Anterograde transneuronal tracers derived from herpes simplex virus 1 (HSV-1) strain 129 (H129) are important tools for mapping neural circuit anatomic and functional connections. It is, therefore, critical to elucidate the transport pattern of H129 within neurons and between neurons. We constructed recombinant H129 variants with genetically encoded fluorescence-labeled capsid protein and/or glycoprotein to visualize viral particle movement in neurons. Both electron microscopy and light microscopy data show that H129 capsids and envelopes move separately, and notably, capsids are enveloped at axonal varicosity and terminals, which are the sites forming synapses to connect with other neurons. Superresolution microscopy-based colocalization analysis and inhibition of H129 particle movement by inhibitors of molecular motors support that kinesin-1 contributes to the anterograde transport of capsids. These results shed light into the mechanisms for anterograde transport of H129-derived tracer in axons and transmission between neurons via synapses, explaining the anterograde labeling of neural circuits by H129-derived tracers.
KEYWORDS: HSV-1 strain H129, anterograde transneuronal tracer, cortical neuron, axonal transport, capsids, virions, anterograde movement, retrograde movement, kinesin-1, dynein
ABSTRACT
The features of herpes simplex virus 1 (HSV-1) strain 129 (H129), including natural neurotropism and anterograde transneuronal trafficking, make it a potential tool for anterograde neural circuitry tracing. Recently anterograde polysynaptic and monosynaptic tracers were developed from H129 and have been applied for the identification of novel connections and functions of different neural circuitries. However, how H129 viral particles are transported in neurons, especially those of the central nervous system, remains unclear. In this study, we constructed recombinant H129 variants with mCherry-labeled capsids and/or green fluorescent protein (GFP)-labeled envelopes and infected the cortical neurons to study axonal transport of H129 viral particles. We found that different types of viral particles were unevenly distributed in the nucleus, cytoplasm of the cell body, and axon. Most H129 progeny particles were unenveloped capsids and were transported as capsids rather than virions in the axon. Notably, capsids acquired envelopes at axonal varicosities and terminals where the sites forming synapses are connected with other neurons. Moreover, viral capsids moved more frequently in the anterograde direction in axons, with an average velocity of 0.62 ± 0.18 μm/s and maximal velocity of 1.80 ± 0.15 μm/s. We also provided evidence that axonal transport of capsids requires the kinesin-1 molecular motor. These findings support that H129-derived tracers map the neural circuit anterogradely and possibly transsynaptically. These data will guide future modifications and improvements of H129-based anterograde viral tracers.
IMPORTANCE Anterograde transneuronal tracers derived from herpes simplex virus 1 (HSV-1) strain 129 (H129) are important tools for mapping neural circuit anatomic and functional connections. It is, therefore, critical to elucidate the transport pattern of H129 within neurons and between neurons. We constructed recombinant H129 variants with genetically encoded fluorescence-labeled capsid protein and/or glycoprotein to visualize viral particle movement in neurons. Both electron microscopy and light microscopy data show that H129 capsids and envelopes move separately, and notably, capsids are enveloped at axonal varicosity and terminals, which are the sites forming synapses to connect with other neurons. Superresolution microscopy-based colocalization analysis and inhibition of H129 particle movement by inhibitors of molecular motors support that kinesin-1 contributes to the anterograde transport of capsids. These results shed light into the mechanisms for anterograde transport of H129-derived tracer in axons and transmission between neurons via synapses, explaining the anterograde labeling of neural circuits by H129-derived tracers.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is a human neurotropic and neuroinvasive pathogen which belongs to the alphaherpesvirus subfamily. HSV-1 traffics as a fully enveloped capsid (i.e., virions, the “married model”) or capsid alone (the “separate model”) in the axon, and the dominant model varies with different HSV-1 strain infections of different neuron types or cell lines (1–6). HSV-1 strains vary in their virulence (7), which is probably due to base substitutions leading to amino acid or cis-regulatory changes (8–12). Most strains of HSV-1 spread primarily in the retrograde direction, i.e., from the neuronal soma to dendrites, while HSV-1 strain H129 (H129) moves primarily anterogradely from the soma to the distal axons (13). Subsequent studies have confirmed the preferential anterograde transport of strain H129 (14–18). H129-derived anterograde transneuronal tracers (19) are mostly applied to studies of the connectivity of central nervous system (16–18). Therefore, further study of H129 axonal transport in cortical neurons is crucial for accurate interpretation of neural circuit mapping results.
HSV-1 capsids exposing the inner tegument but not a naked capsid could associate simultaneously with multiple motor types in vitro (20). It has not been documented yet which motor protein(s) drives anterograde transport of H129 particles in axons. Kinesins and dynein are microtubule-based molecular motors responsible for the plus end-directed and minus end-directed movement of various intracellular cargoes, respectively (21, 22). In neurons, organelles are usually observed to move outward from the cell body in a “fast” transport manner with speeds of up to ∼1 μm/s, while cytoskeletal proteins and some soluble proteins are delivered via a “slow” transport manner at speeds of <0.1 μm/s (20). Motors of the kinesin-1, kinesin-2, and kinesin-3 subfamilies contribute to long-range axonal transport (20, 21).
Studies on the motility of pseudorabies virus (PRV), an alphaherpesvirus member, indicate that the majority of viral particles within axons are enveloped and resident in the lumen of vesicles during the egress phase of infection (23, 24). PRV moves with an average velocity of 1.97 ± 0.06 μm/s for all anterograde runs and a top speed of ≈5 μm/s and an average velocity of 1.28 ± 0.12 μm/s for retrograde runs in chick embryo dorsal root ganglia (DRG) (25). In contrast, progeny HSV-1 particles are less engaged in active transport in vertebrate neurons than those of PRV (3). The velocities of HSV-1 retrograde runs range from 0.5 to 5.0 μm/s, with averages of 2.1 to 2.6 μm/s in vertebrate neurons (3), and at 2.2 ± 0.26 μm/s in squid giant axon (26). These viral particles move in the axon by fast axonal transport at a rate comparable to that of organellar transport in cultured neurons.
In this study, we created single or dual fluorescence-labeled HSV-1 strain H129 recombinant viruses to investigate its axonal transport in mouse cortical neurons cultured in a microfluidic device described previously (19). Both electron microscopy (EM) and live-cell imaging were used to analyze virion components and their movement in cortical neuronal axons. We found that H129 particles were mainly transported as capsids in accordance with a separate model in the anterograde direction in the axon. We also showed that viral capsids were associated with the kinesin-1 family members KIF5A and KIF5B, and their transport required kinesin-1 activity.
RESULTS
H129 viral particles distribute unevenly in the subcellular compartments of cortical neurons.
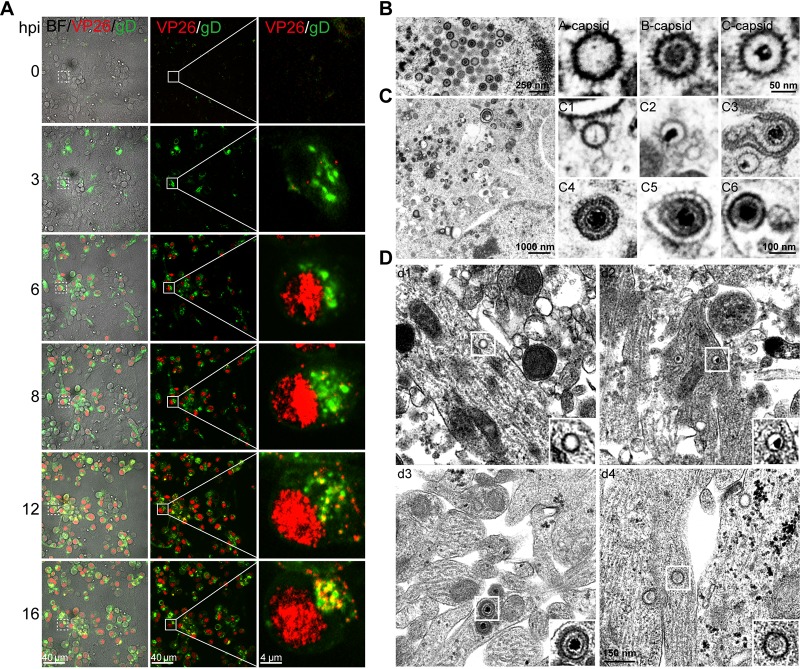
We investigated the infection and life cycle of HSV-1 strain H129 (H129) in cultured cortical neurons. Infection of the neurons was performed with the following genetically modified H129 viruses: H129-G4, which encodes 4 copies of the enhanced green fluorescent protein (EGFP) gene (19); a dually labeled recombinant H129 virus, H129-VP26-mCherry-gD-GFP (H129-G/R), in which VP26, a small capsid protein located on top of the hexon proteins (27), fused with mCherry, and gD, an envelope glycoprotein (28, 29) required for viral entry (30), were fused with GFP; and wild-type H129 virus. The infection progress and life cycle of all three viruses in cortical neurons appear identical. Fig. 1A shows images of live cells from the same viewfield and amplified images that were obtained from the same infected cell at the indicated times postinfection with H129-G/R. Most of capsids and capsid proteins were found in the nucleus, but a few egressed from the nucleus and appeared in the cytoplasm at 8 hours postinfection (hpi); capsids filled up the entire nucleus, and more capsids were observed in the cytoplasm and some at the edge of the cell membrane at 12 and 16 hpi. Thus, the life cycle of H129-G/R is about 12 h, which is consistent with previous reports with wild-type HSV-1 infection (31).
FIG 1.
