Abstract
Detection of early infectious disease may be challenging due to the low copy number of organisms present. To overcome this limitation and rapidly measure low concentrations of the pathogen, we developed a novel technology: Nanotrap particles, which are designed to capture, concentrate, and protect biomarkers from complex biofluids. Nanotrap particles are thermoresponsive hydrogels that are capable of antigen capture through the coupling of affinity baits to the particles. Here, we describe recent findings demonstrating that Nanotrap particles are able to capture live infectious virus, viral RNA, and viral proteins. Capture is possible even in complex mixtures such as serum and allows the concentration and protection of these analytes, providing increased performance of downstream assays. The Nanotrap particles are a versatile sample preparation technology that has far reaching implications for biomarker discovery and diagnostic assays.
Keywords: Rift Valley fever virus, influenza, diagnostics, Venezuelan equine encephalitis virus
The Nanotrap particles are a versatile sample preparation technology that has far reaching implications for biomarker discovery and diagnostic assays.

The need for improved diagnostic assays for biodefense and emerging infectious diseases
Diagnosis of early‐stage infections with viral pathogens is generally very difficult because patients typically present with nonspecific flu‐like symptoms. By the time a definitive diagnosis can be made using conventional methods, the infection will have already advanced to an untreatable, severe, or lethal stage. Thus, there is a substantial need for rapid, accurate, reliable, and safe diagnostic assays to guide physicians to administer treatment options (when available) as well as to notify public health officials if outbreaks are detected. As many pathogens can be transmitted through aerosol routes, the progression to disease will likely be accelerated underscoring the need to have diagnostic assays for early‐stage disease. In addition, some available diagnostic assays require biosafety level (BSL)‐3 or BSL‐4 facilities. While there are currently no specific treatments for some of the viral pathogens to be discussed, it is important that the development of sensitive and specific diagnostic assays is carried out in parallel with therapeutic development to ensure that both needs are met. The concepts presented in this review are widely applicable to viruses; however, we will limit our discussion to Rift Valley fever virus (RVFV), Venezuelan equine encephalitis virus (VEEV), and influenza viruses, as these pathogens are currently being studied in the context of Nanotrap particles.
Pathogens of interest and current state of diagnostics
Rift Valley fever virus
RVFV is a highly pathogenic arthropod‐borne virus that is primarily transmitted by mosquitoes, particularly after heavy rainfall. Although it can infect a wide range of vertebrate hosts, RVFV primarily affects livestock and humans (Mansuroglu et al., 2010). Humans are typically affected when they come in close contact with infected bodily fluids or tissues, but transmission via mosquito bites as well as aerosolization may also occur. However, humans are dead‐end hosts (Wilson, 1994; Paweska et al., 2005a). Mortality rates are dependent on animal species and age. In livestock, mortality rates are as high as 30%. Mortality rates can reach as high as 95% in newborns, and young and abortion rates are as high as 100% (Paweska et al., 2005a). Fever is often observed after a short incubation period. While symptoms in humans are usually mild and include febrile illness resembling the flu, a small percentage may develop serious clinical manifestations such as retinal lesions, meningoencephalitis, hepatitis, severe hemorrhagic fever, coma, and death. In recent years, an increase in mortality amongst humans from 2% to 45% has been reported, which could be due to evolving mechanisms of virulence and mutations or increased surveillance and reporting (Pepin et al., 2010).
Due to its transmission via aerosolization, high pathogenicity, and listing as a group III (bioterrorism potential) Category A emerging infectious disease by the NIAID, work with RVFV requires biosafety level 3 (BSL‐3) containment laboratories. It is also a priority pathogen for the United States Department of Agriculture (USDA) due to its potential impact on livestock, should it ever be introduced into the US. It is highly suggested that laboratory staff working with RVFV be vaccinated. Therefore, diagnosis of RVFV is restricted to a small number of laboratories. This limitation has led to some delay in diagnostics associated with virus isolation and identification techniques that may pose a problem for healthcare authorities in the event of an RVFV epidemic. There is a crucial need for rapid detection and identification of the virus now that the virus has crossed beyond its traditional endemic boundaries (Paweska et al., 2005a; Pepin et al., 2010).
Many times, RVFV infections are recognized late in infection when ocular complications occur. Diagnosis methods include virus isolation, antigen detection, and nucleic acid amplification. The virus can be isolated from serum or whole blood, as well as from other tissues such as the liver, spleen, and brain. However, these virus isolation techniques are lengthy and expensive and are not optimal when facing a potential epidemic. Nucleic acid techniques such as reverse transcription polymerase chain reaction (RT‐PCR), quantitative real‐time PCR (qRT‐PCR), and real‐time reverse‐transcription loop‐mediated isothermal amplification assays (RT‐LAMP) have also been developed (Pepin et al., 2010). However, these tests are not definitive and should be run in parallel with additional tests such as antibody detection techniques. Antibodies primarily directed against the viral glycoproteins, and the nucleoprotein is detectable by day 8 of infection (Pepin et al., 2010). Methods for antibody detection to RVFV include hemagglutination inhibition, complement fixation, indirect immunofluorescence, enzyme‐linked immunosorbent assays (ELISAs), and virus neutralization. These assays pose a health risk to laboratory personal and are restricted for use outside RVFV endemic areas. While known as the gold standard for detection, virus neutralization is time‐consuming, expensive, and lengthy (requiring up to 7 days for completion; Pepin et al., 2010). ELISAs may allow for the rapid detection of antibodies against RVFV, but the technique is cumbersome, expensive, and poses a risk to laboratory personnel (Peeling & Mabey, 2010; WHO, 2013). Sandwich ELISAs for antigen detection (sAG‐ELISAs) provide a safer and faster detection method. However, RVFV viremia remains high for only a short duration, and viral titers may reach undetectable concentrations at both early and late time points postinfection (Pepin et al., 2010). In summary, there are still multiple limitations with the currently available RVFV diagnostics methods that could be improved through a sample preparation technology that is compatible with numerous downstream assays, while increasing their sensitivity and safety.
Venezuelan equine encephalitis virus
VEEV is a mosquito‐borne illness that can cause severe encephalitis in horses and humans, as well in other mammals and birds. VEEV is the subtype I of a complex, which contains six other subtypes (Mosso das Pedras, Cabassou, Everglades, Mucambo, Pixuna, and Rio Negro; Aguilar, 2011). VEEV is further divided into epizootic and enzootic groups and five antigenic variants, AB to F. The epizootic groups, I‐AB and I‐C, are responsible for most epidemics, whereas the enzootic groups normally occur in natural cycles between sylvatic rodents or marsupials (and at time birds) and Culex mosquitoes, and are not usually pathogenic in equines. Enzootic subtypes normally cause mild symptoms. In the epizootic subtype, early symptoms occur within one to 5 days after infection and include fever, depression, abnormal heart rate, weakness, and ataxia. Neurological symptoms appear after 5 days. Death may occur within hours after neurological symptoms. Permanent symptoms in recovered animals may remain. While most enzootic subtypes do not result in serious diseases, the epizootic subtypes have seen a fatality rate of up to 90% in horses. Furthermore, fatal cases have been reported for other mammals, including humans, rabbits, dogs, and sheep. During epidemics, more than 10% of the human population in the affected area can be infected. Humans normally develop a mild, systemic illness, and the illness and its symptoms typically resolve within 2 weeks (Weaver et al., 2004). Neurological disease is caused in < 1% of symptomatic adults (Weaver et al., 2004; Quiroz et al., 2009). However, the virus does have a high fatality rate in the young and elderly (up to 35%). Furthermore, placental damage, abortions, and stillbirths have been reported in pregnant women (Kirsch et al., 2008).
