On 11 February 2003, the Chinese Ministry of Health notified the World Health Organization (WHO) of an outbreak of atypical pneumonia that likely emerged in Guandong Province, China, in November 2002.1 During late February to early March 2003, clusters of atypical pneumonia were recognized in Vietnam, Hong Kong, Canada, and Singapore.2–6 Epidemiological investigations revealed that the index patients for each of these clusters had stayed on the ninth floor of a hotel in Hong Kong on 21–22 February (Figure 1). Further investigation indicated that the likely source of the outbreak was a physician from Guandong Province (Case A) who stayed on the same hotel floor on 21 February. This physician had cared for patients affected by the respiratory disease outbreak and he had been symptomatic with a febrile, respiratory illness since 15 February. This dramatic chain of transmission brought to the world's attention this new respiratory disease, called severe acute respiratory syndrome (SARS), and clearly illustrated the potential for SARS to spread extensively from a single infected person and to rapidly disseminate globally through air travel. The WHO issued an historic global alert7 and, together with its international partners, initiated a rapid and intense response to this global public health emergency. The response led within 2 weeks to the identification of the aetiological agent, SARS-associated coronavirus (SARS-CoV),8–11 and to a series of decisive and effective containment efforts that interrupted the last chain of human transmission in less than 4 months.12 In this article, we review what has been learned about the aetiological agent and its pathogenesis and pathology, clinical manifestations, epidemiology, and diagnosis plus strategies for control of SARS.
Figure 1.
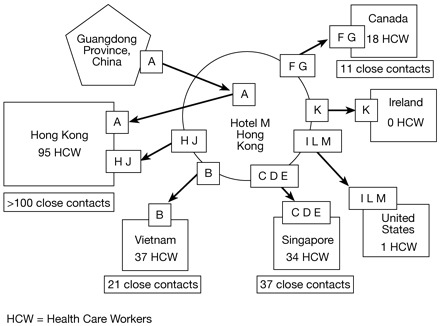
Chain of transmission of severe acute respiratory syndrome (SARS) among guests at Hotel M—Hong Kong, 2003 (Source: MMWR 2003;52:241–48). A represents the index patient (physician from Guandong Province) and B-J represent individual patients that were affected in the outbreak. HCW = health care worker(s)
Aetiological agent
SARS-CoV is an enveloped, positive-stranded, RNA virus in the Coronaviridae family (Figure 2). Coronaviruses are associated with a variety of enteric and respiratory disease syndromes in several animal species. The two recognized human coronaviruses other than SARS-CoV, OC 43, and 229 E, have definitively been associated only with upper respiratory illness. SARS-CoV was first cultured in Vero E6 cells from autopsy tissue, bronchoalveolar lavage, and nasal swab samples obtained from patients hospitalized in Hong Kong, Canada, Germany, and Singapore, with identification by electron microscopy and complete genomic sequencing. The aetiological role of SARS-CoV was confirmed by the replication of an illness similar to that seen among humans in experimentally infected cynomolgus macaques, followed by re-isolation of the virus and detection of a specific immune response in the infected macaques.13,14 The genomic sequence of SARS-CoV is quite distinct from that of other human and animal coronaviruses,15,16 suggesting that the virus has likely circulated in its natural reservoir for a considerable period of time.
Figure 2.
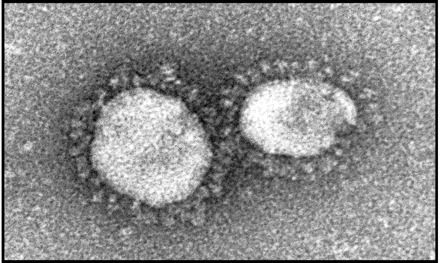
Electron micrograph of SARS-associated coronavirus
The search for the origins of SARS-CoV and its potential reservoir(s) is ongoing. As many as 42% of the early SARS cases in Guandong occurred among people who were involved in animal trade or in food preparation,17 and people involved in animal trade in Guandong were more likely to have antibodies to SARS-CoV than those who did not trade animals or general population controls.18 In addition, a coronavirus with 99% homology with human SARS-CoV isolates was recovered from civet cats and a racoon dog sold live for food in markets in Guangdong.19 Collectively, these observations support the hypothesis that SARS-CoV was first transmitted from wild animals used for food to humans, with subsequent person-to-person transmission.