Subcellular distributions of H129 viral particles in cultured cortical neurons. (A) Primary mouse embryonic cortical neurons were cultured in a microfluidic chamber for 14 days and then infected with H129-G/R at the soma side at an MOI of 1. Confocal images were captured at the indicated time points postinfection. (B) Primary mouse embryonic cortical neurons were cultured in 35-mm dish for 14 days and then were infected with H129-G4 at an MOI of 1. At 24 hours postinfection (hpi), neurons were fixed with 2.5% glutaraldehyde and collected for transmission electron microscopy (EM) analysis. Total viral particles, including capsids, enveloped virions, or other forms of viral particles were observed and analyzed. Capsids in the nuclei: empty A capsid; scaffold-containing B capsid; viral DNA-containing C capsid. (C) Viral particles in the cytoplasm: cytosolic A capsid (C1); cytosolic C capsid (C2); wrapping intermediate: capsids closely associated with cytoplasmic membrane (enveloping viral particles) (C3); complete virion after secondary envelopment (C4); complete virion with an extra membrane (C5); a complete virion along with other cargos wrapped within an extra vesicular structure (virions in MVB-like structure) (C6). (D) Viral particles in the neurites: neurite empty A capsid (d1); a neurite C capsid (d2); virions (d3); noninfectious particles (d4).
To determine the subcellular distribution of different types of viral particles in the nucleus, somatic cytoplasm and neurites (including axons) of cortical neurons cultured in a 35-mm dish for 14 days were infected with H129-G4 or H129-G/R. At 24 hpi when virions are released or ready for release, infected cortical neurons were fixed for further analysis by transmission electron microscopy (EM). Similar results were observed for H129-G4 and H129-G/R infection. Images showed that there were three types of viral capsids in the nuclei, namely, empty A capsids (n = 55, 34.8%), scaffold-containing B capsids (n = 66, 41.8%), and viral genome-containing C capsids (n = 37, 23.4%) (Fig. 1B; Table 1). In the cytoplasm of the soma, in addition to A (Fig. 1C, panel C1) and C capsids (Fig. 1C, panel C2), there were C capsids being wrapped by a secondary envelope (Fig. 1C, panel C3), enveloped virions (Fig. 1C, panel C4), and enveloped virions within vesicles (Fig. 1C, panel C5) and wrapped within multivesicle body (MVB)-like membrane structures (Fig. 1C, panel C6). Approximately 66% (n = 130) of the viral particles in the somatic cytoplasm are cytosolic C capsid (Table 1). In neurites, we observed A capsid (Fig. 1D, panel d1), C capsids (Fig. 1D, panel d2), virions (Fig. 1D, panel d3), and noninfectious particles (NIPs) (Fig. 1D, panel d4). The occurrence probability of viral particles in neurites was 27.9% (n = 12) for A capsids, 46.5% (n = 20) for C capsids, 18.6% (n = 8) for complete virions, and 7% (n = 3) for NIPs (Table 1). These results indicate that there are more C capsids than fully assembled virions in both the somatic cytoplasm and neurites and that the C capsids are the most dominant progeny H129 viral particles in cortical neurons.
TABLE 1.
Distribution of different types of viral particles in cellular compartments of H129-G/R infected mouse cortical neurons at 24 hpia
| Region | No. of viral particles (% of all viral particles) |
|||||
|---|---|---|---|---|---|---|
| A capsidb | B capsidb | C capsidb | Enveloping | Enveloped | NIPsc | |
| Nuclei | 55 (34.8) | 66 (41.8) | 37 (23.4) | |||
| Soma cytoplasm | 9 (4.6) | 0 (0) | 130 (66) | 17 (8.6) | 41 (20.8) | |
| Neurites | 12 (27.9) | 0 (0) | 20 (46.5) | 8 (18.6) | 3 (7.0) | |
EM images from 10 ultrathin sections of H129-G4 infected mouse cortical neurons were analyzed by counting and characterizing the viral particles.
Indicates capsids in the nuclei, cytoplasm, and neurites.
NIP, noninfectious particles.
Transport of H129 viral particles favor a separate model in cortical neuron axons.
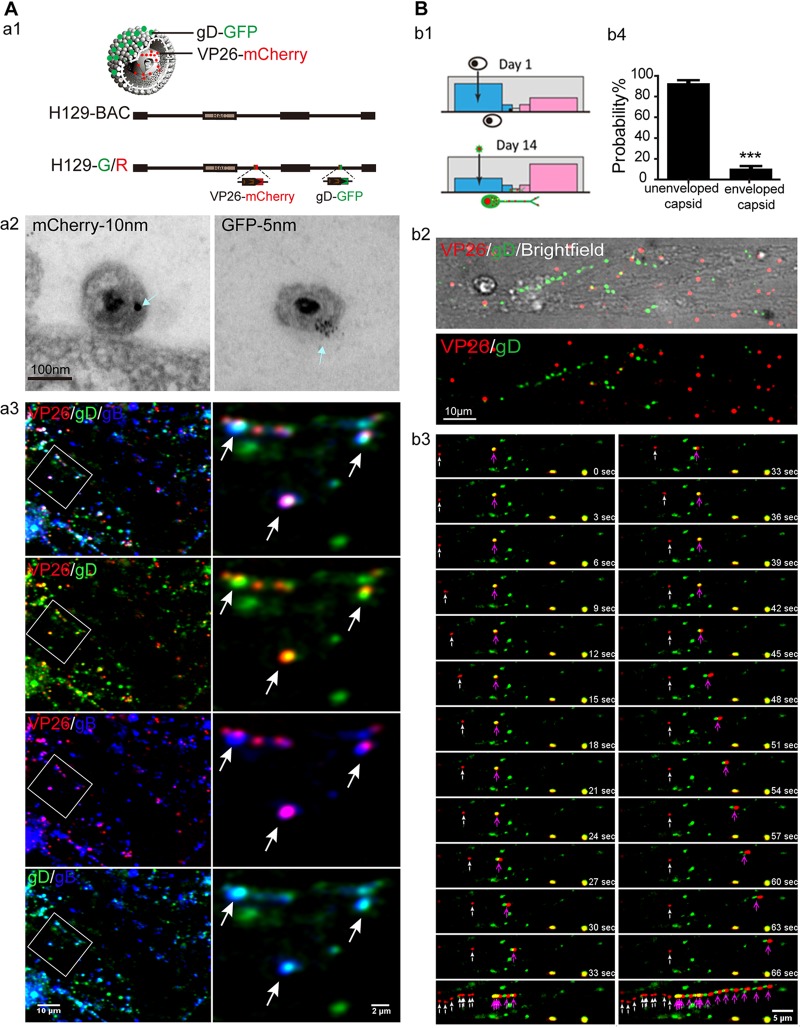
To visualize and trace the movements of viral particles, we used H129G/R, VP26 fused with mCherry, and gD fused with GFP (Fig. 2A, panel a1). An association of VP26-mCherry and gD-GFP with the virion was confirmed by immunoEM with colloidal gold particles of different sizes (Fig. 2A, panel a2). Moreover, at 24 hpi when virions are released or ready for release, most VP26-mCherry+ gD-GFP+ puncta colocalized with gB in the cultured cortical neuron neurites (Fig. 2A, panel a3), indicating that VP26-mCherry and gD-GFP expressed by recombinant H129-G/R could actually label virions.
FIG 2.
H129 viral particles are transported mainly in the form of a capsid in cortical neuron axon by live-imaging analysis. (A) (a1) Schematic of recombinant H129 genome and viral particles; VP26 fused with mCherry and gD fused with green fluorescent protein (GFP), H129-G/R. (a2) EM images for H129-G/R, dually labeled H129 virions. Cell-free viruses in the supernatant were collected at 24 hpi and then processed for immunoEM analysis. Ultrathin sections were probed with antibodies against mCherry or GFP, followed by colloidal gold-conjugated secondary antibodies. Shown are representative images for VP26-mCherry (10 nm gold, left) or gD-GFP (5 nm gold, right) stained on complete virions. Scale bar: 100 nm. (a3) Colocalization of VP26-mCherry, gD-GFP, and gB. Neurons cultured for 14 days were infected with H129-G/R for 24 h and then fixed with 4% paraformaldehyde and immunostained with mouse-anti-gB. Shown are progeny viral particles in neurites. Arrows indicate VP26-mCherry (red) colocalized with both gD-GFP (green) and gB (blue). (B) (b1) The schematic of the microfluidic chamber and culture and infection of cortical neurons in the microfluidic chamber. (b2) Neurons were cultured for 14 days and then infected by adding H129-G/R into the soma (blue) side at an MOI of 1. Movement of viral particles in infected neuron axons was observed by live-cell imaging from 12 hpi to 24 hpi. (b3) Frames of capsid (white arrow) and enveloped capsid (purple arrow) were captured by continuous 1.5-s sequential exposures. Example time-lapse series of a single progeny capsid moving toward axon terminal are shown. Scale bar: 5 μm. (b4) Analysis of the moving viral particles in axons. The numbers of moving unenveloped and enveloped capsids were counted. The proportions of unenveloped and enveloped capsids of the total number of running capsids are shown as mean ± SEM; statistical significance was analyzed by Student’s t test. A total of 591 unenveloped capsids and 38 enveloped capsids were collected from 3 independent experiments. ***, P < 0.001.