VEE is often misdiagnosed with dengue fever, as the two viruses share many of the same symptoms (Aguilar et al., 2011; Pisano et al., 2012). VEEV is typically diagnosed by virus isolation and serological tests. The virus is normally found in the blood, cerebrospinal fluid, or throat swabs, but can also be detected in the pancreas or other tissues (Scherer et al., 1972; Valero‐Fuenmayor et al., 1997). VEEV can also be tested and isolated in guinea pigs, hamsters, mice, embryonated chicken eggs, and cell lines such as Vero, RK‐13, and BHK‐21. Viremia levels detected in infected individuals typically range at about 10^5.7 PFU mL−1 (Aguilar et al., 2011). Other detection methods for VEEV include complement fixation, hemagglutination inhibition, virus neutralization, immunofluorescence assays, and ELISAs. ELISAs are used to detect the viral antigens such as the antigenic determinants that are found on the E2 envelope glycoprotein of VEEV in the blood (Wang et al., 2005). However, antibody detection is not possible early on in infection during the period of viremia. Typically, antibodies against VEEV are made 5–6 days after infection. Immunofluorescence assays, PRN tests, and nucleic acid detection by RT‐PCR can be used to characterize the different VEEV subtypes (Navarro et al., 2005; Pisano et al., 2012).
Influenza virus
The influenza virus is a single‐stranded RNA virus belonging to Orthomyxoviridae family (Baigent & McCauley, 2003; WHO, 2013). There are three types of influenza: A, B, and C. The genome of influenza A and B types consists of eight segments (PB2, PB1, PA, HA, NP, NA, M1/M2, NS1/NS2), whereas type C consists of seven linear segments. Both types A and B encode the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) proteins on separate RNA segments, whereas the HEF protein of type C has the same functional properties as HA and NA combined (Labella & Merel, 2013). The type A and B viruses are transmitted via the respiratory route. The virus binds via surface glycoproteins NA and HA to sialic acid found in the α‐2,3‐linkages of human epithelial cells. This association can cause an extensive range of disease that includes lower respiratory tract infection, pneumonia, and encephalitis. While influenza B is limited to humans and seals and influenza C is limited to humans and swine, influenza A can be spread amongst a variety of species (both avian and mammals; Baigent & McCauley, 2003).
In a 2010 article, it was estimated that acute lower respiratory infections cause 1 million deaths annually in children under the age of 5 years (Peeling & Mabey, 2010). The H1N1 pandemic of 2009 highlighted the need to reassess widely used influenza diagnostic tests for their ability to detect viral antigens in clinical specimens. Within 1 month, there were nearly 9000 confirmed cases and 74 deaths from 40 countries. There is significant evolutionary distance between the HA segment of this novel swine‐origin virus and its closest subtype relative, demonstrating a lack of surveillance and testing in the swine populations over the years (Garten et al., 2009). There are currently multiple rapid influenza diagnostic tests (RIDTs) available to physicians to be used at the bedside. These tests can identify the presence of the influenza A or B viral nucleoprotein in respiratory samples, generally in 15 min or less. RIDTs are a useful screening tool that provides a yes/no answer (not quantitative). The results of RIDTs testing can guide a physician in the prescription of antiviral drugs. They can also be used as screening tool during respiratory outbreaks. However, one of the limitations of the RIDTs is their limited sensitivity (ranging anywhere from 10% to 80%), resulting in high rates of false‐negative results. Findings conducted by the CDC in 2012 on 11 commercially available RIDTs and on 23 influenza virus strains found that while most of the tests were able to detect the viruses at the highest concentrations, many of the kits were unable to detect lower concentrations of virus. Furthermore, results varied based on virus type and subtype. For example, one RIDT tailored for influenza A capture was not able to detect many influenza A strains, including the 2009 H1N1 strain. Therefore, it is important for the patients to be tested within 72 h after onset of illness when the virus concentration is at a peak (Peterson et al., 2013).
While RIDTs for influenza are often used by the physician to provide a qualitative diagnostic for influenza, a definitive diagnosis must be confirmed by RT‐PCR, molecular assays, or viral culture, with viral culture being the gold standard. Viral culturing is critical in providing necessary information for the comparison of various strains and subtypes to one another and to vaccine strains, as well as to monitor emerging strains and mutations amongst the strains (Prevention C.f.D.C.a., 2011). Some FDA‐approved RT‐PCR‐based methods include QuickVue A+B, Directigen EZ Flu A+B, and BinaxNOW Influenza A&B. Currently available RIDTs detected H1N1 virus from respiratory specimens containing high levels of virus; however, sensitivity levels were significantly low (40–69%) and decreased significantly as virus levels decreased (Kramarow & Pastor, 2012), leading to false‐negative results. Furthermore, while molecular methods such as PCR are quickly becoming the golden standard, the risk of cross‐contamination is high (Spackman et al., 2002). Collectively, these studies indicated that there is a need for a sample preparation technology that will be compatible with multiple assay types, will increase sensitivity, and will allow for viral propagation.
Nanotrap particles
Nanotrap particle properties
Nanotrap particles are customizable hydrogel microspheres developed by Ceres Nanosciences for target analyte separation and discovery applications. These particles have demonstrated their utility as innovative tools for the collection, concentration, and preservation of dilute levels of low‐molecular‐weight peptides, proteins, and other biomolecules from biofluid samples (Pelton, 2000). Nanotrap particles are based on cross‐linked N‐isopropylacrylamide (NIPAm) and are appealing as analyte sequestration and concentration devices due to their versatility, reproducibility, and low production costs (Pelton, 2000; Hamidi et al., 2008; Luchini et al., 2010; Patanarut et al., 2010; Douglas et al., 2011; Tamburro et al., 2011). The monomers used to generate the Nanotrap particles also have good colloidal stability in biofluids, thereby creating a large surface area that is ideal for the rapid and complete capture of target analytes in complex aqueous biological matrices (Luchini et al., 2010). The cross‐linked polymeric networks that make up the Nanotrap particles are highly hydrated, making it possible for small molecules to attain access to the interior of the particle (Patanarut et al., 2010). Furthermore, the thermoresponsive nature of the NIPAm monomer imparts on the particles, a significant degree of flexibility and porosity in response to changes in pH and temperature (Patanarut et al., 2010). Thermoresponsive hydrogels have been extensively studied for drug delivery applications, and the knowledge attained from these studies can be applied toward engineering Nanotrap particles for analyte collection devices (Hamidi et al., 2008).
Nanotrap particles can be functionalized with a variety of affinity baits to facilitate the binding and retention of collected target proteins, peptides, post‐translationally modified analytes, lipids and fatty acids, metabolites, nucleic acids, and pathogens (Douglas et al., 2011; Tamburro et al., 2011). This versatile nature of these Nanotrap particles also allows them to be tailored to capture target analytes from a variety of complex biological matrices, including blood, serum, plasma, saliva, and nasopharyngeal fluids (Fig. 1). One method of functionalizing the particles involves polymerizing the NIPAm monomer with another monomer species to generate a copolymer hydrogel possessing the chemical properties of both monomers. For example, poly‐NIPAm (p‐NIPAm) particles with incorporated acrylic acid (AAc) moieties were synthesized via precipitation polymerization to create negatively charged thermoresponsive Nanotrap particles. These particles demonstrated the ability to harvest and concentrate low‐molecular‐weight protein species from serum (Luchini et al., 2008). Another way of functionalizing the particles is to utilize the reversible broad‐spectrum protein binding ability of reactive dyes, which are commonly used as ligands for affinity chromatography applications. These reactive dyes can be covalently immobilized onto the polymer matrix, and their low cost and wide commercial availability make them appealing affinity baits for the Nanotrap particles. The Nanotrap particle can also be encapsulated within a cross‐linked p‐NIPAm shell to further increase the sieving functionality of the core particle architecture. This cross‐linked outer shell of the particle can either be inert or contain chemical moieties such as vinyl sulfonic acid (VSA), which has demonstrated the ability to actively exclude interfering albumin peptides of all sizes while simultaneously excluding large unwanted abundant molecules such as immunoglobulins (Tamburro et al., 2011; Fig. 1b). The inverse effect can also be achieved by varying the degree of cross‐linking in the particles. While Nanotrap particles with a high degree of cross‐linking are compact with decreased thermoresponsiveness, particles with low amounts of cross‐linking exhibit a greater degree of volume‐phase transition in response to changes in temperature. Elimination of cross‐linker from aspects of the particle architecture can also tailor the particles for the capture of larger analytes, like virus particles. For example, interpenetrating polymer networks (IPNs) can be included into the particle microsphere structure to introduce environmentally responsive linear or branched polymer chains to the particle (Park & Choi, 1998). Another method is to synthesize a cross‐linker‐free shell structure around the core particles to generate a particle with a shell capable of expanding without the pore size‐limiting effect of the cross‐linker (Gao & Frisken, 2003).
