Abstract
We examined patterns of genetic variation in Rousettus madagascariensis from Madagascar and R. obliviosus from the Comoros (Grande Comore, Anjouan, and Mohéli). Genetic distances among individuals on the basis of 1,130 base pairs of the mitochondrial cytochrome b (Cytb) locus were estimated from specimens collected from 17 sites on Madagascar, 3 sites on Grande Comore, 3 sites on Anjouan, and 2 sites on Mohéli. We observed little variation in Madagascar and nearshore island samples (maximum 1.1%) and interisland Comoros samples (maximum 1.8%). In contrast, pairwise distances between different sampled sites on Madagascar and the Comoros varied from 8.5% to 13.2%. For 131 Malagasy animals, 69 unique haplotypes were recovered with 86 variable sites, and for 44 Comorian individuals, 17 unique haplotypes were found with 30 variable sites. No haplotype was shared between Madagascar and the Comoros, adding to previous morphological evidence that these 2 populations should be considered separate species. Cytb data showed that Rousettus populations of Madagascar (including nearshore islands) and the Comoros are respectively monophyletic and display no geographic structure in haplotype diversity, and that R. madagascariensis and R. obliviosus are strongly supported as sister to each other relative to other Rousettus species. Genotypic data from 6 microsatellite loci confirm lack of geographic structure in either of the 2 species. In pairwise tests of population differentiation, the only significant values were between samples from the Comoro Islands and Madagascar (including nearshore islands). Estimates of current and historical demographic parameters support population expansion in both the Comoros and Madagascar. These data suggest a more recent and rapid demographic expansion in Madagascar in comparison with greater population stability on the Comoros. On the basis of available evidence, open-water crossings approaching 300 km seem rarely traversed by Rousettus, and, if successful, can result in genetic isolation and subsequent differentiation.
Keywords: Comoros, cytochrome b, Madagascar, microsatellite, phylogeny, phylogeography, Rousettus, western Indian Ocean islands
As currently configured, the pteropodid bat genus Rousettus Gray, 1921 is composed of 10 species distributed from southern Europe and the African continent (including offshore islands) eastward across portions of the Middle East, western Indian Ocean islands, mainland Asia, numerous islands to the east of the Sunda Shelf, Australia, and to the Solomon Islands (Simmons 2005). Across this vast geographical expanse certain taxa have broad distributions and others are localized, such as the western Indian Ocean island endemics R. madagascariensisG. Grandidier, 1928 on Madagascar and R. obliviosusKock, 1978 in the Comoros Archipelago (Fig. 1). The Comoros, which are composed of 4 principal islands (Grande Comore, Anjouan, Mohéli, and Mayotte), have their origin as in situ volcanic islands of relatively recent geological age, with the youngest, Grande Comore, at 0.13−0.5 million years (Myr) and the oldest, Mayotte, at 7.7−15 Myr (Emerick and Duncan 1982; Nougier et al. 1986). Members of this genus are unknown from other islands in the western Indian Ocean such as the Seychelles and Mascarenes, but R. aegyptiacus (E. Geoffroy, 1810) occurs on nearshore and offshore islands of eastern and western Africa and the Arabian Peninsula (Bergmans 1994). On the basis of current taxonomy (Simmons 2005), the only other island endemics within the genus are R. bidens (Jentink, 1879) and R. linduensis Maryanto and Yani, 2003 from Sulawesi. Hence, members of this genus have physical capacity to disperse considerable distances across ocean expanses, which in a few cases has led to island-specific endemics. However, on the basis of an extensive phylogenetic analysis, R. bidens is a member of a different subfamily of pteropodids bats, the Harpyionycterinae (Giannini et al. 2009).
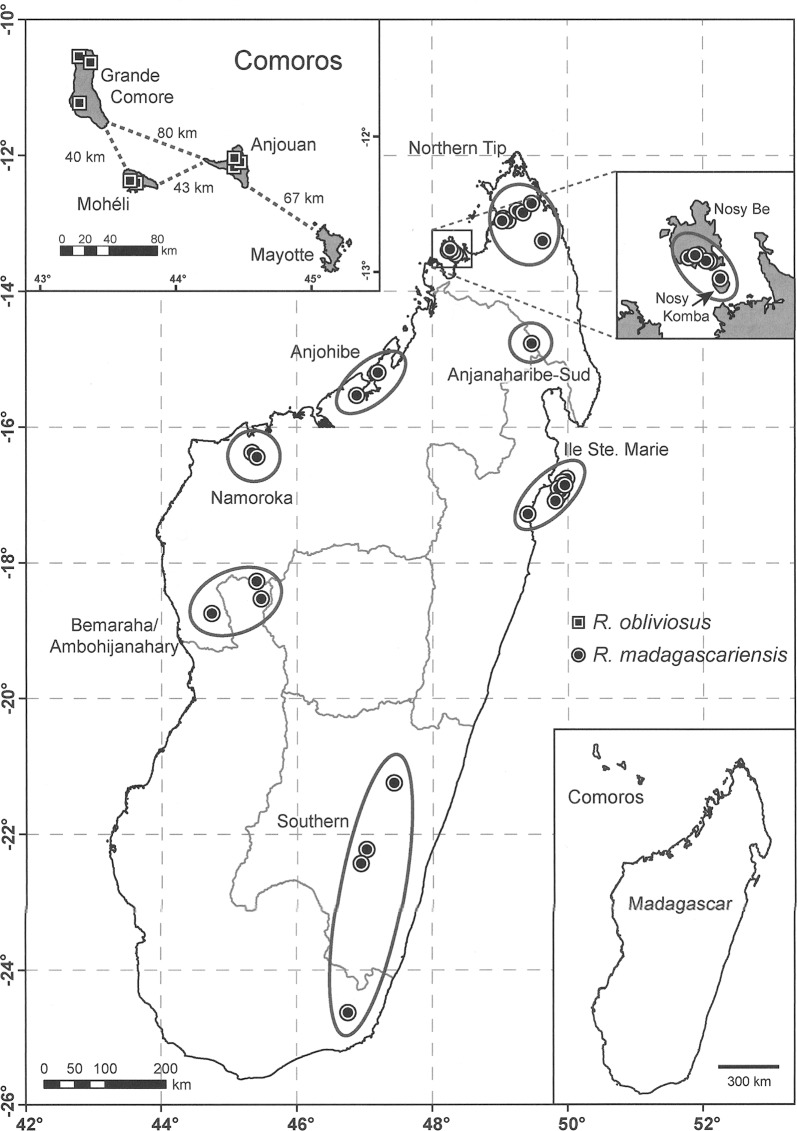
Fig. 1.
Map of principal collection localities on Madagascar and nearshore islands (Ile Sainte Marie, Nosy Komba, and Nosy Be) and in the Comoros Archipelago (Grande Comore, Anjouan, and Mohéli) of Rousettus specimens used in the current study.
Until recently, few details were available on natural history and distribution of R. madagascariensis;Dorst (1947) considered it rare. R. obliviosus was described by Kock (1978) on the basis of material collected in the late 19th century, and even until the early 1980s it was known only from the type series (Bergmans 1994). Subsequently, many aspects of distribution, natural history, and diet of these 40– 75-g, frugivorous, and nonforest-dependent bats have been documented (Andrianaivoarivelo et al. 2009; Goodman et al. 2005, 2010; Louette 2004; MacKinnon et al. 2003; Racey et al. 2010; Razafindrakoto 2006; Sew all et al. 2003). On the basis of recent inventory work, both taxa are common, particularly in portions of Madagascar and the Comoros with caves, lava tubes, and rock crevices where they make their day roosts.
Systematic relationships of R. madagascariensis and R. obliviosus have been unresolved at subgeneric and species levels. R. madagascariensis previously was considered conspecific with R. lanosus Thomas, 1906 of eastern Africa (Hayman and Hill 1971; Kingdon 1974). R. madagascariensis has been shifted between subgenera Rousettus and Stenonycteris (Corbet and Hill 1991; Koopman 1994), and R. obliviosus has been placed in the subgenus Rousettus (Kock 1978). Further, these species have not been included in any explicit morphological phylogeny of the genus or pteropodid bats in general (Springer et al. 1995). Ambiguity of systematic relationships of R. madagascariensis and R. obliviosus is associated with their former rarity in museum collections (Bergmans 1977; Kock 1978) and lack of tissue samples for the latter species in genetic studies (Álvarez et al. 1999; Giannini and Simmons 2003; Juste et al. 1997, 1999; Kirsch et al. 1995).
Given that Madagascar and the Comoros Archipelago are separated by about 300 km, molecular phylogenetic data are useful to decipher whether R. madagascariensis and R. obliviosus are sister taxa, therefore indicating a single continental origin of the 2 species, or show evidence of separate colonization events from continental areas. Further, such data allow for potentially contrasting phylogeographic patterns, with Madagascar being a large single island and the Comoros a series of small islands chained as an archipelago. Hence, this information should provide insight into the evolutionary history of members of this genus and their capacity and constraints to fly across expansive oceanic zones. Finally, Rousettus spp. are known to be reservoirs for a variety of different diseases and ectoparasites that could be important for domestic animals and humans (Calisher et al. 2006; Reeves et al. 2006). To interpret epidemiological patterns, explicit phylogenies and phylogeographic studies of regional members of the genus are needed. The purposes of our study, which uses molecular genetic data, are to determine if R. madagascariensis and R. obliviosus are true species and, if so, to examine possible sister-taxa relationships; unravel phylogeographic patterns within populations of these reputed taxa occurring on Madagascar and the Comoros; and examine patterns of population demographics within and among Rousettus populations in Madagascar and the Comoro Islands.
