Tunneling nanotubes (TNTs) are actin-based intercellular conduits that connect distant cells and allow intercellular transfer of molecular information, including genetic information, proteins, lipids, and even organelles. Besides providing a means of intercellular communication, TNTs may also be hijacked by pathogens, particularly viruses, to facilitate their spread. Viruses of many different families, including retroviruses, herpesviruses, orthomyxoviruses, and several others have been reported to trigger the formation of TNTs or TNT-like structures in infected cells and use these structures to efficiently spread to uninfected cells.
KEYWORDS: TNT, antibodies, intercellular, spread, tunneling nanotubes, virus
ABSTRACT
Tunneling nanotubes (TNTs) are actin-based intercellular conduits that connect distant cells and allow intercellular transfer of molecular information, including genetic information, proteins, lipids, and even organelles. Besides providing a means of intercellular communication, TNTs may also be hijacked by pathogens, particularly viruses, to facilitate their spread. Viruses of many different families, including retroviruses, herpesviruses, orthomyxoviruses, and several others have been reported to trigger the formation of TNTs or TNT-like structures in infected cells and use these structures to efficiently spread to uninfected cells. In the current review, we give an overview of the information that is currently available on viruses and TNT-like structures, and we discuss some of the standing questions in this field.
INTRODUCTION
Cell-to-cell communication is essential to coordinate and maintain physiological activities in organs and tissues. Eukaryotes have developed different strategies for intercellular communication that depend on the nature of the message and the distance across which the message has to be transmitted. For long-distance communications, several strategies can be used, including the secretion of messengers such as cytokines, hormones, or growth factors, or the release of extracellular vesicles, such as exosomes, which may contain different biomolecules (e.g., proteins, lipids, and genetic information) (reviewed in reference 1). An additional type of long-distance intercellular communications was described in 2004, based on the observation of thin membranous nanotubes connecting distant PC12 rat pheochromocytoma cells (2). Rustom et al. described these structures as hovering above the substrate, containing F-actin and enclosed in a lipid bilayer. The term “tunneling nanotube” (TNT) was coined to designate these structures, and it refers to their diameter, originally described as ranging from 50 to 200 nm, and their ability to mediate intercellular transfer of molecular information by forming a “tunnel” between distant and otherwise nonconnected cells. The original report on PC12 cells described TNTs as mediators of intercellular transfer of endocytic vesicles and organelles. Subsequent studies confirmed their presence in a variety of cell types and broadened the range of cargo transported via TNTs, including, for example, cytosolic proteins (3), ions (4), miRNAs (5), and/or organelles such as the endoplasmic reticulum (6), mitochondria (7), Golgi vesicles (6), endosomes (8), or lysosomes (8).
As TNTs started to gain more attention from the scientific community, their heterogeneous nature, e.g., in terms of diameter, length, and cytoskeletal composition, became increasingly clear. All TNTs contain filamentous F-actin as backbone (9). TNTs that are smaller in diameter (<0.7 μm, “thin” TNTs) typically only contain F-actin, whereas “thick” TNTs (>0.7 μm) often contain both actin and microtubules (8). Both types of TNTs may contain nonconventional actin-based myosin motors myosin Va and/or myosin X (MyoX) (2, 10). Myosin Va has been suggested to mediate actomyosin-dependent transport of endocytic vesicles in TNTs (2), while MyoX has been proposed to be a major player in the process of TNT formation (11). In addition to myosin motors, microtubule-containing “thick” TNTs may also harbor microtubule motors kinesin 1 and dynein to drive plus- or minus-end directed movement of cargo, respectively (8, 12). Currently, TNTs are broadly defined as actin-containing intercellular membranous connections that lack substrate adhesion and allow intercellular transfer of molecular information (reviewed in references 9, 13, and 14). TNTs can be open or closed ended, depending on whether they do or do not show cytoplasmic connectivity between donor and recipient cells, respectively (2, 15). A recent study using correlative cryo-electron microscopy to determine the ultrastructure of TNTs in neuronal cell lines (CAD and SH-SY5Y) showed that many of the nanotubes that appeared as a single contiguous structure by fluorescence microscopy in fact constituted a bundle of several thinner individual TNTs (iTNTs) stabilized and held together by N-cadherin (16). The TNTs in this study were open ended and lacked microtubules. It remains to be determined whether other types of TNTs equally may consist of bundles of iTNTs.
In general, two (non-mutually exclusive) models of TNTs formation have been proposed. According to the first model, cells form a filopodium-like protrusion that elongates and docks on a neighboring cell and further differentiates into a TNT (2). In some cases, such as TNTs established by the urothelial cell line T24 or TNTs induced by the US3 protein of pseudorabies virus (PRV), adhesion molecules such as cadherins and β-catenin localize in the contact area between the filopodium/TNT and the recipient cell, stabilizing and anchoring the newly established TNT (17, 18). In the second model, TNTs originate between two joined cells that move away from each other and retain a thin membranous thread upon cell dislodgement (15, 19).
The heterogeneous nature of TNTs suggests that their formation may rely on a variety of molecular pathways, which are still largely unresolved. Nonetheless, some important regulatory factors have been identified over the past years, including M-Sec and the aforementioned MyoX.