Figure 1.

Functionalized Nanotrap particles with core and core–shell architecture. (a) Nanotrap particles are capable of sequestering low‐molecular‐weight biomolecules out of complex solutions, although some high‐abundance, high‐molecular‐weight species may nonspecifically bind to the hydrogel matrix. Note how the low‐molecular‐weight biomolecules are pulled away from carrier proteins (such as albumin) upon binding to the Nanotrap particles. (b) Addition of a cross‐linked shell to the particle architecture increases the sieving performance of the particles, effectively preventing high‐molecular‐weight molecules from binding. (c) Common affinity baits incorporated onto the particle matrix.
While standard pre‐analytical separation techniques often require many days to ‘clean’ a complex biofluid sample, both the core and core–shell Nanotrap particles with incorporated affinity bait are capable of achieving three essential functions within minutes: (1) capturing dilute concentrations of target analytes from complex biofluid samples, (2) polymer network‐limited sieving of interfering species, and (3) protecting captured target analytes from proteolytic degradation (Liotta et al., 2006; Luchini et al., 2008, 2011; Longo et al., 2009; Tamburro et al., 2011). Another benefit afforded using Nanotrap particles is that target classes of analytes sequestered by the particles can be concentrated into a smaller volume to effectively amplify up to 100‐fold the sensitivity of mass spectrometry, Western blotting, and immunoassays with high precision (Fredolini et al., 2008, 2009; Longo et al., 2009; Tamburro et al., 2011).
Previous work with Nanotrap particles
There is a growing interest in the field of biomarker discovery in the pharmaceutical and biotechnological industries. Biomarkers are low‐abundance, low‐molecular‐weight molecules that can serve as an indicator of the presence or stage of specific diseases or some other physiological or pharmacological processes (Patanarut et al., 2010). Complex biological matrices are excellent sources of informative biomarkers, although the presence of high‐molecular‐weight, high‐abundance biomolecules such as serum albumin and immunoglobulins can hinder the detection of the elusive low‐molecular‐weight biomarkers. To further complicate matters, the methods used during sample collection and transportation can compromise the integrity of the collected samples. Fluctuations in temperature and lengthy transportation times can compromise the viability of the target biomarkers in the sample. Furthermore, the presence of proteases in the sample can degrade biomarkers of interest. This can ultimately lead to false‐negative diagnosis of an illness. Currently, mass spectrometry is the favored technique used for candidate biomarker discovery due to its throughput and sensitivity (Merrell et al., 2004; Patanarut et al., 2010). Despite the advantages of mass spectrometry, this method requires small sample sizes consisting of purified target protein, as the presence of large high‐abundance biomolecules can hinder mass spectrometric detection of the biomarkers of interest. This makes it necessary to develop new and innovative tools that can (1) clean collected samples by removing interfering biomolecules, (2) enrich the mixture of only the analytes of interest, and (3) be easily integrated into existing workflows.
Prior studies have demonstrated the ability of Nanotrap particles to protect captured analytes from proteolytic degradation. The analyte preservation ability of the Nanotrap core and core–shell particles functionalized with Cibacron Blue F3G‐A were tested using lysozyme (14.3 kDa) as the model protein and trypsin (24 kDa) as the proteolytic enzyme (Patanarut et al., 2010). While trypsin was small enough to be sequestered by the Nanotrap particles, its enzymatic potency to degrade lysozyme was not retained once captured by the particles (Patanarut et al., 2010). In a similar study, Nanotrap hydrogel particles consisting of a NIPAm and AAc copolymer core encased within an inert cross‐linked shell were capable of protecting platelet‐derived growth factor (PDGF), a small biomarker of c. 14.5 kDa in size, from tryptic degradation (Longo et al., 2009).
Previous research has also employed Nanotrap particles to efficiently concentrate target analytes into smaller volumes to amplify the sensitivity of detection by mass spectrometry or immunoassays (Luchini et al., 2008). By integrating Nanotrap particles into the sample‐processing workflow, low‐molecular‐weight biomarkers were greatly enriched while large interfering biomolecules were removed from the sample. This achieved a 10 000‐fold improvement in the lower limit detection range of mass spectrometry, thereby permitting the discovery of hundreds of candidate biomarkers that were previously undetectable in the nanogram per mL and picogram per mL range (Fredolini et al., 2010; Tamburro et al., 2011). An additional set of low‐abundance peptides not previously included in the PeptideAtlas database was also discovered by mass spectrometry when Nanotrap hydrogel particles were utilized in the sample‐processing workflow (Tamburro et al., 2011). Nanotrap particles with core‐shell architecture demonstrated the ability to protect highly labile proteins such as interleukins and growth factors from enzymatic degradation in blood, sweat, and urine as well as increase the effective detection sensitivity while improving the precision of multiple reaction monitoring (MRM) analysis (Tamburro et al., 2011). In the same study, it was found that incorporation of VSA into the shell matrix of the Nanotrap particles increased the number and dynamic range of sequestered analytes of interest, which allowed for the mass spectrometric identification of proteins known to exist in the picogram per mL range (Tamburro et al., 2011).
Nanotrap particles and infectious diseases
The tests used for infectious disease diagnostics need to be rapid, affordable, and sensitive to allow for timely and effective intervention and treatment (Banoo et al., 2010). The ideal diagnostic test for infectious diseases would (1) utilize noninvasive and/or easily attainable biological fluids such as finger‐stick blood, nasal excretions, saliva, or urine, (2) detect with accuracy and precision early‐stage viral or bacterial disease antigens, living organisms, or pathogen nucleic acids prior to seroconversion, and (3) distinguish active disease processes from immunologic memory of prior exposure (Banoo et al., 2010).
Yet, despite recent technological developments, the challenges that can hinder analyte detection in biomarker discovery can also complicate the discovery, detection, and quantification of pathogen‐related antigens in biofluids and their translation to clinical benefit. For example, dilute concentrations of clinically relevant antigens may exist below the detection limits of mass spectrometry, quantitative PCR, and existing immunoassays. The presence of large interfering biomolecules may also mask the isolation and detection of elusive antigens. Finally, target antigens can also be subjected to proteolytic degradation during sample collection, transportation, and/or storage, which can seriously compromise the validity of test results.
Building on the success of the Nanotrap particles in biomarker discovery, functionalized hydrogel particles consisting of core and core–shell architecture were generated as part of a pioneering effort to develop highly sensitive quantitative assays directed against pathogen antigens and host immune response molecules in biofluids (Douglas et al., 2011). The results of previous research studies have demonstrated the utility of the Nanotrap particles in infectious disease diagnostics. For example, Nanotrap particles with incorporated Acid Black 48 dye were capable of increasing the sensitivity and accuracy of a urinary antigen test for Lyme disease, a bacterial disease caused by the spirochete Borrelia burgdorferi (Douglas et al., 2011). The Acid Black 48 functionalized Nanotrap particles sequestered and enriched the Lyme disease antigen in a 10‐mL urine sample 100‐fold, achieving an immunoassay sensitivity in the pg mL−1 detection range (Douglas et al., 2011).
