Abstract
Influenza is an acute infection of the respiratory tract, which affects each year millions of people. Influenza virus infection is initiated by the surface glycoprotein hemagglutinin (HA) through receptor binding and fusion of viral and endosomal membranes. HA is synthesized as a precursor protein and requires cleavage by host cell proteases to gain its fusion capacity. Although cleavage of HA is crucial for virus infectivity, little was known about relevant proteases in the human airways for a long time. Recent progress in the identification and characterization of HA‐activating host cell proteases has been considerable however and supports the idea of targeting HA cleavage as a novel approach for influenza treatment. Interestingly, certain bacteria have been demonstrated to support HA activation either by secreting proteases that cleave HA or due to activation of cellular proteases and thereby may contribute to virus spread and enhanced pathogenicity. In this review, we give an overview on activation of influenza viruses by proteases from host cells and bacteria with the main focus on recent progress on HA cleavage by proteases HAT and TMPRSS2 in the human airway epithelium. In addition, we outline investigations of HA‐activating proteases as potential drug targets for influenza treatment.
Keywords: influenza virus hemagglutinin, proteolytic cleavage, viral‐bacterial pneumonia, influenza treatment by protease inhibitors, TMPRSS2, HAT
Short abstract
The authors, who are leading experts in this field, present a timely, authoritative review on the proteolytic cleavage of the influenza hemagglutinin (HA), an activation mechanism that is essential for the infectivity of influenza viruses, including the recently emerged H7N9. They also address the potential of host proteases as targets for developing new influenza drugs. This review will be of considerable interest to virologists, microbiologists and pharmaceutical companies alike.
Human influenza and bacterial superinfections
Influenza (flu) is a highly contagious respiratory illness that affects each year millions of people. Of the three genera of influenza viruses (A, B and C), influenza A and influenza B viruses pose a continuous threat to public health. Currently, influenza A viruses of the subtypes H3N2 and H1N1 and influenza B viruses cocirculate with varying predominance in the human population and are responsible for seasonal epidemics that may result in 3–5 million cases of severe respiratory illness and up to 500 000 annual deaths worldwide. The natural reservoir of influenza A viruses are wild aquatic birds from where they are transmitted to a wide range of mammalian and avian hosts including humans, pigs and poultry. The emergence of a new influenza A virus for which there is little or no immunity in the human population may provoke an influenza pandemic, with the most devastating pandemic in 1918, the ‘Spanish flu’, claiming c. 40 million deaths worldwide. In addition, three more influenza pandemics occurred in the 20th and 21st centuries: the ‘Asian flu’ in 1957 (H2N2), the ‘Hong‐Kong flu’ in 1968 (H3N2) and, recently, the 2009 H1N1 pandemic. In contrast, influenza B viruses are predominantly found in humans and are not known to have an animal reservoir from which new virus variants can emerge.
Human influenza viruses spread via aerosols or respiratory droplets due to sneezing and coughing, but may also spread by contact, and infect the respiratory epithelium of the upper or lower airways. Influenza is an acute respiratory illness with a rapid onset of symptoms such as fever, headache, generalized malaise and myalgias that may last for 7–14 days. Complications may be due to bronchitis, viral pneumonia or myocarditis. Moreover, viral‐bacterial pneumonia and secondary bacterial pneumonia due to concomitant or subsequent infection with bacteria, respectively, contribute significantly to morbidity and mortality of influenza infections. Most commonly isolated bacteria are Streptococcus pneumoniae (pneumococcus), Staphylococcus aureus and Haemophilus influenzae. Pathological studies of autopsy specimens and analyses of medical reports revealed that the majority of the deaths of the 1918 influenza pandemic occurred concurrently with bacterial pneumonia predominantly by H. influenzae (McCullers, 2006; Morens et al., 2008). Estimated 70% of severe and fatal cases during the 1957 and 1968 pandemics and c. 30% of influenza‐associated pneumonia during the 2009 H1N1 pandemic likely resulted from pneumonia due to co‐infections by S. aureus or pneumococcus. During seasonal influenza epidemics, estimated 44–57% of hospitalizations and 25% of influenza‐associated deaths are due to secondary bacterial pneumonia (Peltola et al., 2005; McCullers, 2011).
Influenza A virus morphology and viral life cycle
Influenza A viruses belong to the family of Orthomyxoviridae and are enveloped viruses of pleomorphic morphology ranging from spherical to filamentous particles with a diameter of 80–120 nm. The genome consists of eight segments of single‐stranded, negative‐sense RNA, which are associated with the polymerase proteins PB1, PB2 and PA and multiple copies of the nucleoprotein (NP) to form the viral ribonucleoprotein complexes (vRNPs) and together encode up to 15 proteins (Jagger et al., 2012; Muramoto et al., 2013; Fig. 1a). The viral envelope is made up of a lipid bilayer derived from the host cellular plasma membrane and contains the two major spike glycoproteins, hemagglutinin (HA) and neuraminidase (NA), and the proton channel protein M2. The inner side of the envelope is lined by the matrix protein M1. Based on antigenic criteria, influenza A viruses are classified into 17 HA (H1‐H17) and 10 NA (N1‐N10) subtypes to date (Tong et al., 2012). Most subtypes circulate in wild aquatic birds and are sporadically transmitted to other species including humans.
Figure 1.
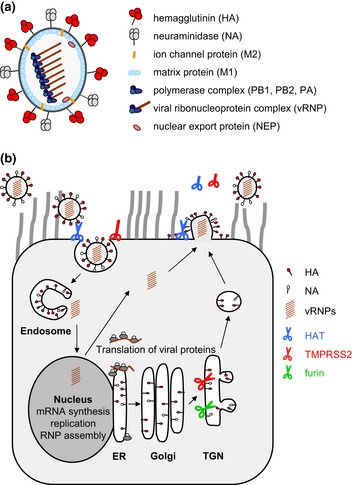
(a) Scheme of the influenza A virus particle. The virion contains a lipid envelope derived from the cellular plasma membrane. The two glycoproteins hemagglutinin (HA) and neuraminidase (NA) and the ion channel protein M2 are embedded in the envelope. The inner side is lined by the matrix protein M1. The genome consists of eight segments of single‐stranded, negative‐sense RNA, which are associated with the nucleoprotein (NP) and the polymerase subunits PB1, PB2 and PA to form viral ribonucleoprotein complexes (vRNPs). The nuclear export protein (NEP) is also present in the virion, while the nonstructural proteins NS1, PB1‐F2, PB1‐N40, PA‐X, PA‐N155 and PA‐N182 are present in the infected cell. (b) Influenza virus replication and proteolytic activation by cellular proteases in human airway epithelial cells. Infection is initiated by the HA through binding to N‐acetyl neuraminic acid‐containing cell surface receptors. Upon receptor‐mediated endocytosis, HA mediates fusion of viral and endosomal membranes at low pH to release the vRNPs into the cytoplasm (uncoating). The vRNPs are imported into the nucleus, where transcription and replication of the viral genome occur. Translation of viral mRNAs is performed by the cellular machinery. HA, NA and M2 are synthesized into the endoplasmic reticulum (ER) and transported along the constitutive secretory pathway to the plasma membrane. The internal viral proteins are synthesized at free ribosomes, and protein components of the vRNPs are then imported into the nucleus, where assembly of new vRNPs occurs. Finally, vRNPs are exported from the nucleus and transported to the plasma membrane, where self‐assembly of viral proteins leads to budding of new virions. The NA cleaves N‐acetyl neuraminic acid from carbohydrate moieties, facilitating release of progeny virions. Proteolytic activation of HA can take place in different compartments and at different time points during viral replication and is indicated by scissors (open scissor: active protease; closed scissor: enzymatically inactive protease; truncated scissor: soluble protease). HA with multibasic cleavage site is cleaved by furin in the TGN. HA containing a monobasic cleavage site is cleaved by TMPRSS2 in the TGN or by HAT on the plasma membrane: either during assembly and budding of progeny virus or during attachment and entry into the cell.
Influenza virus replication is initiated by the HA through binding to N‐acetyl‐neuraminic acid‐containing cell surface receptors, resulting in receptor‐mediated endocytosis and subsequent fusion of the viral envelope with the endosomal membrane to release the vRNPs into the cytoplasm (Fig. 1b). Membrane fusion requires proteolytic cleavage of HA and is induced by the acidic pH in the endosome. The vRNPs are imported into the nucleus where transcription and replication of the genome are catalysed by the viral RNA‐dependent RNA polymerase. Splicing of viral mRNAs and translation into proteins are performed by the cellular machineries. Influenza virus infection causes a shutoff of host protein synthesis, thereby enhancing translation of viral mRNAs. At the later stages of viral infection, newly synthesized vRNPs and viral proteins are transported to the apical membrane of polarized epithelial cells where assembly and budding of new progeny virions take place. Here, NA cleaves N‐acetyl‐neuraminic acid from carbohydrate moieties, facilitating release of progeny virus from the host cell and preventing virions to clump together.