Materials and Methods
Sampling and deoxyribonucleic acid (DNA) extraction.— Since the early 1990s extensive chiropterological surveys have been conducted at numerous localities on Madagascar and nearshore islands (Ile Sainte-Marie, Nosy Be, and Nosy Komba), and in 2006 and 2007 fieldwork was conducted in the Comoros Archipelago (Fig. 1). Specimens referable to R. madagascariensis were collected from 17 sites on Madagascar and those to R. obliviosus from 3 different islands in the Comoro Islands (Grande Comore, Anjouan, and Mohéli). No evidence of the latter species was found on Mayotte. Voucher specimens are deposited in the Field Museum of Natural History (Chicago) and the Université d'Antananarivo, Département de Biologie Animale (Antananarivo, Madagascar). Small muscle samples from each collected individual were preserved in ethylenediaminetetra-acetic acid before specimen preparation. Research involving live animals followed the guidelines for the capture, handling, and care of mammals approved by the American Society of Mammalogists (Gannon et al. 2007). Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, California).
Sequence data collection.—The mitochondrial cytochrome b (Cytb) gene was selected to compare phylogenetic relationships of Comorian and Malagasy Rousettus to other Asiatic and African Rousettus spp. and to characterize the phylogeographic structure among individuals of Rousettus within these 2 island groups. We amplified the entire Cytb region for 44 samples of R. obliviosus and 131 samples of R. madagascariensis using polymerase chain reaction (PCR) with primers L14724 and H15915 (Irwin et al. 1991). PCR was done in a total volume of 20 µl with lx buffer (100 mM Tris-HC1, pH 8.3, 500 mM KCl), 2.0 mM MgCl2, 1 mM deoxynucleotide triphosphate (dNTP), 0.25 µM of each primer, 0.5 U of Taq polymerase, and 1 µl of template DNA. PCR cycles consisted of an initial denaturation at 95°C for 2 min, 30 cycles of 95°C for 30 s, 50°C for 45 s, and 72°C for 1 min 40 s, and a final extension at 72°C for 10 min. Samples were prepared for sequencing reactions by first incubating 5 µl of PCR product with 0.5 µl of ExoSAP-IT (USB Products, Cleveland, Ohio) and 1.5 µl of water at 37°C for 15 min followed by 80°C for 15 min. Cleaned PCR products were sequenced using primers used for amplification in a total volume of 5 µl (l× buffer, 1 µM primer, 0.2 µl of BigDye v3, and 0.5 µl of DNA template) and run on an ABI 3730x1 DNA Analyzer capillary machine. Resulting sequences were checked by eye for errors and contigs were assembled in Sequencher 4.8 (GeneCodes, Ann Arbor, Michigan). Sequences were checked for stop codons and deposited in GenBank (accession numbers GU228597–GU228771; Appendix 1). To test monophyly of Malagasy and Comorian Rousettus, GenBank was searched for Cytb sequences of other members of this genus, which resulted in an additional 57 individuals.
Microsatellite data collection.—We genotyped 193 individ-uals from Madagascar and 43 individuals from the Comoros at 6 microsatellite loci designed for R. leschenaultii (Desmarest, 1820—Hua et al. 2006). For each locus, amplification by PCR using a fluorescently labeled forward primer was done in a total volume of 10 µl with l× buffer, 2.0 mM MgCl2, 0.4 mM dNTP, 0.1 µM each primer, 0.25 U Taq polymerase, and 0.1 µl of template DNA. Cycles consisted of an initial denaturation at 95°C for 2 min, followed by 36 cycles of 95°C for 30 s, 59°C (3 cycles), 56°C (3 cycles), or 50°C (30 cycles) for 30 s and 72°C for 1 min, followed by a final extension at 72°C for 5 min. Fragments were run on an ABI 3730x1 DNA Analyzer, and alleles were called and checked in GeneMarker (Soft-Genetics, State College, Pennsylvania).
Haplotype alignment and phylogenetic analyses.—Haplo-types of Cytb for all samples were aligned by hand in the program MacClade (Maddison and Maddison 2003). Identical haplotypes were condensed using the program Collapse v1.2 (http://darwin.uvigo.es).
We partitioned our data set to account for rate variation among codon positions and used MrModeltest v2.3 (Nylander 2004) to determine the model of nucleotide sequence evolution that best fit each partition according to the Akaike information criterion. We estimated the phylogenetic relationship among haplotypes under a Bayesian framework using the program MrBayes v3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). A SYM+I+G model was applied to first position sites, a HKY+G model to second position sites, and a GTR+G model to third position sites. Analyses consisted of 3 independent runs each with 4 chains sampled every 100 generations for 5,000,000 generations. Convergence was verified by examining the trends in InL scores within and across runs for all parameters. We discarded the first 5,001 trees as burn-in and estimated the 50% majority-rule consensus topology including branch lengths and posterior probabilities (PP) for each node.
Microsatellite data analysis.—We grouped individuals from the Comoros Archipelago by island and divided the Mada-gascar samples into 7 groups for a total of 10 putative populations. We tested for deviation from Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium both within populations and globally using the program Genepop (Ray-mond and Rousset 1995). A Markov chain method (Guo and Thompson 1992) with 10,000 dememorization steps and 1,000 batches of 10,000 iterations per batch was used to determine the significance of each test after Bonferroni correction for multiple comparisons at α = 0.05.
We used Fstat (Goudet 1995) to calculate observed heterozygosity (Ho), gene diversity (HS), and overall FST for Madagascar samples, Comoros samples, and all samples together. We also estimated pairwise FST among population pairs in Fstat using 10,000 permutations of the data to determine significance after Bonferroni correction for multiple comparisons.
We used Bayesian assignment tests in Structure v2.2.3 (Pritchard et al. 2000) as an independent test of genetic structure that does not require prior assumptions of population delineation. Structure runs did not use population of origin as prior information and assumed correlated allelic frequencies among populations. We conducted 5 replicate runs at each K from K =1 to 8 where Kis the number of clusters. Each run consisted of 3.5 million generations sampled every 100 generations with the first 1.0 million steps tossed as burn-in. The best K was determined by calculating the posterior probability of each K and by examining the change in the In L between runs. Additionally, we separately tested for evidence of population structure across individuals within the Comoros.
Parameters for Comoros structure analyses were identical to those used for analysis of the entire data set; we ran 5 replicate runs at each K from K =1 to 5.
Population demographics.—We tested for evidence of demographic expansion in the Comoros and Madagascar clades by examining the distribution of pairwise sequence differences (i.e., mismatch distribution—Rogers and Harpending 1992; Schneider and Excoffier 1999) and by calculating Fu's FS statistic (Fu 1997) using the software Arlequin (Excoffier et al. 2005). For mismatch distribution tests, we used 10,000 simulations of the data to determine the null distribution of pairwise differences under a model of rapid population expansion. Significance of the Fs statistic was determined by simulating 10,000 random samples under a coalescent framework with the model of population equilibrium.
We used the program IM (Hey and Nielsen 2007) to estimate current and historical demographic parameters (current effective population sizes, ancestral effective population size, divergence time, migration rates, and splitting parameter) from the Cytb data for sister clades R. obliviosus and R. madagascariensis. Because we used a single mitochondrial locus, these data conform to assumptions of the IM model. We conducted multiple runs with 4 to 10 heated chains sampling every 10 steps after an initial burn-in of 1,000,000 generations. For each run, we noted effective sample sizes (ESS), mixing rates across heated chains, and plots of parameter trends throughout the run to ensure adequate exploration of likelihood space. We also compared marginal probability densities for parameter estimates across independent runs to verify convergence. The accepted IM run consisted of 8.8 million steps following initial burn-in with ESS greater than 80 for all parameters.
Synonymous and nonsynonymous substitution rates across coding regions of the mammalian mitochondrial genome are substantially different (Pesole et al. 1999). Thus, to approx-imate the mutation rate for the Cytb sequences used in our analyses, we calculated proportion of synonymous and nonsynonymous sites using DNAsp (Rozas et al. 2003) and determined an approximate overall rate for our Cytb data set (25.5% nonsynonymous sites, 74.5% synonymous sites) assuming an average rate of 1.8 x 10−3 substitution site−1myr−1 and 27.4 x 10−3 substitutions site−1 myr−1 for nonsynonymous and synonymous sites, respectively (Pesole et al. 1999).
Results
Sequences results.—We recovered 1,130 base pairs (bp) of the Cytb locus for 175 newly sequenced individuals (Appendix 1). For these sequences, plus 57 Cytb sequences from other Rousettus spp. retrieved from Genbank (for a total of 232 sequences), we found 366 variable sites overall. We did not find any shared haplotypes among samples from the Comoros, Madagascar, or any individuals sampled from Genbank. for 131 animals from Madagascar, we recovered 69 unique haplotypes with 86 variable sites. For the Comoros we found 17 unique haplotypes and 30 variable sites among 44 individuals. Among 32 R. leschenaultii sequences retrieved from Genbank we recovered 27 unique haplotypes with 56 variable sites. The remaining Genbank sequences represented individuals sampled generally as singletons with no reference to geographic location of sampling, and thus similar statistics would not be meaningful.
