Herpesviruses encode an exceptional vesicle formation and scission machinery, which operates at the inner nuclear membrane, translocating the viral nucleocapsid from the nucleus into the perinuclear space. The conserved herpesviral nuclear egress complex (NEC) orchestrates this process. High-resolution imaging approaches as well as the recently solved crystal structures of the NEC provided deep insight into the molecular details of vesicle formation and scission. Nevertheless, the molecular mechanism of nucleocapsid incorporation remained unclear. In accordance with structure-based predictions, a basic amino acid could be pinpointed in the most membrane-distal domain of the NEC (pUL31-K242), indicating that capsid incorporation might depend on a direct electrostatic interaction. Our follow-up study, described here, however, shows that the positive charge is not relevant but that the overall structure matters.
KEYWORDS: herpesvirus, nuclear egress complex, nuclear envelope, pUL31, pUL34, pseudorabies virus
ABSTRACT
Herpesvirus nucleocapsids leave the nucleus by a vesicle-mediated translocation mediated by the viral nuclear egress complex (NEC). The NEC is composed of two conserved viral proteins, designated pUL34 and pUL31 in the alphaherpesvirus pseudorabies virus (PrV). It is required for efficient nuclear egress and is sufficient for vesicle formation and scission from the inner nuclear membrane (INM). Structure-based mutagenesis identified a lysine at position 242 (K242) in pUL31, located in the most membrane distal part of the NEC, to be crucial for efficient nucleocapsid incorporation into budding vesicles. Replacing the lysine by alanine (K242A) resulted in accumulations of empty vesicles in the perinuclear space, despite the presence of excess nucleocapsids in the nucleus. However, it remained unclear whether the defect in capsid incorporation was due to interference with a direct, electrostatic interaction between the capsid and the NEC or structural restrictions. To test this, we replaced K242 with several amino acids, thereby modifying the charge, size, and side chain orientation. In addition, virus recombinants expressing pUL31-K242A were passaged and screened for second-site mutations. Compensatory mutations at different locations in pUL31 or pUL34 were identified, pointing to an inherent flexibility of the NEC. In summary, our data suggest that the amino acid at position 242 does not directly interact with the nucleocapsid but that rearrangements in the NEC coat are required for efficient nucleocapsid envelopment at the INM.
IMPORTANCE Herpesviruses encode an exceptional vesicle formation and scission machinery, which operates at the inner nuclear membrane, translocating the viral nucleocapsid from the nucleus into the perinuclear space. The conserved herpesviral nuclear egress complex (NEC) orchestrates this process. High-resolution imaging approaches as well as the recently solved crystal structures of the NEC provided deep insight into the molecular details of vesicle formation and scission. Nevertheless, the molecular mechanism of nucleocapsid incorporation remained unclear. In accordance with structure-based predictions, a basic amino acid could be pinpointed in the most membrane-distal domain of the NEC (pUL31-K242), indicating that capsid incorporation might depend on a direct electrostatic interaction. Our follow-up study, described here, however, shows that the positive charge is not relevant but that the overall structure matters.
INTRODUCTION
Herpesviruses are large enveloped DNA viruses which replicate in two different cellular compartments. While viral DNA replication, capsid assembly, and genome packaging occur in the nucleus, final virus maturation proceeds in the cytoplasm. To transfer newly assembled nucleocapsids to the cytoplasm, herpesviruses use a unique vesicle-mediated pathway by budding through the inner nuclear membrane (INM) into the perinuclear space (PNS). This is subsequently followed by fusion of the INM-derived primary virion envelope with the outer nuclear membrane (ONM), releasing the nucleocapsids into the cytoplasm (reviewed in references 1, to ,4).
Budding at and scission from the INM are coordinated by the viral nuclear egress complex (NEC), which is conserved throughout the Herpesviridae. The NEC consists of two viral proteins, designated pUL34 and pUL31 in the alphaherpesviruses pseudorabies virus (PrV) and herpes simplex virus (HSV). pUL34 is a tail-anchored membrane protein which is autonomously targeted to the nuclear envelope. pUL31 is diffusely distributed in the nucleus in the absence of pUL34 but is recruited to the INM by interaction with membrane-bound pUL34, forming the NEC (reviewed in references 1 to 4). NEC oligomerization at the INM most likely mediates membrane bending and vesicle scission. The ectopic expression of pUL31 and pUL34 is sufficient for vesicle formation and scission from the INM in eukaryotic cells but also from synthetic lipid bilayers, such as giant unilamellar vesicles, indicating that no other viral or cellular protein(s) is needed for vesiculation (5–7). Despite the simultaneous presence of the NEC components in the nucleus, in infected cells empty vesicles are only rarely observed, while nucleocapsids are apparently selected for translocation (8, 9). How nucleocapsids trigger this process and how they are incorporated into the nascent vesicles remained unclear.
Previous data suggested that pUL31 binds to capsids already in the nucleoplasm and mediates their transport to INM-located pUL34, initiating complex formation and oligomerization (10). The HSV-1 capsid vertex-specific component (CVSC), which is composed of pUL17 and pUL25 and which is enriched on mature capsids (9, 11), was shown to interact with pUL31 (12, 13). In addition, direct contacts between the NEC coat and the CVSC were visualized using cryo-electron tomography on primary enveloped particles (9). Recently, a direct interaction between pUL25 and the NEC was suggested by glutathione S-transferase-pulldown assays (14). In contrast, binding of PrV pUL31 also occurred independently of the pUL25 CVSC component (15), pointing to multiple binding partners or binding sites on the capsids. Due to the transient nature of the nucleocapsid-NEC interaction, which must be formed during envelopment at the INM but which needs to disengage during deenvelopment at the ONM, identification of interaction partners by classical biochemical approaches is difficult. In addition, the observation that preferentially mature nucleocapsids are enveloped at the INM indicates a complex structure-based interaction influenced by subsequent subtle structural changes, ranging from genome packaging and scaffold expulsion to addition of the CVSC.
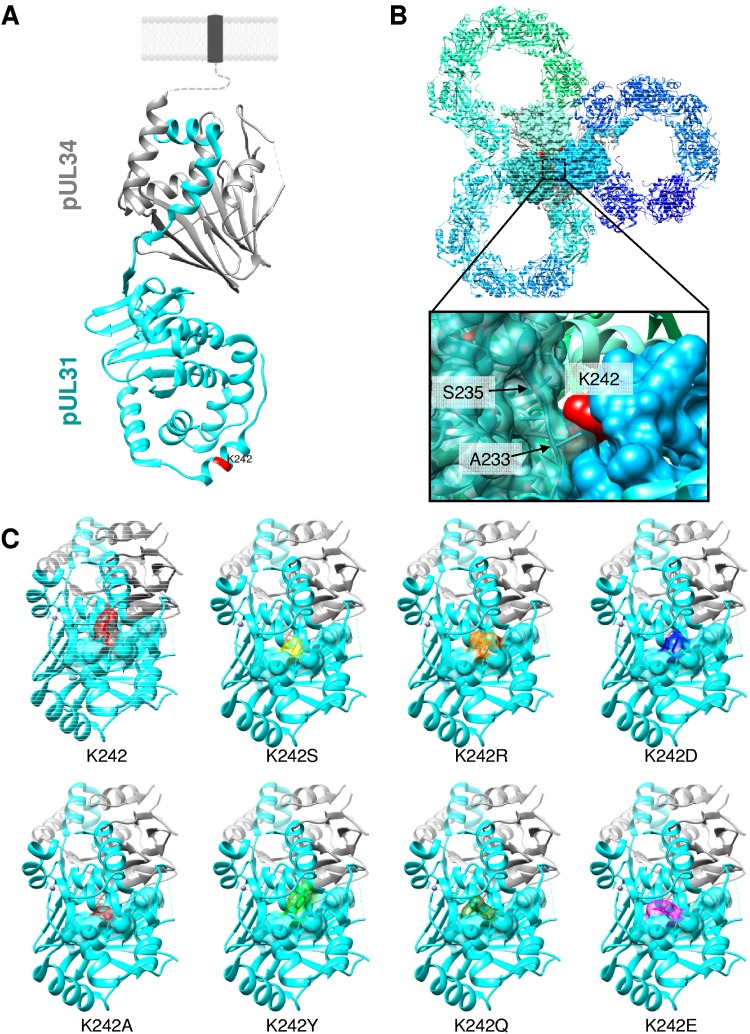
To analyze this process, we modified potential capsid interaction interfaces on the NEC by site-directed mutagenesis (16) based on the crystal structures of the NECs from PrV, HSV-1 (17, 18), and human cytomegalovirus (19, 20), as well as high-resolution imaging of the PrV NEC (21). The NEC is an elongated rod-like structure, with the two, mostly globular components sitting on top of each other (Fig. 1A). Oligomerization of the NEC results in a tightly packed and curved hexameric, honeycomb-like structure (Fig. 1B). Based on these data, only the membrane-distal end of the NEC, formed by pUL31, appears to be accessible for interaction with the nucleocapsid, presumably involving electrostatic interactions (17, 18). To test this hypothesis, we isolated several PrV pUL31 mutants where predicted surface-exposed and charged amino acids were altered to alanine (16). These data resulted in the identification of the relevance of the most membrane-distal alpha-helical (H10) region (17) and especially a conserved lysine at position 242 (K242) as the key residue. Mutation of this lysine or the simultaneous exchange of the triad C241/K242/M243 for alanine (C241-243A) resulted in accumulations of empty membrane vesicles in the PNS, despite the intranuclear presence of numerous nucleocapsids, indicating that this region of pUL31 is crucial for nucleocapsid incorporation (16).