Experimental studies have shown that ferrets and domestic cats are also susceptible to infection by SARS-CoV and that they can efficiently transmit the virus to previously uninfected animals that are housed with them.20 Further clues to the animal reservoir for SARS-CoV might be provided by studies of wild and domesticated animals from animal markets in Guandong and the genomic sequence analysis of any viral isolates obtained from these species as well as from human patients. Studies have also demonstrated the presence of antibody to SARS-CoV or a SARS-CoV-like virus in a small proportion of healthy adults in Hong Kong who had sera banked in May 2001, at least 2 years prior to the SARS outbreak.21 This finding suggests that SARS may have been transmitted periodically in the past from animals to humans and the virus may have evolved and adapted to human infection and transmission22 or that chance events resulting in efficient transmission led to the outbreak in 2003.
Pathogenesis and pathology
The site of initial infection with SARS-CoV is unknown and the pathogenesis of SARS is not completely understood. Angiotensin-converting enzyme 2 (ACE2) was recently identified as a functional SARS-CoV receptor,23 and ACE2 is efficiently expressed in lung, heart, kidney, and gastrointestinal tissue in humans. SARS-CoV has been isolated in cell culture in autopsy specimens from lung, intestinal, and kidney tissue of patients who died of SARS.24,25 SARS-CoV RNA can also be detected by reverse transcription-polymerase chain reaction (RT-PCR) assays in the plasma and serum of more than 50% of patients during the first week of illness.26
Pathological findings in the lungs of SARS patients during the first 10 days of illness include pneumocyte proliferation and desquamation, hyaline membrane formation, mixed inflammatory infiltrate, and intra-alveolar oedema.27,28 Increased numbers of interstitial and alveolar macrophages, with focal haemophagocytosis in interstitial macrophages, have also been described. In cases of longer duration, diffuse alveolar damage, squamous metaplasia, and multinucleated giant cells, of macrophage origin, are observed. Biopsy and autopsy specimens from the colon and terminal ileum of SARS patients show relatively normal architecture with no evidence of villous atrophy or inflammatory infiltrates.29 No distinct histopathological changes have been described in other tissues.
The role of cytokines and immunopathogenic mechanisms in SARS are being investigated. Lymphopaenia is common among patients with SARS. While CD4 T lymphocytes counts are reduced in nearly all SARS patients, reductions in circulating levels of CD8 T lymphocytes, B lymphocytes, and natural killer cells are also common.30,31 CD4 and CD8 lymphocyte counts fall early in the course of illness, and low CD4 and CD8 levels have been associated with more severe illness. The presence of lymphopaenia and pathological manifestations of diffuse alveolar damage, destruction of the white pulp of the spleen, and haemophagocytosis suggest that proinflammatory cytokines released by stimulated alveolar macrophages may have a major role in the pathogenesis of SARS.27
Death from SARS usually occurs late in the course of illness (>1 week after onset) and has been attributed to adult respiratory distress syndrome, multiorgan failure, thromboembolic complications, secondary infections, and septic shock. Co-infection with human metapneumovirus has been reported among patients with laboratory-confirmed SARS;32 however, the role of co-infection in amplifying the pathogenicity of SARS-CoV is unknown.