Next, the axonal transport of H129-G/R progeny viral particles was visualized by live-cell imaging. In order to clearly distinguish axons from the soma and dendritic area of cortical neurons, cortical neurons were plated in the soma compartment of a microfluidic device so that axons would grow and extend through the micrometer-sized grooves to the contralateral side and be separated from the soma by hydrostatic pressure (Fig. 2B, panel b1) (19, 32). Based on the infection progress and life cycle of H129-G/R in cortical neurons described above, visualizing and tracing the axonal movement of viral particles was performed from 12 hpi to 24 hpi, the egress phase of infection (Fig. 2B, panels b2 and b3). Unenveloped capsids (Fig. 2B, panel b3, white arrow) and enveloped capsids (Fig. 2B, panel b3, purple arrow) were both detected by live imaging. The occurrence probability of unenveloped capsid run was 88.6% ± 3.289% in all run puncta (Fig. 2B, panel b4), which is consistent with EM analysis data that unenveloped capsids were predominate.
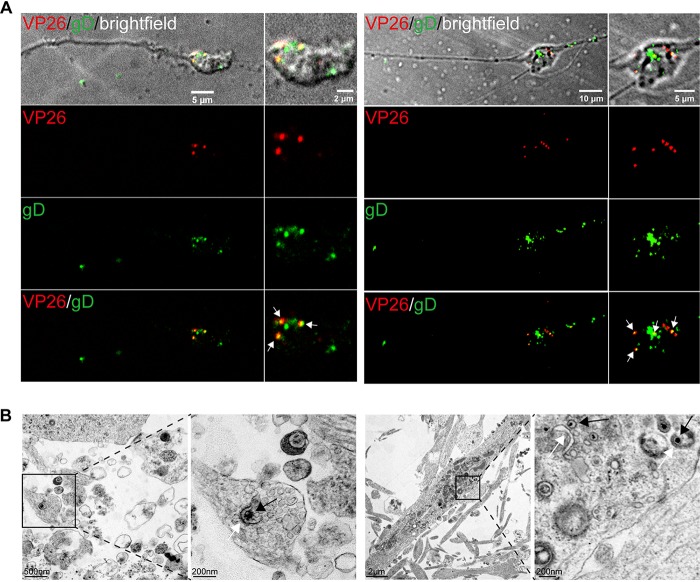
H129 capsids are enveloped in varicosities and terminals of cortical neuron axons.
Most of the released progeny viral particles of HSV-1 are infectious virions. While the H129 capsids and envelopes are transported separately in axons, capsids may become enveloped somewhere in the axon. We observed that the majority of viral particles in growth cones/terminals, VP26-mCherry, and gD-GFP were colocalized with or close to each other, indicating the separately transported capsids become enveloped and form virions (Fig. 3A, left). A similar result was observed in varicosities (Fig. 3A, right). Consistently, enveloping capsids and complete virions were detected by EM analysis of viral particles in growth cones/terminals (Fig. 3B, left) and varicosities (Fig. 3B, right), respectively. Since axonal terminal and varicosity are the sites where synapses are forming and connecting with other neurons, as well as where envelopment of capsids occurs, these data collectively suggest that H129 may transmit to connected neurons via synapses.
FIG 3.
H129 capsids acquire envelopes in the axon terminal and varicosity. (A) Primary mouse embryonic cortical neurons were cultured in the microfluidic chamber for 7 days. When axons grew into the downstream chamber and growth cones were formed, neurons were infected with H129-G/R at an MOI of 1. VP26-mCherry+gD-GFP+ puncta (white arrow) were found in axonal terminal (left panels) and axonal varicosity (right panels) at 24 hpi. (B) Primary mouse embryonic cortical neurons were cultured in a dish for 14 days, then infected with H129-G4 at an MOI of 1, and collected for EM at 24 hpi. (Left) A C capsid being enveloped in axon terminal. (Right) C capsids (black arrows) being closely associated with envelope membrane (white arrows) were found in axon varicosities.
Capsid particles are more motile than enveloped particles in axons.
To assay the motility of viral particles and analyze the movement dynamics of viral particles in the axon, space-time maps (kymographs) were generated by stacking images of either VP26-mCherry-labeled capsids or gD-GFP+ puncta obtained from live-cell imaging of the axons in a 30-μm (width) by 100-μm (length) channel field. Kymographs at time points of 12, 16, 20, and 24 hpi showed that VP26-mCherry-labeled capsids moved more frequently than gD-GFP+ puncta. VP26-mCherry-labeled capsids were more active at 16 hpi and 20 hpi than at other time points (Fig. 4A). The motility of capsids was significantly decreased at 24 hpi compared with 20 hpi. Most of the gD-GFP+ puncta were either stationary or jagged within a limited distance (Fig. 4A). The different dynamics of particle movements further confirmed that the capsid moves separately from the envelope. As the enveloped capsid (virion) is the major form of HSV-1 released from an infected cell, these data suggest that gD-GFP+ puncta are caught up by capsids at either the axon terminal (growth cone) or varicosity, as shown in Fig. 3.
FIG 4.
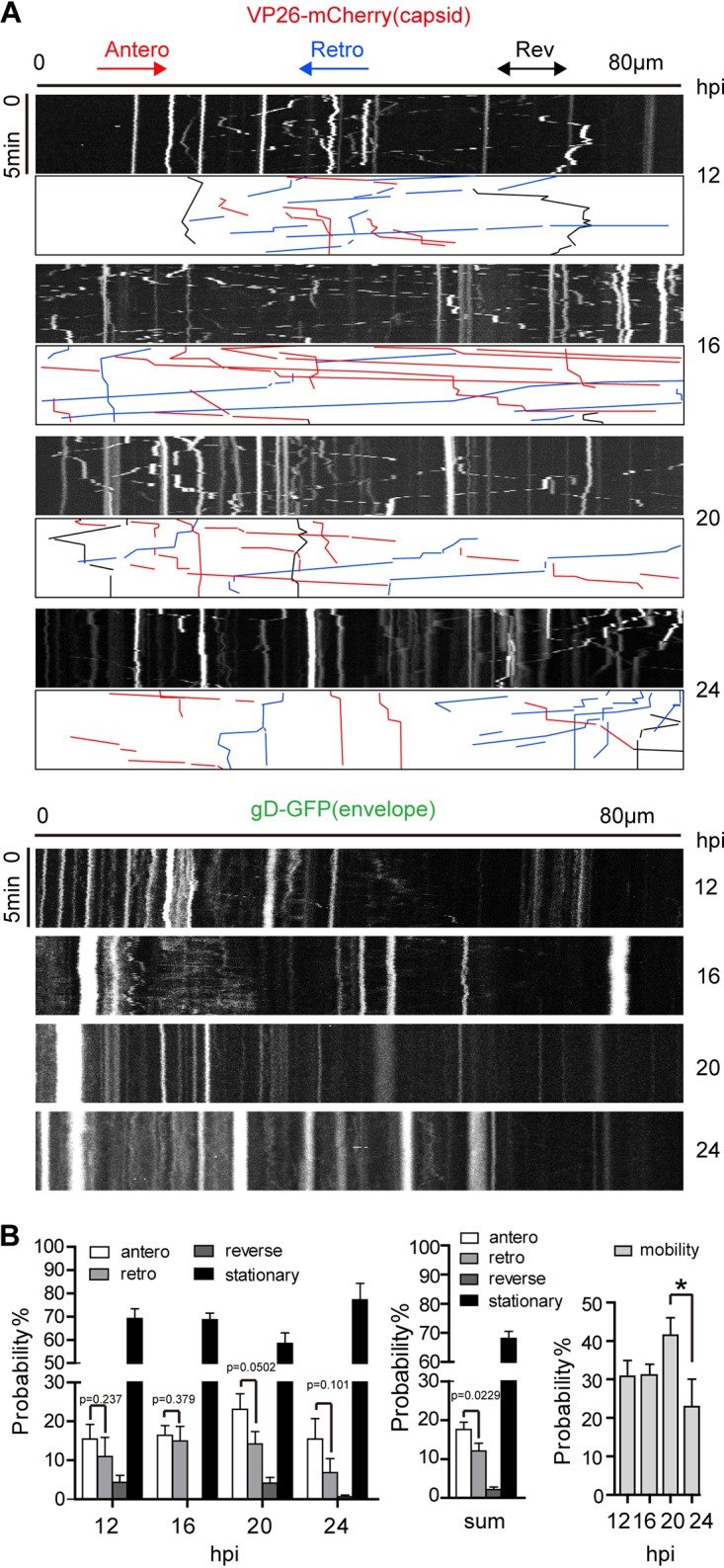
Capsid particles are more motile than enveloped particles in cortical neuron axons. Neurons were cultured for 10 to 14 days and then infected by adding H129-G/R into the soma side at an MOI of 1. The movements of capsids (represented by VP26-mCherry particle) and the envelope particles (represented by gD-GFP particle) were captured by live-cell imaging, and moving characteristics were analyzed. (A) Kymographs for movement of VP26-mCherry puncta (capsids) and gD-GFP puncta (envelopes) in axons. Axon bundle length, 80 μm (horizontal line). Duration of imaging, 5 min (vertical line). Shown are representative kymographs of imaging data obtained at different time points postinfection (12, 16, 20, and 24 hpi). Color-coded traces of anterograde (red), retrograde (blue), and reverse (black) movements were used to quantify puncta transport. (B) Quantification of movement dynamics of VP26-mCherry puncta. The different characteristics of capsid movement were observed, including anterograde movement (antero), retrograde movement (retro), reverse, and stationary state. (Left) Proportions of capsids in the four states at 12, 16, 20, and 24 hpi. (Center) The sum of capsids in the four states at 12, 16, 20, and 24 hpi. (Right) Capsid motility (including movement in anterograde, retrograde, and reverse state) at 12, 16, 20, and 24 hpi. Data were acquired from 3 view fields of each experiment; 3 independent experiments were performed; and 116, 209, 222, and 201 puncta were counted for 12, 16, 20, and 24 hpi, respectively. P value indicates the significant difference between anterograde and retrograde movement, analyzed by Student’s t test. *, P < 0.05; mobility data at 12, 16, 20, and 24 hpi were analyzed by one-way ANOVA followed by Tukey’s test. The results are shown as mean ± SEM.