FIG 1.
Structural prediction of the PrV NEC. (A) pUL34 is shown in gray and pUL31 is shown in cyan in the PrV NEC structure (17). The orientation toward and anchorage in the INM are indicated by the dotted line, and the transmembrane domain in a lipid bilayer is represented by a dark gray box. The location of the amino acid (K242) targeted in this study is indicated in red. (B) Bottom view of the membrane-distal end in the honeycomb array (17), with interacting interhexameric molecules being shown by surface representation with amino acid K242 in red. The zoomed image of the boxed region shows the interaction interface of different NEC heterodimers with one pUL31 in a semitransparent surface presentation, while the neighboring pUL31 is shown with its full surface. Amino acids in close proximity to K242 are indicated by arrows and given with the corresponding position. (C) A bottom view of the membrane-distal end of pUL31 with a partial surface presentation of alpha helix H10 is shown with amino acid K242 in red. Amino acid substitutions at position 242 are modeled into the NEC dimer structure (PDB accession number 5E8C). Molecular graphics and analysis were performed with the UCSF Chimera package (34).
To further explore the role of H10 and K242 in this process, we generated additional mutants by replacing lysine 242 with acidic aspartic acid or glutamic acid with serine, glutamine, or tyrosine, which have smaller or larger side chains than glutamic acid, or with arginine, thereby maintaining the basic character. In addition, virus mutants carrying either the single K242A substitution or the triple C241-243A substitution were passaged in cell culture to select for second-site mutations compensating for the capsid incorporation defect.
The data presented here indicate that capsid incorporation into primary vesicles is not dependent on a specific charge interaction with K242 but may require conformational flexibility within the NEC coat.
RESULTS
Structural predictions of the pUL31-K242 location in the NEC coat.
A lysine residue at position 242 in the PrV pUL31 component of the NEC was found to be crucial for incorporation of nucleocapsids into INM-derived vesicles (16). While in the NEC heterodimer K242 seemed to be freely accessible at the surface (Fig. 1A), modeling of this residue into the hexagonal array showed that this position might indeed be more deeply buried and constrained by residues A233 and S235 in pUL31 of a neighboring hexamer, impairing or even prohibiting direct interaction with the nucleocapsid (Fig. 1B). To further analyze the role of K242 in nucleocapsid incorporation, we replaced this residue by the basic but larger arginine (R), the negatively charged but smaller glutamic acid (E) or aspartic acid (D), the small neutral serine (S), the large aromatic tyrosine (Y), or the medium-sized glutamine (Q). The influence of the amino acid exchanges was calculated and modeled into the heterodimeric NEC structure by using the intrinsic functions of the UCSF Chimera package (Fig. 1C). Mutants were generated by site-specific mutagenesis using the primers shown in Table 1 with pcDNA-UL31 as the template. Correct mutagenesis was verified by sequencing.
TABLE 1.
Primers used for site-directed mutagenesis
| Primer name | Sequence (5′ to 3′)a |
|---|---|
| UL31 K242S | GAC ATT TAT TGC AGC ATG CGG GAC ATC AGC |
| UL31 K242Q | GAC ATT TAT TGC CAG ATG CGG GAC ATC AGC |
| UL31 K242Y | GC GAC ATT TAT TGC TAC ATG CGG GAC ATC AGC |
| UL31 K242R | G AGC GAC ATT TAT TGC AGG ATG CGG GAC ATC AGC |
| UL31 K242E | G AGC GAC ATT TAT TGC GAG ATG CGG GAC ATC AGC |
| UL31 K242D | GCG AGC GAC ATT TAT TGC GAT ATG CGG GAC ATC AGC |
| UL31 A242K | GC GAC ATT TAT TGC AAG ATG CGG GAC ATC AG |
Mismatches are underlined and in bold. Only the forward primer is shown; the reverse complementary primers are not depicted.
Intracellular localization of pUL31 mutants and colocalization with pUL34.
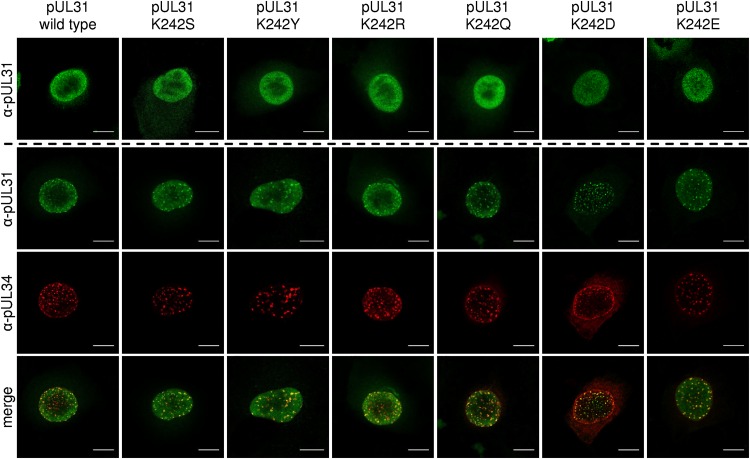
The mutated pUL31 proteins were tested for correct nuclear targeting and interaction with the complex partner pUL34 by either transfection of the corresponding expression plasmids or cotransfection with pcDNA-UL34 (22) into rabbit kidney (RK13) cells and processed for confocal microscopy 2 days later. pUL31 was detected using a monoclonal anti-pUL31 antibody, while pUL34 was visualized by a monospecific rabbit antiserum (Fig. 2) (22). All pUL31 mutants showed the same diffuse distribution in the nucleus as native pUL31 (Fig. 2, top row), indicating stable expression and correct targeting. A punctate pUL31/pUL34-positive staining pattern comparable to that achieved with wild-type pUL31/pUL34 coexpression (Fig. 2, bottom three rows) indicated that complex formation and membrane deformation were not affected by the introduced changes. Only cotransfections of pcDNA-UL31-K242D and pcDNA-UL34 showed the presence of a higher level of pUL34 in the nuclear rim and in cytoplasmic structures.
FIG 2.
Intracellular localization of pUL31 mutants and colocalization with pUL34. Localization was tested after transfection of RK13 cells with the corresponding pUL31 expression plasmids (top row). Colocalization was analyzed after cotransfection with pcDNA-UL34. pUL31 was stained with a pUL31-specific monoclonal antibody (green, second row), while for pUL34, polyclonal anti-pUL34 serum (red, third row) was used. Merged channels are shown in the bottom row. Fluorescence was imaged with a confocal laser-scanning microscope (63× oil immersion objective, single slice; SP5; Leica Germany). Bars, 10 μm.
Functional complementation of PrV-ΔUL31 by the pUL31-K242 substitutions.