Application of Nanotrap particles to viral diagnostics
As reviewed above, viral diagnostics is often achieved through the detection of viral nucleic acids, viral antigens, or viral propagation in tissue culture. Viral propagation is particularly important as it allows the amplification of virus that can then be funneled into additional assays, including next‐generation sequencing to allow subtyping of the virus or identification of novel viruses. Recently, it has been demonstrated that Nanotrap particles are capable of capturing viral antigens, infectious virions and viral RNA, and biomarkers of viral infection [(Shafagati et al., 2013; Narayanan et al., 2014) and N. Shafagati, L. Lundberg, A. Patanarut, K. Fite, B. Lepene and K. Kehn‐Hall, unpublished data]. These data indicate that the Nanotrap particles are a versatile sample preparation technology that can be incorporated upstream of most currently available diagnostic platforms or assays.
Nanotrap particles can capture virions
A recent study by Shafagati et al. (2013) demonstrated that Nanotrap particles are capable of capturing viruses, including RVFV, VEEV, and human immunodeficiency virus (HIV). This was the first report to extend the analyte binding capability of Nanotrap particles to live infectious viruses. Plaque assays confirmed that intact infectious virus was bound by the Nanotrap particles and not merely lysed virus or viral RNA (Shafagati et al., 2013). Control experiments indicated that the Nanotrap particles are not toxic to the host cell (Shafagati et al., 2013). Furthermore, the concentrated virus can be propagated by resuspending the Nanotrap‐bound samples in proper growth medium and adding the sample to relevant cell lines. Additional studies have also demonstrated the capture of influenza A, influenza B, human coronavirus (HCoV), adenovirus, and dengue viruses by Nanotrap particles from viral supernatants (Fig. 2 and unpublished data by Shafagati N, Fite K, Patanarut A, Pinkham C, Baer A, Lepene B, Kehn‐Hall K and Amaya M, Kehn‐Hall K, and Narayanan A). As can be seen in Fig. 2a, all the Nanotrap particles tested were capable of capturing HCoV. NT53 and NT69 demonstrated the greatest enrichment as compared to the no Nanotrap particles control (compare lane 1 with lanes 4 and 6). For influenza A virus, no capture was observed with NT46 and very little with NT71. The greatest capture was observed with NT55, NT69, and NT76. Adenovirus capture was only observed with four of the Nanotrap particles (NT45, NT46, NT53, and NT71). However, this was likely due to the low level of virus present in the starting sample (see lane 1). Importantly, NT45 was capable of a high level of enrichment for adenovirus (compare lanes 1 and 2). These results demonstrate the broad virus capture potential of Nanotrap particles.
Figure 2.

Virus particle capture. (a) Nanotrap particles can capture human coronavirus (HCoV), influenza A, and adenovirus. One hundred microliters of viral supernatants were incubated with 75 µL of each Nanotrap particle for 30 min at room temperature. Nanotrap particles were washed four times with water and viral RNA or DNA extracted and amplified with viral specific primers using either RT‐PCR or PCR. PCR products were separated on 2% agarose gels and visualized with ethidium bromide. NT = no Nanotrap particle control. (b) A proposed model of Nanotrap particle binding to viruses. Virus particles can be separated from biofluid samples via interaction between the affinity bait(s) incorporated into the particle matrix and specific ligands present on the virus particle.
While the exact mechanism of the virus–Nanotrap particle interaction is unknown, it is hypothesized that the viral glycoproteins interact with the Nanotrap particles in a similar fashion as protein biomarkers have been proposed to interact with the Nanotrap particles (Fig. 2b). In the case of RVFV, we hypothesized that the glycoproteins, Gn and Gc, were interacting with the Cibracon Blue affinity bait within the Nanotrap particle. It is interesting to note that the viruses captured to date are all relatively small (c. 100 nM or smaller) and all enveloped RNA viruses, with the exception of adenovirus, which is a nonenveloped double‐stranded DNA virus. The average size of the Nanotrap particles used in the above study was 800 nM; thus, it is unclear whether the viruses are entering inside the core of the Nanotrap particle or binding to the outside of the Nanotrap particles. Preferentially, binding of RVFV with Nanotrap particles containing Cibracon Blue baits has been observed, suggesting that the bait plays at least a partial role in the binding.
The Nanotrap particles have recently been modified to create Nanotrap particles that are more tailored for whole virus capture. These modified Nanotrap particles are larger in size (up to 2 µM) and encased within a non‐cross‐linked shell functionalized with AAc (Fig. 3). Inclusion of this non‐cross‐linked shell onto the particle architecture gives the Nanotrap environmentally responsive ‘arms’ that can capture larger virus particles. Studies are underway to determine whether these environmentally responsive ‘arms’ will further increase the ability of the Nanotrap particles to capture viruses.
Figure 3.

Particles with modified core–shell architecture. The thermoresponsive nature of the Nanotrap particles enables them to expand or shrink in response to changes in pH and temperature, although the species allowed to enter the interior of the particles are limited by the pore sizes of the hydrogel particles. (a) Larger virus particles may not be able to enter the interior of the Nanotrap particles. (b) Inclusion of a non‐cross‐linked shell onto the particle matrix gives the Nanotrap environmentally responsive ‘arms’ that can capture larger virus particles.
The ability of Nanotrap particles to enrich virus is critical early on in RVFV and VEEV infection during which low concentrations of virus can result in a false‐negative result. As demonstrated by several researchers, RVFV titers in the liver and spleen rise from 12 h until about 72 h, after which the titers drop quickly (van Vuren et al., 2011). Similarly, VEEV levels peak at 24–48 h postinfection before dramatically decreasing or becoming virtually undetectable 4–5 days after infection (Wang et al., 2001; Carrara et al., 2007). Presymptomatic diagnosis would be extremely useful during an outbreak (either natural or bioterrorism related), where hundreds to thousands of people and/or livestock may need to be screened for exposure. Nanotrap particles were capable of capturing and enriching RVFV and VEEV in both cell culture supernatants and in serum (Shafagati et al., 2013), demonstrating the ability to utilize the Nanotrap particles in clinically relevant matrices. In addition, viral titers are often low later on in infection, leaving a narrow window for diagnostic assays. Therefore, the concentration of whole virus provided by the Nanotrap particles would resolve a much needed problem to the false‐negative results observed at both early and late time points during the infectious process.
Due to the promiscuity of some of the affinity baits (such as Cibracon Blue), it is likely that Nanotrap particles would be capable of capturing multiple pathogens from a single sample. This extensive binding ability of the particles can be further enhanced by the incorporation of multiple bait chemistries within the core. Alternatively, a cocktail of different Nanotrap particles can also be used during sample processing to capture a wide array of analytes. The ability to capture multiple pathogens within one sample enrichment format is important when the infectious agent is not known at the time of diagnosis. There are many viral infections that result in similar symptoms and thus the causative agent of the illness is not always apparent. For example, respiratory illnesses caused by influenza virus and rhinovirus display similar symptoms (fever, cough, and sore throat) are often misdiagnosed for one another, leading to faulty antiviral treatments (Bellei et al., 2007). Similarly, VEE is difficult to clinically distinguish from other tropical viral diseases such as dengue fever (Quiroz et al., 2009). The composition of the Nanotrap particles allows for the capture of all pathogens present in a sample. Following capture, downstream assays can then be used to determine which pathogen is present in the patient sample. There are also instances where co‐infections occur, and it is important to ensure that multiple pathogens binding to the Nanotrap particles do not interfere with diagnosis and that both pathogens can be captured with the Nanotrap particles. In a mock mixed infection scenario where both HIV and RVFV were spiked into serum, Nanotrap particles were still capable of capturing and enriching RVFV (Shafagati et al., 2013), indicating that Nanotrap particles can be used in the presence of multiple viruses. Similarly, both bacterial and viral co‐infections have been observed for respiratory infections (Freymuth et al., 2005; Bellei et al., 2007; Chertow & Memoli, 2013). Nanotrap particles would function well in these patient samples, as we have demonstrated their ability to capture and enrich influenza A and B viruses, coronaviruses, and adenoviruses (Fig. 2 and N. Shafagati, K. Fite, A. Patanarut, C. Pinkham, A. Baer, B. Lepene and K. Kehn‐Hall, unpublished data), all causative agents of respiratory disease.
