Abstract
Loss-of-function mutations in PARK2, the gene encoding the E3 ubiquitin ligase Parkin, are the most frequent cause of recessive Parkinson's disease (PD). Parkin translocates from the cytosol to depolarized mitochondria, ubiquitinates outer mitochondrial membrane proteins and induces selective autophagy of the damaged mitochondria (mitophagy). Here, we show that ubiquitin-specific protease 15 (USP15), a deubiquitinating enzyme (DUB) widely expressed in brain and other organs, opposes Parkin-mediated mitophagy, while a panel of other DUBs and a catalytically inactive version of USP15 do not. Moreover, knockdown of USP15 rescues the mitophagy defect of PD patient fibroblasts with PARK2 mutations and decreased Parkin levels. USP15 does not affect the ubiquitination status of Parkin or Parkin translocation to mitochondria, but counteracts Parkin-mediated mitochondrial ubiquitination. Knockdown of the DUB CG8334, the closest homolog of USP15 in Drosophila, largely rescues the mitochondrial and behavioral defects of parkin RNAi flies. These data identify USP15 as an antagonist of Parkin and suggest that USP15 inhibition could be a therapeutic strategy for PD cases caused by reduced Parkin levels.
INTRODUCTION
Parkinson's disease (PD) is a disorder in which substantia nigra dopaminergic neurons and several other neuronal subpopulations gradually degenerate, causing a wide range of disabling motor and non-motor problems (1). The lifetime risk of developing PD is 1.5% (1). A subset of PD cases are familial. Mutations in PARK2, the gene encoding the cytosolic E3 ubiquitin ligase Parkin, are the most frequent cause of autosomal recessive PD, probably through a loss-of-function mechanism (2). In addition to its role in familial PD, Parkin deficiency may also contribute to the pathogenesis of sporadic PD, as Parkin is progressively inactivated in the aging human brain as a result of posttranslational modifications (3–6).
The phenotypes of Parkin-null flies (7) and mice (8) and fibroblasts from PD patients with PARK2 mutations (9,10) primarily consist of mitochondrial abnormalities. This has long been puzzling, given the cytosolic localization of Parkin. However, recent work in a variety of experimental models has shown that Parkin translocates from the cytosol to depolarized mitochondria and ubiquitinates a large number of mitochondrial outer membrane (MOM) proteins, thus triggering selective macro-autophagic removal of the damaged mitochondria (mitophagy) (11–19).
The actions of E3 ubiquitin ligases are counteracted by deubiquitinating enzymes (DUBs), which catalyze the removal of ubiquitin from substrates. The human proteome contains ∼80 functional DUBs, belonging to 5 subclasses, the largest of which is the ubiquitin-specific protease (USP) family (20). Whether any DUB antagonizes the enzymatic effects of Parkin is currently unknown. Identification of Parkin's antagonistic DUB could lead to an attractive new therapeutic strategy for PD, namely inhibition of this DUB.
Here, we identify USP15 as a DUB that counteracts Parkin-mediated mitochondrial ubiquitination and mitophagy.
RESULTS
USP15 inhibits Parkin-mediated mitophagy through its DUB activity
We have recently used tandem affinity purification coupled to mass spectrometry to search for Parkin-interacting proteins in HEK293 cells expressing His6-FLAG-Parkin (17). Among the Parkin-associated proteins, we identified two DUBs that were absent in the negative control condition (Supplementary Data): USP11 and USP15, two closely related USPs (46% identity and 62% similarity at the amino acid level). Coimmunoprecipitation and western blotting confirmed the interaction of overexpressed Parkin with USP15 (Supplementary Data), but not USP11. As we were unable to coimmunoprecipitate USP15 with endogenous Parkin in HEK293 cells, we assumed that the binding of Parkin and USP15 is weak and not detectable at endogenous Parkin expression levels.
We then asked if there was a functional interaction between Parkin and USP15. To assay Parkin function, we employed a widely used HeLa cell model of Parkin-mediated mitophagy (11,12,15,17). When HeLa cells, which do not express endogenous Parkin, are transfected with Parkin and treated with mitochondrial uncouplers such as carbonyl cyanide m-chlorophenylhydrazone (CCCP) or valinomycin for 24 h, depolarized mitochondria are eliminated by autophagy in a Parkin-dependent manner (11,12,15,17). To search for an effect of DUBs on Parkin-mediated mitophagy, HeLa cells were transfected with Parkin alone or cotransfected with Parkin and USP15 or a panel of other DUBs: USP11, which was present in our initial proteomic screen; USP4, the closest homolog of USP15 (59% sequence identity and 73% similarity at the amino acid level); USP30, the only DUB that is anchored into the MOM, facing the cytosol with its catalytic domain (21); UCH-L1, a DUB for which a genetic link with PD has been suggested (22); Ataxin-3, a DUB reported to bind with Parkin (23); and A20, a DUB involved in the NF-κB pathway (24), in which Parkin has also been implicated (25) (Fig. 1). Interestingly, USP15 strongly inhibited Parkin-mediated mitophagy, while the other DUBs had no effect (Fig. 1A and B). Western blot showed that the expression levels of transfected USP4, USP11, USP30, UCH-L1, Ataxin-3 and A20 were at least as high as that of USP15, ruling out stronger expression of USP15 as the reason for its unique effect (Fig. 1C). The inhibitory effect of USP15 on Parkin-mediated mitophagy was demonstrated using both the MOM protein Tom20 (Fig. 1A and B) and the matrix protein Hsp60 (Supplementary Data) as mitochondrial markers.
Figure 1.
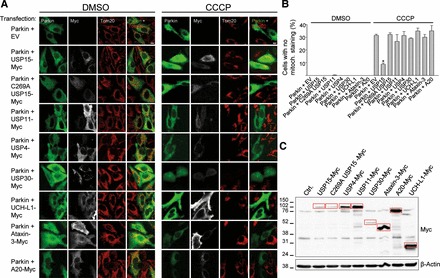
USP15 counteracts Parkin-mediated mitophagy through its deubiquitinating activity. (A) HeLa cells were transfected with Parkin and empty vector (EV) or cotransfected with Parkin and Myc-tagged versions of USP15, the catalytically inactive C269A USP15 mutant, USP11, USP4, USP30, UCH-L1, Ataxin-3 or A20. At 24 h after transfection, cells were treated for 24 h with DMSO or CCCP (10 µm) and immunostained for Parkin, Myc and the mitochondrial marker Tom20. Scale bar, 10 μm. (B) The percentage of Parkin-positive cells (in the condition transfected with Parkin alone) or Parkin- and Myc-positive cells (in the conditions cotransfected with Parkin and DUBs) without detectable Tom20 immunoreactivity was quantified (n ≥ 3). *P < 0.001 compared with CCCP-treated cells transfected with Parkin alone. (C) Anti-Myc WB showing expression levels of the Myc-tagged DUBs (indicated by the red boxes).
USPs are cysteine proteases (20). To determine whether the effect of USP15 on mitophagy depended on its enzymatic activity, we generated a catalytically inactive form of USP15 by replacing the active site cysteine residue at position 269 with alanine (26,27). The C269A mutation abolished the inhibitory effect of USP15 on Parkin-mediated mitophagy, indicating that this effect of USP15 required its deubiquitinating activity (Fig. 1; Supplementary Data).
Knockdown of USP15 enhances mitophagy
Next we used RNAi-mediated knockdown of USP15 (Supplementary Data) to investigate whether endogenous USP15 regulated mitophagy in Parkin-transfected HeLa cells and in two cell models expressing endogenous Parkin: human dopaminergic neuronal SH-SY5Y cells and primary fibroblasts from healthy human subjects (Fig. 2).
Figure 2.