Phylogenetic analysis of Cytb sequence data demonstrates that Rousettus populations from Madagascar display no geographic structure in haplotype diversity with respect to the main island and nearshore islands (Nosy Be, Nosy Komba, and Ile Sainte Marie; Fig. 2). A similar pattern was evident among interland comparisons within the Comoros Archipelago (Grande Comore, Anjouan, and Mohéli). Average pairwise genetic distances within and among putative species indicate a relatively deep divergence between Malagasy and Comorian Rousettus, represented by an average pairwise genetic distance that is at least 15 times larger between Madagascar and the Comoros Archipelago than within each island group (Table 1). Furthermore, within-island genetic distances were only slightly less than average genetic distance among all sequences from R. leschenaultii.
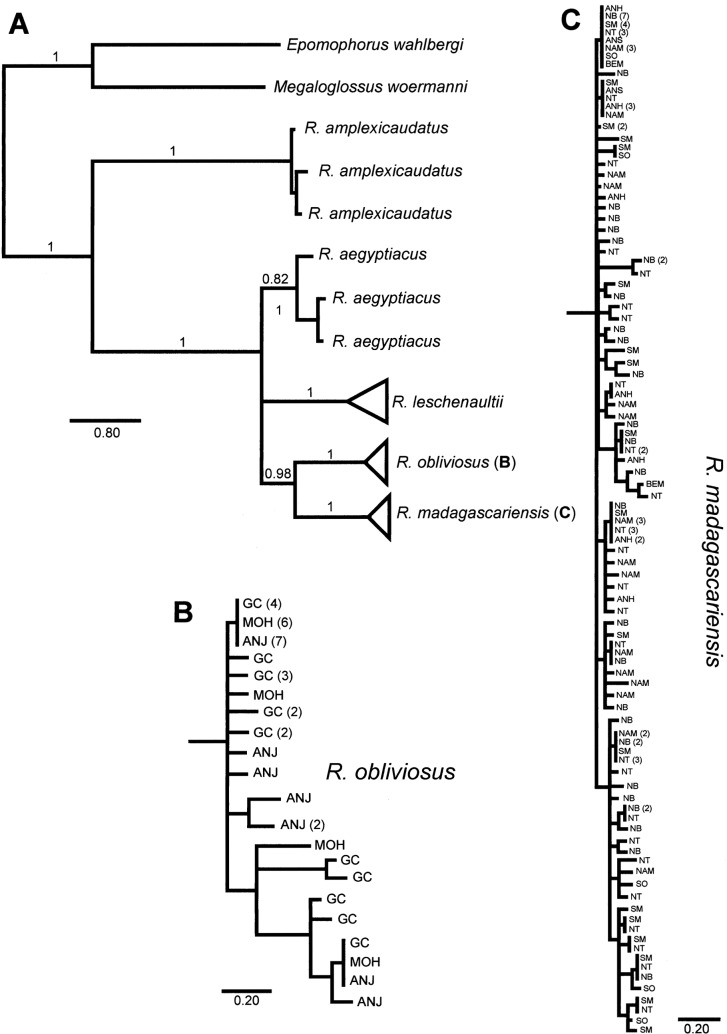
Fig. 2.
A) Bayesian consensus phylogram of Rousettus cytochrome b data. Branches of phylogram are labeled with posterior probability of bipartition, and scale bar for branches is provided below each tree. B) Detail of haplotype tree for samples of R. obliviosus. Tips of tree are labeled according to source islands: ANJ = Anjouan, MOH = Mohéli, and GC = Grande Comore. C) Detail of haplotype tree for samples of R. madagascariensis. Tips of tree are labeled according to source populations: ANH = Anjohibe, ANS = Anjanaharibe-Sud, NT = Northern Tip, NB = Nosy Be and Nosy Komba, SM = IIe Sainte Marie, BEM = Bemaraha/Ambohijanahary, NAM = Namoroka, and SO = Southern. Scale bar for branches is provided below each tree.
Table 1.
Average pairwise distances of cytochrome b sequence data within 3 species of Rousettus from Madagascar, the Comoro Islands, and China.
| Uncorrected P | HKY+I+G | |
|---|---|---|
| Comoros: R. obliviosus | 0.005578 | 0.005775 |
| Madagascar: R. madagascariensis | 0.005382 | 0.005555 |
| China: R. leschenaultii | 0.007654 | 0.007983 |
| Total data set | 0.060776 | 0.102126 |
| Madagascar: Comoros | 0.062075 | 0.086758 |
To test monophyly of Malagasy and Comorian Rousettus we performed a Bayesian phylogenetic analysis of the Cytb haplotype data. The resulting consensus phylogram (Fig. 2) demonstrates strong support for monophyly of all Cytb haplotypes in Madagascar and the Comoros, respectively. As with neighbor-joining analyses, no geographic structure to haplotypes within Madagascar and nearshore islands, or from 3 disjunct islands in the Comoros (Grande Comore, Anjouan, and Mohéli), is suggested.
Monophyly of all different western Indian Ocean Rousettus spp. included in this analysis was strongly supported. Although monophyly of R. aegyptiacus, R. leschenaultii, and the Malagasy/Comorian Rousettus also was strongly supported (i.e., PP = 1.0), R. aegyptiacus haplotypes were unresolved with respect to haplotype clades of sister species. Rousettus madagascariensis and R. obliviosus were strongly supported (i.e., PP = 0.99) as sisters to each other. It was not clear what species of Rousettus is sister to the Malagasy/Comorian clade, but the topology with the monophyletic group of R. leschenaultii haplotypes sister to the Malagasy/Comorian clade was supported by nearly 70% of trees in the posterior distribution.
Microsatellite results.—Microsatellite loci had between 14 and 25 alleles (average 17.5). We did not detect any deviation from H WE or any evidence of linkage after Bonferroni correction. Across all samples observed heterozygosity (H0) was 0.817 and gene diversity (HS) was 0.852. H0 and Hs were higher for R. madagascariensis in comparison with R. obliviosus (Table 2). FST across all samples was 0.050, but only 0.004 and 0.009 within R. madagascariensis and R. obliviosus, respectively. In pairwise tests of population differentiation, the only significant FST values were between samples from the Comoros and Madagascar + nearshore islands (Table 3). Between the Comoros and Madagascar groups the only nonsignificant values of FST involved populations with limited sampling. We did not find any significant genetic differentiation on the basis of FST within the Comoros or among Madagascar populations.
Table 2.
Observed heterozygosity (HO), gene diversity (HS), and FST (with 95% CI) for all individuals and each species individually. Confidence intervals for FST were generated with 10,000 bootstrap replicates.
| Ho | H S | F ST | |
|---|---|---|---|
| All | 0.822 | 0.852 | 0.050 (0.026–0.083) |
| Madagascar (R. madagascariensis) | 0.866 | 0.901 | 0.004 (0–0.009) |
| Comoros (R. obliviosus) | 0.720 | 0.740 | 0.009 (−0.009–0.023) |
Table 3.
F ST pairwise values of Rousettus madagascariensis from Madagascar and R. obliviosus from the Comoros; sample size (n) is indicated in first column and significant values are in bold. The first 7 sites are from Madagascar and include the nearshore island of Ile Sainte Marie in the east and a nearshore island complex of Nosy Be and Nosy Komba in the northwest. The last 3 sites are from 3 different islands in the Comoros Archipelago.
| Southern | Bemaraha/Ambohij anahary | Ile Ste. Marie | Namoroka | Anjohibe | Nosy Be/Nosy Komba | Northern | Grande Comore | Anjouan | |
|---|---|---|---|---|---|---|---|---|---|
| Southern (n = 5) | |||||||||
| Bemaraha/Ambohijanahary (n = 3) | 0.007 | ||||||||
| He Ste. Marie (n = 35) | 0.0133 | −0.025 | |||||||
| Namoroka (n = 26) | 0.0098 | −0.0087 | 0.0121 | ||||||
| Anjohibe (n = 12) | −0.0046 | −0.0078 | 0.0025 | 0.0029 | |||||
| Nosy Be/Nosy Komba (n = 70) | 0.0241 | −0.0104 | 0.0029 | 0.0101 | 0.0027 | ||||
| Northern (n = 40) | 0.0312 | −0.0152 | 0.0002 | 0.0114 | 0.0026 | −0.0002 | |||
| Grande Comore (n = 19) | 0.0968 | 0.1061 | 0.1158 | 0.1096 | 0.105 | 0.1224 | 0.1263 | ||
| Anjouan (n = 13) | 0.0957 | 0.1063 | 0.1169 | 0.1092 | 0.1105 | 0.1274 | 0.1264 | 0.0179 | |
| Mohéli (n = 11) | 0.1154 | 0.1201 | 0.1273 | 0.1193 | 0.1098 | 0.1312 | 0.1322 | 0.0015 | 0.0053 |
Likewise, Bayesian assignment tests across the entire data set found 2 genetic clusters, one corresponding to the Comoros species and the other corresponding to the Madagascar species. Tests focused within the Comoros clade did not recover additional genetic structure among islands, and average likelihood values from structure analyses did not differ across K.