Proteolytic cleavage of HA is a prerequisite for membrane fusion
The HA is the major surface glycoprotein of influenza viruses, mediates receptor binding and membrane fusion and is the main target for neutralizing antibodies. HA is a type I transmembrane protein and is integrated in the viral envelope as a homotrimer. HA is synthesized as a precursor protein, HA0 (75 kDa), that is transported along the constitutive secretory pathway to the plasma membrane (Fig. 1b). During its transport, HA becomes N‐glycosylated, palmitoylated and cleaved into the subunits HA1 (55 kDa) and HA2 (25 kDa) (Fig. 2a and b). Cleavage can take place either in the trans‐Golgi network (TGN) or on the cell surface as discussed below. The amino‐terminal cleavage fragment HA1 contains the receptor‐binding site and remains linked by a disulphide bond to the carboxy‐terminal fragment HA2, which is membrane‐anchored and responsible for fusion (Fig. 2b). HA0 is cleaved at a specific arginine residue (R) to expose the fusion peptide consisting of about 14 hydrophobic amino acids at the N‐terminus of HA2. Cleavage is a prerequisite for conformational changes at low pH in the endosome that trigger membrane fusion and is essential for the infectivity and spread of influenza viruses. The cleavage site is located in a prominent loop that protrudes from the surface (Fig. 2a). The amino acid sequence as well as the structure and susceptibility of the cleavage site are critical for cleavage of HA by different proteases (Klenk & Rott, 1988; Klenk & Garten, 1994; Steinhauer, 1999; Garten & Klenk, 2008).
Figure 2.
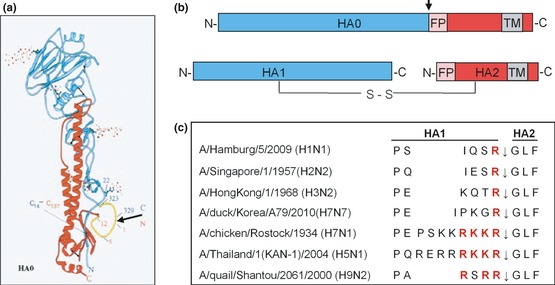
Cleavage of HA0 into HA1 and HA2 at a specific cleavage site. (a) Structure of the monomeric HA0 precursor of A/HongKong/68 containing the mutation R329Q to prevent cleavage determined by X‐ray crystallography (Chen et al., 1998). The cleavage site is located in a prominent surface loop (yellow) highlighted by an arrow. (b) Schematic illustration of the HA0 precursor and the cleaved form consisting of the disulphide‐linked subunits HA1 and HA2. The colours of HA1 (blue) and HA2 (red) are based on the structure shown in Fig. 1a. The cleavage site is indicated by an arrow. FP: fusion peptide, TM: transmembrane domain. (c) Alignment of amino acid sequences at the HA cleavage site of different human and avian influenza viruses. The arrow indicates the cleavage site between HA1 and HA2. Basic amino acids crucial for cleavage by relevant proteases are highlighted in red.
Cleavage of HA is a prime determinant of avian influenza virus pathogenicity
Avian influenza A viruses are responsible for recurrent outbreaks in poultry farms worldwide and lead to serious economic losses in the poultry industry. Avian viruses differ in their pathogenicity and are classified as either low‐ or high‐pathogenic avian influenza viruses (LPAIV or HPAIV, respectively). LPAIV replicate primarily in the intestinal and also in the respiratory tract of birds, cause mild or asymptomatic infections and spread via the faecal‐oral route. In contrast, HPAIV cause systemic infections in poultry with mortality rates up to 100%, known as fowl plague or avian influenza. HPAIV are restricted to the subtypes H5 and H7 under natural settings, but not all H5 and H7 viruses are highly pathogenic.
Already early studies of natural avian influenza virus isolates indicated that the cleavage site sequence connecting HA1 and HA2 is a key determinant for pathogenicity and organ tropism of the viruses. LPAIV possess a single arginine, rarely a single lysine, at the cleavage site, whereas the HA of HPAIV contains a multibasic motif with the consensus sequence R‐X‐R/K‐R (Vey et al., 1992). Cleavage of HA0 into HA1 and HA2 is crucial for virus infectivity. LPAIV were found to require addition of exogenous trypsin to most tissue cultures for proteolytic activation and multicycle replication in vitro, whereas HPAIV were shown to be activated by endogenous proteases (Klenk & Rott, 1988; Klenk & Garten, 1994).
HA with multibasic cleavage site is cleaved by ubiquitous subtilisin‐like endoproteases furin and the proprotein convertase 5/6 (PC5/6), supporting systemic infection (Stieneke‐Gröber et al., 1992; Horimoto et al., 1994; Feldmann et al., 2000). Furin is a type I transmembrane protease that is predominantly localized in the TGN, but is also transported through the constitutive secretory pathway to the plasma membrane, where it can be shed or recycled and targeted back to the TGN (Schäfer et al., 1995). Furin cleaves a large number of cellular protein precursors such as prohormones and growth factors at the C‐terminus of the consensus sequence R‐X‐R/K‐R into biologically active forms. Moreover, furin supports proteolytic activation of a large number of viral glycoproteins (e.g. measles virus, respiratory syncytial virus, yellow fever virus, HIV‐1, Ebola virus) and bacterial toxins (e.g. anthrax toxin, diphtheria toxin) at such basic motifs (Klenk & Garten, 1994; Thomas, 2002). Thus, furin plays important roles in numerous physiological as well as pathogenic processes. Cleavage of HA by furin takes place in the TGN (Fig. 1b) and results in the release of infectious progeny virus containing cleaved HA from the cells, supporting spread of infection.
In contrast, LPAIV possess a monobasic cleavage site and are proteolytically activated by trypsin in vitro (Klenk et al., 1975). Relevant trypsin‐like proteases are present in a restricted number of tissues such as the respiratory or the intestinal tract, limiting spread of infection to these tissues. A protease homologous to blood clotting factor Xa was identified as HA‐activating protease in the chorioallantoic membrane of embryonated chicken eggs (Gotoh et al., 1990), and a number of other trypsin‐like proteases, such as plasmin (Lazarowitz & Choppin, 1975) and tryptase Clara (Kido et al., 1992), have been found to activate HA with a monobasic cleavage site in vitro. The identity of relevant proteases in the respiratory or intestinal tract of avian species, however, remains to be investigated. In 2006, the human proteases HAT and TMPRSS2 present in the human airway epithelium were identified as proteases that activate HA with monobasic cleavage (Böttcher et al., 2006). A chicken protease homologous to human TMPRSS2 has been demonstrated to activate the HA at a monobasic cleavage site in vitro, suggesting that the chTMPRSS2 might be responsible for cleavage of HA also in avian hosts (Bertram et al., 2012).
HPAIV emerge from LPAIV by extension of the cleavage site loop due to insertion of mostly basic amino acids (Fig. 2c). Interestingly, acquisition of a multibasic cleavage site that is activated by furin seems to be restricted to the subtypes H5 and H7 and has not been observed for any other subtype under natural settings (Klenk & Garten, 1994; Garten & Klenk, 2008). Analysis of the cleavage site of different HPAIV demonstrated that extension of the cleavage site can occur by different mechanisms: polymerase slippage at purine‐rich arginine or lysine codons, recombination with viral gene segments (NP or M genes) or 28S ribosomal RNA or still unknown mechanisms and has been reviewed in detail in the study by Garten & Klenk (2008). Already in early studies, it has been demonstrated that substitution of the multibasic cleavage site of a HPAIV by a monobasic motif results in a low‐pathogenic virus. However, insertion of a multibasic cleavage motif into a HA with a monobasic cleavage site does not automatically confer high pathogenicity to the virus, demonstrating that the emergence of a HPAIV is a multifactorial process (Stech et al., 2009; Schrauwen et al., 2011). In a recent study, Veits et al. (2012) introduced a multibasic cleavage site into the HA of several influenza A virus subtypes and generated reassortant viruses in the genetic background of a highly pathogenic H5N1 virus. Interestingly, reassortants with H2, H4, H8 and H14 with multibasic cleavage site caused lethal infections in chickens, while other subtypes did not. These observations demonstrate that a multibasic HA cleavage site can confer high pathogenicity to subtypes other than H5 and H7 in a suitable genetic background. Therefore, the restriction of natural HPAIV isolates to H5 and H7 subtypes seems to be due to their unique ability to acquire a multibasic cleavage site under natural settings; however, underlying mechanisms are not yet understood.