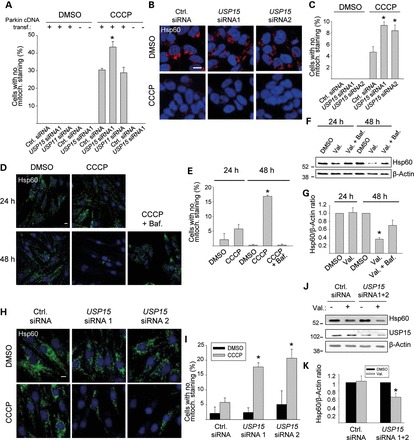
Knockdown of USP15 enhances mitophagy in HeLa cells, SH-SY5Y cells and primary human fibroblasts. (A) HeLa cells were transfected with empty vector or Parkin cDNA and with siRNAs, as indicated, treated with CCCP (10 µm) for 24 h and immunostained for Parkin and Tom20. The percentage of cells (in the conditions that were not transfected with Parkin) or Parkin-positive cells (in the conditions transfected with Parkin) without mitochondrial staining was quantified (n = 3). *P < 0.001 compared with the CCCP-treated condition transfected with control (Ctrl.) siRNA and Parkin cDNA. (B and C) SH-SY5Y cells were transfected with the indicated siRNAs, treated with CCCP (25 µm) for 24 h and immunostained for Hsp60. (C) Quantification of the percentage cells without mitochondrial staining (n = 3). *P < 0.005 compared with the CCCP-treated control siRNA condition. (D–G) Fibroblasts from healthy control 1 were treated with DMSO, CCCP (10 µm), CCCP + bafilomycin (Baf., 100 nm), valinomycin (Val., 1 µm) or valinomycin + bafilomycin for 24 or 48 h, as indicated, followed by anti-Hsp60 immunostaining (D and E) or western blot (F and G). (E) Quantification of the percentage cells without detectable Hsp60 immunoreactivity (n = 3). *P < 0.001 compared with the 48 h DMSO condition. (G) Quantification of the Hsp60/β-Actin ratio on western blot, normalized to this ratio in the DMSO condition (n = 7). *P < 0.005 compared with the 48 h DMSO condition. (H–K) Fibroblasts from control 1 were transfected with the indicated siRNAs and treated with DMSO, CCCP or valinomycin for 24 h, followed by anti-Hsp60 immunostaining (H, I) or western blot (J and K). (I) Quantification of the percentage cells without detectable Hsp60 immunoreactivity (n = 3). * P ≤ 0.01 compared with the CCCP-treated control siRNA condition. (K) Quantification of the Hsp60/β-Actin ratio on western blot, normalized to this ratio in the DMSO condition (n = 5). * P < 0.05 compared with the DMSO condition.
RNAi-mediated reduction of USP15 protein levels in Parkin-transfected HeLa cells enhanced Parkin-mediated mitophagy induced by 24 h exposure to CCCP, while RNAi-mediated knockdown of USP11 had no effect (Fig. 2A; Supplementary Data). Interestingly, siRNA-mediated USP15 knockdown did not result in mitochondrial clearance after CCCP exposure in HeLa cells lacking Parkin (Fig. 2A). Thus, USP15 knockdown enhanced mitophagy but only in the presence of Parkin.
In SH-SY5Y cells, CCCP-induced mitophagy was also markedly promoted by siRNA-mediated knockdown of USP15 (Fig. 2B and C). This mitophagy-promoting effect of USP15 knockdown in SH-SY5Y cells was suppressed by simultaneous siRNA-mediated knockdown of endogenous Parkin protein levels, indicating that the effect of USP15 knockdown on mitophagy was Parkin-dependent (Supplementary Data).
In primary human fibroblasts, exposure to CCCP or valinomycin for 24 h did not induce significant mitochondrial clearance (Fig. 2D–G). However, 48 h treatment with CCCP or valinomycin resulted in substantial loss of mitochondria, as demonstrated both immunocytochemically (Fig. 2D and E) and by western blotting (Fig. 2F and G). The CCCP- and valinomycin-induced loss of mitochondria in fibroblasts was inhibited by the lysosomal inhibitor bafilomycin A1, confirming its autophagic nature (Fig. 2D–G). Mitophagy induced by 48 h exposure to CCCP or valinomycin was also inhibited by RNAi-mediated knockdown of PARK2, indicating that endogenous Parkin mediates mitophagy in these fibroblasts (Supplementary Data). Interestingly, RNAi-mediated knockdown of USP15 in these fibroblasts strongly enhanced mitophagy, so that mitophagy already became demonstrable after 24 h exposure to CCCP or valinomycin (Fig. 2H–K). As in SH-SY5Y cells, this mitophagy-enhancing effect of USP15 knockdown was reversed by simultaneous PARK2 knockdown (Supplementary Data).
USP15 knockdown rescues the mitophagy defect of PARK2 mutant PD patient fibroblasts
As demonstrated above in HeLa cells (Fig. 2A), USP15 knockdown does not enhance mitophagy in the complete absence of Parkin. However, many clinical PARK2 mutations do not completely abolish the E3 ligase activity of Parkin (28–30), so that PD patients with PARK2 mutations often have diminished but non-zero Parkin activity levels. We wondered whether USP15 knockdown would promote Parkin function in PARK2 mutant PD patient cells with reduced but non-zero Parkin activity. We used fibroblasts from a PD patient with compound heterozygous PARK2 mutations (c.8_171del/c.535_871del; p.V3EfsX3/p.G179LfsX) (31). In protein extracts from control human fibroblasts, Parkin exists as a major 52-kDa full-length species and a minor 42-kDa species that lacks the N-terminal ubiquitin-like (UBL) domain and originates from an internal translation initiation site at codon 80 of the PARK2 gene (32) (Fig. 3A). Both full-length Parkin and Parkin lacking the UBL domain are capable of inducing mitophagy (25). In the PARK2 mutant cells, total Parkin protein levels were reduced to 30.3±10.8% of control levels (n = 3), with complete absence of 52-kDa Parkin and residual expression of the 42-kDa, UBL-lacking Parkin species generated through the internal start site from the c.8_171del mutant allele (Fig. 3A). In contrast to control fibroblasts, the PARK2 mutant fibroblasts showed little mitochondrial clearance after exposure to CCCP or valinomycin for 48 h (Fig. 3B–E). Strikingly, however, this mitophagy defect of the PARK2 mutant fibroblasts was rescued by knockdown of USP15 (Fig. 3F–I). This rescue was reversed by simultaneous RNAi-mediated knockdown of residual Parkin in the mutant fibroblasts (Supplementary Data).
Figure 3.
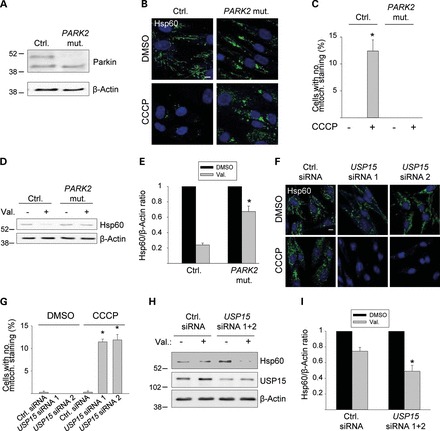
Knockdown of USP15 rescues the mitophagy defect of PARK2 mutant PD patient fibroblasts. (A) Fibroblasts from a PD patient with c.8_171del/c.535_871del PARK2 mutations (PARK2 mut.) and from age-matched healthy control 2 (Ctrl.) were analyzed by western blotting for Parkin expression. (B–E) Mutant and control 2 fibroblasts were treated for 48 h with DMSO, CCCP or valinomycin (Val.), as indicated, followed by anti-Hsp60 immunostaining (B and C) or western blotting (D and E). (C) Quantification of the percentage cells without detectable Hsp60 immunoreactivity (n = 3). * P < 0.001 compared with DMSO-treated control fibroblasts. (E) Quantification of the Hsp60/β-Actin ratio on western blot, normalized to this ratio in the DMSO condition (n = 3). * P < 0.01 compared with valinomycin-treated control fibroblasts. (F–I) PARK2 mutant fibroblasts were transfected with the indicated siRNAs and treated for 48 h with DMSO, CCCP or valinomycin, as indicated, followed by anti-Hsp60 immunostaining (F, G) or western blotting (H, I). (G) Quantification of the percentage cells without detectable Hsp60 immunoreactivity (n = 3). *P < 0.001 compared with the CCCP-treated control siRNA condition. (I) Quantification of the Hsp60/β-Actin ratio on western blot, normalized to this ratio in the DMSO condition (n = 8). *P < 0.001 compared with the valinomycin-treated control siRNA condition. Scale bars, 10 µm.