Comparative demographic estimates of R. madagascariensis on Madagascar and R. obliviosus in the Comoros.—Fu's Fsstatistic was significant for both the Comoros and Madagascar samples, supporting demographic expansion in each population. We also were unable to reject the hypothesis of demographic expansion on the basis of the mismatch distribution tests in either population despite a qualitatively multimodal distribution of pairwise sequence divergence among individuals from the Comoros (Fig. 3). Estimates of x, the mutation-scaled time since expansion (2 µt, where µ is the mutation rate), were larger for Comoros individuals in comparison with Madagascar individuals, but with wide 95% confidence intervals (CIs; τcomoros = 8.908, CI = 0.221– 14.293; τMadagascar = 3.996, CI = 1.893–9.518). Estimates of current θ (2 µN, where N is the effective population size) were 5.748 for the Comoros and nearly 5 times greater for Madagascar (θMadagascar = 26.836).
Fig. 3.
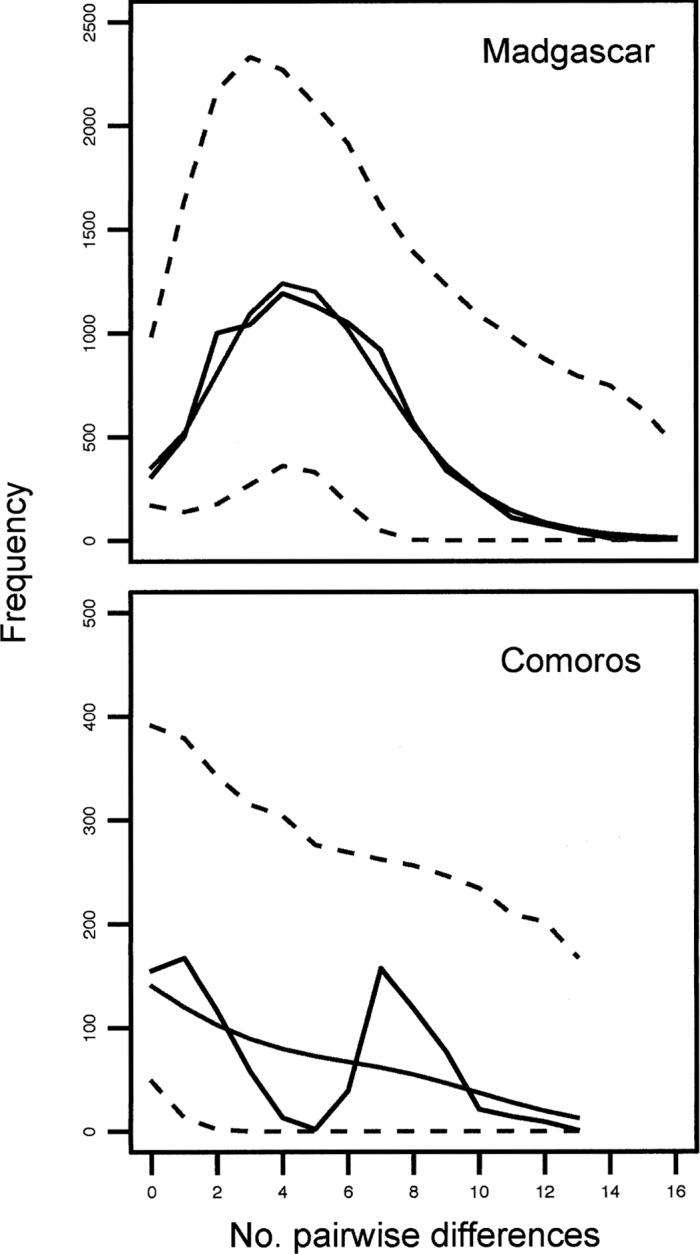
Mismatch distributions for Rousettus madagascariensis from Madagascar (top) and R. obliviosus from the Comoros (bottom). Empirical estimate of pairwise difference in solid black and simulated pairwise differences with 95% CI under a model of rapid demographic expansion in gray.
Analysis of R. obliviosus and R. madagascariensis Cytb data under a model of isolation with migration in the program IM confirmed distinctiveness of each species and also suggested recent demographic expansion in R. madagascariensis. Estimates of migration between species approached zero, indicating no gene flow. The 90% highest probability density for the divergence time parameter ranged from 1.87 to 4.75 with a highest posterior probability at 2.93. Estimates of effective population size largely corroborated results of the mismatch distribution test. We found a greater effective population size in Madagascar (θMadagascar > θComoros, PP = 1.0) and a strong signature of population expansion in R. madagascariensis postdivergence from R. obliviosus (θMadagascar > θAncestral, PP = 0.998). However, evidence for postdivergence population expansion for R. obliviosus was weak (θComoros > θAncestral, PP = 0.068), and analyses indicated demographic stability and suggested that a much larger proportion of the predivergence ancestral population contributed to genetic diversity currently found in the Comoros populations.
Assuming an overall mutation rate of 8.3315 × 10−3substitutions site−1 Myr−1, mean time since expansion for the Comoros was 0.545 Myr (95% CI = 0.013–0.875 Myr) and for Madagascar was 0.245 Myr (95% CI = 0.116–0.583 Myr) on the basis of mismatch distribution analyses. Coalescent-based analyses of divergence time between Madagascar and the Comoros from IM suggested that the split occurred 0.358 Myr (90% HPD = 0.229–0.581 Myr).
Discussion
Specific status of the Comorian Rousettus—In his description of R. obliviosus, Kock (1978) made extensive comparisons with other species of Rousettus, including R. aegyptiacus, R. leschenaultii, R. (=Lissonycteris)angolensis (Bocage, 1898), and R. lanosus and found consistent morphological characters to diagnose this new species. Kock explicitly mentioned that he did not have access to material of R. madagascariensis and thus for many years it was unclear whether R. obliviosus and R. madagascariensis were synonyms. This is particularly important given that the Comoros and Madagascar are known to share a number of bat species (Goodman et al. 2009; Ratrimomanarivo et al. 2008, 2009; Weyeneth et al. 2008). Peterson et al. (1995) made cranial and dental comparisons to address this issue on the basis of measurements and figures in Kock (1978) and concluded that obliviosus was best considered a geographical form of madagascariensis. This was readdressed by Bergmans (1994), who was able to compare specimens of R. madagas-cariensis with R. obliviosus. He found a number of cranial characters that separate these taxa and concluded that R. obliviosus was specifically distinct from R. madagascariensis.
On the basis of molecular results we present herein, populations of Rousettus in Madagascar and Comoros, which are sister taxa, display 6.2–8.7% sequence divergence from one another and clearly form reciprocally monophyletic clades. If one applies the genetic species concept of Baker and Bradley (2006), which proposes that a genetic distance of greater than 5% between 2 populations may warrant their recognition as separate species, R. madagascariensis and R. obliviosus would be considered specifically distinct. Further, these 2 taxa do not share any common haplotypes. Hence, on the basis of these different lines of evidence, we consider R. madagascariensis and R. obliviosus distinct species.
Origin of Malagasy / Comorian Rousettus spp.—Several hypotheses have been proposed about origin of the genus Rousettus on the basis of phylogenetic inference, and centers of diversity pinpoint a southeastern Asian origin (Juste et al. 1999). Three separate routes have been suggested for western expansion of pteropodids into Africa, via a middle Asian-European-Gibraltar route, a middle-Asian, Middle Eastern route, and an Indian subcontinent-western Indian Ocean, east African route (Juste et al. 1999). Phylogenetic relationships presented herein on western Indian Ocean, African, and Asian Rousettus spp. are unresolved and hence do not provide clear support for any of these three hypotheses. In other studies with larger genetic data sets but not including R. obliviosus (Giannini and Simmons 2003, 2005), support exists for the Middle Eastern route with the Afrotropical clade composed of R. aegyptiacus + R. madagascariensis being the sister group to R. leschenaultii. Further geographic and species genetic sampling is needed, with the inclusion of new material of R. obliviosus, to have a greater resolution to phylogeny and patterns of colonization of this genus.
Phylogeography of Malagasy/Comorian Rousettus.—No clear phylogeographic structure was found in populations of R. madagascariensis on Madagascar or R. obliviosus in the Comoros. Across the approximately 1,600-km length of Madagascar individuals of R. madagascariensis from extreme ends of the island and across numerous biomes, and nearshore islands up to 13 km from the main island, demonstrated complete genetic mixing. Nothing is known about dispersal patterns of R. madagascariensis, but on the basis of genetic data presented here, dispersal movements are large scale. A possible explanation for this can be found in the unusual phenological patterns in fruiting of certain native plant genera, such as Ficus (Moraceae), resulting in a local paucity of food for frugivores during certain seasons, forcing obligate fruit-eating species to disperse (Goodman and Ganzhorn 1997). This potentially could explain lack of phylogeographic structure in R. madagascariensis. Even more striking is that a similar pattern of no phylogeographic structure was found in populations of R. obliviosus on Grande Comore, Anjouan, and Mohéli, with open-water distances separating these islands between 40 and 80 km.
