Abstract
Background
Viral infections have been suggested as possible triggers for the onset of ulcerative colitis (UC).
Methods
We employed VirCapSeq-Vert, a high-throughput sequencing virus capture platform, to examine the stool virome of children with newly diagnosed moderate to severe UC. We surveyed fecal samples collected at presentation, after symptom remission, and from a control group diagnosed with irritable bowel syndrome.
Results
Seventy subjects with UC (mean age 13 years, 45 had moderate symptoms, 25 had severe, 69 of 70 had a Mayo endoscopy subscore 2/3) were studied. We detected a wide range of animal viruses that were taxonomically classified into 12 viral families. A virus was present in 50% of fecal samples collected at presentation, 41% of samples collected after remission, and 40% of samples in our control group. The most frequently identified viruses were diet-based gyroviruses. The UC cohort had a significantly higher prevalence of anelloviruses compared with the control cohort. However, we did not identify a single virus that can be implicated in the onset of UC and did not find an association between UC disease severity and viral presence.
Conclusion
Presence of virus in stool was not associated with the onset of pediatric UC.
Keywords: ulcerative colitis, children, virome
We used VirCapSeq-VERT, a high-throughput sequencing virus capture platform, to examine the stool virome of children with newly diagnosed moderate to severe ulcerative colitis. We did not identify a link between a viral infection and the onset of ulcerative colitis.
Introduction
The clinical presentation of ulcerative colitis (UC) in children varies from minimal bleeding to acute severe colitis resulting in hospitalization.1–3 Although most pediatric patients initially respond to standard therapy with corticosteroids and mesalamine, almost half require additional therapy with immunomodulators or biologic agents, and up to 10% are refractory to all medical therapies and require colectomy in the first 1 to 2 years following diagnosis.3–5
The precise pathogenesis of UC is unknown. A wide range of potential factors have been suggested to play a role, including genetics, the microbiome, the mucosal immune response, and epithelial cell abnormalities. What has not been fully assessed is the role, if any, that viruses may play in the pathogenesis of UC. Thus far, the majority of UC virome studies have focused on analyses of bacteriophage populations, and the implications of vertebrate viral infections are unknown.6–8
The Predicting Response to Standardized Pediatric Colitis Therapy (PROTECT) study, initiated in 2012, enrolled treatment-naïve children and adolescents with UC who received initial standardized therapy with mesalamine and corticosteroids. Enrolled patients had stool obtained at diagnosis and weeks 4, 12, and 52 after diagnosis. Clinical observation during the PROTECT study revealed clusters of severe cases suggesting a possible environmental trigger such as viral infection. The aim of this study was to examine the gastrointestinal virome of children with newly diagnosed UC to explore the role of viruses in triggering UC. To enhance the likelihood of successful detection of viral nucleic acid, we employed VirCapSeq-VERT, a viral template enrichment platform that uses ~2 million probes that cover the genomes of 207 viral taxa known to infect vertebrates.9,10 The use of VirCapSeq-VERT results in a 100- to 10,000-fold enrichment of viral sequences over standard metagenomic methods. This method is highly useful in instances where viral sequences represent only a small proportion of overall genetic material present in clinical specimens.
METHODS
Study Population
The PROTECT study was initiated in 2012 to systematically examine the response of children and adolescents newly diagnosed with ulcerative colitis to consensus-defined therapy. After giving informed consent/assent, children aged 4 to 17 years with a clinical history consistent with ulcerative colitis underwent diagnostic ileocolonoscopy and biopsy, and a diagnosis of ulcerative colitis was made using standardized criteria.11 Further characterization of the PROTECT study population has previously been reported.3 Enrolled subjects provided stool samples before diagnostic colonoscopy and before bowel cleansing, and at defined follow-up times including 12 and 52 weeks after therapy started.
Sample Collection
Fecal samples were collected from children enrolled in the PROTECT study. We selected 70 pretreatment samples obtained at presentation (designated as baseline) from “clusters” defined as 2 or more cases seen within 1 week at a study center and random patients in whom no other UC case was identified within a month at a study center. Patients selected had either a moderate Pediatric Ulcerative Colitis Activity Index (PUCAI) score (35 to 60) or a severe PUCAI score ≥65. For the same cohort, we analyzed 70 fecal samples collected after symptom remission (designated as follow-up). These included 64 samples obtained at week 52. For 6 patients, week 52 fecal samples were not available; accordingly, we analyzed samples obtained at week 12. For noninflammatory controls, we examined fecal samples collected from a patient cohort diagnosed with irritable bowel syndrome by standardized clinical criteria.12 Fecal samples were collected at home and kept on ice or frozen until submission to the local investigative site where they were frozen at −80˚C, shipped on dry ice to the biorepository, and then maintained at −80˚C.