The movement of capsids in the axons was further characterized. The movement of capsids was classified into the following four categories: anterograde movement, retrograde movement, reverse motion, and stationary state (Fig. 4B, left). A total of 17.6% ± 1.9% of capsids were moving in the anterograde direction, and 12.1% ± 2% of capsids were moving in the retrograde direction. The anterograde movement was statistically significantly more frequent than the retrograde movement (Fig. 4B, middle). The reverse motion occurred at a lower frequency of <5%, while >50% of the capsids in the axons were in the stationary state (Fig. 4B, left and middle graph). Notably, the motility of capsids reached a peak by 20 hpi and dropped at 24 hpi (Fig. 4B, right graph).
The anterograde mobility of capsids fluctuates with infection stages.
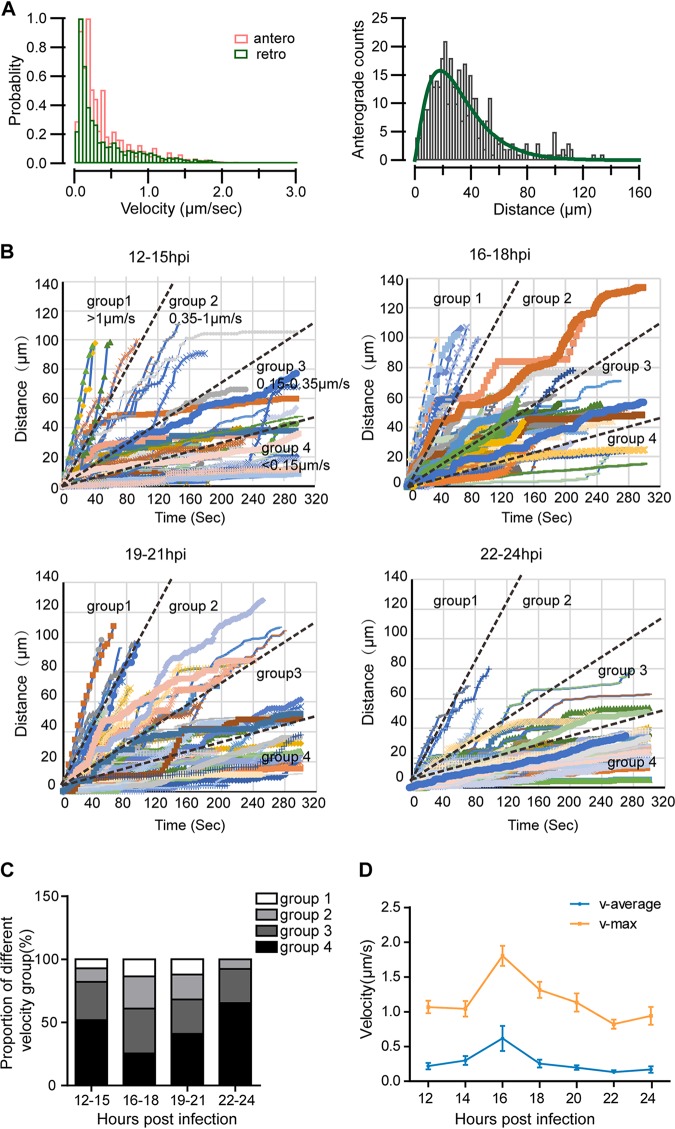
The velocity and run length of anterograde-moving VP26-mCherry+ capsids in axons were analyzed. The instantaneous velocity distribution patterns of anterograde and retrograde capsids were similar during a 12-h period from 12 hpi to 24 hpi, with the largest fraction of puncta moving at a speed below 0.5 μm/s (Fig. 5A, left graph). Most of the anterograde-moving capsids ran a distance between 10 μm and 60 μm during the 5-min imaging period, and the distribution of run length approximated to a gamma distribution was described as f(x) = A*x̂ (alpha−1)*ê (−x/beta) (33), which is consistent with processivity-determined runs (Fig. 5A, right graph).
FIG 5.
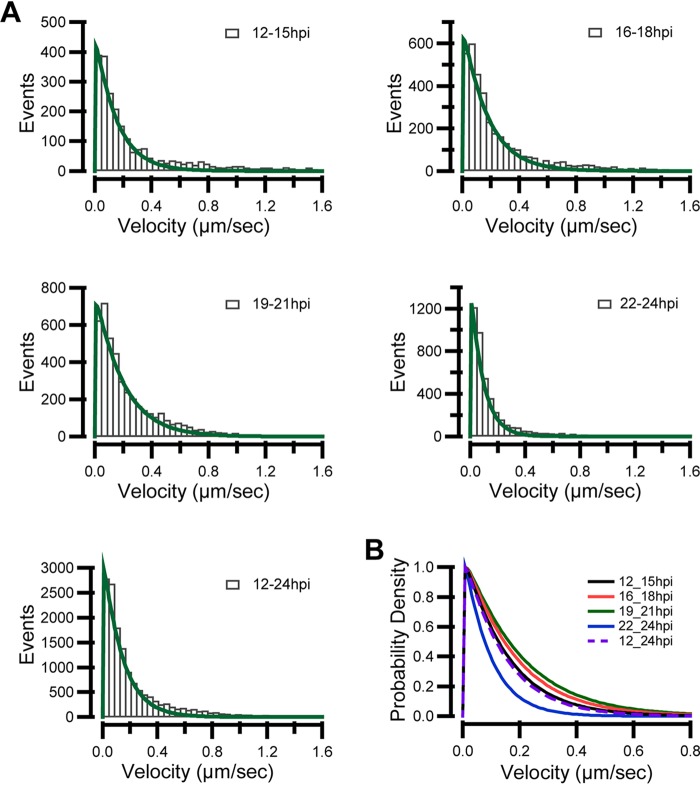
Capsid motility fluctuates with the time of infection. Neurons were cultured for 10 to 14 days and then infected by adding H129-G/R into the soma side at an MOI of 1. The movements of capsids were captured by live-cell imaging, and moving characteristics were analyzed. (A) (Left) Instantaneous velocity distributions of anterogradely (orange histogram) and retrogradely (green histogram) moving capsids. (Right) The histogram of run lengths of anterograde movements and fit using gamma distribution; alpha is 2.2779 ± 0.18, and beta is 14.037 ± 1.52. (B) The time-distance traces of every anterogradely moving capsid during different time periods (12 to 15 hpi, 16 to 18 hpi, 19 to 21 hpi, and 22–24 hpi) were collected. The traces were classified into the following four velocity groups: group 1, fast (>1 μm/s); group 2, medium fast (0.35 to 1 μm/s); group 3, medium (0.15 to 0.35 μm/s); and group 4, slow (<0.15 μm/s). (C) The proportion of the four velocity groups during the indicated periods. (D) Average velocity and maximum velocity of anterograde moving capsids during different periods of 12 to 24 hpi. Data were acquired from 233 anterograde moving capsids from 120 fields of 3 independent experiments.
Next, anterogradely moving capsids which had moved either further than 50 μm or longer than 100 s were analyzed (Fig. 5B). The traces were classified into four groups based on velocity, namely, fast speed (>1 μm/s), medium fast speed (0.35 to 1 μm/s), medium speed (0.15 to 0.35 μm/s), and slow speed (<0.15 μm/s) based on their distribution profile (Fig. 5A). Capsids which have general velocity above 0.15 μm/s occupied a larger proportion at 16 to 18 hpi and 19 to 21 hpi than those at 12 to 15 hpi and 22 to 24 hpi (Fig. 5C). The average velocity (v-average) and maximal velocity (v-max) of anterograde-moving capsids both peaked at 16 hpi, which were 0.62 ± 0.18 μm/s and 1.80 ± 0.15 μm/s, respectively (Fig. 5D). We also plotted modeled velocity distribution for each period using a gamma distribution to calculate the probability of the occurrence of a specific velocity (33). Consistently, there was a higher probability of fast speed during the period of 16 to 21 hpi than earlier (before 16 hpi) and later periods postinfection (after 21 hpi) (Fig. 6A). The velocity distribution also fit a gamma distribution. The shape parameter α ranged from 1.0479 to 1.0699. Thus, a fixed α of 1.05 was used. The values of β were 0.14162 ± 0.0055 from 12 to 15 hpi, 0.16444 ± 0.00553 from 16 to 18 hpi, 0.18309 ± 0.00593 from 19 to 21 hpi, and 0.086236 ± 0.00192 from 22 to 24 hpi, respectively; while the value of β was 0.13191 ± 0.00401 from 12 to 24 hpi. The larger the scale parameter, β, the more spread out the distribution is. The fitting data were normalized and are shown in Fig. 6B. These data indicate that the velocity of anterograde transport capsids varies during the different periods of infection, which is also consistent with the observation shown in Fig. 2B.
FIG 6.