PINK1 is a serine/threonine kinase that, like Parkin, has been linked genetically with autosomal recessive PD (2). PINK1 has also been implicated in mitophagy (12–14) and enhances the E3 ligase activity of Parkin in response to mitochondrial depolarization (13,33–35). Studies in Drosophila and human cells have revealed that overexpression of Parkin can rescue some of the mitochondrial phenotypes caused by PINK1 deficiency, further supporting the view that PINK1 and Parkin function, at least in part, in the same pathway (36–38). To assess whether knockdown of USP15 can also compensate for impaired PINK1 function, we examined fibroblasts from a PD patient with homozygous PINK1 mutations (c.1309 T>G). This mutation results in substitution of tryptophan at position 437 by glycine. This residue is located within the kinase domain of PINK1 and is highly conserved across species. Interestingly, these PINK1mutant fibroblasts showed reduced mitophagy (Supplementary Data), indicating that the W437G mutation impaired PINK1 function. RNAi-mediated USP15 knockdown improved the impaired mitophagy of the PINK1mutant fibroblasts (Supplementary Data).
USP15, like Parkin, is widely expressed across tissues
In view of the antagonistic relationship of Parkin and USP15, we examined their relative expression levels in mouse brain and other tissues. Like Parkin, USP15 was widely expressed across different brain regions (Fig. 4A) and other organs (Fig. 4B). The Parkin/USP15 expression ratio in brain was among the lowest of all organs tested (Fig. 4B and C). Immunostaining of endogenous USP15 in mouse primary midbrain cultures revealed strong expression in neurons, including dopaminergic neurons (Fig. 4D and E). Endogenous USP15 in neurons had a predominantly cytosolic distribution but also partially colocalized with mitochondria (Fig. 4F–I).
Figure 4.
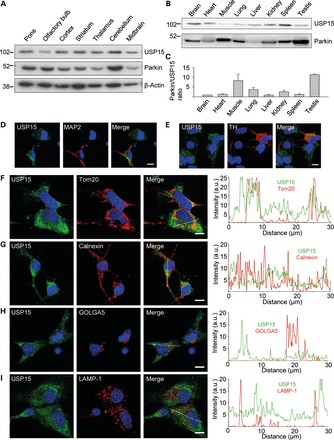
Anatomical, cellular and subcellular distribution of USP15. (A) Extracts from 2- to 3-month-old mouse brain regions (30 μg of total protein per region) were analyzed using WB with the indicated antibodies. (B and C) Extracts from 2- to 3-month-old mouse organs (30 μg of total protein per organ) were analyzed by WB to compare Parkin and USP15 expression levels on the same blot. (C) The Parkin/USP15 expression level ratio was quantified and normalized to the ratio in brain (n = 4). (D–I) Mouse embryonic midbrain cultures were double-labeled for USP15 and either the neuronal marker MAP2 (D), the dopaminergic marker tyrosine hydroxylase (TH) (E), the mitochondrial marker Tom20 (F), the ER marker calnexin (G), the Golgi marker GOLGA5 (H) or the lysosomal marker LAMP-1 (I). Graphs in (F–I) depict the relative intensities of each channel over the drawn lines shown in the merged images. A.U., arbitrary units. Scale bars, 10 µm.
USP15 antagonizes Parkin-mediated mitochondrial ubiquitination
What could be the mechanism for the inhibitory effect of USP15 on Parkin-mediated mitophagy? We considered the possibility that USP15 might modulate Parkin function by deubiquitinating Parkin itself. However, we found no evidence that USP15 deubiquitinates Parkin in basal conditions or after exposure to valinomycin (Fig. 5A and B). We also investigated whether USP15 inhibited the translocation of Parkin from the cytosol to depolarized mitochondria. However, USP15 had no effect on the CCCP-induced translocation of transfected Parkin in HeLa cells (Fig. 5C and D). Moreover, subcellular fractionation and western blotting indicated that USP15 did not prevent the CCCP-induced translocation of endogenous Parkin in HEK293 cells (Fig. 5E). These experiments also showed that a substantial portion of endogenous USP15 in HEK293 cells was present in the mitochondria-enriched fractions in basal conditions (Fig. 5E), consistent with the immunocytochemical colocalization experiments in neurons (Fig. 4F); the amount of USP15 in the mitochondrial fractions did not increase after mitochondrial depolarization (Fig. 5E).
Figure 5.
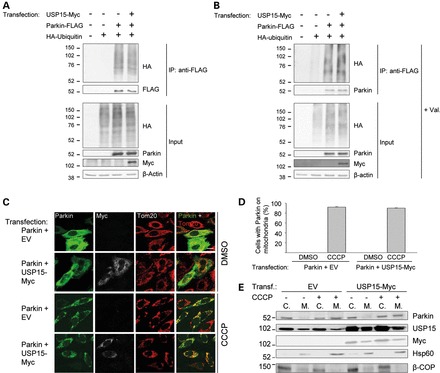
USP15 does not affect the ubiquitination status of Parkin or the translocation of Parkin to depolarized mitochondria. (A and B) HeLa cells were transfected with combinations of HA-tagged ubiquitin, FLAG-tagged Parkin and Myc-tagged USP15, as indicated. At 24 h after transfection, extracts were made in denaturing conditions to dissociate Parkin from its binding partners. After dilution in non-denaturing buffer, immunoprecipitation was performed with anti-FLAG. The immunoprecipitate (IP) and input samples were resolved by SDS–PAGE and western blot with the indicated antibodies. The experiment was performed in basal conditions (A) or after treatment with valinomycin (Val., 1 μm) for 3 h (B). (C and D) HeLa cells were transfected with Parkin and empty vector (EV) or with Parkin and Myc-tagged USP15, as indicated. At 24 h after transfection, cells were treated with DMSO or CCCP (10 µm) for 3 h and immunostained for Parkin, Myc or Tom20. Scale bar, 10 μm. (D) Quantification of the percentage of Parkin-positive cells (in the conditions transfected with Parkin and EV) or Parkin- and Myc-positive cells (in the conditions transfected with Parkin and USP15-Myc) in which Parkin colocalized with mitochondria (n = 3). (E) HEK293 cells were transfected with EV or Myc-tagged USP15 and treated with DMSO or CCCP (10 µm) for 3 h. Cells were fractionated into cytosolic (C.) and mitochondria-enriched (M.) fractions. After loading the same total amount of protein on the gel for each fraction, SDS–PAGE and immunoblotting were performed for Parkin, USP15, Myc, the mitochondrial marker Hsp60 and the non-mitochondrial protein β-COP.
We then determined whether USP15 counteracted Parkin-mediated ubiquitination of depolarized mitochondria. In HeLa cells transfected with Parkin and HA-tagged ubiquitin, ubiquitin had a diffusely cytosolic distribution (Fig. 6A and D). However, CCCP treatment for 6 h induced striking colocalization of ubiquitin with mitochondria (Fig. 6A and D), as previously reported (12,13). This recruitment of ubiquitin to depolarized mitochondria was Parkin-dependent (Fig. 6A and D) (12,13). Importantly, overexpression of USP15, but not the catalytically inactive C269A USP15 mutant or USP4, strongly reduced CCCP-induced clustering of ubiquitin on mitochondria, indicating that USP15 opposed Parkin-mediated mitochondrial ubiquitination (Fig. 6A and D).