Sample Preparation and Nucleic Acid Extraction
Approximately 0.2 grams of feces from each sample was transferred to a 1-mL Eppendorf tube containing 1 mL of sterile phosphate buffered saline. The contents were briefly vortexed and then centrifuged at 5000 relative centrifugal force for 5 minutes to precipitate the particulate material. The supernatant was purified through a 0.45-μm pore-size sterile disk filters (Merck/Millipore, Billerica, MA, USA). Total nucleic acids (TNA) were extracted from 250 μL of the purified supernatant using the easyMag platform (bioMerieux, Marcy-l'Etoile, France). All samples were eluted in 40 μL.
VirCapSeq-VERT Library Preparation and Sequencing
For library preparation, TNA extracts (11 μL) were reverse transcribed using SuperScript III (Thermo, Fisher Waltham, MA, USA) with random hexamers. The cDNA was RNase H treated before second-strand synthesis with Klenow fragment (New England Biolabs, Ipswich, MA, USA). The resulting double-stranded cDNA/DNA mix was mechanically sheared to an average fragment size of 200 bp (Covaris E210 focused ultrasonicator). Sheared product was purified (AxyPrep), and libraries were constructed using KAPA library preparation kits (KAPA) with Roche/NimbleGen (Madison, WI, USA) adapter kits. The quality and quantity of libraries were assessed using a Bioanalyzer (Agilent, Santa Clara, CA, USA).
A total of 10 lanes on Illumina 4000 sequencer were allocated for the sequencing of the libraries generated from the 210 samples. Following library preparations, all samples were randomized before sequencing, with each lane consisting of 21 samples representing 7 baseline, 7 follow-up, and 7 control samples each. Once the samples for each lane were selected, the libraries were mixed with a SeqCap HE universal oligonucleotide, SeqCap HE index oligonucleotides, and COT DNA and vacuum evaporated at 60°C for approximately 40 minutes. Dried samples were mixed with 2× hybridization buffer and hybridization component A (Roche/NimbleGen) before denaturation at 95°C for 10 minutes. The VirCap probe library (4.5 μL) was added and hybridized at 47°C for 12 hours in a standard PCR thermocycler. SeqCap Pure capture beads (Roche/NimbleGen) were washed twice, mixed with the hybridization mix, and kept at 47°C for 45 minutes with vortexing for 10 seconds every 10 to 15 minutes. The streptavidin capture beads complexed with biotinylated VirCapSeq-VERT probes were trapped (DynaMag-2 magnet; Thermo, Fisher) and washed once at 47°C and then twice more at room temperature with wash buffers of increasing stringency. Finally, beads were suspended in 50 µL of water and directly subjected to posthybridization PCR (SeqCap EZ accessory kit V2; Roche/NimbleGen). The PCR products were purified (Agencourt Ampure DNA purification beads; Beckman Coulter, Brea, CA, USA) and quantitated by Bioanalyzer (Agilent) for Illumina sequencing.
Data Analysis and Bioinformatics Pipeline
The demultiplexed FastQ files were adapter trimmed using cutadapt program (v 1.10). Adaptor trimming was followed by generation of quality reports using FastQC software (v 0.11.5); these were used to determine filtering criteria based on the average quality scores of the reads, presence of indeterminate nucleotides, and homopolymeric reads. The reads were quality filtered and end-trimmed with PRINSEQ software (v 0.20.3). Host background levels were determined by mapping filtered reads against a human reference database using Bowtie2 mapper (v 2.2.9). The host-subtracted reads were de novo assembled using MIRA (4.0) assembler. Contigs and unique singletons were subjected to homology search using Megablast against the GenBank nucleotide database. Sequences that showed low or no homology at the nucleotide level were subjected to a BLASTX homology search against the viral GenBank protein database. Viral sequences from BLASTX analysis were subjected to another round of BLASTX homology search against the entire GenBank protein database to correct for biased E values and taxonomic mis-assignments. Based on the contigs identified for different viral strains, GenBank sequences were downloaded and used for mapping the whole data set to recover partial or complete genomes. Viral read numbers were obtained from counting the number of reads mapping to contig sequences and unassembled singletons, and percentages were calculated in relation to the total read number obtained.