Instantaneous velocity distributions of anterograde transporting capsids. Neurons were cultured for 10 to 14 days and then infected by adding H129-G/R into the soma side at an MOI of 1. The movements of capsids were captured by live-cell imaging, and moving characteristics were analyzed. (A) Instantaneous velocity distributions of anterogradely moving capsids in different time periods, namely, 12 to 15 hpi, 16 to 18 hpi, 19 to 21 hpi, and 22 to 24 hpi and the total during 12 to 24 hpi. Data were described using a gamma distribution, shape parameter α ranged from 1.0466 to 1.0699, and a fixed α of 1.05 was used. The values of the scale parameter β were 0.14162 ± 0.0055 during 12 to 15 hpi, 0.16444 ± 0.00553 during 16 to 18 hpi, 0.18309 ± 0.00593 during 19 to 21 hpi, and 0.086236 ± 0.00192 during 22 to 24 hpi, while it was 0.13191 ± 0.00401 during 12 to 24 hpi. The larger the scale parameter of β, the more spread out the distribution is. (B) Modeled probability of velocity distribution was plotted to summarize velocity distributions for each period using normalized gamma distribution. Data were acquired from 347 anterogradely moving capsids from 120 fields of 3 independent experiments.
Anterograde axonal capsid transport requires kinesin-1 motor activity.
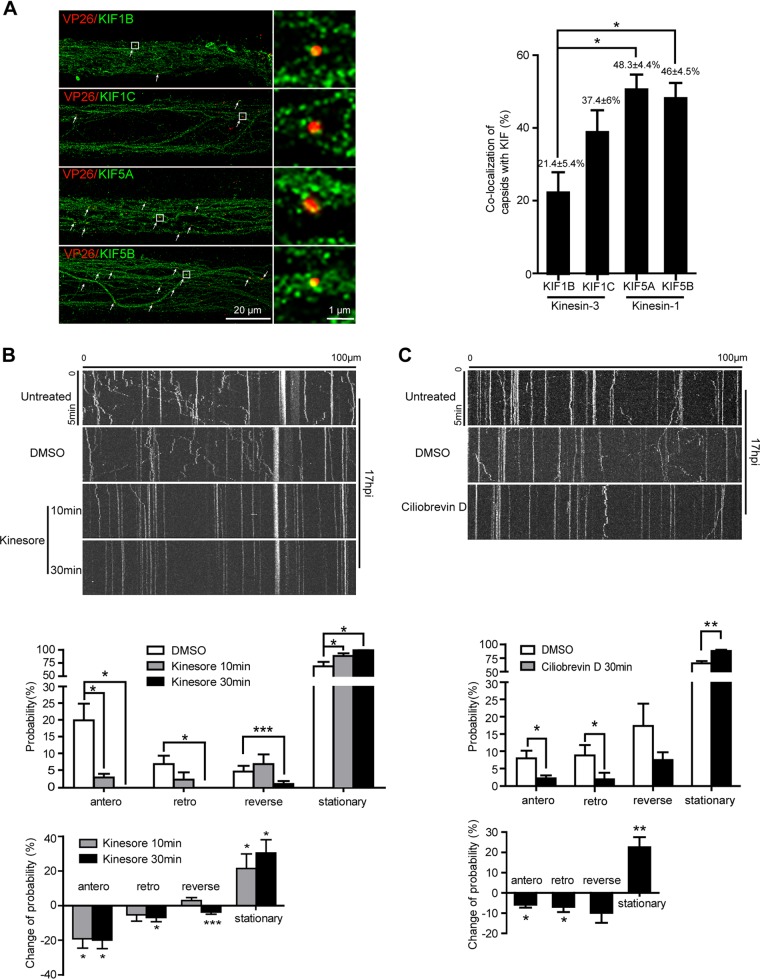
In the axons, microtubules are uniformly oriented with their plus ends pointing toward the distal end. Most motor proteins of the kinesin family move toward the plus ends of the microtubule and are, therefore, capable of driving anterograde cargo transport from the cell body to the axon terminal. Kinesin-1 and kinesin-3 are anterograde motors expressed in the axon. To identify which anterograde motor(s) drives capsid movement in the axon, we infected cortical neurons with H129-VP26-mCherry, and then we analyzed colocalization between VP26-mCherry-labeled capsids and KIF5A and KIF5B of the kinesin-1 family and KIF1B and KIF1C of the kinesin-3 family in the proximal axon bundles (100 μm from the soma chamber) in microchannels by immunostaining and super-resolution microscopy. Quantitative analysis showed that 21.43% ± 5.44% of capsids colocalized with KIF1B, 37.39% ± 5.91% colocalized with KIF1C, 48.34% ± 4.42% colocalized with KIF5A, and 46.00% ± 4.50% colocalized with KIF5B (Fig. 7A). Furthermore, to determine whether axonal transport of H129 capsids requires kinesin activity, we treated neurons with kinesore, a small-molecule inhibitor of kinesin-1 (34), at 17 hpi. Within 10 min, the motility of capsid puncta was inhibited significantly by kinesore, especially anterograde runs; by 30 min, there were barely any moving puncta (Fig. 7B). The effect of ciliobrevin D, an inhibitor of dynein motor activity (35), was also determined. Treatment with ciliobrevin D for 30 min caused significant inhibition of both anterograde and retrograde movement of capsids to a similar extent (Fig. 7C). Taken together, these data indicate that both kinesin-1 and dynein are involved in H129 capsid transport in axons and the kinesin-1 family members are primarily responsible for anterograde transport of capsids toward the axon terminal.
FIG 7.
Kinesin-1 is required for anterograde transport of HSV-1 capsids in axons. (A) Primary mouse embryonic cortical neurons were cultured in the microfluidic chamber for 14 days and then infected with H129-VP26-mCherry at the soma side at an MOI of 1. At 24 hpi, neurons were fixed and stained with indicated kinesin antibodies, followed by superresolution imaging analysis. Shown are representative superresolution images of axon bundle; colocalization of capsids with kinesins is defined as Pearson’s correlation coefficients (PCCs) above 0.5, which is indicated by arrows. Scale bar: 20 μm. Data were collected from at least 18 fields/group of 3 independent experiments and analyzed by one-way ANOVA followed by Tukey’s test. Results are shown as mean ± SEM. *, P < 0.05. (B) Primary mouse embryonic cortical neurons cultured in the microfluidic chamber for 14 days were infected with H129-VP26-mCherry at an MOI of 1. At 17 hpi, neurons were treated with dimethyl sulfoxide (DMSO) for 30 min or kinesore (100 μM) for 10 min or 30 min. Movement of capsids (VP26-mCherry puncta) in axons was observed by live-cell imaging. (Top) Kymographs for movement of capsids without treatment, with DMSO treatment, or with kinesore treatment for 10 min or 30 min in the same field. (Middle) Quantification of the motility of capsids with the treatment of DMSO or kinesore. (Bottom) Quantification of motility changes normalized to the DMSO vehicle control. Data were acquired from 4 view fields containing 20 to 41 VP26 puncta for each group. *, P < 0.05; ***, P < 0.001. (C) Movement of capsids upon ciliobrevin D treatment. (Top) Kymographs for movement of capsids exposed to DMSO or ciliobrevin D (200 μM) at 17 hpi for 30 min; movements of capsids in the same field were captured as described in B. (Middle) Quantification of capsid motility. (Bottom) Quantification of motility changes normalized to the DMSO vehicle control. Data were acquired from 4 view fields containing 23 to 53 capsids. Statistically significant change in probability compared with control was analyzed by Student’s t test. Results are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Our present results are relevant to understanding the features and mechanisms of axonal transport of H129 viral particles. We analyzed the infection progress of the H129-G/R variant, a virus with fluorescence-labeled VP26 and gD. We found that the infection progress was similar to that of H129-G4, which we developed as an anterograde polysynaptic viral tracer from HSV-1 strain H129 (19). Most H129 viral particles are transported as capsids instead of virions in cultured cortical neuron axons. Capsids acquire envelopes in the axon growth cone and varicosity, which may provide the sites for virion egress and transneuronal transmission. More capsids are transported anterogradely than retrogradely, which supports the anterograde transneuronal transmission of H129-derived tracers. These findings are important for the interpretation of H129 tracer-based circuitry mapping, as they provide clarification of the underlying transportation processes.
Our EM analysis revealed that the different types of viral particles were unevenly distributed in different cellular compartments. Notably, capsids were much more frequently observed than virions in axons. These results are consistent with studies on HSV-1 strain F and strain KOS in mouse hippocampal neurons (4). Progeny cytosolic A, B, and C capsids were found in neurons infected with HSV-1 strain F, while only C capsids were found with HSV-1 strain KOS (4). In our study, B capsids were absent in both the somatic cytoplasm and neurites of HSV-1 strain H129-infected cortical neurons. These data indicate that the population distribution of particles may be different with the infections of different strains of HSV-1 in different neuronal cells.
We also investigated axonal transport of H129-G/R in cortical neuron by live-cell imaging. The size of VP26-mCherry puncta is homogeneous, suggesting that capsids regularly assemble as an icosahedron. The size of gD-GFP puncta varies, implying that gD-GFP is embedded in vesicles with different sizes, which is consistent with a previous report (36). Axonal transport analysis found that H129 virus particles were transported separately as unenveloped capsids and envelope-like puncta, in support of the separate model. Later, the capsids acquired an envelope in the axonal growth cone/terminal and varicosity, which might be the eventual exit sites for virions as reported previously (37). Consistent with previous reports that varicosities might be the sites where virions egress (38, 39), we also found that the gD protein accumulated in varicosities. These results indicate that capsids mostly envelop at the axonal terminal or varicosity, and axonal varicosities and terminals are the sites forming synapses connected with other neurons (40, 41). Therefore, H129 might transmit via the synapse, and the H129-derived tracers label neural circuits in a transsynaptic way. In addition, it was reported that during envelopment, the capsid encounters viral glycoproteins on vesicles derived from the trans-Golgi network (42, 43). These findings may explain why the motility of capsids is more frequent than enveloped particles.