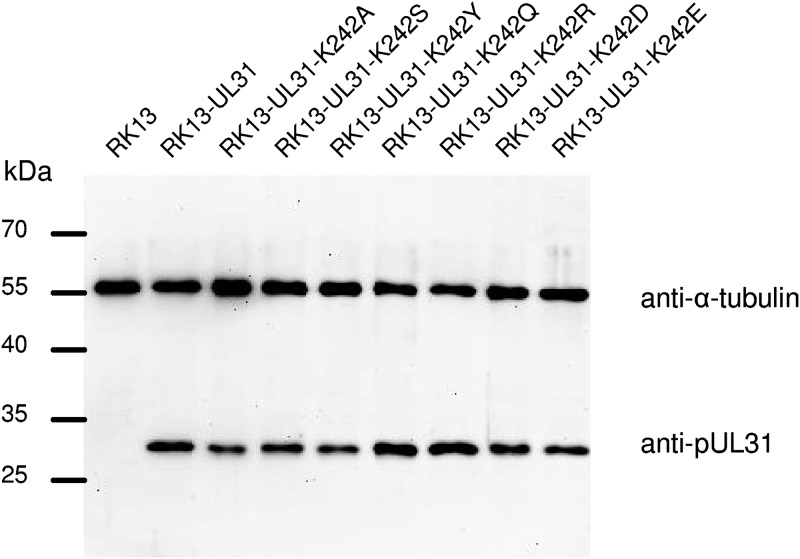
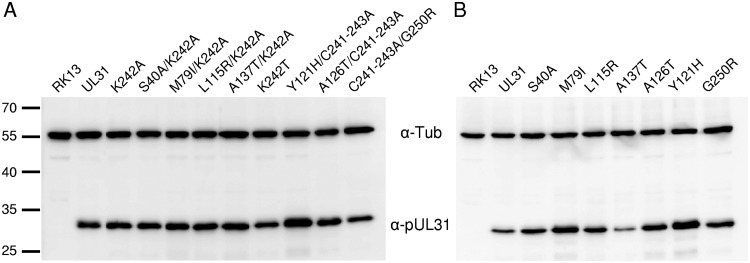
To investigate the ability of the generated mutants to complement the defect of PrV-ΔUL31, cell lines stably expressing the respective pUL31 mutants in trans were generated. Cell clones were isolated and tested by indirect immunofluorescence and immunoblotting, using monospecific anti-pUL31 rabbit serum (23). In contrast to the parental RK13 cells, expression of pUL31 could be detected in all transgenic cell lines. Anti-α-tubulin was included as a loading control (Fig. 3).
FIG 3.
pUL31 expression in RK13 cells. Lysates of cell lines stably expressing native or mutated pUL31 as well as nontransgenic RK13 cells were separated in an SDS-10% polyacrylamide gel. Proteins were detected after transfer to nitrocellulose with the monospecific anti-pUL31 rabbit serum. As loading control, anti-α-tubulin was used. The molecular masses of the marker proteins (in kilodaltons) are indicated on the left.
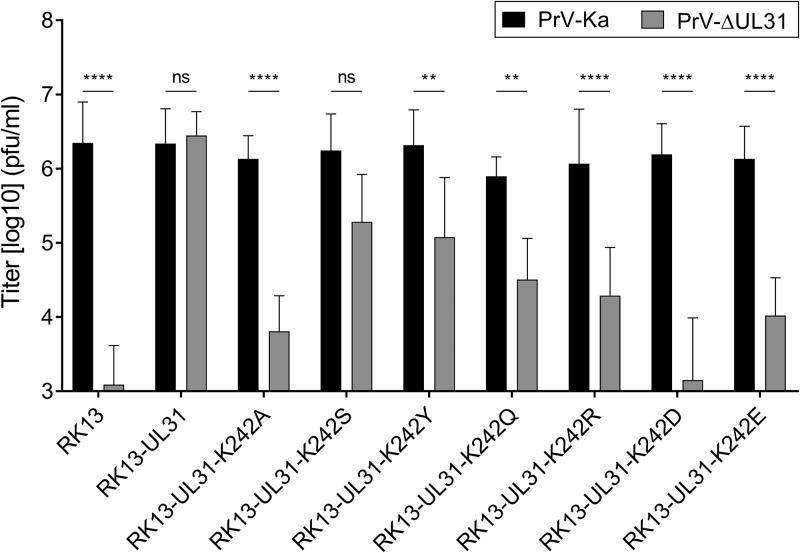
To test for functional complementation, RK13 and the pUL31-expressing RK13 cell lines (RK13-UL31 cells) were infected with PrV wild-type strain Kaplan (PrV-Ka) or PrV-ΔUL31 (23) at a multiplicity of infection (MOI) of 5. At 24 h postinfection (p.i.), the supernatant and cells were harvested and progeny virus titers were determined on RK13-UL31 cells (Fig. 4). Infection with PrV-Ka resulted in progeny titers of approximately 106 PFU/ml in all cell lines, similar to the findings for parental RK13 cells, indicating that none of the pUL31 mutants exerted a dominant negative effect. The titers of PrV-ΔUL31 derived from RK13-UL31-K242S and RK13-UL31-K242Y cells were only slightly reduced compared to those of PrV-Ka derived from the corresponding cells (6- and 10-fold lower, respectively), indicating functional complementation. The titers from RK13-UL31-K242Q cells were approximately 20-fold lower, while the titers resulting from infection of RK13-UL31-K242R cells were reduced 70-fold. No or only very low levels of complementation of PrV-ΔUL31 (reduced more than 100-fold) were found on RK13-UL31-K242E and RK13-UL31-K242D cells, which were in the range of those found on cells expressing pUL31-K242A or for nontransgenic RK13 cells (Fig. 4) (16), indicating that these pUL31 mutants are nonfunctional.
FIG 4.
trans-Complementation assays. Functional complementation by the generated pUL31 mutants was tested after infection of the stably expressing cell lines with PrV-Ka and PrV-ΔUL31 at an MOI of 5. Cells and supernatant were harvested at 24 h p.i., and titers were determined on RK13-UL31 cells. Shown are the mean values from four independent experiments with the corresponding standard deviations. Statistically significant differences are indicated (**, P ≤ 0.01; ****, P ≤ 0.0001; ns, not significant).
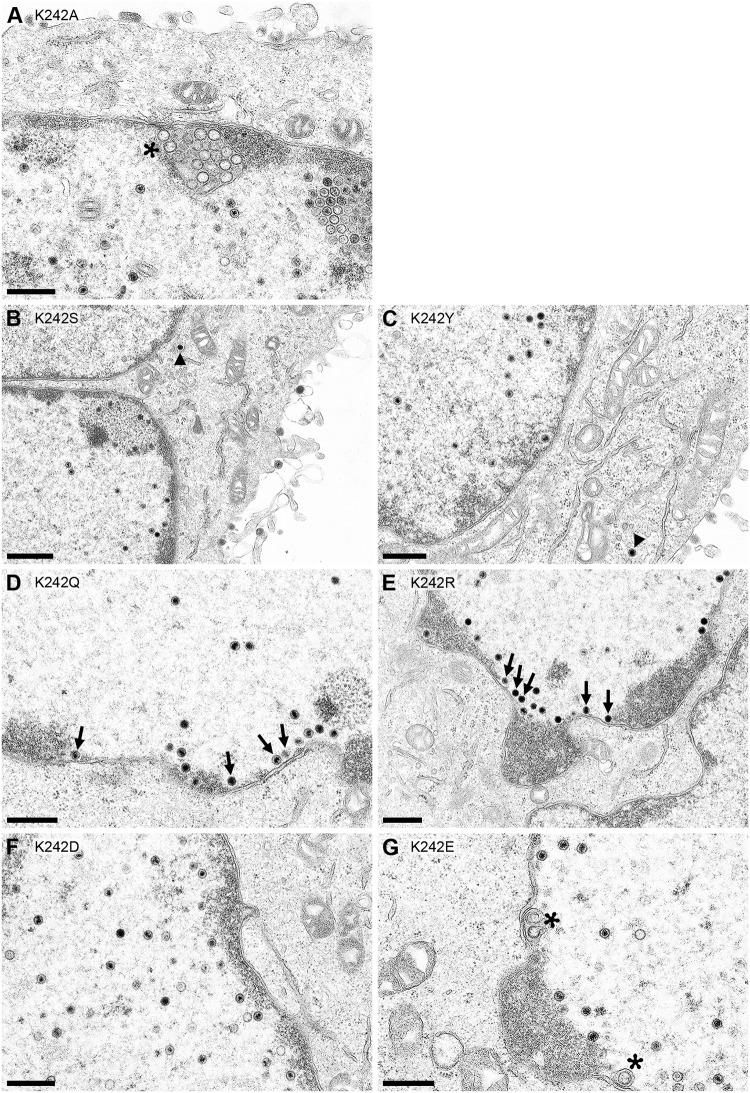
To characterize the effect of the substitutions on nuclear egress, cell lines were infected with PrV-ΔUL31 at an MOI of 1 and processed for electron microscopy. Surprisingly, none of the pUL31 mutant cell lines showed accumulations of empty vesicles within the PNS similar to those observed after infection of RK13-UL31-K242A cells (16) (Fig. 5A). In contrast, PrV-ΔUL31-infected RK13-UL31-K242S cells (Fig. 5B) and RK13-UL31-K242Y cells (Fig. 5C) showed all stages of virion morphogenesis, including nucleocapsids in the cytoplasm and virions at the plasma membrane, paralleling their complementation phenotype (Fig. 4). In PrV-ΔUL31-infected RK13 cells expressing pUL31-K242Q (Fig. 5D) or pUL31-K242R (Fig. 5E), intranuclear nucleocapsids were often detected in close apposition to the inner nuclear membrane, but membrane bending and/or budding was observed only infrequently. Only a few vesicle-like structures in the PNS were present in RK13-UL31-K242E cells (Fig. 5G), while nucleocapsids accumulated in the nucleus in RK13-UL31-K242D cells (Fig. 5F), paralleling the observed lack of complementation (Fig. 4).