Statistical Analysis
To test for the association between each detected virus and UC, we conducted logistic regression model with the binary case status (case vs control) as dependent variable and the presence (vs absence) of each virus as independent variable. In addition, we repeated the logistic regression model replacing the independent variable with the number of viruses present or presence of any virus. Because of the quasi-complete or complete separation phenomenon caused by the low prevalence of certain viruses, we applied a penalized likelihood based method to reduce potential bias.13 To determine virus association with disease severity within cases at baseline, we conducted the same logistic regression models replacing the dependent variable with the binary severity status. Age, sex, and Mayo endoscopy score were adjusted for as potential confounding variables. Furthermore, we examined the association between virus presence and clusters (vs nonclusters) using the same logistic regression models.
We compared the prevalence of each individual virus, any virus, or total virus abundance between the cases at baseline and at follow-up using generalized estimating equation (GEE) method to account for the inter-correlations within the same subjects.14 The choice of the link function in GEE depended on the assumption of the distribution of the variable of interest (ie, for binary presence/absence outcome, logistic link function was used, whereas for the counts of virus richness, we assumed that it followed Poisson distribution, and loglinear link function was applied).
Data analysis was implemented using MatLab and Statistics Toolbox release 2013a (MathWorks, Inc., Natick, MA, USA) and R version 3.5.0 (RStudio, Inc., Boston, MA, USA).
RESULTS
Patients
Seventy children with UC provided stool samples. The mean age was 13 years (range 4 to 17 years), >80% had extensive/pancolitis, 39 of 70 (56%) were male, 45 had moderate disease activity, 25 had severe activity, and 69 of 70 had Mayo Grade 2/3 initial endoscopic activity. No patient had proctitis only. All provided a stool at diagnosis, 64 a stool at 52 weeks, and 6 a stool at 12 weeks following diagnosis. Control stool was obtained from 70 adolescent patients with irritable bowel syndrome who were diagnosed a minimum of 3 months previously participating in an observational study and who were receiving no medications and had no evidence of enteric infections.
VirCapSeq-VERT generated between 295 million to 407 million reads per lane, with an average of 14.0 million to 19.4 million reads per sample (Table 1). Analysis of host-subtracted reads and assembled contigs identified the presence of a wide array of viruses within 30 known viral families (Table 2). These included a broad range of viruses known to infect plants. In total, we identified reads from 18 families of plant viruses. Since these viruses were presumably obtained through the diet and do not infect humans, they were not analyzed further. Our approach also identified viruses that have not yet been classified into any viral families (eg, smacovirus).
Table 1.
Number of Reads Per Pool Generated by VirCapSeq-VERT
| Poola | Average # Reads per Sample |
|---|---|
| 1 | 18,175,993 |
| 2 | 19,409,402 |
| 3 | 19,398,157 |
| 4 | 19,320,585 |
| 5 | 17,989,652 |
| 6 | 16,811,059 |
| 7 | 15,351,970 |
| 8 | 14,048,833 |
| 9 | 14,983,248 |
| 10 | 14,673,411 |
aEach pool consisted of 21 samples
Table 2.
List of Viral Families Represented by the Viruses Identified by High Throughput Sequencing
| Viral family | |
|---|---|
| Vertebrate viruses | Plant viruses |
| Adenoviridae | Alphaflexiviridae |
| Anelloviridae | Baculoviridae |
| Astroviridae | Betaflexiviridae |
| Caliciviridae | Bromoviridae |
| Circoviridae | Chrysoviridae |
| Coronaviridae | Closteroviridae |
| Flaviviridae | Endornaviridae |
| Herpesviridae | Geminiviridae |
| Parvoviridae | Luteoviridae |
| Picobirnaviridae | Mimiviridae |
| Picornaviridae | Partitiviridae |
| Reoviridae | Phycodnaviridae |
| Potyviridae | |
| Secoviridae | |
| Togaviridae | |
| Tombusviridae | |
| Tymoviridae | |
| Virgaviridae |
Virome of Baseline, Follow-up, and Control Cases
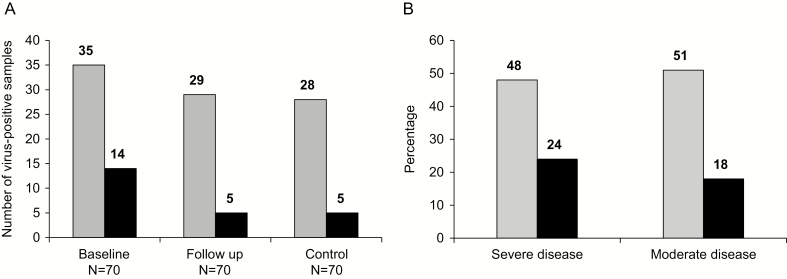
We identified an animal virus in 35 (50%) baseline UC cases (Fig. 1A). Fourteen contained >1 virus; 3 viruses were present in 3 samples, and 2 viruses were present in 11 samples. In the follow-up samples, 29 stools (41%) were virus-positive, with >1 virus present in 5 samples. The total number of viruses was 0.549-fold (95% confidence interval [CI], 0.316–0.956) lower in the follow-up samples compared with baseline (Supplemental Table 1). We did not find any significant difference between virus prevalence in follow-up samples relative to baseline (data not shown). Of the 35 virus-positive baseline patients, 21 had a virus detected in the follow-up samples. In the IBS control samples, 28 (40%) were virus-positive, with 3 viruses present in 2 samples, and 2 viruses present in 2 samples.