Clinical manifestations
SARS-CoV disease has a median incubation period of approximately 4–6 days; most patients become ill within 2–10 days after exposure. The most common initial symptom is fever, often accompanied by headache, myalgias, malaise, chills, and rigor. Respiratory symptoms typically do not begin until 2–7 days after onset of fever, but may be present at onset in up to one-third of patients. Lower respiratory tract symptoms are common and typically include a non-productive cough with later onset of dyspnoea. Upper respiratory symptoms such as rhinorrhoea and sore throat are seen in less than 25% of patients. Diarrhoea has been reported at presentation in approximately 25% of patients, although this symptom was observed in as many as 73% of all patients affected by an outbreak at an apartment complex in Hong Kong that is believed to have resulted from fecal-oral/respiratory transmission of SARS-CoV.33 Respiratory signs such as rales and rhonchi are present in less than one-third of cases, and their severity often does not correlate with the findings seen on chest radiographs. Elderly patients and those with underlying chronic illnesses such as renal failure sometimes present with atypical symptoms, including the absence of fever.34 In general, SARS appears to be milder in children.35,36
Haematological abnormalities include lymphopaenia (70–90% of patients), thrombocytopaenia (30–50%), prolongation of the activated partial thromboplastin time (40–60%), and elevated levels of lactate dehydrogenase (70–90%), alanine or aspartate aminotransferases (20–30%), and creatinine phosphokinase (30–40%). None of these laboratory findings can reliably discriminate between SARS-CoV disease and other atypical pneumonias. Chest radiographs may be normal in up to 30% of patients who are evaluated early in illness, and high-resolution chest computed tomography scans can detect abnormalities in up to two-thirds of patients with normal chest X-rays.37–44 Nearly everyone with SARS-CoV disease develops radiographic evidence of pneumonia by illness day 7–10. The radiographic abnormalities typically begin as isolated, focal, ground glass opacities predominately in a peripheral location, often in the lower lobes, and progress over several days to focal, multifocal, or diffuse consolidation involving additional lobes or both lungs; mediastinal lymphadenopathy, cavitation, atelectasis, and pleural effusions are uncommon.
Some 15–25% of patients with SARS-CoV disease require mechanical ventilation, and about half of these people die despite ventilation therapy. Severity of illness and risk of death from SARS increases with advancing age and in the presence of certain underlying medical conditions, particularly diabetes mellitus.45 The overall case-fatality rate of approximately 10% can increase to >50% in people older than age 60.
Epidemiology
During the 2003 global epidemic, 8098 cases of probable SARS with 774 (9.6%) deaths were reported in 29 countries.46 The countries with the greatest number of reported cases included mainland China (N = 2674), Hong Kong Special Administrative Region (N = 977), Taiwan (N = 218), Singapore (N = 161), and Canada (N = 151). Health care facilities were severely affected and transmission in hospitals was a major factor in the amplification of outbreaks. In addition to health care workers, who accounted for 21% of all cases globally, other patients on the same ward as SARS patients and visitors to the hospital were affected. For example, of 74 cases reported from 15 April to 9 June in Toronto, Canada, 39% occurred among health care workers, 38% resulted from exposure during hospitalization, and 23% occurred among hospital visitors.47 After health care facilities, households of SARS patients were the second most common setting of SARS-CoV transmission. Surveys in Hong Kong and Singapore indicated that 6–8% of people in the households of SARS patients may have been secondarily infected.48,49 The risk of transmission was greatest among those involved in direct patient care or other close contact with the patient. Transmission to casual and social contacts appears to be uncommon, but secondary cases have been documented after exposures in the workplace and on airplanes and other conveyances.50,51
The epidemiology of SARS outbreaks suggests that SARS-CoV is transmitted primarily through droplets and close personal contact, a conclusion that is supported by the finding that surgical masks confer protection against infection.51,52 Studies documenting stability of the virus for days in the environment raise the possibility of fomite transmission, and in a few instances, possible transmission by small-particle aerosols cannot be excluded. In particular, aerosol-generating medical procedures (e.g. endotracheal intubation, bronchoscopy) may be associated with an increased risk of transmission in health care settings.53–56 Given that profuse watery diarrhoea is seen in a significant proportion of patients and SARS-CoV can be shed in large quantities in stool, faeces remain a possible source of virus and fecal-oral or fecal-respiratory spread are the leading hypotheses for a large outbreak affecting more than 300 people at an apartment complex in Hong Kong.9
Mathematical modelling studies indicate that each SARS case infects an average of two to four people.57–59 However, some SARS patients are very efficient in transmitting SARS-CoV to susceptible people under certain circumstances, leading to so-called ‘super-spreading events’.51,53–56,60 Super-spreading events have occurred in a number of settings, including the hotel in Hong Kong, from which the SARS outbreak disseminated globally, and hospitals in many locations. The factors contributing to super-spreading events are not completely understood but may be related to the inherently greater infectiousness of some patients, alternative modes of transmission, or the exposure of large numbers of contacts in an environment conducive to transmission.