FIG 5.
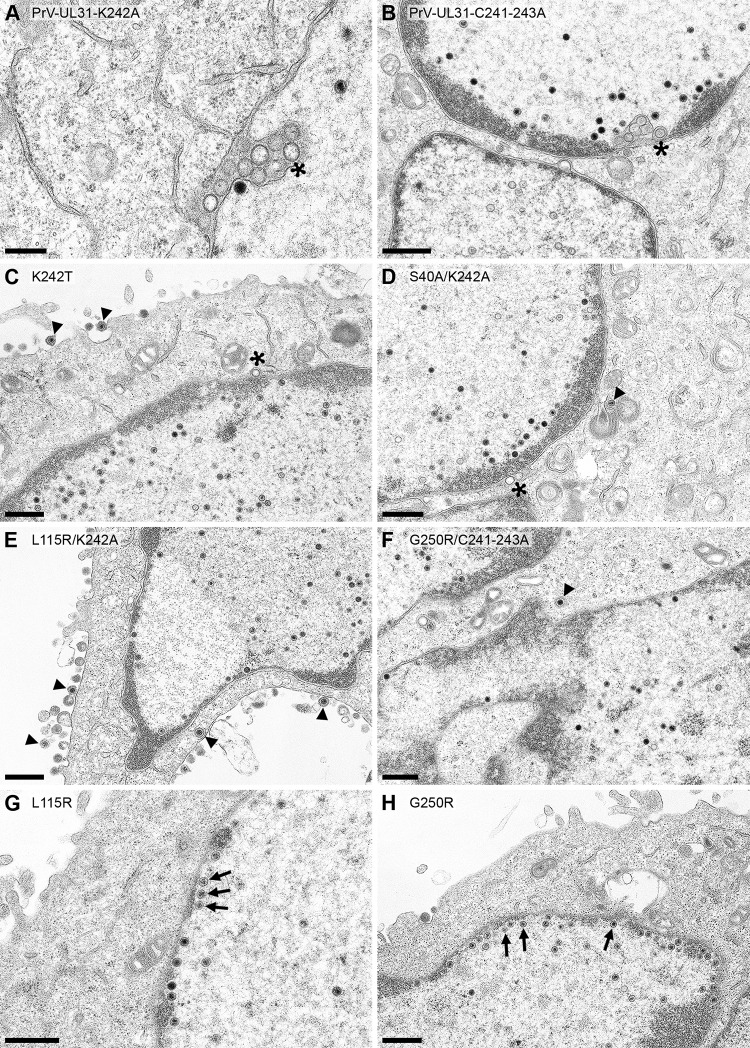
Ultrastructural analyses of cells expressing pUL31 mutants infected with PrV-ΔUL31. RK13-UL31-K242A (A), RK13-UL31-K242S (B), RK13-UL31-K242Y (C), RK13-UL31-K242Q (D), RK13-UL31-K242R (E), RK13-UL31-K242D (F), and RK13-UL31 K242E (G) cells were infected with PrV-ΔUL31 at an MOI of 1 and processed for electron microscopy at 14 h p.i. Representative images are shown. Asterisks mark empty vesicles in the PNS, arrows point to nucleocapsids close to the INM, and arrowheads indicate nucleocapsids or virions in the cytoplasm. Bars, 500 nm (A, C to G) and 800 nm (B).
Serial passaging of virus recombinants.
The results obtained with the various constructs with amino acid substitutions at position K242 were unexpected, arguing against a direct electrostatic interaction between the K242 region within pUL31 alpha helix H10 and nucleocapsids. To analyze the NEC-nucleocapsid interaction further, virus recombinants expressing noncomplementing pUL31-K242A or pUL31-C241-243A were generated by homologous recombination. PrV-UL31-K242A and PrV-UL31-C241-243A exhibited strongly reduced growth properties comparable to those of PrV-ΔUL31 on the corresponding stably expressing cell lines (16), with only low progeny virus titers and striking accumulations of empty vesicles in the PNS. However, the production of a small amount of infectious virus progeny was observed and used for reversion analysis.
To select for compensating second-site mutations, PrV-UL31-K242A and PrV-UL31-C241-243A were passaged in RK13 and Vero cells. Within only a few cell passages, the titers of the supernatants reached up to 106 PFU/ml. Single plaques from several independent assays were isolated and tested. Genomic DNA of revertants which replicated to titers of at least 106 PFU/ml was isolated, and the UL31- and UL34-coding regions were amplified by PCR and sequenced. The mutations identified in pUL31 and pUL34 are summarized in Table 2. In this study, we subsequently focused on the pUL31 mutations.
TABLE 2.
Second-site mutations detected in reversion analyses
| Mutant used for passaging | Cell line used for passaging | Second-site mutation in: |
|
|---|---|---|---|
| UL31 | UL34 | ||
| PrV-UL31-K242A | Vero | S40 | —a |
| PrV-UL31-K242A | RK13 | M79I | — |
| PrV-UL31-K242A | RK13 | L115R | — |
| PrV-UL31-K242A | RK13 | A137T | — |
| PrV-UL31-K242A | Vero | K242T | — |
| PrV-UL31-C241-243A | Vero | Y121H | — |
| PrV-UL31-C241-243A | RK13 + Vero | A126T | — |
| PrV-UL31-C241-243A | RK13 + Vero | G250R | — |
| PrV-UL31-K242A | Vero | — | G15W |
| PrV-UL31-K242A | Vero | — | T25M |
| PrV-UL31-K242A | Vero | — | A26V |
| PrV-UL31-K242A | RK13 + Vero | — | T98A |
| PrV-UL31-K242A | Vero | — | A99V |
—, no mutation.
Characterization of second-site mutations.
To test whether the additional mutations in pUL31 are sufficient to compensate for the replication defect in pUL31-K242A and pUL31-C241-243A (16), the UL31 genes were cloned into pcDNA3. The localization and colocalization with pUL34 after transfection into RK13 cells were comparable to those of the wild-type proteins (data not shown), pointing to correct nuclear targeting and interaction with pUL34.