Capsids move in an anterograde, retrograde, or reverse direction or stay stationary in axons. Neurons use opposite polarity molecular motors of kinesins and dynein to move cargos along microtubules, either anterograde or retrograde. In the “tug-of-war” model, both anterograde and retrograde motors are simultaneously engaged. Thus, the cargo moves in the direction of whichever set of motors exerts a stronger force. In the “exclusionary presence” model, only one set of motors associates with the cargo at any given time. In the “competition-coordination” model, both sets of motors are tethered to the cargo simultaneously; the cargo moves in the direction of whichever set of motors is activated (22, 44). Kinesin-1, kinesin-2, and dynein/dynactin are reported to bind with tegument proteins on cytosolic C capsids in vitro (45). HSV-1 strain H129 is reported to selectively spread in the anterograde direction in neural circuits in vivo (19, 46). The bidirectional movement of H129 capsids in axons of cultured neurons fits well with the “competition-coordination” model for bidirectional movement driven by molecular motors and was confirmed by the inhibitory effects of kinesin-1 and dynein inhibitors on capsid motility. H129 capsid motility, especially the anterograde motility, was severely inhibited by kinesore, a small molecule that targets the cargo-binding interface of the kinesin-1 light chain (34). Interestingly, consistent with previous reports that ciliobrevin D, an inhibitor of dynein ATPase activity (63), inhibits the bidirectional transport of multiple axonal organelles in the sensory neuron (64), it also stopped H129 capsid movement in both directions in cortical axons. These results indicate that both kinesin-1 and dynein are involved in axonal transport of H129 capsids.
We found that 17.6% ± 1.9% of capsids moved anterogradely, which is higher than those observed moving retrogradely (12.1% ± 2%). The preference of capsids for anterograde axonal transport in cultured cortical neurons in vitro is consistent with neural circuitry tracing results in vivo. This evidence further supports the results from anterograde neural circuit labeling by H129-derived tracers. Nevertheless, bidirectional transport of H129 viral particles in axons might reduce the labeling efficiency and direction specificity of H129-derived tracers. Increasing anterograde run and reducing retrograde run might be a strategy to improve the anterograde labeling efficiency and direction specificity of H129 tracers.
An analysis of the dynamics of anterograde capsid transport showed that the motility of anterograde-moving capsids gradually increased from 12 hpi and reached a peak at about 16 hpi, and then most of the capsids appear to reach a quiescent state after 24 hpi. Several details may explain these motility dynamics. First, as the infection progresses, more and more viral particles and proteins are produced and transported in the axon, which leads to steric hindrance and affects motor movement along microtubules (47, 48). Second, HSV-1 induces mitochondrial damage (49, 50) and affects power supply and cell energy. Studies show that alphaherpesviruses, including PRV and HSV-1, hijack the mitochondria transport machinery, as well as elevate neuron activity by increasing intracellular levels of calcium (51). PRV or HSV-1 infection causes disruption of mitochondria axonal transport after 12 hpi, especially from 16 to 18 hpi, raising the possibility that kinesin is hijacked and repurposed for viral particle transport (51). Our results are in line with these findings.
Kinesin-1 family molecular motors of KIF5A, 5B, and 5C have been identified to drive anterograde transport of enveloped HSV virions in axons (52). KIF5A is neuron specific, whereas KIF5B, KIF1B, and KIF1C are ubiquitously expressed in cells (53). We found that more capsids are colocalized with KIF5A and KIF5B than KIF1B and KIF1C, suggesting an interaction between the H129 capsid and these motor proteins. Tegument proteins are involved in capsid trafficking along the microtubule. For example, tegument protein US11 interacts with the ubiquitous kinesin heavy chain (54), and UL36 is required for HSV transport along microtubules (55). However, the viral proteins that interact with kinesin-1 and dynein to mediate axonal transport remain to be identified.
In summary, more capsids are transported anterogradely than retrogradely, which facilitates the anterograde transneuronal tracing for H129-derived tracers. Considering that both kinesin-1 and dynein are required for axonal transport of H129 capsids, viral protein-molecular motor interactions need to be investigated to identify the targets for viral genome manipulation and to improve the anterograde labeling efficiency and direction specificity of H129-derived tracers in the future.
MATERIALS AND METHODS
Ethics statement.
All experiments followed the procedures of the Ethics Committee on Animal Welfares at Wuhan Institute of Virology (WIVA10201502) according to the guidelines of the Care and Use of Laboratory Animals. All efforts have been made to minimize the suffering and the number of animals used.
Construction of recombinant H129 variants.
The bacterial artificial chromosome (BAC) of HSV-1 strain H129 (H129-BAC) was created as described previously (19). A new recombinant H129 with VP26 fused with mCherry and gD fused with GFP was generated by homologous recombination in Escherichia coli SW302 using H129-BAC. The newly resulted H129 variant was designated H129-VP26-mCherry-gD-GFP (H129-G/R). The expression of fluorescent proteins enables visualization of dynamic process of de novo synthesis of viral proteins and transport of H129-G/R viral particles in real time in living cells. Homologous arms were generated by PCR amplification of the upstream and downstream sequences of the VP26 open reading frame and cloning into H129-BAC (H129-VP26-mCherry). The VP26 primers were GCACCTATTCACCGTTTGTCGTTCGAGAACCTTCGACGCCCGGGACCCCGATGGTGAGCAAGGGCGAGGAGGATAAC and GTTGGTGTGGTCTTTTATTGATTAAAACACCCCAGAAGGAACTCCCCGGGCCTGATCGGCACGTAAGAGGTTCCAAC. Similarly gD-GFP was cloned onto H129-VP26-mCherry BAC using the primer pair of CCCACATCCGGGAAGACGACCAGCCGTCC TCGCACCAGCCCTTGTTTTACGGAGGAGGAGTGAGCAAGGGCGAGG and CCATCCCAACCCCGCAGACCTGACCCCCCCGCACCCATTAAGGGGGGGTATGCGGAACCCCTATTTGTTTATTTTTC.
H129-G4 was generated as previously described (19). Recombinant H129 variants of H129-G4, H129-G/R, and H129-VP26-mCherry were used in this study.
Preparation of cell-free virus stock.
Vero-E6 cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Life Technologies, USA) supplemented with 10% of fetal bovine serum (Life Technologies) and 1% of penicillin/streptomycin (Life Technologies) and infected with H129-G4, H129-G/R or H129-VP26-mCherry at a multiplicity of infection (MOI) of 0.5. The culture media were harvested when 100% of the cells displayed severe cytopathic effects (CPEs), the cell debris was discarded, and the supernatant was subjected to high-speed centrifugation (Beckman rotor SW32; 25,000 rpm, 2.5 h, 4°C). Pellets were resuspended in DMEM, aliquots were stored at –80°C. Titers of virus stocks were determined by plaque-forming assay (19).
Primary mouse cortical neuron isolation and culture in a microfluidic device.
Primary mouse forebrain cortical neurons were prepared from embryonic day 16.5 (E16.5) C57BL/6 mouse embryos according to a protocol described previously (56, 57). Briefly, forebrain tissues were dissected from E16.5 C57BL/6 mouse embryos, dissociated with 0.125% trypsin in Hanks’ balanced salt solution without Ca2+ and Mg2+ at 37°C for 15 min, and triturated in serum-free neurobasal (NB) medium supplemented with 2% B27 supplement and GlutaMAX (Life Technologies, USA). The microfluidic chambers were fabricated as described previously (19), with a microgroove length of 500 μm and width of 10 μm. The isolated cortical neurons were plated in microfluidic device at a density of 1 × 106 cells/side. Medium was changed every day.
Transmission electron microscopy.
Primary forebrain cortical neurons cultured on a 35-mm dish for 14 days in vitro (DIV) were infected with H129-G4 at an MOI of 1. At 24 hours postinfection (hpi), neurons were fixed with 2.5% glutaraldehyde, followed by postfixation with 1% osmium tetroxide, dehydration in ethanol of ascending purity (30%, 50%, 70%, 90%, and 100%,), and embedding with an Embed 812 kit (Electron Microscopy Sciences, Fort Washington, PA). Ultrathin sections (90 nm) were sequentially stained with 3.5% aqueous uranyl acetate and 0.2% lead citrate. Images were acquired on FEI Tecnai G2 20 TWIN transmission electron microscope (Hillsboro, OR, USA) (58).
Live-cell imaging.
Cortical neurons (1 × 106) were plated in the soma side of the microfluidic chamber placed on 35-mm glass-bottom dishes (Nest Biotechnology, China) coated with poly-d-lysine (1 mg/ml; Millipore) diluted in ddH2O, and cultured for 14 days to allow axons to extend into the microchannels driven by hydrostatic pressure. Neurons in the somal chamber were infected with H129-G/R at an MOI of 1, with the liquid level in the somal chamber lower than that of the contralateral side to prevent virus leakage through the microchannel, and the virus inoculum was removed and rinsed after incubation for 3 h. Live-cell imaging of axons in microchannels was performed in the 100-μm-wide field in the proximal axon bundle with a 60× oil objective on double disc fluorescence confocal microscope (PerkinElmer, USA) at 37°C with 5% CO2 in an enclosed chamber (EMBL workshop; no. 530010; Cell Biology Trading). Images were acquired for 5 min at 1.5-sec intervals for both channels, and a punctum run was defined as a period of displacement above 1 μm. The images were deconvoluted using the Volocity Demo software package (PerkinElmer, USA). The movement trace of every punctum was manually tracked for 100 frames in 5-min time-lapse imaging for velocity or distance analysis.