Nanotrap particles protect viral RNA, preserve viral infectivity, and eliminate the need for cold chain transport
One of the most important and unique aspects of the Nanotrap particles is the ability to preserve samples. Common preservation methods are often dependent on a cold chain for storage, as is the case for whole virus, or inactivation of the virus for the sake of preserving its genomic material. Most RNA samples can be easily degraded in the presence of various enzymes, making their downstream analyses difficult if the sample is not processed properly. Field conditions may restrict the ability to preserve whole virus. Preserving only the genome limits analysis of viability and pathogenicity. Early studies by the US Army on RVFV confirmed its stability in serum for long term, cryogenic storage at a neutral pH (Klein et al., 1969). Subsequent studies demonstrated blood or serum could preserve virulent RVFV for up to 2 months at 4 °C (Shimshony & Barzilai, 1983). Unfortunately, bypassing the cold chain has not been successful: Nobuto, Whatman FTA, and Whatman 903 filter paper spotted with spiked blood and stored at room temperature did not preserve the RVFV genome or whole virus (Naslund et al., 2011). Attempts to preserve the genome of RVFV and VEEV in mock field conditions have been more promising. RNA from both viruses was preserved in a commercially available RNA extraction buffer for up to 2 days at tropical temperatures (32 °C) prior to placing in the cold chain, but at the cost of viral inactivation (Blow et al., 2008).
Concerns over pandemic influenza have spurred intensive study into field collection and preservation methods of viral samples for genomic typing. The World Health Organization recommends long‐term storage of whole blood and swab samples at −70 °C and found that virus can be preserved up to 7 and 4 days at 4 °C, respectively (WHO, 2006). FTA cards will inactivate virus, but genomic RNA can be extracted after 5 months when stored at room temperature; likewise, Whatman cellulose filter paper will preserve virus for 3 days at room temperature (Abdelwhab et al., 2011). Absolute ethanol will inactivate influenza virus, but will also allow for RNA extraction when stored for 5 months at room temperature (Krafft et al., 2005). Finally, both rayon‐tipped and flocked nylon swabs will preserve viral RNA at 4 °C for up to 14 days (Fereidouni et al., 2012).
Our group has demonstrated that the Nanotrap‐captured RVFV and influenza A RNA are protected from enzymatic degradation, while free‐floating RNA is completely degraded in the presence of RNase A (Shafagati et al., 2013; N. Shafagati, K. Fite, A. Patanarut, C. Pinkham, A. Baer, B. Lepene and K Kehn‐Hall, unpublished data). RNase A is a fairly small protein, 13.7 kDa, and therefore we expect that it will enter the Nanotrap particles. Our working model is that RNase is inactivated when bound to the affinity baits (which mimic natural ligands) incorporated onto the polymer matrix, which serve to indirectly protect the harvested viral RNA from degradation (Fig. 4; Puri & Roskoski, 1994). The ability to protect RVFV RNA from degradation was not observed with RVFV RNA captured by commercially available Bio‐gel HTP hydroxyapatite chromatography beads (Shafagati et al., 2013), suggesting that this is a unique feature of the Nanotrap particles.
Figure 4.
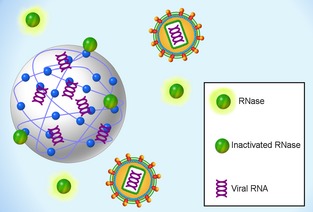
RNA protection. The Nanotrap particles can sequester viral RNA from complex biofluids while protecting them from degradation by RNase.
Viruses are subject to degradation soon after sample collection. This is particularly problematic if the sample contains low viral titers, leading to false negatives during testing. Nanotrap particles are not only capable of capturing and enriching the sample, but also protecting the virus after capture. Specifically, we have demonstrated that Nanotrap particles were capable of preserving viral infectivity (as compared to a sample without Nanotrap particles) at ambient and increased temperatures for up to 3 days (Shafagati et al., 2013). In addition, unpublished work by Shafagati et al. demonstrated that Nanotrap particles with acrylic acid baits preserve viral infectivity of influenza A virus (N. Shafagati, K. Fite, A. Patanarut, C. Pinkham, A. Baer, B. Lepene and K. Kehn‐Hall, unpublished data). After capture of the influenza A 2009 H1N1 virus with Nanotrap particles, the virus can be incubated at ambient temperature for at least 48 h and infectious virus still recovered. The viral protection provided by the Nanotrap particles allows time for unknown samples to be taken from patients in the clinic or cases from the field and transferred to the laboratory for further investigation without the need to utilized cold chain transport.
Conversely, there are instances where an inactivated, noninfectious sample is preferred. BSL3 laboratories are sparse, and work in them is time‐consuming and requires highly trained individuals. Recently, we have demonstrated that Nanotrap particles allow for the binding of inactivated virus. After capture with Nanotrap particles, the infectious virus can be quickly inactivated using two simple methods – heating at 57 °C or addition of NP‐40 detergent or other inactivating agents directly to the sample. Interestingly, Nanotrap particles were more efficient at capturing inactivated virus than Bio‐gel HTP hydroxyapatite beads (Shafagati et al., 2013). The ability to capture inactivated samples is of particular importance when collecting infectious samples in the field, allowing for the safe transport of samples to a BSL2 facility.
Nanotrap particles can capture viral antigens
Viral diagnostics is often achieved with ELISAs for either antibodies against the pathogen or detection of viral antigens. ELISA methodology offers the advantages of being affordable, relatively quick, and highly sensitive and specific technique compared with other molecular techniques. Viral antigens of interest are typically a surface protein or a nucleoprotein with high abundance within the host. These antigens may be virion associated or found in bodily fluids independent of virions having been released from infected cells during cell death.
In the case of RVFV, the nucleoprotein (NP) is the most abundant protein within virions (Ferron et al., 2011; Williams et al., 2011) and the only antigen for which commercial ELISAs are available (offered through Immune Technology Corp and BDSL). Unfortunately to date, none of the RVFV ELISAs are FDA approved. The nucleotide sequence of the NP gene is highly conserved among different isolates, and antibodies against NP are detected early on in infection (Williams et al., 2011). Therefore, it is commonly used in antigen‐capture ELISAs to detect RVFV, and in the last decade, highly sensitive and specific ELISAs for the detection of RVFV NP have been developed (Paweska et al., 2003a, b, 2005a, b, 2008; Fafetine et al., 2007; Jansen van Vuren et al., 2007; Jansen van Vuren & Paweska, 2009; Pepin et al., 2010). One disadvantage of ELISAs as a diagnostic platform is that they pose a biohazard risk due to the use of live virus in an open‐bench system, pipetting, and washing procedures, and the use of OD readings. Furthermore, the reagents are expensive and cumbersome, and the internal antigen controls are not readily available or well characterized (Pepin et al., 2010). In a paper published by Paweskaet al. in 2009, the researchers have addressed at least one of those disadvantages, by developing a sandwich antigen ELISA (sAG‐ELISA) that works with noninfectious samples (Paweska et al., 2005a). The authors first inactivated supernatants from RVFV‐infected Vero cell cultures at 56 °C for 1 h in the presence of 0.5% Tween‐20. They demonstrated that the thermo‐inactivated NP antigen was detected early on after infection (8 h). In human patients infected with RVFV, the sAg‐ELISA had 68% sensitivity and 98% specificity, whereas in experimentally infected sheep, there was 70% sensitivity and 100% specificity (Paweska et al., 2005a). In another paper published by Fukushi et al. in 2012, an antigen‐captured ELISA against RVFV NP using new monoclonal antibodies was developed. The authors tested both histidine‐tagged recombinant NP (His‐rNP) or culture supernatants infected with RVFV and found a detection limit of 0.11 ng per 100 µL in His‐rNP and 7.8 PFU per 100 µL of RVFV in RVFV antigen from culture supernatants of RVFV‐infected cells infected. While this newly developed Ag‐capture ELISA had a detection limit that was ten times higher than that reported for detecting the RVFV antigen, the detection limit was still lower compared with qRT‐PCR assay (Fukushi et al., 2012).