Asymptomatic SARS-CoV infections have been documented,61 but they appear to be uncommon and their epidemiological significance in disease transmission has not been established. The risk of transmission from patients appears to be greatest during the second week of illness,51 which correlates with the timing of peak viral load in respiratory secretions.9 The timing of peak infectiousness might also explain, in part, the predilection for SARS-CoV to spread in health care settings, as most patients seek medical care by the end of the first week of illness. Although SARS-CoV RNA can be detected in fecal specimens by RT-PCR for more than a month after the onset of illness, no transmission has been documented more than 10 days after the resolution of fever.
Genome sequencing analyses indicate that SARS-CoV isolates from the outbreaks in Hong Kong, Vietnam, Singapore, Toronto, and Taiwan are closely related and match the viral isolate obtained from the ill physician from Guandong Province, supporting the epidemiological conclusion that each of these outbreaks was directly or indirectly linked to the ill physician.62 On the other hand, SARS-CoV isolates from Guangdong Province exhibit greater diversity and other viruses belonging to genetic lineages different from the global outbreak strain were introduced into Hong Kong prior to March 2003, but these introductions did not lead to large outbreaks. These data raise the question of whether some viral strains may have an inherently greater epidemic potential; this hypothesis needs further evaluation. Alternatively, differences in transmission unrelated to virus strain (i.e. super-spreading events) could explain the presence or absence of outbreaks associated with independent human infections.
Diagnosis
The clinical manifestations of SARS-CoV disease are not sufficiently distinct from those of other respiratory pathogens to permit a reliable differential diagnosis. However, a combination of clinical and epidemiological features can provide clues to the diagnosis of SARS.63 With no documented person-to-person transmission of SARS-CoV worldwide, the predilection for SARS-CoV to cause unusual clusters of pneumonia in health care settings provides a means to focus on surveillance efforts. If the re-emergence of SARS-CoV is confirmed, then a history of exposure to known SARS case-patients or SARS-affected areas may be helpful in early recognition of patients.
The laboratory diagnosis of SARS-CoV is based on either detection of viral RNA in clinical specimens or the finding of antibodies directed against SARS-CoV in serum. Viral isolation is not recommended for routine diagnosis because of its low sensitivity and the biosafety hazards it poses. Available real-time RT-PCR assays for SARS-CoV are highly specific and can detect as few as 1–10 copies of viral RNA in clinical specimens, but a low viral load in respiratory and fecal specimens during the first week of illness limits the clinical sensitivity and utility of these assays. Limited data suggest that SARS-CoV RNA can be detected in the serum of more than 50% of SARS patients during the first week of illness.26 Because of the potential for contamination, positive RT-PCR results should be confirmed by testing a second specimen in a reference laboratory.64 The identification of serological antibody to SARS-CoV remains the reference standard for confirmation of SARS-CoV infection. However, antibody is usually detectable only after the first week of illness and some patients may not mount an antibody response for up to 21–28 days after illness onset. It is important to note that neither a negative RT-PCR test nor a negative test of serum obtained <28 days after illness onset can reliably exclude infection with SARS-CoV.
Body fluids that should be submitted for testing from patients with suspected SARS include respiratory tract secretions, serum, whole blood, and stool. Lower respiratory tract specimens (e.g. sputum, bronchial alveolar lavage fluid) appear to have higher yield than upper respiratory tract specimens (e.g. nasal or pharyngeal aspirates or swabs). Collection of multiple specimens of different types is likely to increase the overall diagnostic yield, and different specimens will have a greater diagnostic yield at different times in illness (Table 1).
Table 1.
Selection of specimens for SARS-CoV testing, by time from onset of symptoms
|
Specimen
|
<1 week after symptom onset
|
1–3 weeks after symptom onset
|
>3 weeks after symptom onset
|
|---|---|---|---|
| Serum | ++ | ++ | ++ |
| Blood plasma | ++ | + | − |
| Respiratory (BAL, sputum, nasal aspirate and wash, np/op swabs) | + | ++ | + |
| Stool | + | ++ | ++ |
np/op = nasopharyngeal/oropharyngeal; BAL = bronchioalveloar lavage.