Activation of human influenza viruses by host cell proteases TMPRSS2 and HAT
As most avian and mammalian influenza viruses, human influenza viruses possess a monobasic cleavage site and require activation by trypsin‐like proteases. Seasonal and pandemic influenza viruses belong to the subtypes H1, H2 and H3 and influenza B virus, respectively, and are activated C‐terminal of the HA cleavage site motifs I‐Q‐S‐R, I‐E‐S‐R and K‐Q‐T‐R for H1, H2 and H3 subtypes, respectively (Fig. 2c), and L‐K‐E‐R for influenza B viruses. A number of trypsin‐like proteases isolated from rat and swine lung, such as tryptase Clara, mini‐plasmin or tryptase TC30, were shown to support proteolytic activation and multicycle replication of human influenza viruses in vitro (Kido et al., 1992, 2007; Murakami et al., 2001; Sato et al., 2003). Because the genetic identity is unknown for these proteases, it still remains unclear whether they play a role in in vivo infection, and appropriate proteases that cleave HA in the human airway epithelium were unknown until recently.
Activation of HA with monobasic cleavage site by plasmin, tryptase Clara or the factor Xa‐like protease in embryonated chicken eggs occurs outside the cell after release of newly synthesized virions or probably already during assembly and budding when HA is present on the plasma membrane. According to this, the classical dogma of activation of HA with monobasic cleavage site is HA cleavage by soluble proteases outside the cells. A study by Zhirnov et al. (2002), however, demonstrated that cleavage of HA in human airway epithelial cells occurs by cell‐associated proteases. Furthermore, activation of influenza viruses in human intestinal Caco‐2 cells was shown to take place intracellularly (Zhirnov et al., 2003). The relevant proteases remained unknown in these studies, but indicated that cleavage of HA by human proteases may be performed by membrane‐bound proteases.
In search for human HA‐activating proteases, a number of proteases that possess trypsin‐like activity were cloned from human primary tracheobronchial epithelial (HTBE) cells (Böttcher et al., 2006). Among a couple of candidates, the two proteases HAT (human airway trypsin‐like protease; also designated as TMPRSS11D) and TMPRSS2 (transmembrane protease serine S1 member 2; also designated as epitheliasin) were demonstrated to cleave influenza virus HA with a monobasic cleavage site and support multicycle virus replication in vitro (Böttcher et al., 2006). Later on, the TMPRSS2‐related protease TMPRSS4 was shown to activate HA with monobasic site in vitro, too (Chaipan et al., 2009). By contrast, the trypsin‐like proteases prostasin, TMPRSS3, TMPRSS6 and hepsin were not able to activate HA upon co‐expression in mammalian cells (Bertram et al., 2010; Böttcher‐Friebertshäuser et al., 2010), emphasizing that only certain trypsin‐like proteases present in the airways support proteolytic activation of influenza viruses.
TMPRSS2 and HAT belong to the family of type II transmembrane serine proteases (TTSPs) and contain an N‐terminal transmembrane domain, a highly variable stem region and a catalytic domain of the chymotrypsin S1 type (Fig. 3a). The most prominent member of the TTSPs is the digestive enzyme enteropeptidase that originally was isolated as a soluble protease, which later was shown to be the catalytic domain of a membrane‐bound protease (Szabo & Bugge, 2008). TTSPs are synthesized as single‐chain zymogens that are activated by cleavage C‐terminal of a highly conserved arginine or lysine residue into a mature form (Fig. 3a). The catalytic domain is predicted to remain linked to the membrane‐anchored domains by a disulphide bond or can be released as a soluble protease.
Figure 3.
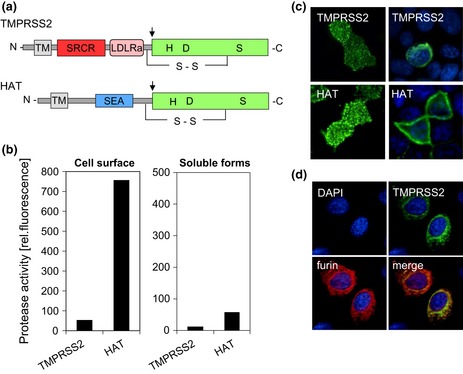
Human proteases TMPRSS2 and HAT cleave HA with monobasic cleavage site. (a) Schematic domain structures. HAT and TMPRSS2 are synthesized as single‐chain zymogens that consist of an N‐terminal transmembrane domain (TM), a stem region containing, for example, sea urchin sperm protein enterokinase; agrin domain (SEA) for HAT or low‐density lipoprotein receptor class A domain (LDLRA); and scavenger receptor cysteine‐rich domain (SRCR) for TMPRSS2; and a C‐terminal trypsin‐like serine (S1) protease domain with the catalytic triad histidine (H), aspartic acid (D) and serine (S). The zymogens undergo autocatalytic cleavage activation (indicated by arrows), and the catalytic domain remains linked to the transmembrane rest of the molecule by a disulphide bond. (b) Enzymatic activity of cell surface‐anchored or soluble HAT and TMPRSS2 in MDCK‐HAT and MDCK‐TMPRSS2 cells. Protease activity on the cell surface and in concentrated protease‐containing supernatants, respectively, was measured by incubation with the fluorogenic peptide substrate Boc‐Gly‐Pro‐Arg‐AMC (Böttcher‐Friebertshäuser et al., 2010). (c) Subcellular localization of HAT and TMPRSS2. HAT and TMPRSS2 on the cell surface (nonpermeabilized cells; left panels) of transient protease expressing MDCK cells and within the cell (permeabilized cells; right panels) were stained using protease‐specific antibodies and FITC‐conjugated secondary antibodies. Cell nuclei were counterstained with DAPI. (d) Colocalization of TMPRSS2 and furin. Huh‐7 cell with transient expression of TMPRSS2 and furin were permeabilized, and protease expression was analysed by TMPRSS2‐ and furin‐specific antibodies, respectively, and FITC‐ or TRITC‐conjugated secondary antibodies. Cell nuclei were counterstained using DAPI.
The physiological roles of HAT and TMPRSS2 are still unknown. Interestingly, mice deficient in TMPRSS2 and HAT, respectively, have been shown to lack a discernible phenotype (Kim et al., 2006; Sales et al., 2011). HAT was originally isolated from patients with chronic airway diseases and among other functions has been demonstrated to cleave fibrinogen, to activate the protease‐activated receptor 2 (PAR‐2) and to increase mucus gene expression and to stimulate bronchial fibroblast proliferation in airway epithelial cells in vitro (Yoshinaga et al., 1998; Chokki et al., 2004; Matsushima et al., 2006). HAT expression is prominent in the trachea and bronchi and was also detected in the gastrointestinal tract, the skin and the brain (Table 1). In the airway epithelium, HAT has been shown to be expressed in ciliated cells, but not in submucosal glands and mast cells (Takahashi et al., 2001). TMPRSS2 is widely expressed in epithelial cells of the respiratory, gastrointestinal and urogenital tract with high expression levels in prostate and colon (Szabo & Bugge, 2008). Moreover, immunohistochemical studies revealed that TMPRSS2 is expressed in cardiac myocytes, suggesting that it might contribute to influenza‐associated myocarditis (Bertram et al., 2012). TMPRSS2 is associated with prostate carcinogenesis in two different ways. The protease has been shown to be overexpressed in prostate cancer tissue. Moreover, fusion of the androgen‐regulated TMPRSS2 promoter to different ETS (E 26) transcription factor genes, resulting in overexpression of the respective transcription factors, is seen in nearly 50% of patients and has established as a prognostic marker of prostate carcinogenesis (Tomlins et al., 2009). The role of TMPRSS2 in the airways remains to be investigated. Expression studies in Xenopus oocytes suggested that TMPRSS2 is involved in regulation of the airway surface liquid (ASL) volume by proteolytic cleavage of epithelial sodium channels (ENaCs) (Donaldson et al., 2002).
Table 1.