Immunofluorescence staining and superresolution imaging.
Cortical neurons were cultured in a microfluidic chamber placed on 35-mm glass-bottom dishes (Nest Biotechnology) for 14 days to allow axon outgrowth and infected with H129-VP26-mCherry at an MOI of 1. Samples were collected at 24 hpi, followed by fixation with 4% paraformaldehyde and permeabilization with 0.4% Triton X-100 (Sigma-Aldrich). After sequential immunofluorescence staining with primary antibodies and secondary antibodies (59), samples were soaked with standard stochastic optical reconstruction microscopy (STORM) buffer (60) and imaged with a 640-nm laser at an intensity of ∼5 kW/cm2. Superresolution images were acquired on an Olympus IX73 inverted optical microscope equipped with a high numerical aperture (NA) oil-immersion objective (60×, NA1.4; Olympus), a high-power 640-nm laser (LWRL640-3W; LaserWave), and a 405-nm laser. The lasers were coupled into a 4 by 1 multimode fiber combiner, and the output lasers were collimated and focused on the sample using the objective with a 100- by 100-μm2 field of view (61). The microscope is also equipped with a sCMOS camera (Flash 4.0 V3; Hamamatsu Photonics) at a frame rate of 100 Hz (exposure time, 10 ms). The samples were soaked with a standard STORM buffer (60) and imaged with the 640-nm laser at an intensity of ∼5 kW/cm2. The primary antibodies used were polyclonal rabbit anti-KIF1B (catalog number A6638; ABclonal), rabbit anti-KIF1C (catalog number A15786; ABclonal), rabbit anti-KIF5A (catalog number A3303; AbClonal), and rabbit anti-KIF5B (catalog number ab167429; Abcam). The secondary antibody was Alexa Fluor 647-conjugated goat-anti-rabbit IgG (H+L) (catalog number A-31573; Invitrogen).
Statistical analysis.
Each experiment was performed in triplicate, and the results were presented as means ± SEM from three independent experiments. Student’s t test and ANOVA were performed to analyze the statistical significance between data groups. Differences were considered to be significant when the P value was <0.05. The fluorescent puncta were manually tracked by using the Volocity Demo software package (PerkinElmer, USA). Kymographs were generated by FIJI (NIH Image) (62). The instantaneous velocity was analyzed by Igor Pro (Wavemetrics, Lake Oswego, OR, USA).
ACKNOWLEDGMENTS
We thank Lin Yang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), Ding Gao, Anna Du, and Pei Zhang (The Core Facility, Wuhan Institute of Virology, CAS) for technical advice on electron microscopy. We also thank the Optical Bioimaging Core Facility of WNLO for support in data acquisition.
This work was supported by grants from the National Nature Science Foundation of China (81620108021, 81427801, 81871660, 81571355, and 31530039), Key Basic Research Project from MOST (2015CB755601), State Key Laboratory of Molecular Developmental Biology (2017-MDB-KF-08, 2018-MDB-KF-09, 2019-MDB-KF-11), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB32030200 and XDB32020100), and NIHRF1 (1RF1MH120020-01).
REFERENCES
- 1.Miranda-Saksena M, Boadle RA, Aggarwal A, Tijono B, Rixon FJ, Diefenbach RJ, Cunningham AL. 2009. Herpes simplex virus utilizes the large secretory vesicle pathway for anterograde transport of tegument and envelope proteins and for viral exocytosis from growth cones of human fetal axons. J Virol 83:3187–3199. doi: 10.1128/JVI.01579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wisner TW, Sugimoto K, Howard PW, Kawaguchi Y, Johnson DC. 2011. Anterograde transport of herpes simplex virus capsids in neurons by both separate and married mechanisms. J Virol 85:5919–5928. doi: 10.1128/JVI.00116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antinone SE, Zaichick SV, Smith GA. 2010. Resolving the assembly state of herpes simplex virus during axon transport by live-cell imaging. J Virol 84:13019–13030. doi: 10.1128/JVI.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ibiricu I, Huiskonen JT, Döhner K, Bradke F, Sodeik B, Grünewald K. 2011. Cryo electron tomography of herpes simplex virus during axonal transport and secondary envelopment in primary neurons. PLoS Pathog 7:e1002406. doi: 10.1371/journal.ppat.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kratchmarov R, Taylor MP, Enquist LW. 2012. Making the case: married versus separate models of alphaherpes virus anterograde transport in axons. Rev Med Virol 22:378–391. doi: 10.1002/rmv.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham A, Miranda-Saksena M, Diefenbach R, Johnson D. 2013. Letter in response to: making the case: married versus Separate models of alphaherpes virus anterograde transport in axons. Rev Med Virol 23:414–418. doi: 10.1002/rmv.1760. [DOI] [PubMed] [Google Scholar]
- 7.Hill JM, Rayfield MA, Haruta Y. 1987. Strain specificity of spontaneous and adrenergically induced HSV-1 ocular reactivation in latently infected rabbits. Curr Eye Res 6:7. [DOI] [PubMed] [Google Scholar]
- 8.Chou J, Roizman B. 1990. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J Virol 64:1014–1020. doi: 10.1128/JVI.64.3.1014-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negatsch A, Mettenleiter TC, Fuchs W. 2011. Herpes simplex virus type 1 strain KOS carries a defective US9 and a mutated US8A gene. J Gen Virol 92:167–172. doi: 10.1099/vir.0.026484-0. [DOI] [PubMed] [Google Scholar]
- 10.Perng G-C, Mott KR, Osorio N, Yukht A, Salina S, Nguyen Q-H, Nesburn AB, Wechsler SL. 2002. Herpes simplex virus type 1 mutants containing the KOS strain ICP34.5 gene in place of the McKrae ICP34.5 gene have McKrae-like spontaneous reactivation but non-McKrae-like virulence. J Gen Virol 83:2933–2942. doi: 10.1099/0022-1317-83-12-2933. [DOI] [PubMed] [Google Scholar]
- 11.Macdonald SJ, Mostafa HH, Morrison LA, Davido DJ. 2012. Genome sequence of herpes simplex virus 1 strain KOS. J Virol 86:6371–6372. doi: 10.1128/JVI.00646-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szpara ML, Parsons L, Enquist LW. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J Virol 84:5303–5313. doi: 10.1128/JVI.00312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zemanick MC, Strick PL, Dix RD. 1991. Direction of transneuronal transport of herpes-simplex virus-1 in the primate motor system is strain-dependent. Proc Natl Acad Sci U S A 88:8048–8051. doi: 10.1073/pnas.88.18.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun N, Cassell MD, Perlman S. 1996. Anterograde, transneuronal transport of herpes simplex virus type 1 strain H129 in the murine visual system. J Virol 70:5405–5413. doi: 10.1128/JVI.70.8.5405-5413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner JA, LaVail JH. 1999. Differential anterograde transport of HSV type 1 viral strains in the murine optic pathway. J Neurovirol 5:140–150. doi: 10.3109/13550289909021996. [DOI] [PubMed] [Google Scholar]
- 16.Zhu HZ, Yan HH, Tang N, Li XY, Pang P, Li H, Chen WT, Guo Y, Shu S, Cai Y, Pei L, Liu D, Luo MH, Man HY, Tian Q, Mu YL, Zhu LQ, Lu YM. 2017. Impairments of spatial memory in an Alzheimer’s disease model via degeneration of hippocampal cholinergic synapses. Nat Commun 8:1676. doi: 10.1038/s41467-017-01943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li YD, Xu JM, Liu YF, Zhu J, Liu N, Zeng WB, Huang N, Rasch MJ, Jiang HF, Gu X, Li X, Luo MH, Li CY, Teng JL, Chen JG, Zeng SQ, Lin LN, Zhang XH. 2017. A distinct entorhinal cortex to hippocampal CA1 direct circuit for olfactory associative learning. Nat Neurosci 20:559–570. doi: 10.1038/nn.4517. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y-Y, Wang Y, Jiang H-F, Liu J-H, Jia J, Wang K, Zhao F, Luo M-H, Luo M-M, Wang X-M. 2018. Impaired glutamatergic projection from the motor cortex to the subthalamic nucleus in 6-hydroxydopamine-lesioned hemi-parkinsonian rats. Exp Neurol 300:135–148. doi: 10.1016/j.expneurol.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Zeng W-B, Jiang H-F, Gang Y-D, Song Y-G, Shen Z-Z, Yang H, Dong X, Tian Y-L, Ni R-J, Liu Y, Tang N, Li X, Jiang X, Gao D, Androulakis M, He X-B, Xia H-M, Ming Y-Z, Lu Y, Zhou J-N, Zhang C, Xia X-S, Shu Y, Zeng S-Q, Xu F, Zhao F, Luo M-H. 2017. Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Mol Neurodegener 12:38. doi: 10.1186/s13024-017-0179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur E. 2014. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirokawa N, Noda Y, Tanaka Y, Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 22.Gross SP. 2004. Hither and yon: a review of bi-directional microtubule-based transport. Phys Biol 1:R1–R11. doi: 10.1088/1478-3967/1/2/R01. [DOI] [PubMed] [Google Scholar]