Figure 1.
Prevalence of viruses identified by VirCapSeq-VERT. A, The number of virus-positive samples (gray columns) and the number of samples with >1 virus (black). B, The percentage of virus-positive samples from baseline patients with severe and moderate disease. Gray columns indicate the percentage of virus-positive samples, and the black columns indicate the percentage of samples with >1 virus.
Virus Association With Case Clustering, Disease Severity
Twelve of the 25 samples originating from patients with severe UC (48%) were virus-positive. Twenty-three of 45 (51%) samples from patients with moderate UC were virus-positive (Fig. 1B). We found no significant association between UC disease severity and the presence of viral sequences (Supplemental Table S2). We also did not find significant association between the presence of a virus, or total viral abundance in baseline cases and disease clusters vs sporadic cases (Supplemental Table S3). Footprints of 2 or more viruses were identified in 6 (24%) of patients with severe UC vs 8 (18%) of patients with moderate UC. We found no significant association between UC and the presence of sequences representing multiple viruses.
Virome Classification
The most frequently detected vertebrate viruses in both UC and IBS were gyroviruses (Gyrovirus genus from the Anelloviridae family) that are presumed to originate from a poultry-based diet.15,16 We detected the presence of a gyrovirus in 35%, 52%, and 40% of baseline UC, follow-up UC, and IBS control cases. We also identified additional vertebrate viruses that presumably originate from food, including porcine anelloviruses and porcine parvoviruses, which are not known to infect humans.
The majority of human viruses detected in our study were human anelloviruses (family Anelloviridae), a diverse group of single-stranded DNA viruses with a short genome of <5 kB (Table 3). Human anelloviruses are characterized into 3 genera including Alphatorquevirus (includes torque teno viruses), Betatorquevirus (torque teno mini virus), and Gammatorquevirus (torque teno midi virus).17 Viruses from all 3 genera were present in the UC and IBS cohorts. We detected the presence of an anellovirus in 25 UC baseline and 25 follow-up UC (36%) fecal samples. Samples from 14 UC patients (20%) were anellovirus-positive at both baseline and follow-up. A total of 13 IBS control samples were anellovirus-positive (19%). The presence of anelloviruses was significantly associated (P < 0.05) with elevated risk of UC relative to IBS controls with an odds ratio of 2.39 (95% CI, 1.10–5.18) (Supplemental Table S4). There was no significant association between the presence of an anellovirus and disease severity: 28% of severe UC cases and 40% of moderate UC cases were anellovirus-positive (Supplemental Table S1). Moreover, the presence of annellovirus in baseline fecal samples was not associated with either week 52 steroid-free remission on mesalamine alone (P = 0.423) or escalation to anti-TNF-α therapy (P = 0.748).
Table 3.