HA‐activating host cell proteases, cleavage specificity and expression in human tissues
| Protease | HA cleavage site sequence | Expression in human tissues | References |
|---|---|---|---|
| TMPRSS2 | R↓ | Nasal epithelium, trachea, bronchi, lung (type II pneumocytes), larynx, tonsil, alveolar macrophages, myocardium, prostate, liver, kidney, small intestine, skin, testis, colon, pancreas |
Vaarala et al. (2001) Böttcher et al. (2006) Lucas et al. (2008) Szabo & Bugge (2008) Bertram et al. (2012) |
| HAT | R↓ | Trachea, bronchi, oesophagus, tongue, nasal epithelium, larynx, epiglottis, tonsil, skin, brain |
Yasuoka et al. (1997) Böttcher et al. (2006) Szabo & Bugge (2008) Bertram et al. (2012) |
| TMPRSS4 | R↓ | Oesophagus, lung, small intestine, stomach, colon, bladder, kidney, pancreas |
Szabo & Bugge (2008) Chaipan et al. (2009) |
| Matriptase | R/K‐X‐X/S‐R↓ | Widespread expression in epithelial tissues: nasal epithelium, trachea, bronchi, salivary gland, oesophagus, kidney, small intestine, stomach, prostate, skin, hair follicle, monocytes, macrophages, mast cells, B cells, spinal neurons |
Oberst et al. (2003) Szabo & Bugge (2008) Baron et al. (2013) |
| TMPRSS13/MSPL (splice isoforms) | R/K‐K‐K‐R↓ | Widespread tissue distribution: lung, brain, kidney, liver, spleen, prostate, pancreas, skin, small intestine, colon, testis, thymus, placenta, endothelial cells, T cells, monocytes |
Szabo & Bugge (2008) Okumura et al. (2010) |
| Furin | R‐X‐R/K‐R↓ | Ubiquitous expression |
Stieneke‐Gröber et al. (1992) Thomas (2002) |
| PC5 (also known as PC6) | R‐X‐R/K‐R↓ | Widespread tissue distribution |
Feldmann et al. (2000) Seidah et al. (2008) |
Cleavage of HA at the indicated amino acid sequence is shown by an arrow. Protease expression in different human tissues is listed; prominent expression levels are highlighted in bold.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
TTSPs are predicted to be situated in the plasma membrane with an extracellular catalytic domain. In addition, soluble forms of HAT and TMPRSS2 have been described. HAT was originally purified as an active soluble protease from sputum of patients with chronic bronchitis and bronchial asthma (Yoshinaga et al., 1998). TMPRSS2 was shown to be released from prostate and prostate cancer cells (Afar et al., 2001). The subcellular localization and enzymatic activity of HAT and TMPRSS2 in airway epithelial cells have not been investigated in more detail so far however, have been studied in Madin‐Darby canine kidney (MDCK) cells with doxycycline‐induced expression of either protease that proved to be a suitable model system (Böttcher et al., 2009).
Initial studies on inhibition of influenza virus activation in MDCK‐HAT and MDCK‐TMPRSS2 cells using natural and synthetic protease inhibitors demonstrated that proteolytic activation of HA by HAT can be efficiently inhibited by exogenous protease inhibitors (Böttcher et al., 2009; Böttcher‐Friebertshäuser et al., 2010; Sielaff et al., 2011). In contrast, inhibition of virus activation by TMPRSS2 required higher inhibitor concentrations. Interestingly, modification of a protease inhibitor by attachment of a fatty acid to improve cellular uptake allowed efficient inhibition of HA cleavage in TMPRSS2‐expressing MDCK cells. These data indicated that cleavage by TMPRSS2 takes place within the cell and inhibition requires cellular uptake of the inhibitor, whereas cleavage of HA by HAT occurs on the plasma membrane and therefore is easy accessible to exogenous inhibitors (Fig. 1b).
HAT and TMPRSS2 were shown to be expressed on the cell surface of MDCK cells both as zymogen and mature form (Böttcher‐Friebertshäuser et al., 2010). Incubation of HAT‐ or TMPRSS2‐expressing cells with fluorogenic peptides to measure the protease activity on the cell surface revealed that HAT is present as an enzymatically active enzyme on the cell surface, while TMPRSS2 shows poor if any protease activity (Fig. 3b). The reasons for the lack of TMPRSS2 activity on the cell surface is unknown but might be related to the presence of protease inhibitors, missing cofactors or lower amounts of TMPRSS2 on the cell surface compared with HAT. Immunofluorescence microscopy of transient TMPRSS2‐ or HAT‐expressing cells demonstrates that both HAT and TMPRSS2 are expressed on the cell surface; however, TMPRSS2 accumulates in the Golgi apparatus and TGN and colocalizes with furin, while HAT is expressed predominantly in the plasma membrane (Fig. 3c and d). These data strongly suggest that TMPRSS2 cleaves HA in the TGN. Moreover, these observations demonstrate that cleavage of HA with monobasic and multibasic cleavage site can be performed in the same compartment but by different proteases (cf. Fig. 1b).
HA cleavage by HAT has been demonstrated to take place on the cell surface (Böttcher‐Friebertshäuser et al., 2010). Interestingly, HAT on the one hand is capable of cleaving newly synthesized HA0, probably during assembly and budding of new virions on the plasma membrane. Thus, infectious virus containing cleaved HA is released from HAT‐expressing cells. Furthermore, HAT can cleave the HA0 of incoming virus at the stage of entry during attachment to the cell, facilitating infection of HAT‐expressing cells with virus containing noncleaved HA0 (Figs 1 and 5). Activation of HA at two different steps during virus replication seems to be redundant at the first few. But considering that different cell types in the airway epithelium express a partially different protease repertoire, activation of incoming virus by HAT may enable proteolytic activation of progeny viruses released from cells that lack expression of appropriate proteases (Fig. 5). This concept is supported by the observation that the WSN virus is activated upon entry into MDBK cells (Boycott et al., 1994).
Figure 5.
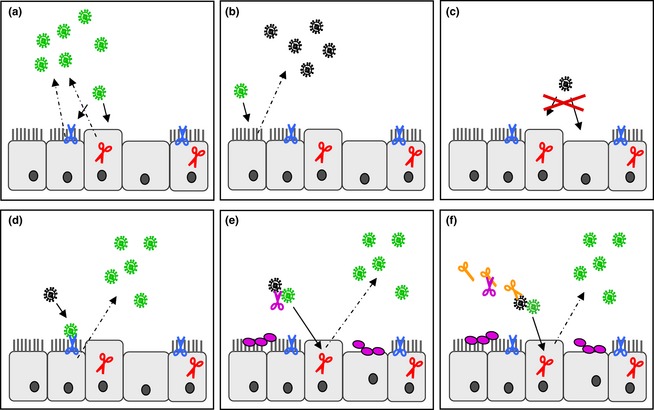
Activation of influenza viruses by host cellular and bacterial proteases in the human airway epithelium. Ciliated and nonciliated epithelial cells are shown. HA‐activating proteases are shown by scissors: HAT (blue), TMPRSS2 (red); continuous arrows indicate infection of cells, and dashed arrows, release of virus progeny. (a) Infection of HAT‐ and/or TMPRSS2‐expressing cells results in release of infectious progeny virus containing cleaved HA (green). (b) Infection of cell without expression of relevant HA‐activating proteases results in progeny virus containing noncleaved HA (black). (c) Virus containing noncleaved HA is not able to infect cells expressing only TMPRSS2 or cells without expression of any relevant protease. (d) HAT‐expressing cells support proteolytic activation of HA of incoming virus on the cell surface, facilitating infection of the cell and release of infectious progeny virus. (e) In co‐infections of influenza virus and certain bacteria, soluble bacterial proteases (purple) may support proteolytic activation and spread of the virus. (f) Bacterial proteases or proteins may activate or augment cellular proteases (orange), which in turn cleave HA.
It is still unknown whether autocatalytic activation of HAT occurs on the plasma membrane or already during its transit through the constitutive secretory pathway to the plasma membrane (Fig. 1b). It therefore remains to be analysed whether HAT could cleave newly synthesized HA0 also within the cell in addition to HA cleavage on the plasma membrane.
As mentioned above, soluble forms of HAT and TMPRSS2 have been described. Shedding of soluble HAT and TMPRSS2 into the cell supernatant has also been observed in MDCK‐HAT and MDCK‐TMPRSS2 cells (Böttcher‐Friebertshäuser et al., 2010). But the soluble forms showed only marginal enzymatic activity (Fig. 3b, right panel) and were not sufficient to support cleavage of influenza virus HA in these cells, indicating that HA cleavage in human airway epithelial cells occurs by cell‐associated and not by soluble proteases. Interestingly, enhanced shedding of TMPRSS2 and HAT from differentiated human nasal epithelial cells (NECs) and hence enhanced influenza virus replication were observed upon exposure to ozone (Kesic et al., 2012). Thus, enhanced shedding of TMPRSS2 and HAT under stress conditions or in chronic airway diseases might play a role in influenza infections in the human airways. However, further studies are needed to prove this hypothesis.