Recent work by our group has demonstrated the capture and enrichment of RVFV NP in viral supernatants and in animal serum (Fig. 5). Seven different Nanotrap particles were analyzed (NT45, 46, 53, 55, 69, 71, and 76) to determine their ability to capture RVFV NP (panel a). NT45 was the most effective at NP capture and showed a robust enrichment as compared to the no NT sample (compare lanes 1 and 2). Interestingly, NT45 is a Nanotrap particle that contains two affinity baits, Reactive Red 120 and Reactive Yellow 86. Experiments were also performed to determine the ability of NT45 to capture RVFV NP from clinically relevant matrices. RVFV NP was spiked into 100% bovine, sheep, goat, or donkey serum, incubated with NT45 and analyzed by Western blot analysis. RVFV NP was detectable in all four sera analyzed when incubated with NT45 (Fig. 5b). In contrast, the same concentration of RVFV NP (lane 2) was not detectable in this Western blot analysis. These results demonstrate that NT45 is capable of capturing and enriching RVFV NP from serum. RVFV NP is found free in serum as well as within virions coating the viral RNA. Thus, Nanotrap particles are capable of capturing both virion associated and free NP, providing a method to capture both pools of NP that can then be further analyzed through downstream assays including the sandwich ELISAs described above. In theory, targeting both pools of NP should increase the sensitivity of ELISAs, with the goal of being comparable to qRT‐PCR detection of RVFV. These studies are currently underway. In addition, as we have shown that Nanotrap particles are capable of capturing inactivated RVFV (Shafagati et al., 2013), these assays could be performed in a BSL‐2 environment.
Figure 5.

Nanotrap particles are capable of capturing and enriching RVFV NP. (a) Nanotrap particle screening for RVFV NP capture. One hundred microliters of purified RVFV His‐NP (0.02 mg mL−1) was incubated with 75 µL of seven different Nanotrap particles (NT45, 46, 53, 55, 69, 71, 76). Samples were incubated at room temperature for 30 mins, washed four times with water, and separated by SDS‐PAGE. Western blot analysis was performed with anti‐His antibody. NP input is 0.02 mg mL−1 as a positive control and had no Nanotrap particles added. (b) NT45 is capable of capturing RVFV NP from serum. RVFV His‐NP (20 µg mL−1) was spiked into 1 mL of 100% serum and incubated with 150 µL of NT45. Samples were incubated at room temperature for 30 mins, washed four times with water, and separated by SDS‐PAGE. Western blot analysis was performed with anti‐His antibody. NP (100 µg mL−1) was included as a positive control.
Influenza diagnostics are much more advanced with multiple assay types available for the physician and the clinical diagnostic laboratory. For viral antigen detection, RIDTs are available, which are lateral flow assays that can identify the presence of the influenza A or B viral nucleoprotein in respiratory samples, generally in 15 min or less. While these tests are very successful at recognizing infection at high titers, false‐negative results are likely to occur at low viral concentrations. This is therefore a key diagnostic area that could benefit from the Nanotrap technology, as a larger volume of sample can be enriched using the Nanotrap particles to provide more accurate detection. Interestingly, many RIDTs (such as the AlereBinaxNOW® Influenza A&B Card) allow the detection of both free and virion‐associated nucleoprotein, as there is a lysis step prior to applying the swab material to the strip. Current research efforts are underway to determine whether Nanotrap particles can enhance the sensitivity of RIDT assays by enriching the virus and its low‐abundance analytes prior to testing with the lateral flow assays.
Use of Nanotrap particles for host biomarker discovery applications to viral diagnostics
Recent interest has emerged over the role of cytokines as potential serum biomarkers in predicting the severity of viral infections. Several papers have demonstrated that these host responses are increased early during infection and, if prolonged, could actually worsen the disease (known as a ‘cytokine storm’; Paquette et al., 2012). Mouse and ferret animal models have shown that virulent strains of influenza A virus have been associated with increased levels of biomarkers such as IL‐6, TNFα, and IFNα. Furthermore, patients' samples show increased IL‐6 and IFNα levels in nasal lavage fluids early on in influenza A infection (Davey et al., 2013). IL‐6, an important feature of the host response that can predict disease and outcome, is found in increased levels in the serum of patient samples, which contains many high‐abundant proteins. A recent paper demonstrated that IL‐6 could be a potential disease severity biomarker for the 2009 pandemic H1N1 influenza A (H1N1pdm09; Paquette et al., 2012). Infection with VEEV and RVFV also results in increased levels of transcripts of interferon, interferon‐regulated factors, and Toll‐like receptors in the brain such as IFNβ and TLR9 (Bouloy et al., 2001; Erwin‐Cohen et al., 2012). In conjunction with high‐RVFV viral titers, the level of serum cytokines (IL‐6, IL‐12, G‐GSF) is significantly increased by 72 h postinfection, but drastically drop to normal cytokine levels at 96 h postinfection (van Vuren et al., 2011). During the course of viral infection, the virus can antagonize these innate and adaptive immune responses, leading to severe disease (Bouloy et al., 2001). Recently, we have demonstrated that Nanotrap particles can help enhance the detection of IL‐8 in cell culture supernatants following RVFV infection (Narayanan et al., 2014). The detection of early host response biomarkers is a promising avenue to allow accurate early diagnostics prior to severe symptoms of disease.
Nanotrap particles have previously been shown to sequester low‐abundance chemokines such as CCL28, CCL24, and CXCL12 (Longo et al., 2009). One core issue with the collection of cytokines is their extremely short half‐life (roughly 10 min) and degradation as soon as blood is drawn (Zhou et al., 2010). Furthermore, cytokines such as IL‐6 begin to decrease significantly at 4 h (Paquette et al., 2012). Nanotrap particles have been shown to protect cytokines from degradation at room temperature (Longo et al., 2009). Another important class of biomarkers is the cell surface proteins (such as the receptors for cytokines), which are important indicators of disease state and clinical outcome. While FACS analysis is the most common method for cell surface protein analysis, it requires at least several hundreds of thousands of proteins per cell for a measurable signal. Unfortunately, protein levels, such as those found for low‐abundant cytokine receptors, can be too low for detection (Joensson et al., 2009). The Nanotrap particles can work to enrich cytokines and cell surface proteins from serum and nasal swab samples into a smaller sample size while simultaneously excluding serum albumin and immunoglobulins, allowing for their downstream analysis with ELISAs and various multiplex assays (Zhou et al., 2010). This will not only enhance early detection but also allow for the comparison of virulent and nonvirulent strains of viruses and the host's immune response.
Scientists and physicians have been trying for years to develop a panel of biomarkers that would predict the development and severity of disease. Individual biomarker candidates cannot reach the level of specificity and sensitivity needed for proper diagnosis of a disease (Petricoin et al., 2006). A determined number of biomarkers will not only allow for proper treatment of the disease before it is too late, but also provide extensive details on host defense. While this concept has been extensively studied in cancer research, it can be extended to viral diagnostics. This could be especially important in influenza detection, where the severity of the disease varies by the subtype of the virus. A study conducted by Davey et al. demonstrated that seven markers were significantly correlated with disease progression in the serum of influenza H1N1pdm09 patients, IL‐6, CD163, IL‐10, LBP, IL‐2, MCP‐1, and IP‐10 (Davey et al., 2013). Influenza infection can predispose an individual to a secondary bacterial infection that is caused by Streptococcus pneumoniae or Streptococcus aureus. There is supporting evidence that the influenza A pandemic of 1918 was caused by a synergy of influenza and pneumococcal infections. The host response and disease progression to pneumonia are variable between individuals. Several biomarkers, such as the inflammatory mediator proteins IP‐10 and CXCL11, are thought to activate other cells that provide a favorable environment for pneumonia infection. While influenza infection cannot be avoided, the progression to pneumonia can be stopped if these biomarkers are detected early enough after viral infection (Brand et al., 2010). However, due to the complexity of serum, the sensitivity of detection for this panel can be too low to make an accurate diagnosis. Due to the composition and promiscuity of the Nanotrap particles, the high‐abundant proteins can be excluded and several of the biomarkers can be captured and tested simultaneously.