Number of Samples From Each Cohort Positive for Every Virus Detected by VirCapSeq-VERT
| Virus | Baseline | Follow-up | Control |
|---|---|---|---|
| Anellovirus | 25 | 25 | 13 |
| Rhinovirus | 8 | 1 | 5 |
| Herpesvirus | 5 | 1 | 2 |
| Picobirnavirus | 4 | 3 | 7 |
| Parvovirus | 2 | 0 | 0 |
| Enterovirus | 1 | 1 | 0 |
| Astrovirus | 1 | 0 | 0 |
| Calicivirus | 1 | 1 | 1 |
| Smacovirus | 1 | 0 | 0 |
| Parainfluenza virus | 1 | 1 | 1 |
| Rotavirus | 1 | 0 | 0 |
| Cardiovirus | 1 | 0 | 0 |
| Pegivirus | 1 | 0 | 0 |
| Bocavirus | 0 | 1 | 1 |
| Coronavirus | 0 | 0 | 1 |
| Adenovirus | 0 | 0 | 1 |
| Parvovirus | 0 | 0 | 1 |
| Kobuvirus | 0 | 0 | 1 |
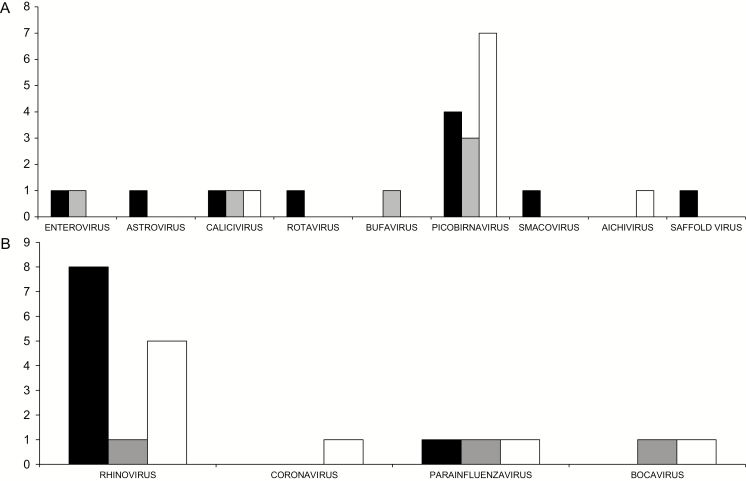
We detected sequences of several viruses associated with outbreaks of gastrointestinal disease, including noroviruses and sapoviruses (family Caliciviridae), astroviruses (Astroviridae), rotaviruses (Reoviridae), and picornaviruses (Picornaviridae).18–23 Although present in baseline UC cases, they were not exclusively associated with UC (Fig. 2A). We detected only 1 baseline UC sample with calicivirus (sapovirus) sequences. This sample also contained sequences of Saffold virus, a picornavirus associated with acute gastroenteritis.24–26 One baseline UC sample each contained sequences of an astrovirus, a rotavirus and an enterovirus. Norovirus sequences were found in 1 follow-up UC and 1 IBS control sample.
Figure 2.
Enteric and respiratory viruses identified by VirCapSeq-VERT. A, The type and number of viruses associated with the gastrointestinal tract. B, The type and number of viruses associated with the respiratory tract. Black bars represent baseline cases, gray bars represent follow-up, and clear bars represent IBS controls.
Another group of viruses often detected in human stool specimens are picobirnaviruses (family Picobirnaviridae).27,28 The association of picobirnaviruses with human disease is unknown, although there have been reports of picobirnavirus-linked gastroenteritis in immunocompromised patients. We identified 4 picobirnavirus-positive samples in the baseline UC cases (Fig. 2A). Sequences of these viruses were present in 3 UC follow-up samples and 7 IBS control samples.
Herpesviruses have been reported in human feces and proposed to have a role in the pathogenesis of UC.29–31 We detected a higher number of herpesvirus-positive samples in UC baseline subjects (n = 5) than follow-up UC subjects (n = 1) or IBS control subjects (n = 2). Between group differences were not statistically significant.
We also detected viruses typically associated with the respiratory tract, which can pass through the digestive system and be present in stool specimens.32–34 Eight baseline UC samples were positive for rhinovirus (HRV) sequences: 5 HRV-A and 3 HRV-B (Fig. 2B). Although only 1 rhinovirus was detected in the follow-up UC samples, 5 control IBS samples also contained a HRV. In addition to HRV, we detected the presence of several other RNA viruses associated with the respiratory tract, including human parainfluenza viruses (1 sample positive in each of the baseline UC, follow-up UC, and IBS control cohorts) and a coronavirus, adenovirus, and bocavirus, each present in a single IBS control.