Data presented herein on phylogeographic structure of R. madagascariensis and R. obliviosus have important implications outside the domain of their evolutionary history. Recent epidemiological work onR. madagascariensis sampled at Ankarana in northern Madagascar revealed presence of Tioman virus (Iehlé et al. 2007). Further, the other Malagasy members of the family Pteropodidae, Eidolon dupreanum (Pollen, 1866) and Pteropus rufus E. Geoffroy, 1803, tested positive for other Paramyxoviridae viruses (e.g., Hendra and Nipah), indicating that these diseases have circulated among these bat species. Given the genetic panmixia of R. madagascariensis across Madagascar, on the basis of the phylogeographic studies presented herein, Tioman virus isolated from this species likely is not restricted to the northern portion of the island. Further, numerous other viruses have been isolated from Rousettus spp. in Asia and Africa, signifying the potential importance of R. madagascariensis and R. obliviosus as reservoirs or vectors of different diseases. These include Rhabdoviridae—European bat lyssavirus 1 and Lagos bat virus; Paramyxoviridae—undetermined Parainfluenzavirus; Togaviridae associated with the Chikungunya virus; Filoviridae—Marburg virus; Flaviviridae—Flavivirus Uganda S; Coronaviridae-severe acute respiratory syndrome Coronavirus; and the unclassified viruses Yogue and Kasokero (Calisher et al. 2006; Kalunda et al. 1986; Kuzmin et al. 2008; Pavri et al. 1971; Towner et al. 2007, 2009; Wellenberg et al. 2002). Further, Rousettus ectoparasites (mites) are known to be vectors of pathogens such as Rickettsia (Reeves et al. 2006).
Dispersal distances and patterns of speciation within Rousettus.—Fruit bats of the family Pteropodidae, and specifically in this case members of the genus Rousettus, are strong fliers (Norberg 1981, 1994) and have broad distributions across a considerable portion of the Old World (Kirsch et al. 1995; Simmons 2005), including notably isolated islands in the western Indian Ocean. As witnessed by 4 of the 10 species in the genus being island endemics (Giannini et al. 2009), these animals have limited capacity to disperse across considerable oceanic distances. In certain cases these events are seemingly rare, having resulted in isolated populations that speciated, and in other cases they remain in contact with other island or continental populations (Bastian et al. 2001). To understand the importance of distance across water crossings as an isolation factor in different populations of Rousettus spp., we present here some examples that do not necessarily rely on the same genetic markers.
On Madagascar panmixia of haplotypes occurs in R. madagascariensis, with no clear phylogeographic structure.
Samples were analyzed from 3 nearshore islands (Nosy Be, Nosy Komba, and Ile Sainte Marie), ranging from 2.5 to 13 km from the main island, and a significant number of shared haplotypes between these islands and the mainland indicate that this species easily traverses these water expanses. The direct distance from Madagascar to the nearest island in the Comoros, Mayotte (where the sister taxa R. obliviosus is unknown to occur), is 300 km and to Anjouan (where R. obliviosus does occur), 390 km. The latter volcanic island formed in situ about 3.7 Myr (Nougier et al. 1986). Given that R. obliviosus and R. madagascariensis are sister taxa and share no common haplotype, this distance of Overwater dispersal is sufficiently great to have been a rare event that subsequently led to speciation. Within Grande Comore, Anjouan, and Mohéli, which are separated by a maximum distance of 80 km, no island-specific genetic structure at mitochondrial or nuclear markers is found, and these animals seemingly cross this water distance with some frequency.
Demographic analysis of mitochondrial data suggests that genetic diversity within each species has a relatively recent origin, within the last million years. Although our results are not conclusive, they also suggest that R. madagascariensis diverged from an established Comoros population. Data from a definitive sister species to this clade would help to test this hypothesis directly.
Distance from the African continent to the nearest of the Comoro Islands is about 300 km, the same distance from Madagascar to Mayotte. Further phylogenetic analyses are needed to determine if the R. madagascariensis/obliviosus group is sister to Asian R. leschenaultii, but if this relationship is upheld it would indicate that African Rousettus were unable to successfully colonize Madagascar or the Comoros across nearly equal distance over the water crossing of the Mozambique Canal.
Outside of western Indian Ocean a few similar comparisons for the genus Rousettus can be presented, such as the oceanic islands in the Gulf of Guinea (São Tomé and Príncipe), which are separated from the African mainland by a maximum of 280 km. As far as we are aware, genetic sequence data are not available from these populations, but on the basis of allozyme variation, populations occurring on São Tomé and Príncipe are different from those on the mainland, and they have been described as endemic subspecies, R. aegyptiacus princepsJuste and Ibañez, 1993 and R. a. tomensisJuste and Ibañez, 1993 (Juste and Ibañez 1993; Juste et al. 1996). For R. amplexicaudatus in the Philippines genetic distances for populations on islands of Luzon and Mindanao, on the basis of Cytb sequence data, were <0.71% across a minimal island-to-island distance of about 215 km (Bastian et al. 2001). In the intermediate area between Luzon and Mindanao other stepping-stone islands house R. amplexicaudatus. Sulawesi holds 3 species of Rousettus, including 2 endemics, and about 120 km of sea separate the coast of New Guinea and northern Sulawesi. Molecular phylogenetic data on the relationships and origins of these taxa are not available.
If the hypothesis presented herein that R. leschenaultii is the sister group to R. madagascariensis/obliviosus is correct and that the ancestor of the latter group arrived in the Madagascar region via stepping-stone islands from the Indian subcontinent region (Juste et al. 1999), this would have involved dispersal across considerable distances: southern India to the Maldives (Male), where no species of Rousettus has been recorded (Bates and Harrison 1997), is about 425 km; from the Maldives to the granitic Seychelles, also where no species of Rousettus has been recorded (Goodman and Gerlach 2007), is about 2,000 km; and then from the granitic Seychelles to northern Madagascar, an additional 800 km. This complete trajectory is over 3,200 km. However, if R. aegyptiacus is the closest species to the madagascariensis!obliviosus group, the distances of 300–400 km that separate the African continent from Madagascar and the Comoros are more reasonable for members of this genus to cross. Clearly, further molecular genetic work is needed, particularly with samples from east of the Sunda Shelf and Australia, to understand the evolutionary and speciation history of this genus across a much broader geographical scale.
Acknowledgments
On Madagascar, we are grateful to the Direction des Eaux et Forêts and Association National pour la Gestion des Aires Protégées, and in the Comoros, to Yahaya Ibrahim of the Centre National de Documentation et de Recherche Scientifique, and Ishaka Said of Action Comores for aid in numerous ways, including permission to collect specimens. We acknowledge Scott G. Cardiff, Zafimahery Rakotomalala, Eddy Rakotonandrasana, Julie Ranivo, Manuel Ruedi, Fanja Ratrimomanarivo, and Nicole Weyeneth for their aid with fieldwork. Teresa Ai, Jonathan Schwartz, and David Weisrock assisted with the collection of sequence data. Conservation International (CABS), John D. and Catherine T. MacArthur Foundation, National Geographic Society (6637–99 and 7402–03), National Science Foundation (DEB 05-16313), and the Volkswagen Foundation have generously supported field research associated with this paper. We are grateful to two anonymous reviewers and Richard D. Stevens for comments on an earlier version of the paper.
Appendix I
Museum number, species, locality, and GenBank numbers of specimens used in this study. Museums holding this material include the Field Museum of Natural History (FMNH) and the Université d'Antananarivo Département de Biologie Animale (UADBA). Abbreviations used in locality names include: PN = Parc National, RNI = Réserve Naturelle Intégrale, RS = Réserve Spéciale, SF = Station Forestière.
Museum number, species, locality, and GenBank numbers of specimens used in this study. Museums holding this material include the Field Museum of Natural History (FMNH) and the Université d'Antananarivo Département de Biologie Animale (UADBA). Abbreviations used in locality names include: PN = Parc National, RNI = Réserve Naturelle Intégrale, RS = Réserve Spéciale, SF = Station Forestière.