Conclusions and future directions
This review provides an overview of the broad utility of Nanotrap particle technology platform for biospecimen sample storage. The initial work presented lays a foundation for the next stage of research and development efforts aimed at the end goal of providing solutions for improved early detection, identification of pathogenic strains or monitoring of disease progression and treatment efficacy. Future directions are focused in three main areas: (1) biospecimen collection and storage, (2) analyte enrichment, and (3) multiplex assay development. Initial results indicated the ability to preserve multiple classes of analytes even at increased temperatures. Further investigations into the fundamental mechanisms of protection will provide valuable insight into this process and potentially extend the ability to prevent degradation to valuable samples and reagents stored at ambient conditions without cold chain storage. We also plan to develop new processes for optional inactivation of pathogenic biospecimens prior to transport or storage. The ability to capture multiple classes of analytes from a single complex biofluid or environmental samples using a combination of Nanotrap particle architectures will provide simple and reliable multiplexed diagnostic workflows and processes.
Acknowledgements
This work was supported by a grant from The National Center for Foreign Animal and Zoonotic Diseases Defense (FAZD Center), Texas A&M/DHS to BL and KK. NS was supported through a graduate student fellowship provided through the FAZD Center, HS‐STEM Career Development Program. This material is based upon work supported by the U.S. Department of Homeland Security under Grant Award Number 20011‐ST‐104‐000002. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Homeland Security. Alexis Patanarut and Ben Lepene are employees of Ceres Nanosciences, Inc. Ceres Nanosciences, Inc. produces and distributes Nanotrap particles.
The manuscript describes an important emerging technology (Nanotrap particle capture) for the diagnosis of important BSL3 viral pathogens. This method offers the potential of improved sensitivity and specificity of virus particle concentration in complex body fluids that is likely to have broad applicability in viral diagnostics.
References
- Abdelwhab EM et al (2011) The use of FTA(R) filter papers for diagnosis of avian influenza virus. J Virol Methods 174: 120–122. [DOI] [PubMed] [Google Scholar]
- Aguilar PV et al (2011) Endemic Venezuelan equine encephalitis in the Americas: hidden under the dengue umbrella. Future Virol 6: 721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigent SJ & McCauley JW (2003) Influenza type A in humans, mammals and birds: determinants of virus virulence, host‐range and interspecies transmission. BioEssays 25: 657–671. [DOI] [PubMed] [Google Scholar]
- Banoo S et al (2010) Evaluation of diagnostic tests for infectious diseases: general principles. Nat Rev Microbiol 8(suppl 12): S17–S29. [PubMed] [Google Scholar]
- Bellei N et al (2007) Influenza and rhinovirus infections among health‐care workers. Respirology 12: 100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JA et al (2008) Viral nucleic acid stabilization by RNA extraction reagent. J Virol Methods 150: 41–44. [DOI] [PubMed] [Google Scholar]
- Bouloy M et al (2001) Genetic evidence for an interferon‐antagonistic function of rift valley fever virus nonstructural protein NSs. J Virol 75: 1371–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand HK, Hermans PW & de Groot R (2010) Host biomarkers and paediatric infectious diseases: from molecular profiles to clinical application. Adv Exp Med Biol 659: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara AS et al (2007) Venezuelan equine encephalitis virus infection of cotton rats. Emerg Infect Dis 13: 1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow DS & Memoli MJ (2013) Bacterial coinfection in Influenza A grand rounds review. JAMA 309: 275–282. [DOI] [PubMed] [Google Scholar]
- Davey RT Jr et al (2013) The association between serum biomarkers and disease outcome in influenza A(H1N1)pdm09 virus infection: results of two international observational cohort studies. PLoS One 8: e57121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas TA et al (2011) The use of hydrogel microparticles to sequester and concentrate bacterial antigens in a urine test for Lyme disease. Biomaterials 32: 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin‐Cohen RA et al (2012) Host responses to live‐attenuated Venezuelan equine encephalitis virus (TC‐83) Comparison of naive, vaccine responder and nonresponder to TC‐83 challenge in human peripheral blood mononuclear cells. Hum Vaccin Immunother 8: 1053–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fafetine JM et al (2007) Cloning and expression of Rift Valley fever virus nucleocapsid (N) protein and evaluation of a N‐protein based indirect ELISA for the detection of specific IgG and IgM antibodies in domestic ruminants. Vet Microbiol 121: 29–38. [DOI] [PubMed] [Google Scholar]
- Fereidouni SR et al (2012) Effect of swab matrix, storage time, and temperature on detection of avian influenza virus RNA in swab samples. Avian Dis 56(suppl 4): 955–958. [DOI] [PubMed] [Google Scholar]
- Ferron F et al (2011) The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog 7: e1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredolini C et al (2008) Concentration and preservation of very low abundance biomarkers in urine, such as human growth hormone (hGH), by Cibacron Blue F3G‐A loaded hydrogel particles. Nano Res 1: 502–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredolini C et al (2009) Nanoparticle technology: amplifying the effective sensitivity of biomarker detection to create a urine test for hGH. Drug Test Anal 1: 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredolini C et al (2010) Investigation of the ovarian and prostate cancer peptidome for candidate early detection markers using a novel nanoparticle biomarker capture technology. AAPS J 12: 504–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymuth F et al (2005) Replication of respiratory viruses, particularly influenza virus, rhinovirus, and coronavirus in HuH7 hepatocarcinoma cell line. J Med Virol 77: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi S et al (2012) Antigen‐capture ELISA for the detection of Rift Valley fever virus nucleoprotein using new monoclonal antibodies. J Virol Methods 180: 68–74. [DOI] [PubMed] [Google Scholar]
- Gao J & Frisken BJ (2003) Cross‐linker‐free N‐isopropylacrylamide gel nanospheres. Langmuir 19: 5212–5216. [Google Scholar]
- Garten RJ et al (2009) Antigenic and genetic characteristics of swine‐origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi M, Azadi A & Rafiei P (2008) Hydrogel nanoparticles in drug delivery. Adv Drug Deliv Rev 60: 1638–1649. [DOI] [PubMed] [Google Scholar]
- Jansen van Vuren P & Paweska JT (2009) Laboratory safe detection of nucleocapsid protein of Rift Valley fever virus in human and animal specimens by a sandwich ELISA. J Virol Methods 157: 15–24. [DOI] [PubMed] [Google Scholar]
- Jansen van Vuren P et al (2007) Preparation and evaluation of a recombinant Rift Valley fever virus N protein for the detection of IgG and IgM antibodies in humans and animals by indirect ELISA. J Virol Methods 140: 106–114. [DOI] [PubMed] [Google Scholar]
- Joensson HN et al (2009) Detection and analysis of low‐abundance cell‐surface biomarkers using enzymatic amplification in microfluidic droplets. Angew Chem Int Ed Engl 48: 2518–2521. [DOI] [PubMed] [Google Scholar]
- Kramarow EA & Pastor PA (2012) Evaluation of 11 commercially available rapid influenza diagnostic tests–United States, 2011–2012. MMWR Morb Mortal Wkly Rep 61: 873–876. [PubMed] [Google Scholar]
- Kirsch MI et al (2008) Development of human antibody fragments using antibody phage display for the detection and diagnosis of Venezuelan equine encephalitis virus (VEEV). BMC Biotechnol 8: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F et al (1969) Interacting factors that influence long‐term storage of live Pasteurella tularensis vaccine and Rift Valley fever virus. Appl Microbiol 17: 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafft AE et al (2005) Evaluation of PCR testing of ethanol‐fixed nasal swab specimens as an augmented surveillance strategy for influenza virus and adenovirus identification. J Clin Microbiol 43: 1768–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella AM & Merel SE (2013) Influenza. Med Clin North Am 97: 621–645, x. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Petricoin EF & D G (2006) Method of isolating analytes from a sample. September 27, 2006.