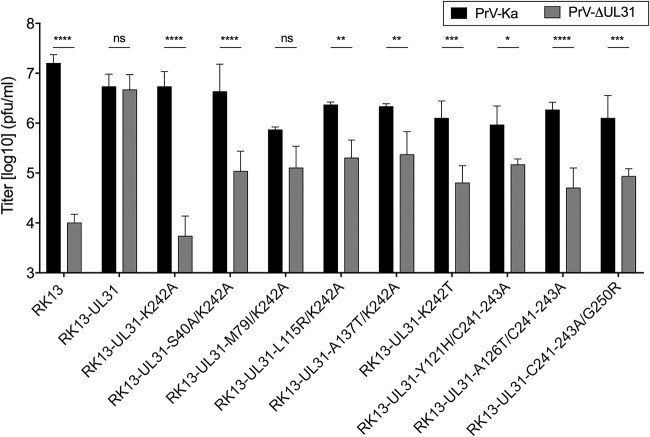
Stably expressing cell lines were isolated, tested for correct pUL31 expression (Fig. 6A), and infected with PrV-ΔUL31 and PrV-Ka as described above. The titers of PrV-Ka reached at least 106 PFU/ml on all cell lines, demonstrating the absence of a dominant negative effect of the second-site mutations (Fig. 7). Only RK13-UL31-M79I/K242A replicated PrV-Ka to slightly lower titers. Nevertheless, complementation of PrV-ΔUL31 with, at the most, 10-fold reduced titers was observed for pUL31-M79I/K242A-, pUL31-A137T/K242A-, pUL31-K242T-, and pUL31-Y121H/C241-243A-expressing cells. RK13-UL31-S40A/K242A, RK13-UL31-L115R/K242A, RK13-UL31-A126T/C241-243A, and RK13-UL31-C241-243A/G250R cells showed a more pronounced deficiency for complementation with 10- to 30-fold lower titers, but these were still significantly above those derived from RK13-UL31-K242A or RK13-UL31-C241-243A cells (Fig. 7), pointing to at least partial compensation of the K242A/C241-243A defect.
FIG 6.
Immunoblot analysis of RK13 cells expressing pUL31 with second-site mutations. RK13 cells expressing UL31-K242A with the second-site mutations (A) or pUL31 carrying only these mutations (B) were harvested and tested for pUL31 expression with the monospecific anti-pUL31 serum. Anti-α-tubulin (α-Tub) was used as a loading control.
FIG 7.
Complementation of PrV-ΔUL31 by second-site-mutated pUL31-K242A and pUL31-C241-243A. Complementation by pUL31-K242A and pUL31-C241-243A carrying second-site mutations was tested after infection of the stably expressing cell lines with PrV-Ka and PrV-ΔUL31 at an MOI of 5. Cells and supernatant were harvested at 24 h p.i., and titers were determined on RK13-UL31 cells. Shown are the mean values from three independent experiments with the corresponding standard deviations. Statistically significant differences were evaluated using GraphPad Prism software and are indicated (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; ns, not significant).
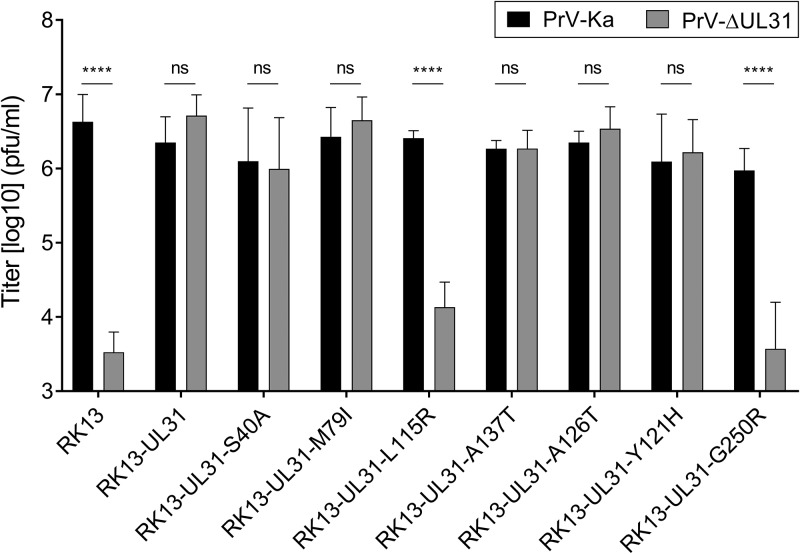
To test whether the second-site mutations affect virus replication in the absence of the K242A or C241-243A substitutions, the latter were reverted by site-directed mutagenesis, resulting in pcDNA-UL31-S40A, pcDNA-UL31-M79I, pcDNA-UL31-L115R, pcDNA-UL31-Y121H, pcDNA-UL31-A126T, pcDNA-UL31-A137T, and pcDNA-UL31-G250R. Since the localization of the mutated pUL31 and colocalization with pUL34 were comparable to those of wild-type pUL31 (data not shown), stably expressing RK13 cells were generated and tested for pUL31 expression (Fig. 6B) and for functional complementation of PrV-ΔUL31 (Fig. 8). While pUL31-S40A, pUL31-M79I, pUL31-A126T, and pUL31-Y121H complementation of PrV-ΔUL31 produced wild-type-like titers with no detectable effect, expression of pUL31-L115R and pUL31-G250R resulted in progeny titers only in the range of those derived from nontransgenic RK13 cells, indicating that these mutations significantly impaired the pUL31 function in the absence of the K242A/C241-243A substitutions (Fig. 8).
FIG 8.
Effect of second-site mutations in the absence of the K242A mutation or the C241-243A mutations. The K242A and C241-243A mutations in plasmids expressing the pUL31 genes derived from the passaged mutants were repaired to the wild type by site-directed mutagenesis. Stably expressing cells were infected with PrV-Ka and PrV-ΔUL31 as described in the Fig. 7 legend. Shown are mean values from three independent experiments with the corresponding standard deviations (****, P ≤ 0.0001; ns, not significant).
In ultrastructural analyses, all stages of nuclear egress and virion formation could be observed in cells expressing pUL31 with substitutions in H10 in the presence of the second-site mutations, as shown for PrV-ΔUL31-infected RK13-UL31-K242T, RK13-UL31-S40A/K242A, RK13-UL31-L115R/K242A, and RK13-UL31-C241-243A/G250R cells (Fig. 9C to F). These data point to an at least partial reversion of the nucleocapsid incorporation defect observed in PrV-ΔUL31-infected RK13-UL31-K242A and RK13-UL31-C241-243A cells (16) or RK13 cells infected with PrV-UL31-K242A and PrV-UL31-C241-243A (Fig. 9A and B). Nevertheless, single empty vesicles in the PNS were still evident, indicating that capsid uptake into the nascent primary virion envelope may not be as efficient as that with the wild-type NEC. Nucleocapsids in close proximity to the INM could be observed in PrV-ΔUL31-infected cells expressing pUL31-L115R and pUL31-G250R, while membrane bending did not ensue (Fig. 9G and H).
FIG 9.
Ultrastructural analysis. RK13 cells were infected with PrV-UL31-K242A (A) or PrV-UL31-C241-243A (B). (C to H) RK13 cells expressing pUL31-K242T (C), pUL31-S40A/K242A (D), pUL31-L115R/K242A (E), pUL31-G250R/C241-243A (F), pUL31-L115R (G), or pUL31-G250R (H), infected with PrV-ΔUL31 (MOI, 1), fixed, and processed for electron microscopy at 14 h p.i. Asterisks mark empty vesicles in the PNS, nucleocapsids close to the INM are indicated by arrows, and nucleocapsids or virions in the cytoplasm or at the plasma membrane are highlighted by arrowheads. Bars, 300 nm (A) and 600 nm (B to H).
DISCUSSION
Elucidation of the structures of several NECs by X-ray diffraction (17–20) as well as a multimodal imaging approach (21) shed light on this exceptional viral vesicle formation and scission machinery. Identification of a clear polarity in the NEC with a membrane-proximal and a membrane-distal face revealed that the major portion of the NEC components is involved in oligomerization and vesicle coat formation, leaving only the most membrane-distal part as a potential interaction interface with the nucleocapsid (21). In line with this, mutation of lysine 242 to alanine in the most membrane-distal helical region (H10) in PrV pUL31 (16) or a R281A/D282A exchange in HSV-1 pUL31 (14) (corresponding to amino acids R244 and D245, respectively, in PrV pUL31) resulted in accumulations of empty membrane vesicles in the PNS, indicating that capsid uptake into the nascent vesicles was impaired. However, in contrast to the findings for HSV-1, a direct physical interaction between H10 and the nucleocapsid cargo was not shown for PrV.
Although H10 and, in particular, K242 appeared to be surface exposed in the crystal structure of the PrV NEC heterodimer, modeling of this position into the hexagonal array showed that it might be more deeply buried in the interior and not available for a direct capsid interaction. To investigate this in more detail, we replaced the lysine K242 with either the smaller amino acid serine (S) or glutamine (Q), the negatively charged aspartate (D) or glutamate (E), the bulky but also basic arginine (R), or the large aromatic tyrosine (Y).