Figure 6.
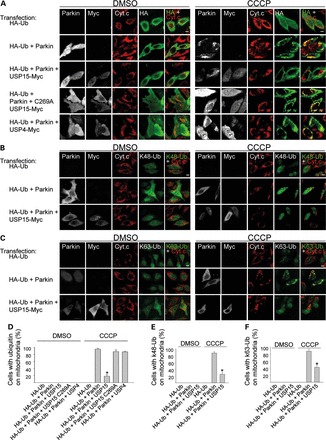
USP15 counteracts Parkin-mediated mitochondrial ubiquitination. (A–C) HeLa cells were transfected with combinations of HA-tagged ubiquitin (HA-Ub), Parkin and Myc-tagged DUBs. After 24 h, cells were treated with DMSO or CCCP (10 µm) for 6 h, followed by quadruple immunostaining for Parkin, Myc, the mitochondrial marker cytochrome c (Cyt. c) and either HA (A), K48-linked ubiquitin chains (K48-Ub) (B) or K63-linked ubiquitin chains (K63-Ub) (C). (D–F) The percentage cells in which HA-Ub (D), K48-Ub (E) and K63-Ub (F) colocalized with mitochondria was quantified among the HA-positive cells (in conditions transfected with HA-Ub alone), the HA- and Parkin-positive cells (in conditions cotransfected with HA-Ub and Parkin) and the HA-, Parkin- and Myc-positive cells (in conditions cotransfected with HA-Ub, Parkin and Myc-tagged DUBs) (n ≥ 3). *P < 0.001 compared with CCCP-treated cells transfected with HA-Ub and Parkin (D–F), with HA-Ub, Parkin and USP15 C269A (D), and with HA-Ub, Parkin and USP4 (D).
Parkin has been shown to catalyze formation of several distinct types of poly-ubiquitin linkages on depolarized mitochondria, including K48- and K63-linked ubiquitin chains (12,15,39,40). We used antibodies that specifically recognize either K48- or K63-linked ubiquitin chains (39,40). Parkin indeed mediated accumulation of K48- and K63-linked ubiquitin chains on mitochondria after CCCP treatment for 6 h (Fig. 6B, C, E, F). Overexpression of USP15 antagonized Parkin-mediated clustering of both K48- and K63-linked ubiquitin chains on depolarized mitochondria (Fig. 6B, C, E, F).
We also examined the effect of USP15 on mitochondrial ubiquitination at endogenous expression levels of USP15 and ubiquitin using subcellular fractionation and western blotting (Fig. 7A–D). HeLa cells were treated with valinomycin for 0–6 h, followed by isolation of mitochondrial fractions and analysis of the mitochondria by immunoblotting for endogenous ubiquitin. Treatment with valinomycin for 1–6 h clearly induced Parkin-dependent accumulation of endogenous ubiquitin in the mitochondrial fractions (Fig. 7A and B). Strikingly, Parkin-dependent mitochondrial ubiquitination was strongly potentiated after RNAi-mediated knockdown of USP15 (Fig. 7C and D). In contrast, knockdown of USP11 had no effect on mitochondrial ubiquitination (Fig. 7E and F).
Figure 7.
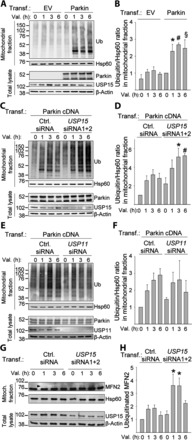
Knockdown of USP15 enhances Parkin-mediated mitochondrial ubiquitination. (A and B) HeLa cells were transfected with empty vector (EV) or Parkin cDNA and treated with valinomycin (Val., 1 µm) for 0, 1, 3 or 6 h, followed by subcellular fractionation and WB of mitochondrial fractions and total lysates with the indicated antibodies. (B) Quantification of the ubiquitin/Hsp60 ratio in the mitochondrial fractions, normalized to this ratio in the Parkin-transfected condition after 0 h of valinomycin (n = 5–9). *P<0.05, #P<0.05 and §P<0.05, compared with valinomycin treatment for 1, 3 or 6 h, respectively, in the EV-transfected cells. (C and D) HeLa cells were transfected with Parkin cDNA and either control (Ctrl.) siRNA or a combination of USP15 siRNAs 1 and 2, and treated with valinomycin for 0, 1, 3 or 6 h, followed by WB of mitochondrial fractions and total lysates. (D) Quantification of the ubiquitin/Hsp60 ratio in the mitochondrial fractions, normalized to this ratio in cells transfected with control siRNA after 0 h of valinomycin (n = 3). *P<0.05 and #P<0.01 versus valinomycin treatment for 3 or 6 h, respectively, in the control siRNA-transfected cells. (E and F) HeLa cells were transfected with Parkin cDNA and either control siRNA or USP11 siRNA, and treated with valinomycin for 0, 1, 3 or 6 h, followed by WB of mitochondrial fractions and total lysates. (F) Quantification of the ubiquitin/Hsp60 ratio in the mitochondrial fractions, normalized to this ratio in cells transfected with control siRNA after 0 h of valinomycin (n = 6). (G and H) Fibroblasts from control 1 were transfected with either control siRNA or a combination of USP15 siRNAs 1 and 2, and treated with valinomycin (1 µm) together with MG132 (20 μm) for 0, 1, 3 or 6 h, followed by WB of mitochondrial fractions and total lysates for mitofusin-2 (MFN2) and other indicated antibodies. Arrowhead indicates the position of the ubiquitinated MFN2 band. (H) Quantification of the amount of ubiquitinated MFN2, normalized to this amount in cells transfected with control siRNA after 0 h of valinomycin (n = 9). *P<0.001 compared with control siRNA-transfected cells after 0 h of valinomycin treatment.
We then used the same mitochondrial fractionation and western blot assay to assess the ubiquitination status of depolarized mitochondria in control human fibroblasts expressing endogenous Parkin (Supplementary Data). After exposure to valinomycin, we observed a time-dependent increase in the ubiquitin content of the mitochondrial fractions. In contrast, in PARK2 mutant fibroblasts there was no increase in mitochondrial ubiquitination after valinomycin treatment (Supplementary Data). However, knockdown of USP15 in the PARK2 mutant fibroblasts resulted in increased ubiquitination of mitochondria after depolarization (Supplementary Data).
Next, we assessed the effect of USP15 on the ubiquitination status of mitofusin-2, a substrate of Parkin in the MOM (15,19,41), in fibroblasts (Fig. 7G and H). As ubiquitinated mitofusin-2 is also degraded by the proteasome (15), we performed these experiments in the presence of the proteasome inhibitor MG132. USP15 knockdown enhanced the amount of ubiquitinated endogenous mitofusin-2 in response to mitochondrial depolarization (Fig. 7G and H).
Parkin interacts genetically with the USP15 homolog CG8334 in Drosophila
We then asked whether USP15 and Parkin also had an antagonistic relationship in vivo. To address this, we utilized the parkin knockdown Drosophila model of PD. Parkin-deficient flies have been reported to display locomotor defects, flight muscle degeneration and accumulation of swollen and dysfunctional mitochondria (7,42). Indeed, RNAi-mediated parkin knockdown in flies expressing a GFP version that specifically localizes to mitochondria (mitoGFP) resulted in the appearance of large clumps of intense mitoGFP signal in adult indirect flight muscles (Fig. 8A–C), consistent with the previous work (42). EM analysis revealed vacuolization of adult flight muscle cells and severely swollen mitochondria with disrupted cristae (Fig. 8D) (7,42). Mitochondrial membrane potential in parkin RNAi flies was reduced, as shown by labeling of neuromuscular junction synapses with the potentiometric dye JC-1 (Fig. 8E and F) (43). Moreover, adult parkin RNAi flies showed a defect in climbing ability, in line with previous studies (Fig. 8G) (7).