Recent studies demonstrated that HAT and TMPRSS2 support proteolytic activation of influenza B viruses, the SARS coronavirus and the human metapneumovirus at single arginines in vitro and therefore may play a role in activation and spread of different respiratory viruses in the human airways (Shirogane et al., 2008; Matsuyama et al., 2010; Shulla et al., 2011; Böttcher‐Friebertshäuser et al., 2012).
Activation of HA at unusual cleavage site sequences
Commonly, influenza virus HA is activated either at a single arginine by trypsin‐like proteases such as TMPRSS2 or HAT or at a multibasic motif by ubiquitous furin and PC5/6. However, a number of studies demonstrated that such a strict separation does not apply to all influenza isolates and that some influenza viruses can possess uncommon cleavage site motifs that may facilitate activation by other or additional proteases.
One of the best studied exceptions is the influenza virus A/WSN/33 (H1N1), which is neurotropic in mice. The HA of A/WSN/33 contains the unusual HA cleavage site I‐Q‐Y‐R instead of I‐Q‐S‐R (cf. Fig. 2c) with a tyrosine (Y) in P2 that was shown to facilitate efficient cleavage by plasmin, supporting proteolytic activation and virus spread to other tissues including the brain (Lazarowitz & Choppin, 1975; Goto & Kawaoka, 1998; Sun et al., 2010).
During a H5N2 avian influenza virus outbreak in Pennsylvania in 1983, loss of a carbohydrate side chain of HA1 due to a single point mutation was shown to be responsible for the appearance of high virulence in chickens. The influenza virus A/chick/Pennsylvania/1/83 (H5N2) possessed the multibasic cleavage site motif K‐K‐K‐R, but the low‐pathogenic isolates contained an oligosaccharide at Asn‐11 of HA1 that was demonstrated to interfere with cleavage of HA by steric hindrance (Kawaoka et al., 1984). Interestingly, upon serial passage of a nonpathogenic H5N2 isolate in chicken embryo cells, pathogenic mutants emerged that had retained the carbohydrate side chain, but instead possessed an increased number of basic amino acids at the cleavage site (Ohuchi et al., 1989). Thus, masking of the cleavage site by an oligosaccharide can be overcome by extension of the cleavage site. The multibasic motif K‐K‐K‐R, however, is not cleaved by furin because of the lysine (K) in P4. In a recent study, the type II transmembrane protease TMPRSS13/MSPL that is widely expressed in several tissues (Table 1) has been shown to cleave the HA at such a motif and to enable systemic infection independent of furin (Okumura et al., 2010).
Another example for an unusual HA cleavage has recently been described for influenza A viruses of the subtype H9N2 that circulate worldwide and have become highly prevalent in poultry in many countries. H9N2 viruses are associated with repeated outbreaks of severe illness in poultry farms in the Middle East and are occasionally transmitted to pigs and humans, raising concern about their pandemic potential. In contrast to most other subtypes, H9N2 viruses vary remarkably in the amino acid sequence at the HA cleavage site. While H9N2 viruses in America, Europe and Africa contain diverse monobasic HA cleavage site motifs, many recent H9N2 isolates from Asia and the Middle East possess di‐ or tribasic HA cleavage sites of the sequence R‐S‐S‐R or R‐S‐R‐R. Importantly, the di‐ or tribasic cleavage sites of H9N2 viruses evolved by substitution and not by insertion of basic amino acids as in case of HPAIV of subtypes H5 and H7 (Fig. 2c) and are not cleaved by ubiquitous furin or PC5/6 (Gohrbandt et al., 2011; Soda et al., 2011). HAT and TMPRSS2 have been demonstrated to cleave H9 with mono‐, di‐ and tribasic cleavage site motifs (Baron et al., 2013). Moreover, the TTSP matriptase was shown to cleave H9 with R‐S‐S‐R and R‐S‐R‐R cleavage sites (Baron et al., 2013). Matriptase is expressed in a wide range of tissues with high expression levels in the kidney (Table 1). Interestingly, nephrotropism has been observed in H9N2 outbreaks in chickens. According to this, H9N2 viruses with R‐S‐S‐R and R‐S‐R‐R cleavage sites were able to undergo multicycle replication in primary chicken embryo kidney (CEK) cells due to proteolytic activation of HA, while H9N2 isolates containing a single R were not. HA activation in CEK cells was inhibited by matriptase inhibitors, suggesting that matriptase may contribute to the nephrotropism of H9N2 viruses in chickens (Baron et al., 2013). However, it remains to be investigated whether expression of matriptase in a wide range of tissues affects organ tropism and pathogenicity of H9N2 viruses in vivo.
Bacterial proteases can cleave HA during co‐infections
Bacterial co‐infections contribute significantly to morbidity and mortality of influenza infections during seasonal epidemics and pandemic outbreaks (McCullers & Rehg, 2002; McCullers, 2006; Morens et al., 2008; Metersky et al., 2012). Bacterial infections can occur concomitant or secondary to an influenza infection. Influenza virus has been shown to increase the susceptibility to secondary bacterial infections by multiple factors: exposure of bacterial attachment sites and receptors due to virus‐induced tissue damage and removal of sialic acids by the viral NA; decreased mucociliary clearance of bacteria; and modulation of the local immune response. The synergism between influenza virus and bacteria has been reviewed in detail (McCullers, 2006, 2011).
In 1987, Tashiro et al. (1987a, b) demonstrated that bacterial infection may also be for the benefit of the virus by producing proteases that facilitate cleavage of HA. Some strains of Staphylococcus aureus (S. aureus) were shown to secrete proteases that are capable of cleaving the HA of certain influenza strains in vitro (Fig. 4a). Interestingly, activation of HA was demonstrated to be specific for both bacteria and virus strains. Thus, for example, a protease secreted by S. aureus Wood 46 was able to cleave the HA of A/swine/1976/31 (H1N1), but not HA of A/chicken/Germany/N/49 (H10N7) (Fig. 4a). By contrast, a protease produced by S. aureus M/86/86 was not capable of activating the HA of A/swine/1976/31 (H1N1), but cleaved the HA of several other influenza A virus isolates. Remarkably, intranasal co‐infection of mice with S. aureus Wood 46 and A/swine/1976/31 (H1N1) resulted in fatal disease with extended lesions in the lungs (Fig. 4b and c left diagram), while infection with either the virus or S. aureus Wood 46 alone did not cause significant symptoms of disease and relevant pathological alterations in the lung (Fig. 4b). Low virus titres were found in lungs of mice infected with A/Swine/1976/31 (H1N1) alone, whereas high titres were present in lung homogenates of mice co‐infected with S. aureus Wood 46 (Fig. 4c, left diagram). Importantly, the infectivity of virus from lung homogenates of mice infected only with virus was markedly enhanced by trypsin treatment prior to plaque titration, indicating inefficient HA cleavage in the mice. In contrast, the infectivity of progeny virus from mice co‐infected with A/swine/1976/31 (H1N1) and S. aureus Wood 46 could not be increased by treatment with trypsin, suggesting that HA was already efficiently cleaved. Thus, co‐infection with influenza virus and S. aureus strains secreting appropriate HA‐cleaving proteases may promote severe pneumonia due to enhanced activation and spread of the virus. Accordingly, co‐infection of mice with influenza viruses and S. aureus strains producing proteases that failed to cleave the HA of the virus in vitro did not lead to significant symptoms of disease in the animals (Fig. 4a and c). Low virus titres were present in lungs of mice co‐infected with virus and bacteria or infected with virus alone, and infectivity of the progeny virus was increased by trypsin treatment. In agreement with the studies by Tashiro et al., a protease secreted by Aerococcus viridans was demonstrated to cleave the HA of several influenza A virus isolates, and co‐infection of mice resulted in severe pneumonia with a fatal outcome (Scheiblauer et al., 1992). However, the HA‐activating proteases from S. aureus and A. viridans have not been identified yet, and mechanisms underlying strain‐specific differences in HA activation remain unknown. In addition, HA‐activating proteases from H. influenzae or S. pneumoniae strains have not been described so far. Attempts to isolate HA‐activating proteases from bacteria present in the respiratory tract of pigs or in the lower digestive tract of waterfowl failed (Callan et al., 1997; King et al., 2009).
Figure 4.
