whether the putative genomic signatures of spider sociality we found (expansion and rapid evolution of genes with transport, metabolic, and behavioral functions, and overall elevated rate of molecular evolution) are consistently found across independent origins of sociality and can provide further evidence concerning the putative candidate genes we identified.
Supplementary Material
Acknowledgments
We would like to thank Yiyong Zhao of Fudan University for generously providing scripts for phylogenomic analysis. We thank Dr Weitao Chen of Chinese Academy of Fishery Sciences for assistance in molecular clock analysis. We thank the Linksvayer lab members for helpful comments and discussion on an early version of the article. This work was funded by the National Institutes of Health (Grant No. GM115509).
Author Contributions
CT and TAL conceived and designed the experiments. GMN, NPW and JNP collected the Stegodyphus dumicola samples for RNA sequencing and GMN dissected the samples. CT performed all transcriptomic and comparative genomic analyses, created all figures, and wrote the first draft of the manuscript. CT and TAL finalized the manuscript.
Data deposition: The sequencing reads have been deposited at NCBI SRA under the NCBI BioProject accession PRJNA575239.
Literature Cited
- Agnarsson I, Avilés L, Coddington JA, Maddison WP.. 2006. Sociality in theridiid spiders: repeated origins of an evolutionary dead end. Evolution 60(11):2342–2351. [PubMed] [Google Scholar]
- Amdam GV, Csondes A, Fondrk MK, Page RE Jr.. 2006. Complex social behaviour derived from maternal reproductive traits. Nature 439(7072):76–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilés L. 1997. Causes and consequences of cooperation and permanent-sociality in spiders. The Evolution of Social Behavior in Insects and Arachnids. Cambridge: Cambridge University Press. . 476–498. [Google Scholar]
- Babb PL, et al. 2017. The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat Genet. 49(6):895–903. [DOI] [PubMed] [Google Scholar]
- Bechsgaard J, et al. 2019. Evidence for faster X chromosome evolution in spiders. Mol Biol Evol. 36(6):1281–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Tal R, Tuni C, Lubin Y, Smith D, Bilde T.. 2014. Fitness consequences of outcrossing in a social spider with an inbreeding mating system. Evolution 68(2):343–351. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, et al. 2019. Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science 364(6446):eaav6202. [DOI] [PubMed] [Google Scholar]
- Cornwell W, Nakagawa S.. 2017. Phylogenetic comparative methods. Curr Biol. 27(9):R333–R336. [DOI] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW.. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22(10):1269–1271. [DOI] [PubMed] [Google Scholar]
- Ebersberger I, Strauss S, von Haeseler A.. 2009. HaMStR: profile hidden Markov model based search for orthologs in ESTs. BMC Evol Biol. 9(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Alonso M, et al. 2018. Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana. Nat Ecol Evol. 2(2):352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaither MR, et al. 2018. Genomics of habitat choice and adaptive evolution in a deep-sea fish. Nat Ecol Evol. 2(4):680–687. [DOI] [PubMed] [Google Scholar]
- Galtier N, et al. 2018. Codon usage bias in animals: disentangling the effects of natural selection, effective population size, and GC-biased gene conversion. Mol Biol Evol. 35(5):1092–1103. [DOI] [PubMed] [Google Scholar]
- Garamszegi LZ. ed. 2014. Modern phylogenetic comparative methods and their application in evolutionary biology: concepts and practice. Heidelberg: Springer publisher.