Discussion
We employed VirCapSeq-VERT to examine the fecal virome of children with UC and IBS with the objective of investigating whether viral infection could trigger the onset of UC or increase the severity of illness. We identified sequences of viruses that represented 30 viral families and taxonomically unclassified viruses. The vertebrate virus sequences detected most frequently represented gyroviruses (genus Gyrovirus, family Anelloviridae). Gyroviruses include chicken anemia virus (CAV) and several novel viruses (designated Gyrovirus 1 through Gyrovirus 4) identified in chickens and humans.15,16,35 Domestic birds are the presumed hosts of gyroviruses, and CAV is ubiquitous in poultry. CAV sequences have been reported in 2% and 51% of human fecal specimens.36–38 Chicken anemia virus can cause immunosuppression in chickens. There is no evidence that it can infect humans. Accordingly, we and others posit that the presence of CAV sequences in human feces is due to ingestion of CAV-infected poultry.39–41
Although we did not identify a single virus as a candidate trigger for UC, we identified a higher prevalence of anellovirus sequences in feces of children with UC relative to IBS controls. Our results are similar to those reported by Norman et al, where an increase in anellovirus sequences was detected in patients with IBD.6 To what extent anelloviruses can trigger or influence the course of human disease is unclear. Anelloviruses are ubiquitous in humans, although their exact prevalence is difficult to estimate because their extremely high sequence divergence impedes epidemiological surveys.42 It is estimated that more than 90% of the human worldwide population is infected with at least 1 type of anellovirus.42, 43 Although they have been detected in fluid and tissue samples representing a wide range of human diseases, they are also found in in healthy individuals.43–49 Some studies suggest that anelloviruses may impact the severity of inflammation through the regulation of inflammatory cytokine homeostasis. It has been proposed that a higher prevalence of anelloviruses may alter the balance of proinflamatory cytokines and that anelloviruses may indirectly augment disease.43 Further studies will be required to determine whether anelloviruses impact the course of UC.
We enrolled UC patients at diagnosis and before treatment, avoiding potential confounding effects of therapy upon the stool virome. However, enrollment was limited to only 70 UC patients with moderate to severe disease activity. We do not know, therefore, whether our findings apply to those with mild disease. We do not have detailed dietary history on the UC subjects because their inclusion in PROTECT only began at their diagnostic colonoscopy. We recognize other limitations in our study. Adolescents with IBS—and not healthy children—were used as controls. Though there are no data suggesting alterations in stool virome in IBS, an altered microbiome has been reported in IBS.50 We only analyzed stool specimens and not rectal biopsy tissue, potentially limiting viral detection. A potential limitation of our capture approach is that it is based on probes designed from viral sequences that have been deposited in Genbank. While we cannot exclude the possibility that yet uncharacterized viruses with extreme genetic diversity would escape detection, VirCapSeq-VERT can detect novel viruses with as little as 60% nucleotide identity to viruses present in the probe set. We also acknowledge that this pilot survey represents a limited sample set and is not a prospective full scale epidemiologic study. Nonetheless, our findings do not support a role of an active viral infection in the onset of UC and may shift the focus toward other potential infectious, genetic, or environmental triggers.
Supplementary Material
Acknowledgments
The authors wish to thank the PROTECT investigators, research coordinators, and patients and families for their help in completing this study.
Supported by: NIDDK 5U01DK095745.
References
- 1. Gower-Rousseau C, Dauchet L, Vernier-Massouille G, et al. The natural history of pediatric ulcerative colitis: a population-based cohort study. Am J Gastroenterol. 2009;104:2080–2088. [DOI] [PubMed] [Google Scholar]
- 2. Fumery M, Duricova D, Gower-Rousseau C, et al. Review article: the natural history of paediatric-onset ulcerative colitis in population-based studies. Aliment Pharmacol Ther. 2016;43:346–355. [DOI] [PubMed] [Google Scholar]