SARS-CoV test results should always be considered in the context of clinical and epidemiological findings. In the setting of no SARS human-to-human transmission worldwide, the positive predictive value of a SARS-CoV test is extremely low, and thus any positive laboratory result must be interpreted with extreme caution because of its implications for global public health.
Diagnostic assays for other respiratory pathogens may be helpful in differentiating SARS-CoV disease from other illnesses, but SARS patients may be simultaneously infected with SARS-CoV and another respiratory pathogen. Factors that should be considered in the evaluation of patients with dual infections include the strength of the epidemiological exposure criteria for SARS-CoV disease, the specificity of the alternate diagnostic test, and the compatibility of the clinical presentation and course of illness with the alternative diagnosis.
Strategies for control of SARS
In the absence of a vaccine, effective drugs, or natural immunity to SARS-CoV, the key to controlling SARS is the classic public health control strategy of case identification and containment. These measures proved effective and were associated with cessation of transmission throughout the world by July 2003.
The key to containment efforts is a surveillance system that provides ready access to timely information on the number of new cases, the likely source of exposure for cases, the number of cases not previously identified as contacts, and the number of contacts with high-risk exposures to known cases (potential prospective cases). Since SARS anywhere has implications locally, nationally, and globally, it is essential that the health care and public health communities exchange information on individual SARS cases and the status of SARS transmission in the community and globally.
Once a potential case of SARS is detected, appropriate infection control measures must be implemented immediately to prevent transmission. SARS patients often require hospitalization because of the severity of their illness; those with less severe illness are sometimes hospitalized to ensure that strict isolation procedures are followed. In other settings, patients have been managed in residential settings after evaluation of their suitability for this purpose. Because the possibility of airborne transmission cannot be excluded, patients who require hospitalization should preferably be admitted to an airborne infection isolation room or specially adapted SARS unit or ward where they can be treated safely and appropriately. In some settings, a lack of isolation rooms and/or a need to concentrate infection control efforts and resources may lead to a strategy of grouping patients in individual rooms on the same floor, which proved effective in many settings in the 2003 outbreak. Health care workers should strictly adhere to use of appropriate personal protective equipment, and the potential for transmission to other non-SARS patients and hospital visitors should be minimized through administrative controls.
Contact tracing, the identification of people potentially exposed to a case of SARS, is essential to prevent spread. This provides a means to focus control efforts on people who are at high risk of SARS, to identify people early in the course of their illness, and to implement control measures before they can spread the virus to others. Contact tracing, evaluation for illness, and monitoring should be an immediate high priority to maximize the chance to rapidly control an outbreak. During the 2003 outbreak, quarantine of exposed people was one of the contact management strategies used to prevent inadvertent SARS-CoV exposures by separating those exposed from the unexposed. This potentially decreased the interval between the onset of symptoms and the institution of control measures.
Other key components of an effective SARS control strategy include the following: (1) systems for rapid and frequent communication of crucial information about SARS; (2) education and training of public health and health care workers about the clinical and epidemiological aspects of SARS, appropriate use and interpretation of laboratory tests, and best practices for effective use of infection control strategies; and (3) efficient information technology systems that provide a means to link clinical, epidemiological, and laboratory data on SARS cases and to disseminate this information locally, nationally, and globally, and systems that allow rapid identification, tracking, evaluation, and monitoring of contacts of SARS cases.
Conclusions
The emergence of SARS-CoV dramatically illustrates the potential for a new disease to suddenly appear and spread, leading to widespread health, social, and economic consequences. Fortunately, the experience also demonstrates the power of traditional public health measures—including surveillance, infection control, isolation, and quarantine—to contain and control a SARS outbreak. It is not possible to predict when and where SARS-CoV will reappear and whether it will cause similar outbreaks in the future. Possible sources of virus for a re-emergence of SARS-CoV include its original animal reservoir, persistent infection in humans, or the laboratory. The recently detected SARS-CoV cases in China are hypothesized to represent animal-to-human transmission,65,66 and each of the laboratory-acquired cases in Singapore and Taiwan highlight the need for stringent biosafety precautions.67,68 To achieve the type of swift and decisive response that is required to control a SARS outbreak, we must be prepared and have learned from the many lessons of the 2003 SARS outbreak. Preparation for a response to an outbreak of SARS requires co-ordination and co-operation among public health, health care, and other emergency response entities at all levels of government. Investments in SARS preparedness are likely to yield additional dividends in preparedness to battle other emerging infectious diseases and other threats to public health.