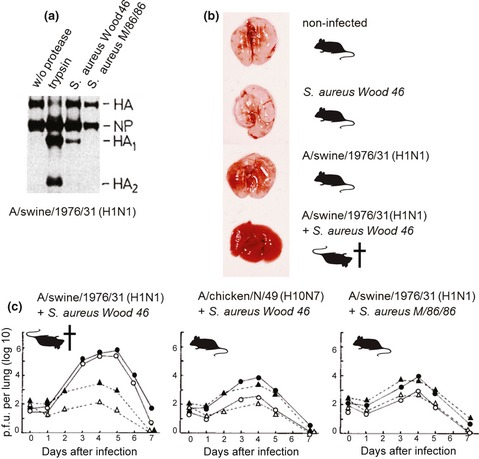
Activation of influenza viruses by proteases from bacteria. (a) Cleavage of HA of A/swine/1976/31 (H1N1) by proteases from S. aureus. Metabolically S35‐labelled A/swine/1976/31 (H1N1) containing noncleaved HA0 was treated with the indicated proteases or remained untreated (w/o protease). Proteins were separated by SDS‐PAGE and visualized by autoradiography (Tashiro et al., 1987a). (b) Pathological alterations in the lungs of mice. Mice were infected with either S. aureus Wood 46 or influenza virus A/swine/1976/31 (H1N1) or co‐infected with both pathogens. Noninfected mice were used as a control. Lungs were taken at 5 days p.i. (Tashiro et al., 1987a). Mice indicate survival or fatal infection of the animals. (c) Virus titres in mice lungs. Virus titres in lung homogenates of mice infected with certain influenza viruses and S. aureus strains are shown as plaque‐forming units (p.f.u.) per lung. Lung homogenates were either treated with trypsin or remained untreated before plaque titration. Infections with virus alone without (▵) and with (▲) trypsin treatment; co‐infections without (○) and with (●) trypsin treatment (Tashiro et al., 1987a, 1987b, 1987c). Mice indicate survival and fatal infection, respectively.
Interestingly, indirect mechanisms of influenza virus activation upon co‐infection with bacteria have been proposed to contribute to secondary bacterial pneumonia. Scheiblauer et al. (1992) observed that Pseudomonas aeruginosa, a frequent complication in patients with cystic fibrosis, secretes a protease that is not capable of cleaving HA, but simultaneous administration of the protease and influenza virus to mice resulted in severe infections with high virus titres in the lungs and a mortality rate of 50%. Analysis of bronchoalveolar lavage (BAL) fluid of the mice revealed an increased trypsin‐like protease activity compared with BAL of mice infected in the absence of P. aeruginosa protease. The increase in protease activity was assumed to be a result of inflammatory‐induced release of trypsin‐like (host) proteases, but remains to be analysed in more detail.
Moreover, generation of plasmin from its inactive zymogen plasminogen by plasminogen activators from bacteria such as staphylokinase and streptokinase, from S. aureus and various streptococci, respectively, has been suggested to facilitate enhanced HA activation in co‐infections with influenza virus (Scheiblauer et al., 1992; Tse et al., 2013). Infection of mice with influenza strains that can be activated by plasmin and simultaneously treatment with plasminogen and either staphylokinase or streptokinase developed severe pneumonia with high virus titres in the lung (Scheiblauer et al., 1992).
The contribution of certain bacteria to proteolytic activation of influenza viruses in the human airways and the development of pneumonia still remain to be demonstrated. However, bacterial proteases might provide novel drug targets for the treatment of viral‐bacterial co‐infections, and further effort in their identification and characterization might support the development of appropriate inhibitors.
Outlook: HA‐activating proteases provide potential drug targets
Currently available measures for the control of influenza in humans are vaccination and antiviral medications, which target the viral NA or the M2 protein. But treatment options become more limited due to the development of drug‐resistant viruses for both M2 and NA inhibitors. Furthermore, the production of influenza vaccines against a newly emerged virus so far requires 4–6 months, highlighting the urgent need for novel drug targets.
HA‐activating host cell proteases provide potential drug targets due to their crucial role for virus infectivity. Early studies using the broad‐range serine protease inhibitor aprotinin from bovine lungs demonstrated that influenza virus replication in embryonated chicken eggs and mice can be markedly suppressed by inhibiting HA cleavage (Zhirnov et al., 1982, 2011). Aprotinin aerosol treatment was shown to protect mice from an otherwise lethal influenza infection and significantly reduced virus titres and lesions in the lung. Moreover, in a clinical trial, inhalations of aerosolized aprotinin in patients with seasonal influenza and parainfluenza markedly reduced the duration of symptoms without causing side effects (Zhirnov et al., 2011).
Inhibition of host factors such as HA‐activating proteases is a novel approach for influenza treatment, and besides a lack of knowledge about relevant proteases until recently, concerns about possible side effects or toxicity have limited the development of drugs that target them. The identification of HAT and TMPRSS2 as HA‐activating proteases from human airways, however, provided potential drug targets, and recent progress in the development of protease inhibitors is considerable. Interestingly, mice deficient in TMPRSS2 and HAT activity, respectively, have been shown to lack a discernible phenotype, indicating functional redundancy or compensation of physiological functions by other host proteases (Kim et al., 2006; Sales et al., 2011). Knockdown of TMPRSS2 expression using an antisense peptide‐conjugated morpholino oligomer (PPMO) strongly suppressed influenza virus replication in human airway epithelial cells without affecting cell viability (Böttcher‐Friebertshäuser et al., 2011). Substrate analogue peptide mimetic inhibitors of HAT containing a 4‐amidinobenzylamide moiety as the P1 residue have been demonstrated to efficiently suppress influenza virus replication in HAT‐expressing cells (Böttcher et al., 2009; Böttcher‐Friebertshäuser et al., 2010; Sielaff et al., 2011). Moreover, a peptide mimetic inhibitor of TMPRSS2 was shown to drastically reduce virus titres and to delay influenza virus propagation by 24–48 h in airway epithelial cells in vitro (Böttcher‐Friebertshäuser et al., 2012). Remarkably, the combination of the protease inhibitor and the current NA inhibitor oseltamivir carboxylate was highly synergistic and efficiently blocked influenza virus propagation in human airway epithelial cells. The further development of abovementioned protease inhibitors may lead to novel drugs that should be considered for influenza treatment – as monotherapy or in combination with NA inhibitors. It should be mentioned that specific inhibitors of furin have recently been developed and were demonstrated to efficiently suppress virus replication of HPAIV of subtype H7 in cell culture. Inhibition of HA cleavage therefore provides a promising concept for treatment of HPAIV infections in humans, most likely in combination with NA inhibitors, for example oseltamivir (Garten et al., 1989; Becker et al., 2010; Y. Lu, W. Garten, T. Steinmetzer, in preparation).
The therapy of viral‐bacterial pneumonia and secondary bacterial pneumonia following influenza has been considered by both antimicrobial drugs and antivirals such as NA inhibitors and has been reviewed in detail (McCullers, 2011; Metersky et al., 2012). Co‐infection studies in animal models suggest that the best option to prevent secondary bacterial pneumonia is the prevention of influenza infection by influenza vaccination (Huber et al., 2010; McCullers, 2011). Moreover, vaccination against both influenza virus and superinfecting bacteria is considered reasonable, but is limited by the availability of vaccines against certain superinfecting bacteria or different strains. Interestingly, Tashiro et al. (1987c) demonstrated that HA‐activating proteases secreted from different S. aureus strains are inhibited by the natural broad‐range protease inhibitor leupeptin. Treatment of mice co‐infected with A/swine/1979/31 (H1N1) and S. aureus Wood 46 with leupeptin led to reduced virus titres in the lungs and survival of the animals, whereas nontreated mice developed fatal pneumonia (cf. Fig. 4).
In summary, HA‐activating proteases have been shown to represent potential drug targets for influenza treatment. The combination of appropriate protease inhibitors with current antivirals and/or antimicrobial drugs provides a novel and promising approach that should be considered for the treatment of both influenza and viral‐bacterial pneumonia.
Conclusion
Proteolytic activation of influenza virus HA is essential for virus infectivity and spread. The identification of relevant proteases in the human airways has demonstrated that HA cleavage occurs by membrane‐bound proteases and can take place in different cellular compartments: either in the TGN or on the cell surface. In addition, HA activation can occur at different time points during the viral life cycle: during transport of HA to the plasma membrane, during budding and release of progeny virus or at a very late time point upon attachment and entry into a new cell. Remarkably, bacteria such as S. aureus and S. pneumoniae have been demonstrated to secrete proteases that cleave HA directly or to produce proteins that augment and activate relevant host cell proteases and thereby may facilitate HA activation upon co‐infection. Thus, influenza viruses can be activated by different proteases and mechanisms in the human airway epithelium (Fig. 5). At the same time, cleavage of HA is a potent drug target for the treatment of influenza infections due to its crucial role for virus infectivity. Further identification and characterization of relevant proteases provide the basis for the development of specific protease inhibitors as novel influenza drugs.