- Garb JE, et al. 2019. The transcriptome of Darwin’s bark spider silk glands predicts proteins contributing to dragline silk toughness. Commun Biol. 2(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garb JE, Sharma PP, Ayoub NA.. 2018. Recent progress and prospects for advancing arachnid genomics. Curr Opin Insect Sci. 25:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison NL, et al. 2016. Spider phylogenomics: untangling the spider tree of life. PeerJ 4:e1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, et al. 2009. Phylogenomic analyses reveal convergent patterns of adaptive evolution in elephant and human ancestries. Proc Natl Acad Sci U S A. 106(49):20824–20829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, et al. 2014. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 24(8):1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinsted L, Pruitt JN, Settepani V, Bilde T.. 2013. Individual personalities shape task differentiation in a social spider. Proc R Soc B. 280(1767):20131407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Harpur BA, Zayed A.. 2013. Accelerated evolution of innate immunity proteins in social insects: adaptive evolution or relaxed constraint? Mol Biol Evol. 30(7):1665–1674. [DOI] [PubMed] [Google Scholar]
- Harrison MC, et al. 2018. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat Ecol Evol. 2(3):557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L-Y, et al. 2018. Doublesex evolution is correlated with social complexity in ants. Genome Biol Evol. 10(12):3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesen J, Lubin Y, Smith DR, Bilde T, Schneider JM.. 2007. The age and evolution of sociality in Stegodyphus spiders: a molecular phylogenetic perspective. Proc R Soc B. 274(1607):231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BR, Linksvayer TA.. 2010. Deconstructing the superorganism: social physiology, groundplans, and sociogenomics. Q Rev Biol. 85(1):57–79. [DOI] [PubMed] [Google Scholar]
- Kapheim KM, et al. 2015. Social evolution. Genomic signatures of evolutionary transitions from solitary to group living. Science 348(6239):1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser CN, Jones DK, Modlmeier AP, Pruitt JN.. 2014. Exploring the effects of individual traits and within-colony variation on task differentiation and collective behavior in a desert social spider. Behav Ecol Sociobiol. 68(5):839–850. [Google Scholar]
- Kriventseva EV, et al. 2015. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 43(D1):D250–D256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulmuni J, Wurm Y, Pamilo P.. 2013. Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 110(6):538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt SD, Menzel F, Nehring V, Schmitt T.. 2016. Ecology and evolution of communication in social insects. Cell 164(6):1277–1287. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer TA, Johnson BR.. 2019. Re-thinking the social ladder approach for elucidating the evolution and molecular basis of insect societies. Curr Opin Insect Sci. 34:123–129. [DOI] [PubMed] [Google Scholar]
- Linksvayer TA, Wade MJ.. 2005. The evolutionary origin and elaboration of sociality in the aculeate Hymenoptera: maternal effects, sib-social effects, and heterochrony. Q Rev Biol. 80(3):317–336. [DOI] [PubMed] [Google Scholar]
- Liu S, Aageaard A, Bechsgaard J, Bilde T.. 2019. DNA methylation patterns in the social spider, Stegodyphus dumicola. Genes 10(2):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N.. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 102:10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin Y, Bilde T.. 2007. The evolution of sociality in spiders In: Brockmann HJ, Roper TJ, Naguib M, Wynne-Edwards KE, Barnard C, Mitani J, editors. Advances in the study of behavior. Vol. 37. Cambridge, MA: Academic Press; p. 83–145. [Google Scholar]
- McKenzie SK, Fetter-Pruneda I, Ruta V, Kronauer D.. 2016. Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proc Natl Acad Sci U S A. 113(49):14091–14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie SK, Oxley PR, Kronauer D.. 2014. Comparative genomics and transcriptomics in ants provide new insights into the evolution and function of odorant binding and chemosensory proteins. BMC Genomics 15(1):718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. 2013. Ion channel reconstitution. Boston, MA: Springer Science & Business Media.
- Mirarab S, et al. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30(17):i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annu Rev Genet. 39(1):197–218. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA.. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14(9):817–818. [DOI] [PubMed] [Google Scholar]
- Privman E, et al. 2018. Positive selection on sociobiological traits in invasive fire ants. Mol Ecol. 27(15):3116–3130. [DOI] [PubMed] [Google Scholar]
- Pruitt JN, et al. 2018. Selection for collective aggressiveness favors social susceptibility in social spiders. Curr Biol. 28(1):100–105.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt JN, Keiser CN.. 2014. The personality types of key catalytic individuals shape colonies’ collective behaviour and success. Anim Behav. 93:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt JN, McEwen BL, Cassidy ST, Najm GM, Pinter-Wollman N.. 2019. Experimental evidence of frequency-dependent selection on group behaviour. Nat Ecol Evol. 3(4):702–707. [DOI] [PubMed] [Google Scholar]
- Ranson H, et al. 2002. Evolution of supergene families associated with insecticide resistance. Science 298(5591):179–181. [DOI] [PubMed] [Google Scholar]