Figure 8.
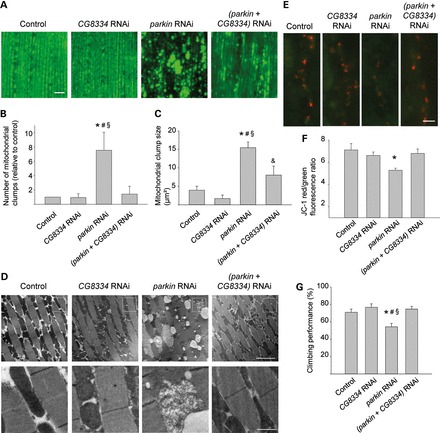
CG8334 knockdown rescues parkin RNAi phenotypes in Drosophila. (A–D) Morphological analysis of mitoGFP-expressing indirect flight muscle from 1-week-old control (w1118; UAS-mitoGFP/+; mef-2-GAL4/+), CG8334 RNAi (w1118; UAS-mitoGFP/+; CG8334 RNAi/mef-2-GAL4), parkin RNAi (w1118; UAS-mitoGFP/parkin RNAi; mef-2-GAL4/+) and (parkin + CG8334) RNAi (w1118; UAS-mitoGFP/parkin RNAi; mef-2-GAL4/CG8334 RNAi) flies. (A) Confocal images of mitoGFP in indirect flight muscle. Scale bar, 10 µm. (B) Quantification of the number of mitochondrial clumps larger than 5 µm² in mitoGFP-expressing indirect flight muscle, normalized to control (n = 6 flies per genotype). *P < 0.05 compared with Control; #P < 0.05 compared with CG8334 RNAi; §P < 0.05 compared with (parkin + CG8334) RNAi. (C) Quantification of mitochondrial clump size in mitoGFP-expressing muscle (n = 6 flies per genotype). *P < 0.001 compared with Control; #P < 0.001, &P < 0.05 compared with CG8334 RNAi; §P < 0.01 compared with (parkin + CG8334) RNAi. (D) EM images of indirect flight muscle. The bottom row shows magnifications of the boxed areas in the row above. Scale bars, first row: 5 µm; second row: 1 µm. (E and F) Mitochondrial membrane potential (Δψm) at third instar larval boutons of control (w1118; tub-GAL4/+), CG8334 RNAi (w1118; ; CG8334 RNAi/tub-GAL4), parkin RNAi (w1118; parkin RNAi/+; tub-GAL4/+) and (parkin + CG8334) RNAi (w1118; parkin RNAi/+; CG8334 RNAi/tub-GAL4) flies was imaged using JC-1, a potentiometric green fluorescent dye that shifts to red fluorescence within mitochondria with a normal negative Δψm (red, JC-1 aggregates; green, JC-1 monomers). Scale bar, 5 µm. (F) Quantification of red over green fluorescence intensity in synaptic mitochondria normalized to control (n ≥ 15 synapses per genotype). *P < 0.005 compared with control. (G) Analysis of negative geotaxis in 2-week-old flies with the same genotypes as in (E) and (F) (n = 10 batches of 10 flies per genotype). The percentage flies that cross a 4 cm line in 15 s after tapping the flies down is indicated. *P < 0.05 compared with Control; #P < 0.01 compared with CG8334 RNAi; §P < 0.005 compared with parkin + CG8334 RNAi.
The Drosophila genome encodes ∼40 different DUBs (44). Of all Drosophila DUBs, CG8334 bears the strongest similarity to USP15 (amino acid sequence identity and similarity 43 and 63%, respectively; E value <10−63). If CG8334 counteracts the effects of Parkin, we would expect silencing of CG8334 to suppress the parkin knockdown phenotypes. Importantly, all these phenotypes were indeed largely rescued by RNAi-mediated knockdown of CG8334, indicating that CG8334 and Parkin have opposite effects on fly mitochondrial morphology and function in vivo (Fig. 8A–G).
DISCUSSION
Based on our findings in human cells, we propose a model in which USP15 opposes mitophagy by antagonizing Parkin-mediated mitochondrial ubiquitination (Fig. 9). Parkin-mediated mitophagy appears to be tightly regulated, and Parkin must overcome the antagonistic effect of USP15 to induce mitochondrial clearance. The inhibitory effect on mitophagy is remarkably specific to USP15, as this effect was not observed with the closely related USPs, USP4 and USP11.
Figure 9.
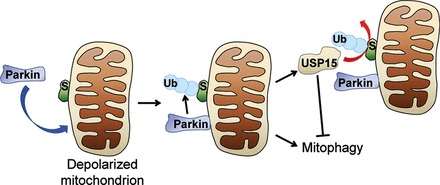
Model of antagonistic control of mitochondrial ubiquitination and mitophagy by Parkin and USP15. S, substrate. Ub, ubiquitin.
A recent proteomic study in human cells showed that Parkin ubiquitinates over 100 sites in more than 30 different MOM proteins, including mitofusin-2, in response to mitochondrial depolarization (19). We show that USP15 knockdown increases the amount of ubiquitinated mitofusin-2 after mitochondrial depolarization. It remains to be investigated which other mitochondrial Parkin substrates besides mitofusin-2 are deubiquinated by USP15 and which mitochondrial ubiquitination events are most critical for the induction of mitophagy.
Parkin has been reported to mediate the formation of K48-, K63- and even K27-linked poly-ubiquitin chains on depolarized mitochondria (12,15,39,40). We show that USP15 antagonizes the Parkin-mediated accumulation of K48- and K63-linked chains on mitochondria. Thus, USP15, like Parkin, does not appear to display strict chain-type specificity. The observed lack of ubiquitin linkage specificity of USP15 is consistent with the properties of other characterized members of the USP family, which are also promiscuous in their ability to hydrolyze multiple ubiquitin linkage types (45).
Our model of antagonistic control of mitochondrial fate by Parkin and USP15 is further supported by our in vivo findings in the parkin RNAi Drosophila model of PD. Knockdown of CG8334, the closest homolog of USP15 in the fly, largely rescues the mitochondrial defects of parkin RNAi flies. Although the cellular mechanisms underlying the mitochondrial abnormalities of Parkin-deficient flies have not been fully delineated, impairment of mitophagy probably contributes to this phenotype (46). If CG8334 inhibits mitophagy in flies, similar to USP15 in human cells, suppression of CG8334 levels would allow residual Parkin in parkin RNAi flies to target damaged mitochondria for removal.
Parkin regulates mitochondrial homeostasis also through pathways unrelated to MOM protein ubiquitination and mitophagy. For example, Parkin-mediated ubiquitination controls the levels of PARIS, a transcriptional repressor of PGC1-α, a regulator of mitochondrial biogenesis (47). Also, Parkin promotes the LUBAC-mediated ubiquitination of NEMO, thereby inducing transcriptional upregulation of OPA1, a mitochondrial GTPase involved in the control of cristae structure, inner membrane fusion and apoptosis (25). Future work will address whether USP15 also affects these pathways.
USP15 has previously been shown to activate the TGF-β pathway through deubiquitination of the type I TGF-β receptor and receptor-activated SMAD proteins (26,27). By enhancing TGF-β signaling, USP15 promotes oncogenesis (27). USP15 gene amplifications and elevated USP15 activity have been found in glioblastomas and several other human malignancies (27). Whether Parkin also affects the TGF-β pathway is currently unknown. Intriguingly, considerable evidence suggests that Parkin is a tumor suppressor (48,49), although the mechanisms for this effect are not well understood. Somatic inactivating PARK2 mutations and diminished Parkin levels have been reported in glioblastomas and other human cancers (48,49). Thus, the antagonistic relationship of Parkin and USP15 may be relevant not only for mitophagy and PD but also for oncogenesis.