Conflict of interest
The authors declare that they have no conflict of interest.
The authors, who are leading experts in this field, present a timely, authoritative review on the proteolytic cleavage of the influenza hemagglutinin (HA), an activation mechanism that is essential for the infectivity of influenza viruses, including the recently emerged H7N9. They also address the potential of host proteases as targets for developing new influenza drugs. This review will be of considerable interest to virologists, microbiologists and pharmaceutical companies alike.
References
- Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB & Jakobovits A (2001) Catalytic cleavage of the androgen‐regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res 61: 1686–1692. [PubMed] [Google Scholar]
- Baron J, Tarnow C, Mayoli‐Nüssle D et al (2013) Matriptase, HAT, and TMPRSS2 Activate the Hemagglutinin of H9N2 Influenza A Viruses. J Virol 87: 1811–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker GL, Sielaff F, Than ME, Lindberg I, Routhier S, Day R, Lu Y, Garten W & Steinmetzer T (2010) Potent inhibitors of furin and furin‐like proprotein convertases containing decarboxylated P1 arginine mimetics. J Med Chem 53: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S, Glowacka I, Blazejewska P et al (2010) TMPRSS2 and TMPRSS4 facilitate trypsin‐independent spread of influenza virus in Caco‐2 cells. J Virol 84: 10016–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, Lucas JM, Nelson PS, Pöhlmann S & Soilleux EJ (2012) Influenza and SARS‐coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE 7: e35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W & Matrosovich M (2006) Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol 80: 9896–9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher E, Freuer C, Steinmetzer T, Klenk HD & Garten W (2009) MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine 27: 6324–6329. [DOI] [PubMed] [Google Scholar]
- Böttcher‐Friebertshäuser E, Freuer C, Sielaff F, Schmidt S, Eickmann M, Uhlendorff J, Steinmetzer T, Klenk HD & Garten W (2010) Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J Virol 84: 5605–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher‐Friebertshäuser E, Stein DA, Klenk HD & Garten W (2011) Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide‐conjugated morpholino oligomer targeting the hemagglutinin‐activating protease TMPRSS2. J Virol 85: 1554–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher‐Friebertshäuser E, Lu Y, Meyer D, Sielaff F, Steinmetzer T, Klenk HD & Garten W (2012) Hemagglutinin activating host cell proteases provide promising drug targets for the treatment of influenza A and B virus infections. Vaccine 30: 7374–7380. [DOI] [PubMed] [Google Scholar]
- Boycott R, Klenk HD & Ohuchi M (1994) Cell tropism of influenza virus mediated by hemagglutinin activation at the stage of virus entry. Virology 203: 313–319. [DOI] [PubMed] [Google Scholar]
- Callan RJ, Hartmann FA, West SE & Hinshaw VS (1997) Cleavage of influenza A virus H1 hemagglutinin by swine respiratory bacterial proteases. J Virol 71: 7579–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaipan C, Kobasa D, Bertram S et al (2009) Proteolytic activation of the 1918 influenza virus hemagglutinin. J Virol 83: 3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ & Wiley DC (1998) Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95: 409–417. [DOI] [PubMed] [Google Scholar]
- Chokki M, Yamamura S, Eguchi H, Masegi T, Horiuchi H, Tanabe H, Kamimura T & Yasuoka S (2004) Human airway trypsin‐like protease increases mucin gene expression in airway epithelial cells. Am J Respir Cell Mol Biol 30: 470–478. [DOI] [PubMed] [Google Scholar]
- Donaldson SH, Hirsh A, Li DC, Holloway G, Chao J, Boucher RC & Gabriel SE (2002) Regulation of the epithelial sodium channel by serine proteases in human airways. J Biol Chem 277: 8338–8345. [DOI] [PubMed] [Google Scholar]
- Feldmann A, Schäfer MK, Garten W & Klenk HD (2000) Targeted infection of endothelial cells by avian influenza virus A/FPV/Rostock/34 (H7N1) in chicken embryos. J Virol 74: 8018–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten W & Klenk HD (2008) Cleavage activation of the influenza virus hemagglutinin and its role in pathogenesis. Avian Influenza: Monogr in Virol 27: 156–167. [Google Scholar]
- Garten W, Stieneke A, Shaw E, Wikstrom P & Klenk HD (1989) Inhibition of proteolytic activation of influenza virus hemagglutinin by specific peptidyl chloroalkyl ketones. Virology 172: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohrbandt S, Veits J, Breithaupt A, Hundt J, Teifke JP, Stech O, Mettenleiter TC & Stech J (2011) H9 avian influenza reassortant with engineered polybasic cleavage site displays a highly pathogenic phenotype in chicken. J Gen Virol 92: 1843–1853. [DOI] [PubMed] [Google Scholar]
- Goto H & Kawaoka Y (1998) A novel mechanism for the acquisition of virulence by a human influenza A virus. P Natl Acad Sci USA 95: 10224–10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh B, Ogasawara T, Toyoda T, Inocencio NM, Hamaguchi M & Nagai Y (1990) An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo. EMBO J 9: 4189–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T, Nakayama K, Smeekens SP & Kawaoka Y (1994) Proprotein‐processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J Virol 68: 6074–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber VC, Peltola V, Iverson AR & McCullers JA (2010) Contribution of vaccine‐induced immunity toward either the HA or the NA component of influenza viruses limits secondary bacterial complications. J Virol 84: 4105–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger BW, Wise HM, Kash JC et al (2012) An overlapping protein‐coding region in influenza A virus segment 3 modulates the host response. Science 337: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y, Naeve CW & Webster RG (1984) Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology 139: 303–316. [DOI] [PubMed] [Google Scholar]
- Kesic MJ, Meyer M, Bauer R & Jaspers I (2012) Exposure to ozone modulates human airway protease/antiprotease balance contributing to increased influenza A infection. PLoS ONE 7: e35108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido H, Yokogoshi Y, Sakai K, Tashiro M, Kishino Y, Fukutomi A & Katunuma N (1992) Isolation and characterization of a novel trypsin‐like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J Biol Chem 267: 13573–13579. [PubMed] [Google Scholar]
- Kido H, Okumura Y, Yamada H, Le TQ & Yano M (2007) Proteases essential for human influenza virus entry into cells and their inhibitors as potential therapeutic agents. Curr Pharm Des 13: 405–414. Review. [DOI] [PubMed] [Google Scholar]
- Kim TS, Heinlein C, Hackman RC & Nelson PS (2006) Phenotypic analysis of mice lacking the Tmprss2‐encoded protease. Mol Cell Biol 26: 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MD, Guentzel MN, Arulanandam BP, Lupiani B & Chambers JP (2009) Proteolytic bacteria in the lower digestive tract of poultry may affect avian influenza virus pathogenicity. Poult Sci 88: 1388–1393. [DOI] [PubMed] [Google Scholar]
- Klenk HD & Garten W (1994) Host cell proteases controlling virus pathogenicity. Trends Microbiol 2: 39–43. Review. [DOI] [PubMed] [Google Scholar]
- Klenk HD & Rott R (1988) The molecular biology of influenza virus pathogenicity. Adv Virus Res 34: 247–281. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk HD, Rott R, Orlich M & Blödorn J (1975) Activation of influenza A viruses by trypsin treatment. Virology 68: 426–439. [DOI] [PubMed] [Google Scholar]
- Lazarowitz SG & Choppin PW (1975) Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68: 440–454. [DOI] [PubMed] [Google Scholar]
- Lucas JM, True L, Hawley S, Matsumura M, Morrissey C, Vessella R & Nelson PS (2008) The androgen‐regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J Pathol 215: 118–125. [DOI] [PubMed] [Google Scholar]
- Matsushima R, Takahashi A, Nakaya Y, Maezawa H, Miki M, Nakamura Y, Ohgushi F & Yasuoka S (2006) Human airway trypsin‐like protease stimulates human bronchial fibroblast proliferation in a protease‐activated receptor‐2‐dependent pathway. Am J Physiol Lung Cell Mol Physiol 290: L385–L395. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M & Taguchi F (2010) Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84: 12658–12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA (2006) Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev 19: 571–582. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA (2011) Preventing and treating secondary bacterial infections with antiviral agents. Antivir Ther 16: 123–135. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA & Rehg JE (2002) Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet‐activating factor receptor. J Infect Dis 186: 341–350. [DOI] [PubMed] [Google Scholar]