Modeling the different residues into the PrV NEC dimer (Fig. 1C) revealed that the side chains of the substituted amino acids occupy the available space differently. While the original lysine contains the longest extension pointing toward the neighboring pUL31 molecules (Fig. 1B and C), alanine is small and compact, leaving some empty space. The positions of serine and, especially, tyrosine with its aromatic ring and aspartic acid partly mimicked the position of lysine. In contrast, side chains of arginine, glutamine, and glutamic acid were kinked and directed inwards but not toward the neighboring pUL31 in the hexameric ring.
All pUL31 mutants translocated to the nucleus indistinguishably from the wild-type protein and interacted with coexpressed pUL34 to deform the nuclear membrane, resulting in the typical punctate speckled pattern observed for the wild-type proteins (7). Only the coexpression of pUL31-K242D slightly affected the localization of pUL34 and showed reduced colocalization. Nevertheless, a double-positive fluorescent punctate pattern was also evident. Modeling of aspartic acid into the NEC structure indicates that it fits into the position of the original lysine, but the negative charge seems to influence the binding affinity to the NEC partner or the NEC oligomer.
The function of the mutated pUL31 was tested by complementation assays and ultrastructural analyses using stably expressing cell lines. Surprisingly, none of the novel mutations resulted in prominent accumulations of empty vesicles, as observed for pUL31-K242A or pUL31-C241-243A (16). Surprisingly, maintaining the basic character by alteration of lysine to arginine led to significantly reduced titers. Thus, the presence of a basic amino acid at this position does not per se result in the proper function of the NEC, arguing against a simple charge interaction. Only intranuclear nucleocapsids were observed, and these were often in close proximity to the INM, suggesting that capsid transport to and docking at the INM occurs but that membrane bending is impaired. A similar phenotype could be found in virus mutants lacking the CVSC component pUL25 (24) or expressing pUL31 with mutations in the neighboring alpha-helical region, H11, which affect the nuclear export signal (pUL31-NESPM) (16, 25). These data indicate that alpha-helical regions H10 and H11 may indeed cooperate in the structural rearrangements during nuclear egress, leading from a flat NEC patch to a curved shape. A similar phenotype was also found in cells expressing pUL31-K242Q.
In contrast, the replacement of K242 by the small and neutral serine (K242S) and by the large and aromatic tyrosine (K242Y) had only a limited effect on the production of infectious progeny, with the titers being approximately 5- to 10-fold lower than those of PrV-Ka. Electron microscopic images showed all stages of herpesvirus assembly and egress, although nucleocapsids and virions were only rarely detected in the cytoplasm or in the extracellular space, which is in line with the reduced titers and the relatively low sensitivity of electron microscopy. In contrast to pUL31-K242A expression, no empty vesicles were observed in the PNS.
Cells expressing pUL31-K242D or pUL31-K242E, which reverted the net charge at position 242, were unable to complement the defect of PrV-ΔUL31, and the titers were comparable to those derived from nontransgenic RK13 cells. Only intranuclear nucleocapsids were observed, in line with the severe drop in viral progeny titers. Very rarely, single empty vesicles in the PNS were found, but these were in striking contrast to the abundant vesiculation in cells expressing pUL31-K242A (Fig. 5A) or pUL31-C241-243A (16).
Thus, the question remains why K242A resulted in these massive accumulations of membrane vesicles in the PNS. Alanine is the only amino acid tested with an apolar side chain, in contrast to the wild-type lysine and all other amino acids substituted. In addition, modeling shows that the side chain does not fill the space between the pUL31 molecules in the dimeric interface of the hexamers (Fig. 1B and C), with less restriction and/or higher flexibility probably resulting in deregulated vesicle budding and scission. A similar dysregulation of vesicle formation from the INM was reported for HSV-1 when a basic patch in pUL34 was mutated to alanine (R158A/R161A, CL13) (26). These two mutations are located in the interaction surface between pUL34 and pUL31 within the core NEC and close to an intramolecular salt bridge (18). It is feasible that both changes, the alanine substitution in PrV pUL31-K242A and the double alanine exchange in HSV-1 pUL34, resulted in a greater flexibility and an enhanced propensity for oligomerization, resulting in increased capsid-independent budding and vesicle scission.
The defect in HSV-1 pUL34 could be compensated for by inter- or intramolecular second-site mutations (26). Thus, we were interested in whether this might also be the case for PrV pUL31-K242A and pUL31-C241-243A. For this, recombinant viruses expressing pUL31-K242A or pUL31-C241-243A were generated and passaged in RK13 and Vero cells. Although severely impaired in viral replication, infectious progeny was formed in nontransgenic cells infected with these mutants, which served as a basis for reversion analysis. Within less than 10 passages, viral titers increased to wild-type-like values of 106 PFU/ml. Surprisingly, second-site mutations occurred at different amino acid positions in pUL31 as well as in pUL34, pointing to higher flexibility of the NEC than expected.
The relevance of mutations uncovered in pUL34 is difficult to test in our trans-complementation assays since the native pUL34 was still present. Therefore, in this study, we focused on the mutations found in pUL31. Cell lines expressing the second-site pUL31 mutants in the presence of the H10 substitutions all replicated PrV-ΔUL31 to significantly higher titers than the parental cell lines RK13-UL31-K242A and RK13-UL31-C241-243A or nontransgenic RK13 cells. Titers only 6- to 12-fold lower than those of PrV-Ka were achieved with pUL31-K242T, pUL31-M79I/K242A, pUL31-L115R/K242A, pUL31-A137T/K242A, pUL31-Y121H/C241-243A, and pUL31-C241-243A/G250R, pointing to at least a partial functional complementation of the nuclear egress defect and infectious virion maturation. Approximately 30-fold-lower titers were obtained on RK13-UL31-S40A/K242A and RK13-UL31-A126T/C241-243A cells than were obtained for PrV-Ka, still pointing to a partial functional rescue of the defect induced by the K242A/C241-243A mutations.
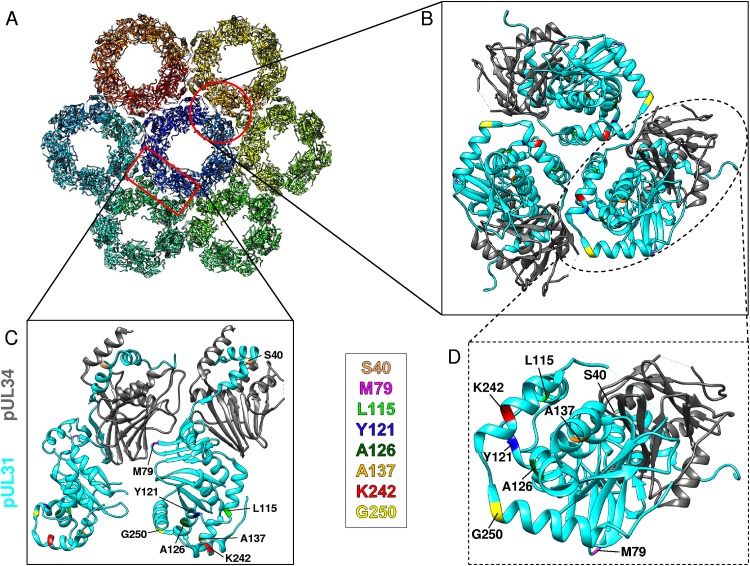
Modeling the second-site mutations into the NEC structure showed that the substitutions L115R, Y121H, A126T, A137T, and G250R were located in close proximity to the alpha-helical region H10 or a direct reversion of K242A (K242T) within the membrane-distal part of pUL31 (Fig. 10), indicating that these mutations compensate for the structural defects imposed by the amino acid changes in H10.
FIG 10.
Location of the pUL31 mutations in the NEC dimer and in the hexagonal lattice. (A) Top view of the NEC hexameric lattice (PDB accession number 5FKI) (17, 21). The chains are rainbow colored. (B to D) Close-up views of NEC heterodimer interfaces of the hexameric lattice, with the mutations being marked in color. pUL31 is shown in cyan, while pUL34 is shown in gray. The interfaces indicated in panel A are represented by a red circle for panel B and a red rectangle for panel C. (B) Top view of the trimer interface between hexamers. (C) Side view of the dimer-dimer interface. (D) Zoom of the membrane-distal part of the NEC shown in panel B. The amino acids that were mutated are highlighted by different colors and labeled. Molecular graphics and analysis were performed with the UCSF Chimera package (34).