- 23.del Rio T, Ch'ng TH, Flood EA, Gross SP, Enquist LW. 2005. Heterogeneity of a fluorescent tegument component in single pseudorabies virus virions and enveloped axonal assemblies. J Virol 79:3903–3919. doi: 10.1128/JVI.79.7.3903-3919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maresch C, Granzow H, Negatsch A, Klupp BG, Fuchs W, Teifke JP, Mettenleiter TC. 2010. Ultrastructural analysis of virion formation and anterograde intraaxonal transport of the alphaherpesvirus pseudorabies virus in primary neurons. J Virol 84:5528–5539. doi: 10.1128/JVI.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith GA, Gross SP, Enquist LW. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci U S A 98:3466–3470. doi: 10.1073/pnas.061029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bearer EL, Breakefield XO, Schuback D, Reese TS, LaVail JH. 2000. Retrograde axonal transport of herpes simplex virus: evidence for a single mechanism and a role for tegument. Proc Natl Acad Sci U S A 97:8146–8150. doi: 10.1073/pnas.97.14.8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen DH, Jakana J, McNab D, Mitchell J, Zhou ZH, Dougherty M, Chiu W, Rixon FJ. 2001. The pattern of tegument-capsid interaction in the herpes simplex virus type 1 virion is not influenced by the small hexon-associated protein VP26. J Virol 75:11863–11867. doi: 10.1128/JVI.75.23.11863-11867.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handler CG, Eisenberg RJ, Cohen GH. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol 70:6067–6075. doi: 10.1128/JVI.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elton DM, Halliburton IW, Killington RA, Meredith DM, Bonass WA. 1992. Identification of the equine herpesvirus type-1 glycoprotein-17/18 as a homolog of herpes-simplex virus glycoprotein-D. J Gen Virol 73:1227–1233. doi: 10.1099/0022-1317-73-5-1227. [DOI] [PubMed] [Google Scholar]
- 30.Sari TK, Pritchard SM, Cunha CW, Wudiri GA, Laws EI, Aguilar HC, Taus NS, Nicola AV. 2013. Contributions of herpes simplex virus 1 envelope proteins to entry by endocytosis. J Virol 87:13922–13926. doi: 10.1128/JVI.02500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1840 In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 1. LWW Press, Philadelphia, PA. [Google Scholar]
- 32.Taylor AM, Rhee SW, Jeon NL. 2006. Microfluidic chambers for cell migration and neuroscience research. Methods Mol Biol 321:167–177. doi: 10.1385/1-59259-997-4:167. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, Schnitzbauer J, Grimes S, Jardine PJ, Bustamante C. 2014. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell 157:702–713. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randall TS, Yip YY, Wallock-Richards DJ, Pfisterer K, Sanger A, Ficek W, Steiner RA, Beavil AJ, Parsons M, Dodding MP. 2017. A small-molecule activator of kinesin-1 drives remodeling of the microtubule network. Proc Natl Acad Sci U S A 114:13738–13743. doi: 10.1073/pnas.1715115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roossien DH, Lamoureux P, Miller KE. 2014. Cytoplasmic dynein pushes the cytoskeletal meshwork forward during axonal elongation. J Cell Sci 127:3593–3602. doi: 10.1242/jcs.152611. [DOI] [PubMed] [Google Scholar]
- 36.Mingo RM, Han J, Newcomb WW, Brown JC. 2012. Replication of herpes simplex virus: egress of progeny virus at specialized cell membrane sites. J Virol 86:7084–7097. doi: 10.1128/JVI.00463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saksena MM, Wakisaka H, Tijono B, Boadle RA, Rixon F, Takahashi H, Cunningham AL. 2006. Herpes simplex virus type 1 accumulation, envelopment, and exit in growth cones and varicosities in mid-distal regions of axons. J Virol 80:3592–3606. doi: 10.1128/JVI.80.7.3592-3606.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Regge N, Nauwynck HJ, Geenen K, Krummenacher C, Cohen GH, Eisenberg RJ, Mettenleiter TC, Favoreel HW. 2006. Alpha-herpesvirus glycoprotein D interaction with sensory neurons triggers formation of varicosities that serve as virus exit sites. J Cell Biol 174:267–275. doi: 10.1083/jcb.200510156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diefenbach RJ, Miranda-Saksena M, Douglas MW, Cunningham AL. 2008. Transport and egress of herpes simplex virus in neurons. Rev Med Virol 18:35–51. doi: 10.1002/rmv.560. [DOI] [PubMed] [Google Scholar]
- 40.Shepherd GM, Raastad M, Andersen P. 2002. General and variable features of varicosity spacing along unmyelinated axons in the hippocampus and cerebellum. Proc Natl Acad Sci U S A 99:6340–6345. doi: 10.1073/pnas.052151299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters A, Harriman KM. 1990. Different kinds of axon terminals forming symmetric synapses with the cell bodies and initial axon segments of layer II/III pyramidal cells. I. Morphometric analysis. J Neurocytol 19:154–174. doi: 10.1007/bf01217295. [DOI] [PubMed] [Google Scholar]
- 42.Turcotte S, Letellier J, Lippé R. 2005. Herpes simplex virus type 1 capsids transit by the trans-golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol 79:8847–8860. doi: 10.1128/JVI.79.14.8847-8860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMillan TN, Johnson DC. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J Virol 75:1928–1940. doi: 10.1128/JVI.75.4.1928-1940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rezaul K, Gupta D, Semenova I, Ikeda K, Kraikivski P, Yu J, Cowan A, Zaliapin I, Rodionov V. 2016. Engineered tug-of-war between kinesin and dynein controls direction of microtubule based transport in vivo. Traffic 17:475–486. doi: 10.1111/tra.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog 6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wojaczynski GJ, Engel EA, Steren KE, Enquist LW, Card JP. 2015. The neuroinvasive profiles of H129 (herpes simplex virus type 1) recombinants with putative anterograde-only transneuronal spread properties. Brain Struct Funct 220:1395–1420. doi: 10.1007/s00429-014-0733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rank M, Frey E. 2018. Crowding and pausing strongly affect dynamics of kinesin-1 motors along microtubules. Biophys J 115:1068–1081. doi: 10.1016/j.bpj.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nam W, Epureanu BI. 2016. Effects of obstacles on the dynamics of kinesins, including velocity and run length, predicted by a model of two dimensional motion. PLoS One 11:e0147676. doi: 10.1371/journal.pone.0147676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR. 2007. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep 8:188–193. doi: 10.1038/sj.embor.7400878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wnęk M, Ressel L, Ricci E, Rodriguez-Martinez C, Guerrero JCV, Ismail Z, Smith C, Kipar A, Sodeik B, Chinnery PF, Solomon T, Griffiths MJ. 2016. Herpes simplex encephalitis is linked with selective mitochondrial damage; a post-mortem and in vitro study. Acta Neuropathol 132:433–451. doi: 10.1007/s00401-016-1597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kramer T, Enquist LW. 2012. Alphaherpesvirus infection disrupts mitochondrial transport in neurons. Cell Host Microbe 11:504–514. doi: 10.1016/j.chom.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DuRaine G, Wisner TW, Howard P, Johnson DC. 2018. Kinesin-1 proteins KIF5A, -5B, and -5C promote anterograde transport of herpes simplex virus enveloped virions in axons. J Virol 92:e01269-18. doi: 10.1128/JVI.01269-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanai Y, Okada Y, Tanaka Y, Harada A, Terada S, Hirokawa N. 2000. KIF5C, a novel neuronal kinesin enriched in motor neurons. J Neurosci 20:6374–6384. doi: 10.1523/JNEUROSCI.20-17-06374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diefenbach RJ, Miranda-Saksena M, Diefenbach E, Holland DJ, Boadle RA, Armati PJ, Cunningham AL. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J Virol 76:3282–3291. doi: 10.1128/jvi.76.7.3282-3291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shanda SK, Wilson DW. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol 82:7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, Chen J, Guo Z, Deng S, Du X, Zhu S, Ye C, Shi YS, Liu JJ. 2018. Endophilin A1 promotes actin polymerization in dendritic spines required for synaptic potentiation. Front Mol Neurosci 11:177. doi: 10.3389/fnmol.2018.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang G, Dong X, Huang X, Huang XJ, Liu H, Wang Y, Ye WC, Shi L. 2015. A natural diarylheptanoid promotes neuronal differentiation via activating ERK and PI3K-Akt dependent pathways. Neuroscience 303:389–401. doi: 10.1016/j.neuroscience.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 58.Yang B, Liu X-J, Yao YX, Jiang X, Wang XZ, Yang H, Sun JY, Miao Y, Wang W, Huang Z-L, Wang YY, Tang QY, Rayner S, Britt WJ, McVoy MA, Luo M-H, Zhao F. 2018. WDR5 facilitates human cytomegalovirus replication by promoting capsid nuclear egress. J Virol 92:e00207-18. doi: 10.1128/JVI.00207-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niu Y, Zhang C, Sun Z, Hong Z, Li K, Sun DM, Yang YR, Tian CL, Gong WM, Liu JJ. 2013. PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nat Cell Biol 15:417–429. doi: 10.1038/ncb2710. [DOI] [PubMed] [Google Scholar]
- 60.van de Linde S, Loschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. 2011. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc 6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Z, Xin B, Li L, Huang ZL. 2017. High-power homogeneous illumination for super-resolution localization microscopy with large field-of-view. Opt Express 25:13382–13395. doi: 10.1364/OE.25.013382. [DOI] [PubMed] [Google Scholar]
- 62.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, O'Donnell M, Gelfand VI, Kapoor TM, Chen JK. 2012. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature 484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sainath R, Gallo G. 2015. The dynein inhibitor Ciliobrevin D inhibits the bidirectional transport of organelles along sensory axons and impairs NGF-mediated regulation of growth cones and axon branches. Dev Neurobiol 75:757–777. doi: 10.1002/dneu.22246. [DOI] [PMC free article] [PubMed] [Google Scholar]