References
- 1.Zhong NS, Zheng BJ, Li YM et al Epidemiological and aetiological studies of patients with severe acute respiratory syndrome (SARS) from Guangdong in February 2003. Lancet 2003;362:1353–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N, Hui D, Wu A et al A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003;348:1986–94. [DOI] [PubMed] [Google Scholar]
- 3.Tsang KW, Ho PL, Ooi GC et al A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003;348:1977–85. [DOI] [PubMed] [Google Scholar]
- 4.Poutanen SM, Low DE, Henry B et al Identification of severe acute respiratory syndrome in Canada. N Engl J Med 2003;348:1995–2005. [DOI] [PubMed] [Google Scholar]
- 5.Booth CM, Matukas LM, Tomlinson GA et al Clinical features and short-term outcomes of 144 patients with SARS in the Greater Toronto area. JAMA 2003;289:2801–09. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Severe acute respiratory syndrome—Singapore, 2003. MMWR 2003;52:405–11.12807088 [Google Scholar]
- 7.World Health Organization. WHO issues a global alert about cases of atypical pneumonia [Press release, 12 March 2003]. Available at http://www.who.int/csr/sars/archive/2003_03_12/en/
- 8.Stohr K, World Health Organization Multicentre Collaborative Network for SARS Diagnosis. A multicentre collaboration to investigate the cause of severe acute respiratory syndrome. Lancet 2003;361:1730–33.12767752 [Google Scholar]
- 9.Peiris JSM, Lai ST, Poon LLM et al Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drosten C, Gunther S, Preiser W et al Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967–76. [DOI] [PubMed] [Google Scholar]
- 11.Ksiazek TG, Erdman D, Goldsmith C et al A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953–66. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. Update 96—Taiwan, China: SARS transmission interrupted in last outbreak area. http://www.who.int/csr/don/2003_07_05/en/
- 13.Fouchier RAM, Kuiken T, Schutten M et al Koch's postulates fulfilled for SARS virus. Nature 2003;423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuiken T, Fouchier RAM, Schutten M et al Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 2003;362:263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marra MA, Jones SJM, Astell CR et al The genome sequence of the SARS-associated coronavirus. Science 2003;300:1399–404. [DOI] [PubMed] [Google Scholar]
- 16.Rota PA, Oberste MS, Monroe SS et al Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003;300:1394–99. [DOI] [PubMed] [Google Scholar]
- 17.Breiman RF, Evans MR, Priser W et al Role of China in the quest to define and control severe acute respiratory syndrome. Emerg Infect Dis 2003;9:1037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention. Prevalence of IgG antibody to SARS-associated coronavirus in animal traders—Guangdong province, China. MMWR 2003;52:986–87. [PubMed] [Google Scholar]
- 19.Guan Y, Zheng BJ, He YQ et al Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science 2003;302:276–78. [DOI] [PubMed] [Google Scholar]
- 20.Martina BE, Haagmans BL, Kuiken T et al SARS virus infection of cats and ferrets. Nature 2003;425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng BJ, Guan Y, Wong KH et al SARS-related virus predating SARS outbreak, Hong Kong. Emerg Infect Dis [serial online] 2004 Feb [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol10no2/03-0533.htm [DOI] [PMC free article] [PubMed]
- 22.The Chinese SARS Molecular Epidemiology Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Published online January 29 Science 2004; 10.1126/science.1092002 (Science Express Reports). [DOI] [PubMed]
- 23.Li W, Moore MJ, Vasilieva N et al Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003;426:450–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farcas GA, Mazzuli T, Willey BM et al Fatal severe acute respiratory syndrome is associated with multiorgan involvement by coronavirus (SARS-CoV). Presented at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, 2013.