The second-site mutations resulting in amino acid substitutions L115R und G250R both introduce a positive charge, which was lost in pUL31-K242A and pUL31-C241-243A. L115 is located at the end of H5, which is supposed to form the trimeric interface in hexamers (17). Addition of this positive charge might rearrange and stabilize the hexamers forming the trimeric interaction interface. G250R is located in H11, which also carries the nuclear export signal sequence (25). The simultaneous substitutions L252A and L254A, which are part of the predicted NES (pUL31-NESPM), resulted in a nonfunctional protein (25), further supporting the suggestion that H10 and H11 may collaborate for membrane deformation and budding during nuclear egress. L115R and G250R, however, resulted in a nonfunctional pUL31 in the absence of the K242A/C241-243A mutation. In ultrastructural analyses, a phenotype similar to that achieved with pUL31-ΔNES, i.e., nucleocapsids lined up at the INM but no detectable membrane deformation or budding events, was observed (25), indicating that H5, in addition to H10 and H11, facilitates membrane deformation and budding.
The second-site mutations S40A and M97I are located distantly from H10 and the membrane-distal part of the NEC (Fig. 10). S40 is located in the second of the two alpha helices forming the hinge region of the pUL31 N-terminal arm, which reaches around and inserts into a groove of the pUL34 core. This arm is one of the major interaction domains between the complex partners in the NEC dimer (17–20). In PrV pUL31, the neighboring D41 forms an intramolecular hydrogen bond with Y34, stabilizing the kink at an angle of ca. 80° (17). It is conceivable that the exchange of the polar serine in the wild-type protein for the nonpolar alanine (S40A) allows more flexibility of this arm and, thus, orientation between the complex partners. In addition, S40 is predicted to be a putative phosphorylation site (http://www.cbs.dtu.dk/services/NetPhos), which may play a regulatory role in assembly and/or disassembly of the NEC during nuclear egress and which is lost by the S40A substitution.
The region comprising amino acids 77 to 79 in PrV pUL31 forms a loop between two β sheets (17) (Fig. 10). This loop, together with the zinc finger (ZNF) motif coordinated by amino acids C73, C89, C92, and H188, is anticipated to form an important intrahexamer interface connecting to a loop comprising amino acids 88 to 93 in pUL34 of a neighboring NEC dimer (17) (Fig. 10). Mutations in or close to the loop as well as mutations in the ZNF motif (L76A, S77A, G80A, C73S, C89S, C92S, and H188A) resulted in nonfunctional proteins (17), highlighting the importance of this region for higher-order complex formation and NEC function. The exchange of methionine at position 79 for the more hydrophobic isoleucine might impose a higher flexibility on the dimer-dimer interaction interface.
None of the revertants revealed a mutation back to K242. However, in one of the revertants, which efficiently replicated in cell culture, the alanine substitution in pUL31-K242A was changed to threonine (pUL31-K242T). Cell lines expressing pUL31-K242T partially complemented the defect of PrV-ΔUL31, and the titers derived from these cells were similar to those observed for RK13-UL31-K242S and RK13-UL31-K242Y cells (Fig. 4), adding another example of a functional amino acid exchange of the lysine at position 242. It is noteworthy that all amino acid substitutions at this position which complemented PrV-ΔUL31 could serve as a potential phosphorylation site. The phosphomimetic amino acids aspartic acid (D) and glutamic acid (E), however, were nonfunctional at this position (Fig. 4). Lysine, besides adding a positive charge, is also a major target for posttranslational modifications, such as ubiquitination, acetylation, sumoylation, and methylation, which play critical roles in regulating biological processes (27). Further studies are needed to uncover putative protein modifications in the NEC, especially at this particular site.
Several K242A second-site revertants had no additional mutations in pUL31 but in pUL34 (Table 2). All these mutations were located in predicted loop regions (G15W, T25M, A26V, T98A, A99V) (17). The loop region comprising amino acids 22 to 26 of PrV pUL34 is thought to form the dimeric interface in the hexagonal lattice (17). In addition, in HSV-1 the dominant negative mutation D35A/E37A in pUL34 (corresponding to amino acids D22/E24 in PrV pUL34) (28, 29) impairs curvature formation of the coat (5). The functional relevance of these mutations will be analyzed in the future.
In summary, our data indicate that K242 in H10 in PrV pUL31 may, in fact, not mediate a direct interaction with the nucleocapsid cargo but indicate that this part of the NEC is important for rearrangements leading to the correct curvature of the coated vesicle, allowing for uptake of the nucleocapsid into the nascent primary virion envelope.
MATERIALS AND METHODS
Cells and viruses.
Rabbit kidney (RK13) cells, RK13-UL31 cells, and Vero cells were cultivated in Dulbeccós modified Eaglés minimum essential medium supplemented with 10% or 5% (Vero) fetal calf serum. PrV laboratory strain Kaplan (PrV-Ka) (30) was propagated in RK13 cells, while PrV-ΔUL31 was grown in RK13-UL31 cells (23).
Site-specific mutagenesis of pUL31.
pUL31 mutants were generated using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) as described previously (16) and the primers listed in Table 1 with the respective UL31 genes cloned in pcDNA3 (Invitrogen) as the template. Correct mutagenesis was verified by sequencing.
Laser-scanning confocal microscopy.
For localization and colocalization studies, RK13 cells were transfected either with the corresponding pUL31 expression plasmids singly or with the corresponding pUL31 expression plasmids cotransfected with pcDNA-UL34 (22) by calcium phosphate coprecipitation (31). At 2 days posttransfection, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS), and incubated with a recently generated anti-pUL31 monoclonal antibody (1:50) and with the monospecific rabbit anti-pUL34 serum (1:500) (22). For detection, Alexa Fluor 488-conjugated goat anti-mouse IgG and Alexa 568-conjugated goat anti-rabbit IgG (Invitrogen) were used. Images were acquired with a confocal laser-scanning microscope (63× oil immersion objective, single slice; SP5; Leica, Germany) and processed using ImageJ software (32).
Generation of stably expressing RK13 cell lines.
To generate stably expressing RK13 cell lines, calcium phosphate coprecipitation was used (31). At 2 days posttransfection, the cells were split and transferred into selection medium containing 500 μg/ml G418 (Invitrogen). After 10 to 14 days, resistant cell colonies were picked by aspiration and screened by indirect immunofluorescence for pUL31 expression using the polyclonal rabbit anti-pUL31 serum (23). Cell clones homogeneously expressing wild-type or mutated pUL31 were used.
Immunoblot analysis.
Cells were cultivated in 24-well plates for 2 days, scraped into medium, and pelleted by centrifugation at 2,000 × g for 5 min. The pellets were resuspended after washing with phosphate-buffered saline (PBS) in sample buffer (0.13 M Tris-HCl, pH 6.8, 4% SDS, 20% glycerin, 0.01% bromophenol blue, 10% β-mercaptoethanol). Lysates were sonicated and boiled for 3 min before separation of proteins in an SDS-10% polyacrylamide gel. Proteins were transferred onto a nitrocellulose membrane and incubated with the polyclonal rabbit anti-pUL31 serum (23) and a monoclonal anti-α-tubulin antibody (Sigma) as a loading control. Bound antibody was detected by peroxidase-coupled goat anti-rabbit and goat anti-mouse immunoglobulin antibodies and visualized by enhanced chemiluminescence (Bio-Rad Clarity Western ECL blotting substrate), recorded in an image analyzer (Bio-Rad).
Generation of virus recombinants expressing pUL31-K242A and pUL31-C241-243A.
Virus recombinants PrV-UL31-K242A and PrV-UL31-C241-243A were generated by homologous recombination. For this, the corresponding mutations were introduced into cloned genomic 3.3-kb SalI fragment 1C, comprising the UL31 gene region (23). Mutations were introduced using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) and the primers described previously (16). Recombination plasmids were cotransfected with PrV-ΔUL31 (green fluorescent protein-positive) genomic DNA (7), followed by purification of nonfluorescing plaques on RK13-UL31 cells. Correct mutagenesis and recombination were tested after PCR amplification by sequencing of the corresponding gene region.
Serial passaging of virus recombinants.