| Museum number | Species | Province/ island | Locality | GenBank accession number |
|---|---|---|---|---|
| FMNH 179237 | R. madagascariensis | Antsiranana | Andlavakarano (Andavadrano) | GU228684 |
| FMNH 179319 | R. madagascariensis | Antsiranana | Andlavakarano (Andavadrano) | GU228685 |
| FMNH 172678 | R. madagascariensis | Antsiranana | Antsahabe River, near village of Ankijabe | GU228639 |
| FMNH 172679 | R. madagascariensis | Antsiranana | Antsahabe River, near village of Ankijabe | GU228640 |
| FMNH 179320 | R. madagascariensis | Antsiranana | Campement Matsaborymadio | GU228686 |
| FMNH 188555 | R. madagascariensis | Antsiranana | Nosy Be | GU228693 |
| FMNH 188556 | R. madagascariensis | Antsiranana | Nosy Be | GU228694 |
| FMNH 188558 | R. madagascariensis | Antsiranana | Nosy Be | GU228695 |
| FMNH 188559 | R. madagascariensis | Antsiranana | Nosy Be | GU228696 |
| UADBA SMG-15185 | R. madagascariensis | Antsiranana | Nosy Be | GU228697 |
| UADBA SMG-15186 | R. madagascariensis | Antsiranana | Nosy Be | GU228698 |
| UADBA SMG-15187 | R. madagascariensis | Antsiranana | Nosy Be | GU228699 |
| UADBA SMG-15188 | R. madagascariensis | Antsiranana | Nosy Be | GU228700 |
| FMNH 188561 | R. madagascariensis | Antsiranana | Nosy Be | GU228701 |
| FMNH 188562 | R. madagascariensis | Antsiranana | Nosy Be | GU228702 |
| FMNH 188564 | R. madagascariensis | Antsiranana | Nosy Be | GU228703 |
| FMNH 188613 | R. madagascariensis | Antsiranana | Nosy Be | GU228704 |
| FMNH 188614 | R. madagascariensis | Antsiranana | Nosy Be | GU228705 |
| FMNH 188615 | R. madagascariensis | Antsiranana | Nosy Be | GU228706 |
| UADBA SMG-15201 | R. madagascariensis | Antsiranana | Nosy Be | GU228707 |
| FMNH 187670 | R. madagascariensis | Antsiranana | Nosy Be | GU228597 |
| FMNH 187675 | R. madagascariensis | Antsiranana | Nosy Be | GU228598 |
| FMNH 187676 | R. madagascariensis | Antsiranana | Nosy Be | GU228599 |
| FMNH 187684 | R. madagascariensis | Antsiranana | Nosy Be | GU228600 |
| FMNH 187690 | R. madagascariensis | Antsiranana | Nosy Be | GU228601 |
| FMNH 187693 | R. madagascariensis | Antsiranana | Nosy Be | GU228602 |
| FMNH 187698 | R. madagascariensis | Antsiranana | Nosy Be | GU228603 |
| FMNH 187699 | R. madagascariensis | Antsiranana | Nosy Be | GU228604 |
| FMNH 187701 | R. madagascariensis | Antsiranana | Nosy Be | GU228605 |
| FMNH 187702 | R. madagascariensis | Antsiranana | Nosy Be | GU228606 |
| FMNH 187703 | R. madagascariensis | Antsiranana | Nosy Be | GU228607 |
| UADBA SMG-15239 | R. madagascariensis | Antsiranana | Nosy Komba | GU228708 |
| UADBA SMG-15240 | R. madagascariensis | Antsiranana | Nosy Komba | GU228709 |
| UADBA SMG-15241 | R. madagascariensis | Antsiranana | Nosy Komba | GU228710 |
| FMNH 188629 | R. madagascariensis | Antsiranana | Nosy Komba | GU228711 |
| FMNH 188630 | R. madagascariensis | Antsiranana | Nosy Komba | GU228712 |
| FMNH 188631 | R. madagascariensis | Antsiranana | Nosy Komba | GU228713 |
| FMNH 188632 | R. madagascariensis | Antsiranana | Nosy Komba | GU228714 |
| UADBA SMG-15250 | R. madagascariensis | Antsiranana | Nosy Komba | GU228715 |
| FMNH 188622 | R. madagascariensis | Antsiranana | Nosy Komba | GU228716 |
| FMNH 188623 | R. madagascariensis | Antsiranana | Nosy Komba | GU228717 |
| FMNH 188624 | R. madagascariensis | Antsiranana | Nosy Komba | GU228718 |
| FMNH 178790 | R. madagascariensis | Antsiranana | RS d'Analamerana, Grotte de Bazaribe | GU228673 |
| FMNH 178791 | R. madagascariensis | Antsiranana | RS d'Analamerana, Grotte de Bazaribe | GU228674 |
| FMNH 178792 | R. madagascariensis | Antsiranana | RS d'Analamerana, Grotte de Bazaribe | GU228675 |
| FMNH 178793 | R. madagascariensis | Antsiranana | RS d'Analamerana, Grotte de Bazaribe | GU228676 |
| FMNH 178795 | R. madagascariensis | Antsiranana | RS d'Analamerana, Grotte de Bazaribe | GU228677 |
| FMNH 178796 | R. madagascariensis | Antsiranana | RS d'Analamerana, Grotte de Bazaribe | GU228678 |
| FMNH 178797 | R. madagascariensis | Antsiranana | RS d'Analamerana, Grotte de Bazaribe | GU228679 |
| FMNH 154287 | R. madagascariensis | Antsiranana | RS d'Anjanaharibe-Sud, Marolakana River | GU228633 |
| FMNH 176268 | R. madagascariensis | Antsiranana | RS d'Ankarana, 3.5 km SE Andrafiabe (village) | GU228672 |
| FMNH 176262 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228668 |
| FMNH 176263 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228669 |
| FMNH 176264 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228670 |
| FMNH 176265 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228671 |
| FMNH 172997 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228641 |
| FMNH 172998 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228642 |
| FMNH 172999 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228643 |
| FMNH 173000 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228644 |
| FMNH 173001 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228645 |
| FMNH 177383 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228629 |
| FMNH 177384 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte des Chauves-souris | GU228630 |
| FMNH 183863 | R. madagascariensis | Antsiranana | RS d'Ankarana, Grotte Milaintety | GU228631 |
| FMNH 169703 | R. madagascariensis | Antsiranana | RS d'Ankarana, near Andrafiabe Cave | GU228637 |
| FMNH 162064 | R. madagascariensis | Fianarantsoa | 9 km NE Ivohibe, 6.5 km ESE Angodongodona | GU228636 |
| FMNH 187600 | R. madagascariensis | Fianarantsoa | Ambodiamontana | GU228692 |
| FMNH 151705 | R. madagascariensis | Fianarantsoa | PN d'Andringitra | GU228632 |
| FMNH 179289 | R. madagascariensis | Mahajanga | Grotte d'Anjohibe | GU228681 |
| FMNH 179290 | R. madagascariensis | Mahajanga | Grotte d'Anjohibe | GU228682 |
| FMNH 179294 | R. madagascariensis | Mahajanga | Grotte d'Anjohibe | GU228683 |
| FMNH 184008 | R. madagascariensis | Mahajanga | Grotte d'Ankelimahogo | GU228687 |
| FMNH 184009 | R. madagascariensis | Mahajanga | Grotte d'Ankelimahogo | GU228688 |
| FMNH 184010 | R. madagascariensis | Mahajanga | Grotte d'Ankelimahogo | GU228689 |
| FMNH 184011 | R. madagascariensis | Mahajanga | Grotte d'Ankelimahogo | GU228690 |
| FMNH 184012 | R. madagascariensis | Mahajanga | Grotte d'Ankelimahogo | GU228691 |
| FMNH 169679 | R. madagascariensis | Mahajanga | PN de Bemaraha, Foret d'Andranogidro | GU228638 |
| FMNH 175759 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228646 |
| FMNH 175760 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228647 |
| FMNH 175761 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228648 |
| FMNH 175762 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228649 |
| FMNH 175763 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228650 |
| FMNH 175765 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228651 |
| FMNH 175766 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228652 |
| FMNH 175897 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228653 |
| FMNH 175898 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228654 |
| FMNH 175899 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228655 |
| FMNH 175900 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228656 |
| FMNH 175901 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228657 |
| FMNH 175903 | R. madagascariensis | Mahajanga | RNI de Namoroka, along Ampandra River | GU228658 |
| FMNH 175767 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228659 |
| FMNH 175768 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228660 |
| FMNH 175769 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228661 |
| FMNH 175770 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228662 |
| FMNH 175771 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228663 |
| FMNH 175772 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228664 |
| FMNH 175773 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228665 |
| FMNH 175774 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228666 |
| FMNH 175775 | R. madagascariensis | Mahajanga | RNI de Namoroka, near source of Mandevy River | GU228667 |
| FMNH 187720 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228608 |
| FMNH 187721 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228609 |
| FMNH 187722 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228610 |
| FMNH 187723 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228611 |
| FMNH 187724 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228612 |
| FMNH 187725 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228613 |
| FMNH 187727 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228614 |
| FMNH 187728 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228615 |
| FMNH 187729 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228616 |
| FMNH 187730 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228617 |
| FMNH 187731 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228618 |
| FMNH 187732 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228619 |
| FMNH 187733 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228620 |
| FMNH 187735 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228621 |
| FMNH 187736 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228622 |