USPs are ‘druggable’ targets, as illustrated by the recent development of a selective small-molecule inhibitor of USP14 (50). An important implication of our data is that inhibition of USP15 may have therapeutic potential for PD cases caused by reduced Parkin levels. Diminished Parkin activity in PD can result from PARK2 mutations (2), but probably also occurs in sporadic forms of PD, as active Parkin becomes progressively depleted in the aging human brain due to oxidative, nitrosative and dopaminergic damage (3–6). In PD cases with diminished but non-zero Parkin activity, USP15 inhibition could enable residual Parkin to exert its neuroprotective function more effectively.
MATERIALS AND METHODS
Antibodies
The following primary antibodies were used for immunofluorescence (IF) or western blot (WB): mouse anti-Parkin (IF, 1:500; WB, 1:4000; clone PRK8, Millipore) (3), mouse anti-USP15 (IF, 1:100; WB, 1:1000; Abnova) (26), mouse anti-FLAG (WB, 1:5000; clone M2, Sigma), rabbit anti-FLAG (WB, 1:1000; Sigma), rabbit anti-USP11 (WB, 1:1000; Bethyl), mouse anti-β-actin (WB, 1:5000; clone AC-15, Sigma), rabbit anti-calnexin (IF, 1:500; Enzo), rabbit anti-GOLGA5 (IF, 1:500; Sigma), rabbit anti-LAMP-1 (IF, 1:200; Sigma), rabbit anti-MAP2 (IF, 1:200; clone H300, Santa Cruz), rabbit anti-tyrosine hydroxylase (IF,1:200; Millipore), sheep anti-cytochrome c (IF, 1:200; Sigma), rabbit anti-Tom20 (IF, 1:500; WB, 1:1000; Santa Cruz), rabbit anti-HA (IF, 1:100; WB, 1:2000; Bethyl), mouse anti-HA (WB, 1:1000; Covance), goat anti-c-myc (IF, 1:250; WB, 1:1000; Bethyl), chicken anti-c-myc (IF, 1:250; Bethyl), goat anti-Hsp60 (IF, 1:500; WB 1:1000; Santa Cruz), mouse anti-ubiquitin (WB 1:1000; Santa Cruz), rabbit anti-ubiquitin, K48-specific (IF, 1:500; Merck Millipore, clone Apu2), rabbit anti-ubiquitin, K63-specific (IF, 1:500; Merck Millipore, clone Apu3), mouse anti-mitofusin-2 (WB, 1:1000; Abcam) and mouse anti-β-COP (WB 1:1000; Sigma). Peroxidase-linked secondary antibodies for WB were from GE Healthcare. Secondary antibodies for IF were Dylight 405 donkey anti-rabbit (Jackson Immunoresearch), Cy5 donkey anti-chicken (Millipore), donkey anti-mouse Alexa Fluor-488 and -647, anti-rabbit and anti-sheep Alexa Fluor-488 and -555, and anti-goat Alexa Fluor-647 (Invitrogen).
cDNAs and siRNAs
The constructs encoding untagged wild-type human Parkin, Parkin with an N-terminal His6-FLAG tag and HA-tagged ubiquitin were described previously (4,17). pCMV6–Entry vectors containing cDNAs for human USP15, USP30 and UCH-L1 with C-terminal Myc-FLAG tags were purchased from Origene. The USP15 C269A mutant construct was generated using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) using sense primer 5′-CCTAAGTAACTTGGGAAATACGGCTTTCATGAACTCAGCTATTCAG-3′ and antisense primer 3′-CTGAATAGCTGAGTTCATGAAAGCCGTATTTCCCAAGTTACTTAGG-5′. Mutagenesis was verified by sequencing. cDNAs encoding Myc-tagged forms of USP4, USP11, Ataxin-3 and A20 were gifts from Dr P.J. Lehner (University of Cambridge, UK) (51), Dr J. Yang (Baylor College of Medicine, Houston, USA) (52), Dr E.A. Fon (Montreal Neurological Institute, Canada) (23) and Dr K. Li (University of Texas Medical Branch, Galveston, USA) (53), respectively. The target sequences of USP15 siRNA 1, USP15 siRNA 2, USP11 siRNA and control siRNA were 5′-GCACGTGATTATTCCTGTT-3′, 5′-GGTTGGAATAAACTTGTCA-3′, 5′-ACCGATTCTATTGGCCTAGTA-3′ and 5′-AATTCTCCGAACGTGTCACGT-3′, respectively (Qiagen) (17,26,54). The siRNAs targeted against human PARK2 (siRNA1, HSS107593; siRNA2, HSS107594) were from Invitrogen.
Skin fibroblasts, cell cultures and transfection
HEK293, HeLa and SH-SY5Y cells were grown and transfected with cDNA or siRNA as described (17). Primary embryonic mouse midbrain neurons were cultured as described (17) and immunostained after 5 days in vitro. Human dermal fibroblasts were obtained by skin biopsy from the medial aspect of the upper limb after written informed consent. All procedures were approved by the local ethical committee. Control 1 was a healthy male born in 1949; control 2 was a healthy female born in 1967; control 3 was a healthy male born in 1944; control 4 was a healthy female born in 1938. The clinical history of the PD patient with compound heterozygous PARK2 mutations, a female born in 1970, has been published (31). The patient with PINK1 mutations was a female born in 1941 who developed early-onset PD at the age of 38, presenting initially with a right-sided arm and leg tremor, bradykinesia and rigidity. She responded well to levodopa. As the disease progressed, she developed motor fluctuations. Twenty years after disease onset, she required bilateral pallidotomy to treat her severe dyskinesias. She was the eldest of seven children, born to consanguineous parents. She has one other sister who developed PD at the age of 46 years. Her twin brother and sister died in early childhood and another sister and two brothers remain unaffected. PINK1 mutations were identified by DNA extraction from blood using JET Quick Spin Columns according to the manufacturer's instructions (Astral Scientific Australia). The purified DNA was then subjected to a PCR cycle to incorporate the dye labeled terminators (Applied Biosystems, Australia). The PCR conditions were as detailed: 25 cycles at 96°C 10 s, 50°C 5 s and 60°C 4 min. The PCR reactions were then cleaned and final reaction samples were sequenced using an ABI 3730 capillary sequencer (Applied Biosystems, Australia). Human fibroblasts were grown in DMEM F12 (Invitrogen) supplemented with fetal bovine serum (10%, Greiner Bio-One), non-essential amino acids (1%) penicillin (100 U/ml), streptomycin (100 µg/ml) and sodium bicarbonate (0.12%) at 37°C in a 5% CO2 humidified atmosphere. Human fibroblasts were transfected with 50 nm siRNA using the Neon Transfection System (Invitrogen) according to the manufacturer's instructions. CCCP (Calbiochem), valinomycin (Sigma), bafilomycin A1 (Sigma) or DMSO were added 24 or 48 h after transfection.
Immunofluorescence, confocal microscopy and image analysis
Immunostaining of cultured cells was performed as described (17). For experiments measuring effects of Parkin and myc-tagged DUBs on colocalization of HA-tagged ubiquitin with mitochondria, quadruple immunofluorescence was performed using mouse anti-Parkin, chicken anti-myc, rabbit anti-HA and sheep anti-cytochrome c as primary antibodies and donkey secondary antibodies anti-mouse Alexa Fluor-488, Cy5 anti-chicken, Dylight 405 anti-rabbit and anti-sheep Alexa Fluor-555. Nuclei were stained with TOTO-3 iodide (1 µm in the secondary antibody solution; Invitrogen). Confocal images with 1 µm slice thickness were acquired at room temperature either with an Axioskop 2 microscope (LSM510 META; Carl Zeiss), a 63X NA 1.4 oil Plan Apochromat objective and an AxioCam HR camera (Carl Zeiss), or (for analysis of quadruple immunostaining) with a Nikon Eclipse E800 microscope equipped with Biorad Radiance 2100 laser scanning hardware and a 60X NA 1.4 oil Plan Apochromat objective. Brightness and contrast were adjusted with NIH ImageJ software. In translocation, mitophagy and ubiquitination experiments, random images were captured and analyzed by an investigator blinded to the experimental condition. A minimum of 100 cells per condition was analyzed per experiment. Colocalization was assessed by line scan analysis over a distance of 30 µm using the Plot Profile function in ImageJ.