- Ren D. 2011. Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron 72(6):899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittschof CC, et al. 2014. Neuromolecular responses to social challenge: common mechanisms across mouse, stickleback fish, and honey bee. Proc Natl Acad Sci U S A. 111(50):17929–17934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittschof CC, Robinson GE.. 2016. Behavioral genetic toolkits: toward the evolutionary origins of complex phenotypes. Curr Top Dev Biol. 119:157–204. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Fernald RD, Clayton DF.. 2008. Genes and social behavior. Science 322(5903):896–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, et al. 2014. Population genomics of eusocial insects: the costs of a vertebrate-like effective population size. J Evol Biol. 27(3):593–603. [DOI] [PubMed] [Google Scholar]
- Roux J, et al. 2014. Patterns of positive selection in seven ant genomes. Mol Biol Evol. 31(7):1661–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein DR, Abbot P.. 2017. Comparative social evolution. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Sadd BM, Kleinlogel Y, Schmid-Hempel R, Schmid-Hempel P.. 2005. Trans-generational immune priming in a social insect. Biol Lett. 1(4):386–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanggaard KW, et al. 2014. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 5:3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul MC, et al. 2019. Cross-species systems analysis of evolutionary toolkits of neurogenomic response to social challenge. Genes Brain Behav. 18(1):e12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager EE, et al. 2017. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 15(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settepani V, Bechsgaard J, Bilde T.. 2014. Low genetic diversity and strong but shallow population differentiation suggests genetic homogenization by metapopulation dynamics in a social spider. J Evol Biol. 27(12):2850–2855. [DOI] [PubMed] [Google Scholar]
- Settepani V, Bechsgaard J, Bilde T.. 2016. Phylogenetic analysis suggests that sociality is associated with reduced effectiveness of selection. Ecol Evol. 6(2):469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settepani V, et al. 2017. Evolution of sociality in spiders leads to depleted genomic diversity at both population and species levels. Mol Ecol. 26(16):4197–4210. [DOI] [PubMed] [Google Scholar]
- Simola DF, et al. 2013. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 23(8):1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, Fei T, Zhang C, Zhao K.. 2017. Comprehensive transcriptomic analysis of Tibetan Schizothoracinae fish Gymnocypris przewalskii reveals how it adapts to a high altitude aquatic life. BMC Evol Biol. 17(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, Tian F, Zhao K.. 2017. Genomic signature of highland adaptation in fish: a case study in Tibetan Schizothoracinae species. BMC Genomics 18(1):948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AL, Robinson GE.. 2007. Evo-devo and the evolution of social behavior. Trends Genet. 23(7):334–341. [DOI] [PubMed] [Google Scholar]
- Vander Meer RK, Breed MD, Espelie KE, Winston ML.. 1998. Pheromone communication in social insects. Ants, wasps, bees and termites. Boulder (CO): Westview. p. 162.
- Vizueta J, et al. 2017. Evolution of chemosensory gene families in arthropods: insight from the first inclusive comparative transcriptome analysis across spider appendages. Genome Biol Evol. 9(1):178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizueta J, Rozas J, Sánchez-Gracia A.. 2018. Comparative genomics reveals thousands of novel chemosensory genes and massive changes in chemoreceptor repertories across chelicerates. Genome Biol Evol. 10(5):1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojvodic S, et al. 2015. The transcriptomic and evolutionary signature of social interactions regulating honey bee caste development. Ecol Evol. 5(21):4795–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, et al. 2019. Morphology and genome of a snailfish from the Mariana Trench provide insights into deep-sea adaptation. Nat Ecol Evol. 3(5):823–833. [DOI] [PubMed] [Google Scholar]
- Warner MR, Qiu L, Holmes MJ, Mikheyev AS, Linksvayer TA.. 2019. Convergent eusocial evolution is based on a shared reproductive groundplan plus lineage-specific plastic genes. Nat Commun. 10(1):2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard SH, et al. 2011. Genes involved in convergent evolution of eusociality in bees. Proc Natl Acad Sci U S A. 108(18):7472–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CM, Keiser CN, Pruitt JN.. 2015. Personality and morphology shape task participation, collective foraging and escape behaviour in the social spider Stegodyphus dumicola. Anim Behav. 105:47–54. [Google Scholar]
- Wu H, et al. 2014. Camelid genomes reveal evolution and adaptation to desert environments. Nat Commun. 5:5188. [DOI] [PubMed] [Google Scholar]
- Wurm Y, et al. 2011. The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci U S A. 108(14):5679–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, et al. 2017. Genomic basis of adaptive evolution: the survival of Amur Ide (Leuciscus waleckii) in an extremely alkaline environment. Mol Biol Evol. 34(1):145–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, et al. 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21(5):650–659. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP.. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 15(12):496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346(6215):1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2015. Chemoreceptor evolution in hymenoptera and its implications for the evolution of eusociality. Genome Biol Evol. 7(8):2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman A. 2007. Assessing the costs of group living: comparing metabolic physiology and growth in social and solitary spiders [Honors Thesis]. Cornell University.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.