| FMNH 187737 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228623 |
| FMNH 187738 | R. madagascariensis | Toamasina | He Sainte Marie | GU228624 |
| FMNH 187739 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228625 |
| FMNH 187741 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228626 |
| FMNH 187742 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228627 |
| FMNH 187743 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228628 |
| FMNH 188656 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228719 |
| FMNH 188657 | R. madagascariensis | Toamasina | He Sainte Marie | GU228720 |
| FMNH 188658 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228721 |
| FMNH 188659 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228722 |
| FMNH 188660 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228723 |
| FMNH 188661 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228724 |
| FMNH 188662 | R. madagascariensis | Toamasina | Ile Sainte Marie | GU228725 |
| FMNH 179197 | R. madagascariensis | Toamasina | SF de Tampolo | GU228680 |
| UADBA ZR-157 | R. madagascariensis | Toliara | Ambohijanahary Mountain | GU228726 |
| FMNH 194598 | R. madagascariensis | Toliara | Ambohijanahary Mountain | GU228727 |
| FMNH 156610 | R. madagascariensis | Toliara | PN d'Andohahela, 8 km NW Eminiminy | GU228634 |
| FMNH 156611 | R. madagascariensis | Toliara | PN d'Andohahela, 8 km NW Eminiminy | GU228635 |
| FMNH 194230 | R. obliviosus | Grande Comore | Panga Milembeni, near village of Dimadjou | GU228753 |
| FMNH 194231 | R. obliviosus | Grande Comore | Nyamaoui Panga [=Panga Chilamouinani], near Fassi | GU228728 |
| FMNH 194232 | R. obliviosus | Grande Comore | Nyamaoui Panga [=Panga Chilamouinani], near Fassi | GU228754 |
| FMNH 194233 | R. obliviosus | Grande Comore | Nyamaoui Panga [=Panga Chilamouinani], near Fassi | GU228755 |
| FMNH 194234 | R. obliviosus | Grande Comore | Nyamaoui Panga [=Panga Chilamouinani], near Fassi | GU228729 |
| FMNH 194236 | R. obliviosus | Grande Comore | Nyamaoui Panga [=Panga Chilamouinani], near Fassi | GU228730 |
| FMNH 194238 | R. obliviosus | Grande Comore | Nyamaoui Panga [=Panga Chilamouinani], near Fassi | GU228731 |
| FMNH 194240 | R. obliviosus | Grande Comore | Nyamaoui Panga [=Panga Chilamouinani], near Fassi | GU228732 |
| FMNH 194241 | R. obliviosus | Grande Comore | Nyamaoui Panga [=Panga Chilamouinani], near Fassi | GU228733 |
| FMNH 194305 | R. obliviosus | Grande Comore | above Boboni, on trail towards La Convalescence | GU228734 |
| FMNH 194306 | R. obliviosus | Grande Comore | above Boboni, on trail towards La Convalescence | GU228735 |
| FMNH 194307 | R. obliviosus | Grande Comore | above Boboni, on trail towards La Convalescence | GU228756 |
| FMNH 194308 | R. obliviosus | Grande Comore | above Boboni, on trail towards La Convalescence | GU228757 |
| FMNH 194313 | R. obliviosus | Anjouan | Grotte de Mangamitsano, between Bambao and Col de Patsi | GU228758 |
| FMNH 194314 | R. obliviosus | Anjouan | Grotte de Mangamitsano, between Bambao and Col de Patsi | GU228759 |
| FMNH 194315 | R. obliviosus | Anjouan | Grotte de Mangamitsano, between Bambao and Col de Patsi | GU228760 |
| FMNH 194316 | R. obliviosus | Anjouan | Grotte de Mangamitsano, between Bambao and Col de Patsi | GU228761 |
| FMNH 194317 | R. obliviosus | Anjouan | Grotte de Mangamitsano, between Bambao and Col de Patsi | GU228736 |
| FMNH 194318 | R. obliviosus | Anjouan | Grotte de Mangamitsano, between Bambao and Col de Patsi | GU228737 |
| FMNH 194320 | R. obliviosus | Anjouan | Grotte de Hapira, near Limbi along Trondroni River | GU228762 |
| FMNH 194322 | R. obliviosus | Anjouan | Grotte de Hapira, near Limbi along Trondroni River | GU228763 |
| FMNH 194438 | R. obliviosus | Anjouan | Lac de Dzialande | GU228738 |
| FMNH 194439 | R. obliviosus | Anjouan | Lac de Dzialande | GU228739 |
| FMNH 194440 | R. obliviosus | Anjouan | Lac de Dzialande | GU228740 |
| FMNH 194441 | R. obliviosus | Anjouan | Lac de Dzialande | GU228741 |
| FMNH 194442 | R. obliviosus | Anjouan | Lac de Dzialande | GU228742 |
| FMNH 194443 | R. obliviosus | Anjouan | Lac de Dzialande | GU228764 |
| FMNH 194444 | R. obliviosus | Anjouan | Lac de Dzialande | GU228765 |
| FMNH 194458 | R. obliviosus | Mohéli | Ouallah I | GU228743 |
| FMNH 194459 | R. obliviosus | Mohéli | Ouallah I | GU228766 |
| FMNH 194460 | R. obliviosus | Mohéli | Ouallah I | GU228744 |
| FMNH 194461 | R. obliviosus | Mohéli | Ouallah I | GU228767 |
| FMNH 194462 | R. obliviosus | Mohéli | Ouallah I | GU228768 |
| FMNH 194463 | R. obliviosus | Mohéli | Ouallah I | GU228769 |
| FMNH 194530 | R. obliviosus | Mohéli | Ouallah I | GU228770 |
| FMNH 194464 | R. obliviosus | Mohéli | near Ouallah I, along Akomodjou River | GU228745 |
| FMNH 194465 | R. obliviosus | Mohéli | near Ouallah I, along Akomodjou River | GU228746 |
| FMNH 194466 | R. obliviosus | Mohéli | near Ouallah I, along Akomodjou River | GU228747 |
| FMNH 194539 | R. obliviosus | Grande Comore | Panga Milembeni, near village of Dimadjou | GU228748 |
| FMNH 194540 | R. obliviosus | Grande Comore | Panga Milembeni, near village of Dimadjou | GU228749 |
| FMNH 194541 | R. obliviosus | Grande Comore | Panga Milembeni, near village of Dimadjou | GU228750 |
| FMNH 194542 | R. obliviosus | Grande Comore | Panga Milembeni, near village of Dimadjou | GU228751 |
| FMNH 194543 | R. obliviosus | Grande Comore | Panga Milembeni, near village of Dimadjou | GU228752 |
| FMNH 194544 | R. obliviosus | Grande Comore | Panga Milembeni, near village of Dimadjou | GU228771 |
Literature Cited
- Álvarez Y., Juste J. B., Tabares E., Garrido-Pertierra A., Ibáñez C., Bautista J. M. 1999. Molecular phylogeny and morphological homoplasy in fruitbats. Molecular Biology and Evolution 16:1061–1067. [DOI] [PubMed] [Google Scholar]
- Andrianaivoarivelo A. R., et al. 2009. Characterization of 22 microsatellite marker loci in the Madagascar rousette (Rousettus madagascariensis). Conservation Genetics 10:1025–1028. [Google Scholar]
- Baker R. J., Bradley R. D. 2006. Speciation in mammals and the genetic species concept. Journal of Mammalogy 87:643–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian S. T., Jr., Tanaka K., Anunciado R. V. P., Natural N. G., Sumalde A. C., Namikawa T. 2001. Phylogenetic relationships among megachiropteran species from the two major islands of the Philippines, deduced from DNA sequences of the cytochrome b gene. Canadian Journal of Zoology 79:1671–1677. [Google Scholar]
- Bates P. J. J., Harrison D. L. 1997. Bats of the Indian subcontinent. Harrison Zoological Museum, Sevenoaks, Kent, United Kingdom. [Google Scholar]
- Bergmans W. 1977. Notes on new material of Rousettus madagascariensis Grandidier, 1829 (Mammalia, Megachiroptera). Mammalia 41:67–74. [Google Scholar]
- Bergmans W. 1994. Taxonomy and biogeography of African fruit bats (Mammalia, Megachiroptera). 4. The genus Rousettus Gray, 1821. Beaufortia 44:79–126. [Google Scholar]
- Calisher C. H., Childs J. E., Field H. E., Holmes K. V., Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clinical Microbiology Reviews 19:531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbet G. B., Hill J. E. 1991. A world list of mammalian species. 3rd ed. Oxford University Press, New York. [Google Scholar]
- Dorst J. 1947. Les chauves-souris de la faune Malgache. Bulletin du Muséum National d'Histoire Naturelle, série 2, 19:306–313. [Google Scholar]
- Emerick C. M., Duncan R. A. 1982. Age progressive volcanism in the Comores Archipelago, western Indian Ocean and implications for Somali plate tectonics. Earth and Planetary Science Letters 60:415–428. [Google Scholar]
- Excoffier L., Laval G., Schneider S. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon W. L., Sikes R. S., the Animal Care,Use Committee of the American Society Of Mammalogists 2007. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy 88:809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GiAnnini N. P., Almeida F. C., Simmons N. B. 2009. Phylogenetic relationships of Harpyionycterine megabats (Chiroptera: Pteropodidae). Bulletin of the American Museum of Natural History 331:183–204. [Google Scholar]
- Gi Annini N. P., Simmons N. B. 2003. A phylogeny of megachiropteran bats (Mammalia: Chiroptera: Pteropodidae) based on direct optimization analysis of one nuclear and four mitochondrial genes. Cladistics 19:496–511. [DOI] [PubMed] [Google Scholar]
- Giannini N. P., Simmons N. B. 2005. Conflict and congruence in a combined DNA morphology analysis of megachiropteran bat relationships (Mammalia: Chiroptera: Pteropodidae). Cladistics 21:411–437. [DOI] [PubMed] [Google Scholar]
- Goodman S. M., et al. 2005. The distribution and conservation of bats in the dry regions of Madagascar. Animal Conservation 8:153–165. [Google Scholar]
- Goodman S. M., Ganzhorn J. 1997. Rarity of figs (Ficus) on Madagascar and its relationship to a depauperate frugivore community. Revue d'Ecologie 52:321–329. [Google Scholar]
- Goodman S. M., Gerlach J. 2007. Chiroptera. Pp. 105–109 in Terrestrial and freshwater vertebrates of the Seychelles Islands (Gerlach J., ed.). Backhuys Publishers, Leiden, The Netherlands. [Google Scholar]