- 25.Nakajima N, Asahi-Ozaki Y, Nagata N et al SARS coronavirus-infected cells in lung detected by new in situ hybridization technique. Jpn J Infect Dis 2003;56:139–41. [PubMed] [Google Scholar]
- 26.Grant PR, Garson JA, Tedder RS, Chan PK, Tam JS, Sung JJ. Detection of SARS coronavirus in plasma by real-time RT-PCR. N Engl J Med 2003;349:2468–69. [DOI] [PubMed] [Google Scholar]
- 27.Nicholls JM, Poon LL, Lee KC et al Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003;361:1773–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franks TJ, Chong PY, Chui P et al Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum Pathol 2003;34:743–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung WK, To KF, Chan PK et al Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 2003;125:1011–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui W, Fan Y, Wu W, Zhang F, Wang JY, Ni AP. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis 2003;37:857–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong RS, Wu A, To KF et al Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ 2003;326:1358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan PKS, Tam JS, Lam CW et al Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg Infect Dis 2003;9:1058–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris JSM, Chu CM, Cheng VCC et al Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 2003;361:1767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tee AKH, Oh HML, Hui KP et al Atypical SARS in geriatric patient. Emerg Infect Dis [serial online] 2004 Feb [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol10no2/03-0322.htm [DOI] [PMC free article] [PubMed]
- 35.Bitnun A, Allen U, Heurter H et al Children hospitalized with severe acute respiratory syndrome-related illness in Toronto. Pediatrics 2003;112:e261. [DOI] [PubMed] [Google Scholar]
- 36.Leung TF, Wong GW, Hon KL, Fok TF. Severe acute respiratory syndrome (SARS) in children: epidemiology, presentation and management. Paediatr Respir Rev 2003;4:334–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong KT, Antonio GE, Hui DS et al Thin-section CT of severe acute respiratory syndrome: evaluation of 73 patients exposed to or with the disease. Radiology 2003;228:395–400. [DOI] [PubMed] [Google Scholar]
- 38.Wong KT, Antonio GE, Hui DS et al Severe acute respiratory syndrome: radiographic appearances and pattern of progression in 138 patients. Radiology 2003;228:401–06. [DOI] [PubMed] [Google Scholar]
- 39.Antonio GE, Wong KT, Hui DS et al Thin-section CT in patients with severe acute respiratory syndrome following hospital discharge: preliminary experience. Radiology 2003;228:810–15. [DOI] [PubMed] [Google Scholar]
- 40.Nicolaou S, Al-Nakshabandi NA, Muller NL. Radiologic manifestations of severe acute respiratory syndrome. N Engl J Med 2003;348:2006. [DOI] [PubMed] [Google Scholar]
- 41.Nicolaou S, Al-Nakshabandi NA, Muller NL. SARS: imaging of severe acute respiratory syndrome. Am J Roentgenol 2003;180:1247–49. [DOI] [PubMed] [Google Scholar]
- 42.Muller NL, Ooi GC, Khong PL, Nicolaou S. Severe acute respiratory syndrome: radiographic and CT findings. Am J Roentgenol 2003;181:3–8. [DOI] [PubMed] [Google Scholar]
- 43.Grinblat L, Shulman H, Glickman A, Matukas L, Paul N. Severe acute respiratory syndrome: radiographic review of 40 probable cases in Toronto, Canada. Radiology 2003;228:802–09. [DOI] [PubMed] [Google Scholar]
- 44.Antonio GE, Wong KT, Hui DS et al Imaging of severe acute respiratory syndrome in Hong Kong. Am J Roentgenol 2003;181:11–17. [DOI] [PubMed] [Google Scholar]
- 45.Choi KW, Chau TN, Tsang O et al Outcomes and prognostic factors in 267 patients with severe acute respiratory syndrome in Hong Kong. Ann Intern Med 2003;139:715–23. [DOI] [PubMed] [Google Scholar]
- 46.World Health Organization. Cumulative number of reported probable cases of severe acute respiratory syndrome (SARS). Available at http://www.who.int/csr/sars/country/en/.