Coimmunoprecipitation and western blot
Coimmunoprecipitation was performed as described (17). In brief, HeLa cells were washed with ice-cold PBS, removed with a scraper, and resuspended in lysis buffer (20 mm Tris–HCl and 1% Triton X-100, pH 7.4). After solubilization for 30 min at 4°C, insoluble material was removed by centrifugation at 20 000g for 5 min. Protein concentrations were determined using the Bio-Rad Protein assay. Equal protein amounts were incubated with anti-FLAG M2 Affinity Gel (Sigma) for 2 h at 4°C and washed three times with lysis buffer. Bound proteins were eluted in lysis buffer containing FLAG peptides (0.1 µg/µl; Sigma) for 30 min at 4°C, followed by SDS–PAGE, blotting onto PVDF membranes and incubation with blocking solution, primary and secondary antibodies. Immunoreaction was visualized with ECL (Pierce) or ECL Prime (GE Healthcare) and Amersham Hyperfilm ECL (GE Healthcare) or a Fujifilm LAS-3000 Imager. The density of scanned signals was measured with UN-SCAN-IT gel 6.1 (Silk Scientific).
Isolation of mitochondria
Mitochondria were isolated as described (17), with minor modifications. Cells were harvested with a scraper, washed in PBS and homogenized with a glass pestle in isolation medium (250 mm sucrose, 1 mm EDTA and 10 mm Tris–MOPS, pH 7.4). Nuclei and undisrupted cells were removed by centrifugation (600g) for 10 min at 4°C. The supernatant was centrifuged at 7000g for 10 min at 4°C and the resulting supernatant (containing cytosol) was separated from the pellet. The pellet was resuspended in isolation medium and centrifuged at 7000g for 10 min at 4°C. This step was repeated three times and the final pellet (enriched in mitochondria) was analyzed by SDS–PAGE and western blot. In experiments assaying mitochondrial and mitofusin-2 ubiquitination, N-ethylmaleimide (10 mm, Sigma) was added to the isolation medium to prevent deubiquitination of mitochondria after cell lysis.
Ubiquitination status of Parkin
Ubiquitination of Parkin was assayed as described (17,23). One day after transfection of HeLa cells with FLAG-Parkin and HA-ubiquitin, proteins were extracted in denaturating lysis buffer [50 mm Tris–HCl, pH 7.4, 5 mm EDTA, 1% SDS, 15 U/ml DNase I and protease inhibitor cocktail (Roche)] to dissociate Parkin from its binding partners, and boiled for 5 min. Extracts were then diluted 1:10 in non-denaturating lysis buffer [50 mm Tris–HCl, pH 7.4, 300 mm NaCl, 5 mm EDTA, 1% Triton X-100 and protease inhibitor cocktail], followed by anti-FLAG immunoprecipitation as described above and western blot.
Drosophila stocks
Flies were kept on standard cornmeal and molasses medium. parkin (CG10523) and CG8334 transgenic UAS-RNAi lines were obtained from the Vienna Stock Center (VDRC) and were expressed using w1118; tub-GAL4 and w1118; mef-2-GAL4 at 28°C. Controls used were w1118; tub-GAL4/+ and w1118; mef-2-GAL4/+ at 28°C. w1118; UAS-mitoGFP was obtained from Bloomington stock center (Indiana, USA). To quantify parkin and CG8334 mRNA levels, RNA was isolated from adult thoraces and real-time RT-PCR was performed as previously described (55) using primers 5′-CCAGCAATGTCACCATCAAAG-3′ and 5′-GCGTGTCCACTCAGTCTG-3′ for parkin and 5′-GGAGTGACGCATCTTGAG-3′ and 5′-TTCTTTGGTATGGGTGGACTG-3′ for CG8334. The data were normalized using RP-49, a ribosomal gene (55). Real-time PCR showed that parkin mRNA levels in parkin RNAi lines were 47.6±1.8% of control parkin levels (n = 12), and CG8334 mRNA levels in CG8334 RNAi lines were 59.0±3.3% of control CG8334 levels (n = 11).
Mitochondrial morphology in Drosophila flight muscles
For fluorescence imaging of mitoGFP in adult thorax muscles, 1-week-old adult flies were fixed in PBS with 5% formaldehyde and 0.4% Triton for 3 h. After fixation, dissected thoraces were mounted in Vectashield (Vector Laboratories) and imaged on a Zeiss LSM 510 META confocal microscope using a 63x oil NA 1.4 objective. MitoGFP was imaged using a 488 nm laser and a band pass emission filter (500–530). Number of mitochondrial clumps was determined in an automated manner using the ‘analyze particles’ plugin in NIH ImageJ software, with the following settings: size >5 µm2; circularity >0.2. Mitochondrial clump size was analyzed with the following settings: size >1.5 µm2; circularity: >0.2. For electron microscopy, 1-week-old adult fly thoraces were first fixed in paraformaldehyde/glutaraldehyde, then postfixed in osmium tetroxide and finally dehydrated and embedded in Epon as described (55). For TEM analysis, different sections (80 nm thick) were stained with uranyl acetate and lead citrate.
JC-1 labeling
Dissected third instar Drosophila larvae were incubated for 10 min in 4 µm JC-1 (Molecular Probes) in HL-3 solution and then washed for 1 min in HL-3 solution before imaging. Mitochondria at the neuromuscular junctions of muscle 6 and 7 in segments A2 and A3 were imaged using the following filters (Semrock) for green and red emission, respectively: ex: 500/24 nm; em: 542/27 nm; dichroic: 520 nm and ex: 543/22 nm; em: 593/40 nm; dichroic: 562 nm. Mitochondria in neuromuscular boutons were easily distinguished by their shape and morphology and by the green JC-1 labeling that labels boutons differently from muscle cells (43). Images were captured on a Nikon FN1 microscope with a 60X NA 1 water immersion objective. Data were analyzed in NIS-Elements 3.0 (Nikon) by measuring the ratios of red to green JC-1 fluorescence.
Negative geotaxis
Ten flies per batch were loaded into an empty vial (2 cm diameter, 10 cm height) and the number of flies that crosses a 4 cm line in 15 s after tapping the flies down was counted. Assays were performed on 10 batches of 10 flies each at 28°C.
Statistics
Significance of differences was analyzed with two-tailed Student's t-test for comparison between two groups and with one-way ANOVA and post hoc Holm–Sidak test for comparison between more than two groups (SigmaStat 3.5, Systat). Values and error bars represent mean±SEM. NCBI BLAST (blastp) was used to search for USP15 homologs and determine E values and amino acid sequence identity and similarity.