- Goodman S. M., et al. 2009. The use of molecular and morphological characters to resolve the taxonomic identity of cryptic species: the case of Miniopterus manavi (Chiroptera: Miniopteridae). Zoologica Scripta 38:339–363. [Google Scholar]
- Goodman S. M., Weyeneth N., Ibrahim Y., Saïd I., Ruedi M. 2010. A review of the bat fauna of the Comoro Archipelago. Acta Chiropterologica 12:117–141. [Google Scholar]
- Goudet J. 1995. Fstat (version 1.2): a computer program to calculate F-statistics. Journal of Heredity 86:485–486. [Google Scholar]
- Guo S. W., Thompson E. A. 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361–372. [PubMed] [Google Scholar]
- Hayman R. W., Hill J. E. 1971. The mammals of Africa; an identification manual. Part 2 Order Chiroptera. Smithsonian Institution Press, Washington, D.C. [Google Scholar]
- Hey J., Nielsen R. 2007. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proceedings of the National Academy of Sciences 104:2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua P. Y, Chen J. P., Sun M., Liang B., Zhang S. Y., Wu D. H. 2006. Characterization of microsatellite loci in fulvous fruit bat Rousettus leschenaulti. Molecular Ecology Notes 6:939–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. [DOI] [PubMed] [Google Scholar]
- Iehle C, et al. 2007. Henipavirus and tioman virus antibodies in pteropodid bats, Madagascar. Emerging and Infectious Diseases 13:159–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin D. M., Kocher T. D., Wilson A. C. 1991. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution 32:128–144. [DOI] [PubMed] [Google Scholar]
- Juste J., Álvarez Y., Tabares E., Garrido-Pertierra A., Ibáñez C., Bautista J. M. 1999. Phylogeography of African fruitbats (Megachiroptera). Molecular Phylogenetics and Evolution 13:596–604. [DOI] [PubMed] [Google Scholar]
- Juste J., Ibáñez C. 1993. Geographic variation and taxonomy of Rousettus aegyptiacus (Mammalia: Megachiroptera) in the islands of the Gulf of Guinea. Zoological Journal of the Linnean Society 107:117–129. [Google Scholar]
- Juste J. B., Ibáñez C., Machordom A. 1997. Evolutionary relationships among the African fruit bats: Rousettus egyptiacus, R. angolensis, and Myonycteris. Journal of Mammalogy 78:766–774. [Google Scholar]
- Juste J. B., Machordom A., Ibáñez C. 1996. Allozyme variation of the Egyptian Rousette (Rousettus egyptiacus; Chiroptera Pteropodidae) in the Gulf of Guinea (West-Central Africa). Biochemical Systematics and Ecology 24:499–508. [Google Scholar]
- Kalunda M., et al. 1986. Kasokero virus: a new human pathogen from bats (Rousettus aegyptiacus) in Uganda. American Journal of Tropical Medicine and Hygiene 35:387–392. [DOI] [PubMed] [Google Scholar]
- Kingdon J. 1974. East African mammals: an atlas of evolution in Africa. Vol. IIPart A (insectivores and bats) Academic Press, London, United Kingdom. [Google Scholar]
- Kirsch J. A., Flannery T. F., Springer M. S., Lapointe F. J. 1995. Phylogeny of the Pteropodidae (Mammalia: Chiroptera) based on DNA hybridisation, with evidence for bat monophyly. Australian Journal of Zoology 43:395–428. [Google Scholar]
- Kock D. 1978 A new fruit bat of the genus Rousettus Gray 1821, from the Comoro Islands, western Indian Ocean (Mammalia: Chiroptera). Pp. 205–216 in Proceedings of the Fourth International Bat Research Conference (Olembo R. J., Castelino J. B., Mutere F. A., eds.). Kenya Literature Bureau, Nairobi, Kenya. [Google Scholar]
- Koopman K. F. 1994. Chiroptera: systematics. Pp. 1–217 in Handbook of zoology (Niethammer J., Schliemann H., Starck D., eds.). Walter de Gruyter, Berlin, Germany: Vol. 8. [Google Scholar]
- Kuzmin I. V., et al. 2008. Lagos bat virus in Kenya. Journal of Clinical Microbiology 46:1451–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louette M. 2004. Mammifères. Pp. 65–87 in La faune terrestre de l'archipel des Comores (Louette M., Meitre D., Locqué R., eds.). Musée royal de l'Afrique centrale, Tervuren, Belgium. [Google Scholar]
- MacKinnon J. L., Hawkins C. E., Racey P. A. 2003. Pteropodidae, fruit bats. Pp. 1299–1302 in The natural history of Madagascar (Goodman S. M., Benstead J. P., eds.). University of Chicago Press, Chicago, Illinois. [Google Scholar]
- Maddison D. R., Maddison W. P. 2003. MacClade 4: analysis of phylogeny and character evolution. Version 4.06. Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Norberg U. M. 1981. Allometry of bat wings and legs and comparison with bird wings. Philosophical Transactions of the Royal Society London, B. Biological Sciences 292:359–398. [Google Scholar]
- Norberg U. M. 1994. Wing design, flight performance and habitat use in bats. Pp. 205–239 in Ecological morphology: integrative organismal biology (Wainwright P. C., Reilly S. M., eds.). University of Chicago Press, Chicago, Illinois. [Google Scholar]
- Nougier J., Cantagrel J. M., Karche J. P. 1986. The Comores Archipelago in the western Indian Ocean: volcanology, geochronology and geodynamic setting. Journal of African Earth Sciences 5:135–145. [Google Scholar]
- Nylander J. A. A. 2004. MrModeltest v2. Program distributed by the author Evolutionary Biology Centre, Uppsala University, Sweden. [Google Scholar]
- Pavri K. M., Singh K. R. P., Hollinger F. B. 1971. Isolation of a new parainfluenza virus from a frugivorous bat, Rousettus leschenaulti, collected at Poona, India. American Journal of Tropical Medicine and Hygiene 20:125–130. [DOI] [PubMed] [Google Scholar]
- Pesole G., Gissi C., De Chirico A., Saccone C. 1999. Nucleotide substitution rate of mammalian mitochondrial genomes. Journal of Molecular Evolution 48:427–434. [DOI] [PubMed] [Google Scholar]
- Peterson R. L., Eger J. L., Mitchell L. 1995. Chiroptères. Faune de Madagascar 84:1–204. [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racey P. A., Goodman S. M., Jenkins R. K. B. 2010. The ecology and conservation of Malagasy bats. Pp. 369–404 in Island bats (Fleming T. H., Racey P. A., eds). University of Chicago Press, Chicago, Illinois. [Google Scholar]
- Ratrimomanarivo F. H., Goodman S. M., Stanley W. T., Hoosen N., Taylor P. J., Lamb J. 2008. Morphological and molecular variation in Mops leucostigma (Chiroptera: Molossidae) of Madagascar and the Comoros: phylogeny, phylogeography and geographic variation. Mitteilungen aus dem Hamburgischen Zoologischen Museum 105:57–101. [Google Scholar]
- Ratrimomanarivo F. H., Goodman S. M., Stanley W. T., Naidoo T., Taylor P. J., Lamb J. 2009. Geographie and phylogeographic variation in Chaerephon leueogaster (Chiroptera: Molossidae) of Madagascar and the western Indian Ocean islands of Mayotte and Pemba. Acta Chiropterologica 11:25–52. [Google Scholar]
- Raymond M., Rousset F. 1995. Genepop (Version 1.2)— population-genetics software for exact tests and ecumenicism. Journal of Heredity 86:248–249. [Google Scholar]
- Razafindrakoto N. 2006. Etude comparative du régime alimentaire de Pteropus rufus Tiedemann, 1808 et de Rousettus madagascariensis Grandidier, 1928 (Pteropodidae) dans le district de Moramanga. Mémoire Diplôme d'tudes Approfondies, Département de Biologie Animale, Université d'Antananarivo, Antananarivo, Madagascar. [Google Scholar]
- Reeves W., Dowling A., Dasch G. 2006. Rickettsial agents from parasitic Dermanyssoidea (Acari: Mesostigmata). Experimental and Applied Acarology 38:181–188. [DOI] [PubMed] [Google Scholar]
- Rogers A. R., Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Molecuar Biology and Evolution 9:552–569. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Rozas J., Sánchez-DelBarrio J. C., Messeguer X., Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497. [DOI] [PubMed] [Google Scholar]
- Schneider S., Excoffier L. 1999. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sew all B. J., Granek E. F., Trewhella W. J. 2003. The endemic Comoros Islands fruit bat Rousettus obliviosus: ecology, conservation, and Red List status. Oryx 37:344–352. [Google Scholar]
- Simmons N. B. 2005. Order Chiroptera. Pp. 312–529 in Mammal species of the world: a taxonomic and geographic reference, 3rd ed. (Wilson D. E., Reeder D. M., eds.). Johns Hopkins University Press, Baltimore, Maryland. [Google Scholar]
- Springer M. S., Hollar L. J., Kirsch J. A. 1995. Phylogeny, molecules versus morphology, and rates of character evolution among fruitbats (Chiroptera: Megachiroptera). Australian Journal of Zoology 43:557–582. [Google Scholar]
- Towner J. S., et al. 2007. Marburg virus infection detected in a common African bat. PLoS ONE 2(8):e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J. S., et al. 2009. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog 5(7):e 1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenberg G. J., Audry L., Rønsholt L., van der Poel W. H. M., Bruschke C. J. M., Bourhy H. 2002. Presence of European bat lyssavirus RNAs in apparently healthy Rousettus aegyptiacus bats. Archives of Virology 147:349–361. [DOI] [PubMed] [Google Scholar]
- Weyeneth N., Goodman S. M., Stanley W. T., Ruedi M. 2008The biogeography of Miniopterus bats (Chiroptera: Miniopteridae) from the Comoro Archipelago inferred from mitochondrial DNA. Molecular Ecology 17:5205–5219. [DOI] [PubMed] [Google Scholar]