- Longo C et al (2009) Core‐shell hydrogel particles harvest, concentrate and preserve labile low abundance biomarkers. PLoS One 4: e4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini A et al (2008) Smart hydrogel particles: biomarker harvesting: one‐step affinity purification, size exclusion, and protection against degradation. Nano Lett 8: 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini A et al (2010) Nanoparticle technology: addressing the fundamental roadblocks to protein biomarker discovery. Curr Mol Med 10: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini A et al (2011) Smart hydrogel particles for biomarker harvesting. United States.
- Mansuroglu Z et al (2010) Nonstructural NSs protein of Rift Valley fever virus interacts with pericentromeric DNA sequences of the host cell, inducing chromosome cohesion and segregation defects. J Virol 84: 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell K et al (2004) Analysis of low‐abundance, low‐molecular‐weight serum proteins using mass spectrometry. J Biomol Tech 15: 238–248. [PMC free article] [PubMed] [Google Scholar]
- Narayanan A et al (2014) Reactive oxygen species activate NFκB (p65) and p53 and induce apoptosis in RVFV infected liver cells. Virology 449: 270–286. [DOI] [PubMed] [Google Scholar]
- Naslund J et al (2011) Detection of Puumala and Rift Valley fever virus by quantitative RT‐PCR and virus viability tests in samples of blood dried and stored on filter paper. J Virol Methods 178: 186–190. [DOI] [PubMed] [Google Scholar]
- Navarro JC et al (2005) Postepizootic persistence of Venezuelan equine encephalitis virus, Venezuela. Emerg Infect Dis 11: 1907–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paessler S & Weaver SC (2009) Vaccines for Venezuelan equine encephalitis. Vaccine 27(suppl 4): D80–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette SG et al (2012) Interleukin‐6 is a potential biomarker for severe pandemic H1N1 influenza A infection. PLoS One 7: e38214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TG & Choi HK (1998) Thermally induced core‐shell type hydrogel beads having interpenetrating polymer network (IPN) structure. Macromol Rapid Commun 19: 167–172. [Google Scholar]
- Patanarut A et al (2010) Synthesis and characterization of hydrogel particles containing Cibacron Blue F3G‐A. Colloids Surf A Physicochem Eng Asp 362: 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paweska JT et al (2003a) IgG‐sandwich and IgM‐capture enzyme‐linked immunosorbent assay for the detection of antibody to Rift Valley fever virus in domestic ruminants. J Virol Methods 113: 103–112. [DOI] [PubMed] [Google Scholar]
- Paweska JT et al (2003b) Indirect enzyme‐linked immunosorbent assay for the detection of antibody against Rift Valley fever virus in domestic and wild ruminant sera. Onderstepoort J Vet Res 70: 49–64. [PubMed] [Google Scholar]
- Paweska JT et al (2005a) An inhibition enzyme‐linked immunosorbent assay for the detection of antibody to Rift Valley fever virus in humans, domestic and wild ruminants. J Virol Methods 127: 10–18. [DOI] [PubMed] [Google Scholar]
- Paweska JT, Burt FJ & Swanepoel R (2005b) Validation of IgG‐sandwich and IgM‐capture ELISA for the detection of antibody to Rift Valley fever virus in humans. J Virol Methods 124: 173–181. [DOI] [PubMed] [Google Scholar]
- Paweska JT et al (2008) Recombinant nucleocapsid‐based ELISA for detection of IgG antibody to Rift Valley fever virus in African buffalo. Vet Microbiol 127: 21–28. [DOI] [PubMed] [Google Scholar]
- Peeling RW & Mabey D (2010) Point‐of‐care tests for diagnosing infections in the developing world. Clin Microbiol Infect 16: 1062–1069. [DOI] [PubMed] [Google Scholar]
- Pelton R (2000) Temperature‐sensitive aqueous microgels. Adv Colloid Interface Sci 85: 1–33. [DOI] [PubMed] [Google Scholar]
- Pepin M et al (2010) Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res 41: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S, Dugas AF & Rothman RE (2013) Evaluation of 11 commercially available rapid influenza diagnostic tests‐United States, 2011–2012. Ann Emerg Med 61: 573–577. [DOI] [PubMed] [Google Scholar]
- Petricoin EF et al (2006) The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nat Rev Cancer 6: 961–967. [DOI] [PubMed] [Google Scholar]
- Pisano MB et al (2012) Specific detection of all members of the Venezuelan Equine Encephalitis complex: development of a RT‐Nested PCR. J Virol Methods 186: 203–206. [DOI] [PubMed] [Google Scholar]
- Prevention C.f.D.C.a. (2011) Influenza Symptoms and the Role of Laboratory Diagnostics. Seasonal Influenza (Flu) 2011 December 9, 2011; Available from: http://www.cdc.gov/flu/professionals/diagnosis/labrolesprocedures.htm.
- Puri RN & Roskoski R Jr (1994) Inactivation of yeast hexokinase by Cibacron Blue 3G‐A: spectral, kinetic and structural investigations. Biochem J 300(Pt 1): 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz E et al (2009) Venezuelan equine encephalitis in Panama: fatal endemic disease and genetic diversity of etiologic viral strains. PLoS Negl Trop Dis 3: e472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer WF et al (1972) Observations of equines, humans and domestic and wild vertebrates during the 1969 equine epizootic and epidemic of Venezuelan encephalitis in Guatemala. Am J Epidemiol 95: 255–266. [DOI] [PubMed] [Google Scholar]
- Shafagati N et al (2013) The use of nanotrap particles as a sample enrichment method to enhance the detection of Rift Valley fever virus. PLoS Negl Trop Dis 7: e2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimshony A & Barzilai R (1983) Rift Valley fever. Adv Vet Sci Comp Med 27: 347–425. [PubMed] [Google Scholar]
- Spackman E et al (2002) Development of a real‐time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 40: 3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburro D et al (2011) Multifunctional core‐shell nanoparticles: discovery of previously invisible biomarkers. J Am Chem Soc 133: 19178–19188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero‐Fuenmayor N et al (1997) Importance of transmission electron microscopy in the diagnosis of a Venezuelan equine encephalitis outbreak in 1995 in the Venezuelan Guajira. Invest Clin 38: 73–82. [PubMed] [Google Scholar]
- van Vuren PJ, Tiemessen CT & Paweska JT (2011) Anti‐nucleocapsid protein immune responses counteract pathogenic effects of Rift Valley fever virus infection in mice. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang EY et al (2001) Virulence and viremia characteristics of 1992 epizootic subtype IC Venezuelan equine encephalitis viruses and closely related enzootic subtype ID strains. Am J Trop Med Hyg 65: 64–69. [DOI] [PubMed] [Google Scholar]
- Wang E et al (2005) A novel, rapid assay for detection and differentiation of serotype‐specific antibodies to Venezuelan equine encephalitis complex alphaviruses. Am J Trop Med Hyg 72: 805–810. [PubMed] [Google Scholar]
- Weaver SC et al (2004) Venezuelan equine encephalitis. Annu Rev Entomol 49: 141–174. [DOI] [PubMed] [Google Scholar]
- WHO (2006) Collecting, preserving and shipping specimens for the diagnosis of avian influenza A (H5N1) virus infection. Guide for Field Operations, pp. 1–80.
- WHO (2013) Influenza. WHO. [Google Scholar]
- Williams R et al (2011) Validation of an IgM antibody capture ELISA based on a recombinant nucleoprotein for identification of domestic ruminants infected with Rift Valley fever virus. J Virol Methods 177: 140–146. [DOI] [PubMed] [Google Scholar]
- Wilson ML (1994) Rift Valley fever virus ecology and the epidemiology of disease emergence. Ann NY Acad Sci 740: 169–180. [DOI] [PubMed] [Google Scholar]
- Zhou X et al. (2010) Conceptual and methodological issues relevant to cytokine and inflammatory marker measurements in clinical research. Curr Opin Clin Nutr Metab Care 13: 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]