For serial passaging, monolayers of RK13 and Vero cells were infected with either PrV-UL31-K242A or PrV-UL31-C241-243A in 24-well cell culture plates in dilutions of 10−1 to 10−6. Infected cells were incubated for 3 to 5 days. The contents of the well with the highest virus dilution showing a 100% cytopathic effect were harvested and centrifuged at 15,000 × g for 5 min. The supernatant was again titrated on 24-well culture plates. After 10 and 30 passages, the supernatants were titrated under plaque assay conditions and single virus plaques were picked. After a second round of plaque purification, virus stocks were prepared and the titer was determined on RK13 cells. Plaque isolates which reached titers of at least 106 PFU/ml were further investigated. For this, viral DNA was isolated, and the UL31 and UL34 genes were amplified by PCR (23) and sequenced. UL31 PCR products carrying additional mutations were cloned into pcDNA3, and stably expressing cells were generated as described above.
In vitro complementation studies.
To test for functional complementation, stably expressing cell lines were infected on ice with PrV-Ka or PrV-ΔUL31 at a multiplicity of infection (MOI) of 5 for 1 h to allow for attachment of viruses to the cells. Subsequently, the inoculum was replaced by prewarmed medium to initiate infection and incubated at 37°C for 1 h. Thereafter, the remaining extracellular virus was inactivated by low-pH treatment (33), and cells were kept for an additional 24 h at 37°C. Cells and supernatant were harvested, frozen, and thawed. After the removal of cellular debris by centrifugation, progeny virus in the supernatant was titrated on RK13-UL31 cells. The mean values from at least three independent experiments were calculated. Statistical significance was determined by a two-way analysis of variance, followed by Sidak’s multiple-comparison test. All statistical tests were performed using GraphPad Prism (version 8.1.0) software (GraphPad Software, La Jolla, CA, USA). Significant differences between PrV-Ka and PrV-ΔUL31 infection are indicated by asterisks in the appropriate figures.
Electron microscopy.
Stably expressing cell lines infected with PrV-ΔUL31 or RK13 cells infected with PrV-UL31-K242A or PrV-UL31-C241-243A at an MOI of 1 were processed for electron microscopy at 14 h p.i. as described previously (22).
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft (grant DFG ME 854/12-2). Molecular graphics and analyses were performed with the UCSF Chimera package, which was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS grant P41-GM103311).
We thank Cindy Krüper, Karla Günther, and Petra Meyer for technical help and Mandy Jörn for photographic assistance. Molecular graphics and analyses were performed with the UCSF Chimera package.
REFERENCES
- 1.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 2.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Mettenleiter TC, Muller F, Granzow H, Klupp BG. 2013. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 4.Mettenleiter TC. 2002. Herpesvirus assembly and egress. J Virol 76:1537–1547. doi: 10.1128/jvi.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigalke JM, Heuser T, Nicastro D, Heldwein EE. 2014. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun 5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorenz M, Vollmer B, Unsay JD, Klupp BG, Garcia-Saez AJ, Mettenleiter TC, Antonin W. 2015. A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J Biol Chem 290:6962–6974. doi: 10.1074/jbc.M114.627521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc Natl Acad Sci U S A 104:7241–7246. doi: 10.1073/pnas.0701757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klupp BG, Granzow H, Mettenleiter TC. 2011. Nuclear envelope breakdown can substitute for primary envelopment-mediated nuclear egress of herpesviruses. J Virol 85:8285–8292. doi: 10.1128/JVI.00741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newcomb WW, Fontana J, Winkler DC, Cheng N, Heymann JB, Steven AC. 2017. The primary enveloped virion of herpes simplex virus 1: its role in nuclear egress. mBio 8:e00825-17. doi: 10.1128/mBio.00825-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funk C, Ott M, Raschbichler V, Nagel CH, Binz A, Sodeik B, Bauerfeind R, Bailer SM. 2015. The herpes simplex virus protein pUL31 escorts nucleocapsids to sites of nuclear egress, a process coordinated by its N-terminal domain. PLoS Pathog 11:e1004957. doi: 10.1371/journal.ppat.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A 108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang K, Wills E, Lim HY, Zhou ZH, Baines JD. 2014. Association of herpes simplex virus pUL31 with capsid vertices and components of the capsid vertex-specific complex. J Virol 88:3815–3825. doi: 10.1128/JVI.03175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeshima K, Arii J, Maruzuru Y, Koyanagi N, Kato A, Kawaguchi Y. 2019. Identification of the capsid binding site in the herpes simplex virus 1 nuclear egress complex and its role in viral primary envelopment and replication. J Virol 93:e01290-19. doi: 10.1128/JVI.01290-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leelawong M, Guo D, Smith GA. 2011. A physical link between the pseudorabies virus capsid and the nuclear egress complex. J Virol 85:11675–11684. doi: 10.1128/JVI.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronfeldt S, Klupp BG, Franzke K, Mettenleiter TC. 2017. Lysine 242 within helix 10 of the pseudorabies virus nuclear egress complex pUL31 component is critical for primary envelopment of nucleocapsids. J Virol 91:e01182-17. doi: 10.1128/JVI.01182-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeev-Ben-Mordehai T, Weberruß M, Lorenz M, Cheleski J, Hellberg T, Whittle C, El Omari K, Vasishtan D, Dent KC, Harlos K, Franzke K, Hagen C, Klupp BG, Antonin W, Mettenleiter TC, Grünewald K. 2015. Crystal structure of the herpesvirus nuclear egress complex provides insights into inner nuclear membrane remodeling. Cell Rep 13:2645–2652. doi: 10.1016/j.celrep.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bigalke JM, Heldwein EE. 2015. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J 34:2921–2936. doi: 10.15252/embj.201592359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lye MF, Sharma M, El Omari K, Filman DJ, Schuermann JP, Hogle JM, Coen DM. 2015. Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex. EMBO J 34:2937–2952. doi: 10.15252/embj.201592651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walzer SA, Egerer-Sieber C, Sticht H, Sevvana M, Hohl K, Milbradt J, Muller YA, Marschall M. 2015. Crystal structure of the human cytomegalovirus pUL50-pUL53 core nuclear egress complex provides insight into a unique assembly scaffold for virus-host protein interactions. J Biol Chem 290:27452–27458. doi: 10.1074/jbc.C115.686527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, Bäuerlein FJB, Cheleski J, Werner S, Guttmann P, Rehbein S, Henzler K, Demmerle J, Adler B, Koszinowski U, Schermelleh L, Schneider G, Enquist LW, Plitzko JM, Mettenleiter TC, Grünewald K. 2015. Structural basis of vesicle formation at the inner nuclear membrane. Cell 163:1692–1701. doi: 10.1016/j.cell.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol 74:10063–10073. doi: 10.1128/jvi.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J Virol 76:364–378. doi: 10.1128/jvi.76.1.364-378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klupp BG, Granzow H, Keil GM, Mettenleiter TC. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J Virol 80:6235–6246. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paßvogel L, Klupp BG, Granzow H, Fuchs W, Mettenleiter TC. 2015. Functional characterization of nuclear trafficking signals in pseudorabies virus pUL31. J Virol 89:2002–2012. doi: 10.1128/JVI.03143-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roller RJ, Haugo AC, Kopping NJ. 2011. Intragenic and extragenic suppression of a mutation in herpes simplex virus 1 UL34 that affects both nuclear envelope targeting and membrane budding. J Virol 85:11615–11625. doi: 10.1128/JVI.05730-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu HD, Zhou JQ, Lin SF, Deng WK, Zhang Y, Xue Y. 2017. PLMD: an updated data resource of protein lysine modifications. J Genet Genomics 44:243–250. doi: 10.1016/j.jgg.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Bjerke SL, Cowan JM, Kerr JK, Reynolds AE, Baines JD, Roller RJ. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 U(L)34 protein. J Virol 77:7601–7610. doi: 10.1128/jvi.77.13.7601-7610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roller RJ, Bjerke SL, Haugo AC, Hanson S. 2010. Analysis of a charge cluster mutation of herpes simplex virus type 1 UL34 and its extragenic suppressor suggests a novel interaction between pUL34 and pUL31 that is necessary for membrane curvature around capsids. J Virol 84:3921–3934. doi: 10.1128/JVI.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan AS, Vatter AE. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 31.Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 32.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mettenleiter TC. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623–625. doi: 10.1016/0042-6822(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]