- 3. Hyams JS, Davis S, Mack DR, et al. Factors associated with early outcomes following standardised therapy in children with ulcerative colitis (PROTECT): a multicentre inception cohort study. Lancet Gastroenterol Hepatol. 2017;2:855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schechter A, Griffiths C, Gana JC, et al. Early endoscopic, laboratory and clinical predictors of poor disease course in paediatric ulcerative colitis. Gut. 2015;64:580–588. [DOI] [PubMed] [Google Scholar]
- 5. Turner D, Mack D, Leleiko N, et al. Severe pediatric ulcerative colitis: a prospective multicenter study of outcomes and predictors of response. Gastroenterology. 2010;138:2282–2291. [DOI] [PubMed] [Google Scholar]
- 6. Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fernandes MA, Verstraete SG, Phan TG, et al. Enteric virome and bacterial microbiota in children with ulcerative colitis and Crohn disease. J Pediatr Gastroenterol Nutr. 2019;68:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nusbaum DJ, Sun F, Ren J, et al. Gut microbial and metabolomic profiles after fecal microbiota transplantation in pediatric ulcerative colitis patients. FEMS Microbiol Ecol. 2018;94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Briese T, Kapoor A, Mishra N, et al. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. Mbio. 2015;6:e01491–e01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cummings MJ, Tokarz R, Bakamutumaho B, et al. Precision surveillance for viral respiratory pathogens: virome capture sequencing for the detection and genomic characterization of severe acute respiratory infection in Uganda. Clin Infect Dis. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bousvaros A, Antonioli DA, Colletti RB, et al. Differentiating ulcerative colitis from Crohn disease in children and young adults: report of a working group of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the Crohn’s and Colitis Foundation of America. J Pediatr Gastroenterol Nutr. 2007;44:653–74. [DOI] [PubMed] [Google Scholar]
- 12. Hyams JS, Di Lorenzo C, Saps M, et al. Functional disorders: children and adolescents. Gastroenterology. 2016. [DOI] [PubMed] [Google Scholar]
- 13. Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 14. Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 15. Jiménez-Melsió A, Parés S, Segalés J, et al. Detection of porcine anelloviruses in pork meat and human faeces. Virus Res. 2013;178:522–524. [DOI] [PubMed] [Google Scholar]
- 16. Zhou Y, Wylie KM, El Feghaly RE, et al. Metagenomic approach for identification of the pathogens associated with diarrhea in stool specimens. J Clin Microbiol. 2016;54:368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Family—Anelloviridae. In: King AMQ, Adams M, Carstens EB, Lefkowitz EJ, eds. Virus Taxonomy. San Diego: Elsevier; 2012:331–341. [Google Scholar]
- 18. Boros Á, Raáb M, Károly É, et al. A cluster of salivirus A1 (picornaviridae) infections in newborn babies with acute gastroenteritis in a neonatal hospital unit in Hungary. Arch Virol. 2016;161:1671–1677. [DOI] [PubMed] [Google Scholar]
- 19. Finkbeiner SR, Li Y, Ruone S, et al. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol. 2009;83:10836–10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCarthy KS, Guntapong R, Thattiyaphong A, et al. Outbreak of norovirus gastroenteritis infection, Thailand. Southeast Asian J Trop Med Public Health. 2013;44:409–416. [PubMed] [Google Scholar]
- 21. Monica B, Ramani S, Banerjee I, et al. Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, South India. J Med Virol. 2007;79:544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pacilli M, Cortese MM, Smith S, et al. Outbreak of gastroenteritis in adults due to rotavirus genotype G12P[8]. Clin Infect Dis. 2015;61:e20–e25. [DOI] [PubMed] [Google Scholar]
- 23. Verma H, Chitambar SD, Gopalkrishna V. Circulation of aichi virus genotype B strains in children with acute gastroenteritis in India. Epidemiol Infect. 2011;139:1687–1691. [DOI] [PubMed] [Google Scholar]
- 24. Khamrin P, Thongprachum A, Kikuta H, et al. Three clusters of saffold viruses circulating in children with diarrhea in Japan. Infect Genet Evol. 2013;13:339–343. [DOI] [PubMed] [Google Scholar]
- 25. Nielsen AC, Gyhrs ML, Nielsen LP, et al. Gastroenteritis and the novel picornaviruses aichi virus, cosavirus, saffold virus, and salivirus in young children. J Clin Virol. 2013;57:239–242. [DOI] [PubMed] [Google Scholar]
- 26. Ren L, Gonzalez R, Xiao Y, et al. Saffold cardiovirus in children with acute gastroenteritis, Beijing, China. Emerg Infect Dis. 2009;15:1509–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun G, Zang Q, Gu Y, et al. Viral metagenomics analysis of picobirnavirus-positive feces from children with sporadic diarrhea in China. Arch Virol. 2016;161:971–975. [DOI] [PubMed] [Google Scholar]