SUPPLEMENTARY MATERIAL
FUNDING
W.V. is a Senior Clinical Investigator of the Research Foundation Flanders (FWO). C.M.S is supported by a National Health and Medical Research Council Clinical Practitioner Fellowship (#1008433). This work was supported by FWO (G.0583.12N, G074709, G053913N), University of Leuven (GOA/13/017), European Research Council (260678), Interuniversity Attraction Poles (P7/16), a Methusalem grant (University of Leuven and Flanders government to B.D.S.) and Vlaams Instituut voor Biotechnologie.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to E.A. Fon, P.J. Lehner, K. Li, K. Winklhofer and J. Yang for providing constructs. We thank S. Van Meensel and J. Swerts for technical support and V. Morais and G. Esposito for discussion.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lees A.J., Hardy J., Revesz T. Parkinson's disease. Lancet. 2009;373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 2.Corti O., Lesage A., Brice A. What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol. Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- 3.Pawlyk A.C., Giasson B.I., Sampathu D.M., Perez F.A., Lim K.L., Dawson V.L., Dawson T.M., Palmiter R.D., Trojanowski J.Q., Lee V.M. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J. Biol. Chem. 2003;278:48120–48128. doi: 10.1074/jbc.M306889200. [DOI] [PubMed] [Google Scholar]
- 4.Winklhofer K.F., Henn I.H., Kay-Jackson P.C., Heller U., Tatzelt J. Inactivation of parkin by oxidative stress and C-terminal truncations: a protective role of molecular chaperones. J. Biol. Chem. 2003;278:47199–47208. doi: 10.1074/jbc.M306769200. [DOI] [PubMed] [Google Scholar]
- 5.Chung K.K., Thomas B., Li X., Pletnikova O., Troncoso J.C., Marsh L., Dawson V.L., Dawson T.M. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 6.Lavoie M.J., Ostaszewski B.L., Weihofen A., Schlossmacher M.G., Selkoe D.J. Dopamine covalently modifies and functionally inactivates parkin. Nat. Med. 2005;11:1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- 7.Greene J.C., Whitworth A.J., Kuo I., Andrews L.A., Feany M.B., Pallanck L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl Acad. Sci. USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palacino J.J., Sagi D., Goldberg M.S., Krauss S., Motz C., Wacker M., Klose J., Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 9.Mortiboys H., Thomas K.J., Koopman W.J., Klaffke S., Abou-Sleiman P., Olpin S., Wood N.W., Willems P.H., Smeitink J.A., Cookson M.R., et al. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann. Neurol. 2008;64:555–565. doi: 10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grünewald A., Voges L., Rakovic A., Kasten M., Vandebona H., Hemmelmann C., Lohmann K., Orolicki S., Ramirez A., Schapira A.H., et al. Mutant Parkin impairs mitochondrial function and morphology in human fibroblasts. PLoS ONE. 2010;5:e12962. doi: 10.1371/journal.pone.0012962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L., Hess S., Chan D.C. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C., Andres A.M., Ratliff E.P., Hernandez G., Lee P., Gottlieb R.A. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS ONE. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Humbeeck C., Cornelissen T., Hofkens H., Mandemakers W., Gevaert K., De Strooper B., Vandenberghe W. Parkin interacts with Ambra1 to induce mitophagy. J. Neurosci. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Q., Zakaria H.M., Simone A., Sheng Z.H. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 2012;22:545–552. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarraf S.A., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P., Harper J.W. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komander D., Clague M.J., Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura N., Hirose S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol. Biol. Cell. 2008;19:1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leroy E., Boyer R., Auburger G., Leube B., Ulm G., Mezey E., Harta G., Brownstein M.J., Jonnalagada S., Chernova T., et al. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 23.Durcan T.M., Kontogiannea M., Thorarinsdottir T., Fallon L., Williams A.J., Djarmati A., Fantaneanu T., Paulson H.L., Fon E.A. The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum. Mol. Genet. 2011;20:141–154. doi: 10.1093/hmg/ddq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wertz I.E., O'Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 25.Müller-Rischart A.K., Pilsl A., Beaudette P., Patra M., Hadian K., Funke M., Peis R., Deinlein A., Schweimer C., Kuhn P.H., et al. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol. Cell. 2013;49:908–921. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- 26.Inui M., Manfrin A., Mamidi A., Martello G., Morsut L., Soligo S., Enzo E., Moro S., Polo S., Dupont S., et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat. Cell Biol. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 27.Eichhorn P.J., Rodon L., Gonzalez-Junca A., Dirac A., Gili M., Martinez-Saez E., Aura C., Barba I., Peg V., Prat A., et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda N., Kitami T., Suzuki T., Mizuno Y., Hattori N., Tanaka K. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 2006;281:3204–3209. doi: 10.1074/jbc.M510393200. [DOI] [PubMed] [Google Scholar]
- 29.Sriram S.R., Li X., Ko H.S., Chung K.K., Wong E., Lim K.L., Dawson V.L., Dawson T.M. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum. Mol. Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- 30.Hampe C., Ardila-Osorio H., Fournier M., Brice A., Corti O. Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum. Mol. Genet. 2006;15:2059–2075. doi: 10.1093/hmg/ddl131. [DOI] [PubMed] [Google Scholar]
- 31.Koentjoro B., Park J.S., Ha A.D., Sue C.M. Phenotypic variability of parkin mutations in single kindred. Mov. Disord. 2012;27:1299–1303. doi: 10.1002/mds.25041. [DOI] [PubMed] [Google Scholar]
- 32.Henn I.H., Gostner J.M., Lackner P., Tatzelt J., Winklhofer K.F. Pathogenic mutations inactivate parkin by distinct mechanisms. J. Neurochem. 2005;92:114–122. doi: 10.1111/j.1471-4159.2004.02854.x. [DOI] [PubMed] [Google Scholar]
- 33.Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., Alessi D.R., Knebel A., Trost M., Muqit M.M. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014 doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 35.Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., Youle R.J. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J., et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 37.Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with Parkin. Nature. Vol. 441. 1162–1166: 2006. [DOI] [PubMed] [Google Scholar]
- 38.Exner N., Treske B., Paquet D., Holmström K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H., Gasser T., et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by Parkin. J. Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y.S., Kimura M., Sato S., Hattori N., Komatsu M., et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H., Taanman J.W. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng H., Dodson M.W., Huang H., Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl Acad. Sci. USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haddad D.M., Vilain S., Vos M., Esposito G., Matta S., Kalscheuer V.M., Craessaerts K., Leyssen M., Nascimento R.M., Vianna-Morgante A.M., et al. Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair Parkin-dependent mitophagy. Mol. Cell. 2013;50:831–843. doi: 10.1016/j.molcel.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Tsou W.L., Sheedlo M.J., Morrow M.E., Blount J.R., McGregor K.M., Das C., Todi S.V. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS ONE. 2012;7:e43112. doi: 10.1371/journal.pone.0043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faesen A.C., Luna-Vargas M.P., Geurink P.P., Clerici M., Merkx R., van Dijk W.J., Hameed D.S., El Oualid F., Ovaa H., Sixma T.K. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 46.Vincow E.S., Merrihew G., Thomas R.E., Shulman N.J., Beyer R.P., MacCoss M.J., Pallanck L.J. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl Acad. Sci. USA. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin J.H., Ko H.S., Kang H., Lee Y., Lee Y.I., Pletinkova O., Troconso J.C., Dawson V.L., Dawson T.M. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poulogiannis G., McIntyre R.E., Dimitradi M., Apps J.R., Wilson C.H., Ichimura K., Luo F., Cantley L.C., Wyllie A.H., Adams D.J., et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc. Natl Acad. Sci. USA. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veeriah S., Taylor B.S., Meng S., Fang F., Yilmaz E., Vivanco I., Janakiraman M., Schultz N., Hanrahan A.J., Pao W., et al. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat. Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., Gartner C., Dimova N., Hanna J., Gygi S.P., et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nathan J.A., Sengupta S., Wood S.A., Admon A., Markson G., Sanderson C., Lehner P.J. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9:1130–1145. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun W., Tan X., Shi Y., Xu G., Mao R., Gu X., Fan Y., Yu Y., Burlingame S., Zhang H., et al. USP11 negatively regulates TNFα-induced NF-κB activation by targeting on IκBα. Cell. Signal. 2010;22:386–394. doi: 10.1016/j.cellsig.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T., Baker S.C., et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiltshire T.D., Lovejoy C.A., Wang T., Xia F., O'Connor M.J., Cortez D. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J. Biol. Chem. 2011;285:14565–14571. doi: 10.1074/jbc.M110.104745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esposito G., Vos M., Vilain S., Swerts J., De Sousa Valadas J., Van Meensel S., Schaap O., Verstreken P. Aconitase causes iron toxicity in Drosophila pink1 mutants. PLoS Genet. 2013;9:e1003478. doi: 10.1371/journal.pgen.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