- 47.Centers for Disease Control and Prevention. Update: severe acute respiratory syndrome—Toronto, Canada, 2003. MMWR 2003;52:547–50. [PubMed] [Google Scholar]
- 48.Goh DL-M, Lee BW, Chia KS et al Secondary household transmission of SARS, Singapore. Emerg Infect Dis [serial online] 2004 Feb [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol10no2/03-0676.htm [DOI] [PMC free article] [PubMed]
- 49.Lau JTF, Lau M, Kim JH et al Probable secondary infections in households of SARS patients in Hong Kong. Emerg Infect Dis [serial online] 2004 Feb [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol10no2/03-0626.htm [DOI] [PMC free article] [PubMed]
- 50.Olsen SJ, Chang HL, Cheung TY et al Transmission of the severe acute respiratory syndrome on aircraft. N Engl J Med 2003;349:2416–22. [DOI] [PubMed] [Google Scholar]
- 51.Consensus document on the epidemiology of severe acute respiratory syndrome (SARS) WHO/CDS/CSR/GAR/2003.11. Geneva: World Health Organization, 2003. (Accessed 2 February 2004, at http://www.who.int/csr/sars/en/WHOconsensus.pdf.)
- 52.Seto WH, Tsang D, Yung RWH et al Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS). Lancet 2003;361:1519–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varia M, Wilson S, Sarwal S et al Investigation of a nosocomial outbreak of severe acute respiratory syndrome (SARS) in Toronto, Canada. C Med Assoc J 2003;169:285–92. [PMC free article] [PubMed] [Google Scholar]
- 54.Scales DC, Green K, Chan AK et al Illness in intensive care staff after brief exposure to severe acute respiratory syndrome. Emerg Infect Dis 2003;9:1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong T-W, Lee C-K, Tam W et al Cluster of SARS among medical students exposed to single patient, Hong Kong. Emerg Infect Dis [serial online] 2004 Feb [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol10no2/03-0452.htm [DOI] [PMC free article] [PubMed]
- 56.Christian MD, Loutfy M, McDonald LC et al Possible SARS coronavirus transmission during cardiopulmonary resuscitation. Emerg Infect Dis [serial online] 2004 Feb [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol10no2/03-0700.htm [DOI] [PMC free article] [PubMed]
- 57.Lipsitch M, Cohen T, Cooper B et al Transmission dynamics and control of severe acute respiratory syndrome. Science 2003;300:1966–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riley S, Fraser C, Donnelly CA et al Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science 2003;300:1961–66. [DOI] [PubMed] [Google Scholar]
- 59.Dye C, Gay N. Modeling the SARS epidemic. Science 2003;300:1884–85. [DOI] [PubMed] [Google Scholar]
- 60.Shen Z, Ning F, Zhou W et al Superspreading SARS events, Beijing, 2003. Emerg Infect Dis [serial online] 2004 Feb [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol10no2/03-0732.htm [DOI] [PMC free article] [PubMed]
- 61.Lee HKK, Tso EYK, Chau TN, Tsang OTY, Choi KW, Lai TST. Asymptomatic severe acute respiratory syndrome–associated coronavirus infection. Emerg Infect Dis (in press). (Also available at http://www.cdc.gov/ncidod/EID/vol9no11/03-0401.htm.) [DOI] [PMC free article] [PubMed]
- 62.Guan Y, Peiris JS, Zheng B et al Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet 2004;363:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jernigan JA, Low DE, Helfand RF. Combining clinical and epidemiologic features for early recognition of SARS. Emerg Infect Dis [serial online] 2004 Feb [date cited]. Available from: URL: http://www.cdc.gov/ncidod/EID/vol10no2/03-0741.htm [DOI] [PMC free article] [PubMed]
- 64.World Health Organization. WHO SARS International Reference and Verification Laboratory Network: Policy and Procedures in the Inter-Epidemic Period. http://www.who.int/csr/sars/guidelines/en/WHOSARSReferenceLab.pdf
- 65.World Health Organization. Update 4: Review of probable and laboratory-confirmed SARS cases in southern China, 27 January 2004. http://www.who.int/csr/don/2004_01_27/en/
- 66.World Health Organization. New case of laboratory-confirmed SARS in Guangdong, China—update 5, 31 January 2004. http://www.who.int/csr/don/2004_01_31/en/
- 67.World Health Organization. Severe acute respiratory syndrome (SARS) in Singapore, 10 September 2003. http://www.who.int/csr/don/2003_09_10/en/
- 68.World Health Organization. Severe Acute Respiratory Syndrome (SARS) in Taiwan, China, 17 December 2003 http://www.who.int/csr/don/2003_12_17/en/