- 28. Wilhelmi I, Roman E, Sánchez-Fauquier A. Viruses causing gastroenteritis. Clin Microbiol Infect. 2003;9:247–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopes S, Andrade P, Conde S, et al. Looking into enteric virome in patients with IBD: defining guilty or innocence? Inflamm Bowel Dis. 2017;23:1278–1284. [DOI] [PubMed] [Google Scholar]
- 30. Magro F, Santos-Antunes J, Albuquerque A, et al. Epstein-barr virus in inflammatory bowel disease-correlation with different therapeutic regimens. Inflamm Bowel Dis. 2013;19:1710–1716. [DOI] [PubMed] [Google Scholar]
- 31. Verdonk RC, Haagsma EB, Kleibeuker JH, et al. Cytomegalovirus infection increases the risk for inflammatory bowel disease. Am J Pathol. 2010;176:3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harvala H, McIntyre CL, McLeish NJ, et al. High detection frequency and viral loads of human rhinovirus species A to C in fecal samples; diagnostic and clinical implications. J Med Virol. 2012;84:536–542. [DOI] [PubMed] [Google Scholar]
- 33. Paloniemi M, Lappalainen S, Salminen M, et al. Human bocaviruses are commonly found in stools of hospitalized children without causal association to acute gastroenteritis. Eur J Pediatr. 2014;173:1051–1057. [DOI] [PubMed] [Google Scholar]
- 34. Wootton SH, Aguilera EA, Wanger A, et al. Detection of NH1N1 influenza virus in nonrespiratory sites among children. Pediatr Infect Dis J. 2014;33:95–96. [DOI] [PubMed] [Google Scholar]
- 35. Cavanaugh CE, Muscat PL, Telford SR III, et al. Fatal deer tick virus infection in maine. Clin Infect Dis. 2017;65:1043–1046. [DOI] [PubMed] [Google Scholar]
- 36. Phan TG, Li L, O’Ryan MG, et al. A third gyrovirus species in human faeces. J Gen Virol. 2012;93:1356–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang MB, Chang HM, Wu WC, et al. First detection of chicken anemia virus and norovirus genogroup II in stool of children with acute gastroenteritis in Taiwan. Southeast Asian J Trop Med Public Health. 2016;47:416–423. [PubMed] [Google Scholar]
- 38. Chu DK, Poon LL, Chiu SS, et al. Characterization of a novel gyrovirus in human stool and chicken meat. J Clin Virol. 2012;55:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeurissen SH, Janse ME, Van Roozelaar DJ, et al. Susceptibility of thymocytes for infection by chicken anemia virus is related to pre- and posthatching development. Dev Immunol. 1992;2:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ragland WL, Novak R, El-Attrache J, et al. Chicken anemia virus and infectious bursal disease virus interfere with transcription of chicken IFN-alpha and IFN-gamma mRNA. J Interferon Cytokine Res. 2002;22:437–441. [DOI] [PubMed] [Google Scholar]
- 41. McConnell CD, Adair BM, McNulty MS. Effects of chicken anemia virus on cell-mediated immune function in chickens exposed to the virus by a natural route. Avian Dis. 1993;37:366–374. [PubMed] [Google Scholar]
- 42. Spandole S, Cimponeriu D, Berca LM, et al. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol. 2015;160:893–908. [DOI] [PubMed] [Google Scholar]
- 43. Shulman LM, Davidson I. Viruses with circular single-stranded DNA genomes are everywhere! Annu Rev Virol. 2017;4:159–180. [DOI] [PubMed] [Google Scholar]
- 44. Sherman KE, Rouster SD, Feinberg J. Prevalence and genotypic variability of TTV in HIV-infected patients. Dig Dis Sci. 2001;46:2401–2407. [DOI] [PubMed] [Google Scholar]
- 45. Touinssi M, Gallian P, Biagini P, et al. TT virus infection: prevalence of elevated viraemia and arguments for the immune control of viral load. J Clin Virol. 2001;21:135–141. [DOI] [PubMed] [Google Scholar]
- 46. Zhong S, Yeo W, Tang MW, et al. Gross elevation of TT virus genome load in the peripheral blood mononuclear cells of cancer patients. Ann N Y Acad Sci. 2001;945:84–92. [DOI] [PubMed] [Google Scholar]
- 47. Girard C, Ottomani L, Ducos J, et al. High prevalence of torque teno (TT) virus in classical kaposi’s sarcoma. Acta Derm Venereol. 2007;87:14–17. [DOI] [PubMed] [Google Scholar]
- 48. Li L, Deng X, Da Costa AC, et al. Virome analysis of antiretroviral-treated HIV patients shows no correlation between T-cell activation and anelloviruses levels. J Clin Virol. 2015;72:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Young JC, Chehoud C, Bittinger K, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am J Transplant. 2015;15:200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vich Vila A, Imhann F, Collij V, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.