- Metersky ML, Masterton RG, Lode H, File TM Jr & Babinchak T (2012) Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. Int J Infect Dis 16: e321–e331. Review. [DOI] [PubMed] [Google Scholar]
- Morens DM, Taubenberger JK & Fauci AS (2008) Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 198: 962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Towatari T, Ohuchi M, Shiota M, Akao M, Okumura Y, Parry MA & Kido H (2001) Mini‐plasmin found in the epithelial cells of bronchioles triggers infection by broad‐spectrum influenza A viruses and Sendai virus. Eur J Biochem 268: 2847–2855. [DOI] [PubMed] [Google Scholar]
- Muramoto Y, Noda T, Kawakami E, Akkina R & Kawaoka Y (2013) Identification of novel influenza A virus proteins translated from PA mRNA. J Virol 87: 2455–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst MD, Singh B, Ozdemirli M, Dickson RB, Johnson MD & Lin CY (2003) Characterization of matriptase expression in normal human tissues. J Histochem Cytochem 51: 1017–1025. [DOI] [PubMed] [Google Scholar]
- Ohuchi M, Orlich M, Ohuchi R, Simpson BE, Garten W, Klenk HD & Rott R (1989) Mutations at the cleavage site of the hemagglutinin alter the pathogenicity of influenza virus A/chick/Penn/83 (H5N2). Virology 168: 274–280. [DOI] [PubMed] [Google Scholar]
- Okumura Y, Takahashi E, Yano M, Ohuchi M, Daidoji T, Nakaya T, Böttcher E, Garten W, Klenk HD & Kido H (2010) Novel type II transmembrane serine proteases, MSPL and TMPRSS13, Proteolytically activate membrane fusion activity of the hemagglutinin of highly pathogenic avian influenza viruses and induce their multicycle replication. J Virol 84: 5089–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola VT, Murti KG & McCullers JA (2005) Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J Infect Dis 192: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales KU, Hobson JP, Wagenaar‐Miller R, Szabo R, Rasmussen AL, Bey A, Shah MF, Molinolo AA & Bugge TH (2011) Expression and genetic loss of function analysis of the HAT/DESC cluster proteases TMPRSS11A and HAT. PLoS ONE 6: e23261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Yoshida S, Iida K, Tomozawa T, Kido H & Yamashita M (2003) A novel influenza A virus activating enzyme from porcine lung: purification and characterization. Biol Chem 384: 219–227. [DOI] [PubMed] [Google Scholar]
- Schäfer W, Stroh A, Berghöfer S, Seiler J, Vey M, Kruse ML, Kern HF, Klenk HD & Garten W (1995) Two independent targeting signals in the cytoplasmic domain determine trans‐Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J 14: 2424–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiblauer H, Reinacher M, Tashiro M & Rott R (1992) Interactions between bacteria and influenza A virus in the development of influenza pneumonia. J Infect Dis 166: 783–791. [DOI] [PubMed] [Google Scholar]
- Schrauwen EJ, Bestebroer TM, Munster VJ, de Wit E, Herfst S, Rimmelzwaan GF, Osterhaus AD & Fouchier RA (2011) Insertion of a multibasic cleavage site in the haemagglutinin of human influenza H3N2 virus does not increase pathogenicity in ferrets. J Gen Virol 92: 1410–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Mayer G, Zaid A, Rousselet E, Nassoury N, Poirier S, Essalmani R & Prat A (2008) The activation and physiological functions of the proprotein convertases. Int J Biochem Cell Biol 40: 1111–1125. [DOI] [PubMed] [Google Scholar]
- Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, Tahara M, Kikuta H & Yanagi Y (2008) Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J Virol 82: 8942–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A, Heald‐Sargent T, Subramanya G, Zhao J, Perlman S & Gallagher T (2011) A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85: 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sielaff F, Böttcher‐Friebertshäuser E, Meyer D, Saupe SM, Volk IM, Garten W & Steinmetzer T (2011) Development of substrate analogue inhibitors for the human airway trypsin‐like protease HAT. Bioorg Med Chem Lett 21: 4860–4864. [DOI] [PubMed] [Google Scholar]
- Soda K, Asakura S, Okamatsu M, Sakoda Y & Kida H (2011) H9N2 influenza virus acquires intravenous pathogenicity on the introduction of a pair of di‐basic amino acid residues at the cleavage site of the hemagglutinin and consecutive passages in chickens. Virol J 8: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stech O, Veits J, Weber S, Deckers D, Schröer D, Vahlenkamp TW, Breithaupt A, Teifke J, Mettenleiter TC & Stech J (2009) Acquisition of a polybasic hemagglutinin cleavage site by a low‐pathogenic avian influenza virus is not sufficient for immediate transformation into a highly pathogenic strain. J Virol 83: 5864–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA (1999) Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258: 1–20. Review. [DOI] [PubMed] [Google Scholar]
- Stieneke‐Gröber A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk HD & Garten W (1992) Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin‐like endoprotease. EMBO J 11: 2407–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Tse LV, Ferguson AD & Whittaker GR (2010) Modifications to the hemagglutinin cleavage site control the virulence of a neurotropic H1N1 influenza virus. J Virol 84: 8683–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo R & Bugge TH (2008) Type II transmembrane serine proteases in development and disease. Int J Biochem Cell Biol 40: 1297–1316. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Sano T, Yamaoka K, Kamimura T, Umemoto N, Nishitani H & Yasuoka S (2001) Localization of human airway trypsin‐like protease in the airway: an immunohistochemical study. Histochem Cell Biol 115: 181–187. [DOI] [PubMed] [Google Scholar]
- Tashiro M, Ciborowski P, Klenk HD, Pulverer G & Rott R (1987a) Role of Staphylococcus protease in the development of influenza pneumonia. Nature 325: 536–537. [DOI] [PubMed] [Google Scholar]
- Tashiro M, Ciborowski P, Reinacher M, Pulverer G, Klenk HD & Rott R (1987b) Synergistic role of staphylococcal proteases in the induction of influenza virus pathogenicity. Virology 157: 421–430. [DOI] [PubMed] [Google Scholar]
- Tashiro M, Klenk HD & Rott R (1987c) Inhibitory effect of a protease inhibitor, leupeptin, on the development of influenza pneumonia, mediated by concomitant bacteria. J Gen Virol 68: 2039–2041. [DOI] [PubMed] [Google Scholar]
- Thomas G (2002) Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol 3: 753–766. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA & Schalken JA (2009) ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol 56: 275–286. Review. [DOI] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P et al (2012) A distinct lineage of influenza A virus from bats. P Natl Acad Sci USA 109: 4269–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse LV, Marcano VC, Huang W, Pocwierz MS & Whittaker GR (2013) Plasmin‐mediated activation of pandemic H1N1 influenza virus hemagglutinin independent of the viral neuraminidase. J Virol 87: 5161–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarala MH, Porvari KS, Kellokumpu S, Kyllönen AP & Vihko PT (2001) Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol 193: 134–140. [DOI] [PubMed] [Google Scholar]
- Veits J, Weber S, Stech O et al (2012) Avian influenza virus hemagglutinins H2, H4, H8, and H14 support a highly pathogenic phenotype. P Natl Acad Sci USA 109: 2579–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey M, Orlich M, Adler S, Klenk HD, Rott R & Garten W (1992) Hemagglutinin activation of pathogenic avian influenza viruses of serotype H7 requires the protease recognition motif R‐X‐K/R‐R. Virology 188: 408–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuoka S, Ohnishi T, Kawano S, Tsuchihashi S, Ogawara M, Masuda K, Yamaoka K, Takahashi M & Sano T (1997) Purification, characterization, and localization of a novel trypsin‐like protease found in the human airway. Am J Respir Cell Mol Biol 16: 300–308. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S, Nakahori Y & Yasuoka S (1998) Fibrinogenolytic activity of a novel trypsin‐like enzyme found in human airway. J Med Invest 45: 77–86. [PubMed] [Google Scholar]
- Zhirnov OP, Ovcharenko AV & Bukrinskaya AG (1982) Protective effect of protease inhibitors in influenza virus infected animals. Arch Virol 73: 263–272. [DOI] [PubMed] [Google Scholar]
- Zhirnov OP, Ikizler MR & Wright PF (2002) Cleavage of influenza a virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases. J Virol 76: 8682–8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhirnov OP, Vorobjeva IV, Ovcharenko AV & Klenk HD (2003) Intracellular cleavage of human influenza a virus hemagglutinin and its inhibition. Biochemistry (Mosc) 68: 1020–1026. [DOI] [PubMed] [Google Scholar]
- Zhirnov OP, Klenk HD & Wright PF (2011) Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res 92: 27–36. [DOI] [PubMed] [Google Scholar]