In 2009, Hase et al. showed that the 73-kDa cytosolic protein M-Sec is present in TNTs connecting Raw264.7 macrophage cells and that knockdown of M-Sec via RNA interference suppressed the formation of TNTs (20). M-Sec contributes to TNT formation via its association with the Ras-like small GTPase Ral-A and the exocyst complex (20). The LST-1 (leukocyte-specific transcript 1) protein appears to act as a scaffold to recruit Ral-A, the exocyst complex and likely M-Sec to the plasma membrane (21). Once the complex is assembled, Ral-A binds to filamin, MyoX, myoferlin, and Ral binding protein 1 (RalBP1), resulting in the activation of Cdc42, which subsequently leads to actin remodeling and filopodium formation (20–22). Nevertheless, the fact that TNTs can be established between cells that do not express M-Sec, i.e., neurons (11, 23), indicates that different mechanisms of TNT formation may exist.
MyoX is a widespread unconventional myosin motor protein with an established role in filopodium formation and elongation (24, 25). In the central nervous system cell line CAD, overexpression of MyoX results in TNT formation, and knockdown of MyoX via shRNA impairs endogenous TNT formation (11). In this particular model, MyoX-induced TNTs emerge from the extension and docking of filopodia. MyoX is recruited to the plasma membrane of a subset of filopodia through its binding with phosphatidyl inositol moieties [PtdIns(3-5)P3], where it contributes to TNT formation (11). In support of this, in a recent study, expression of the HIV protein Nef was found to correlate with an increase in MyoX expression, and this in turn resulted in MyoX-dependent TNT formation (26).
Although TNTs have been described to occur under physiological conditions, several stress stimuli may also trigger their formation, including oxidative stress, serum starvation, hyperglycemic conditions, inflammation, UV radiation, low pH, and hypoxia, among others (6, 27–29). These data suggest that, under specific circumstances, TNT formation may be part of a stress response mechanism to facilitate biomolecule and/or energy transfer between damaged and healthy cells.
There is increasing evidence that TNT-mediated intercellular communication is involved in several pathologies, including cancer, neurodegenerative diseases, and infectious diseases. TNTs can facilitate cell-to-cell signaling and transfer of cellular contents between tumor cells and their microenvironment (5, 30, 31) and have been observed in human primary tumors (29, 32, 33). TNTs may contribute to cancer progression and therapy resistance through the transfer of, e.g., oncogenic miRNAs (5), multidrug resistance protein 1 (34, 35), mitochondria (36), and intercellular calcium waves (33). For more information on the role of TNTs in cancer pathobiology, we refer the reader to different reviews on the topic (see, for example, references 37, 38, and 39). TNTs may also play a role in the progression of several neurodegenerative diseases since they have been reported to contribute to intercellular transfer of misfolded proteins from affected to healthy neurons, including transfer of amyloid β and tau protein (Alzheimer’s disease), α-synuclein (Parkinson’s disease), huntingtin (Huntington’s disease), superoxide dismutase-1 (amyotrophic lateral sclerosis), and prion protein (PrPSc; transmissible spongiform encephalopathies) (reviewed in reference 40). TNTs may also contribute to intercellular spread of pathogens, primarily viruses—the topic of the present review—but in some cases also bacteria. For example, recently, it has been demonstrated that Mycoplasma hyorhinis induces TNT formation in infected cells and exploits these structures for efficient and rapid cell-to-cell dissemination (41).
INDUCTION AND USE OF TNTs AND TNT-LIKE STRUCTURES BY VIRUSES
Viruses from different families have been described to trigger formation of TNTs or TNT-like structures (Table 1), which these viruses may use to efficiently spread infection and molecular information to neighboring cells. For some viruses, particularly retroviruses and alphaherpesviruses, viral proteins that are responsible for TNT formation have been identified, as well as (part of) the signaling axes that connect the expression of these viral proteins to TNT formation.
TABLE 1.
Overview of TNTs and TNT-like structures triggered by different virusesa
| Virus | Viral protein involved | Infected cells | Cell type(s) | Microtubules | Spread in presence of virus-neutralizing antibodies | Virus particles visualized in or on TNT-like structures by EM | Reference(s) |
|---|---|---|---|---|---|---|---|
| Retroviruses | |||||||
| HIV-1 | Nef | Yes | Macrophages | Yes | Yes | Yes (TEM) | 42, 43, 45 |
| HIV-1 and MLV | Env | Yes | Cos-1, XC and HEK293 | No | No | Yes (SEM) | 53 |
| HTLV-1 | p8 | Yes | p8-overexpressing Jurkat, MT-2 and primary T cells | No | NDb | Yes (TEM) | 51 |
| Herpesviruses | |||||||
| Alphaherpesviruses | |||||||
| PRV | US3 | Yes | ST, RK13, MEF | Yes | Yes | Yes (TEM) | 17, 56, 62 |
| HSV-2 | US3 | ND | Vero | Yes | ND | ND | 66 |
| BoHV-1 | US3 (in transduced cells) | Yes | MDBK, KOP, primary bovine fibroblasts | Yes | Yes | ND | 65, 71 |
| BoHV-5 | US3 | ND | ST, Vero | ND | ND | ND | 67 |
| Gammaherpesviruses | |||||||
| MHV-68 | gp48/ORF58 | Yes | NIH 3T3, BHK-21 | No | ND | ND | 73 |
| EBV | BDLF2/BMRF2 | ND | Cos7, 293T, HeLa | No | ND | ND | 72 |
| Alphaviruses | |||||||
| SINV, SFV, CHIKV | Structural proteins (SFV) | Yes | Vero, primary HUVEC | Yes | Yes | Possibly (SEM) | 75 |
| Pneumoviruses | |||||||
| HMPV | P (transfected cells) | Yes | BEAS-2B, A549 | Yes | Yes | ND | 76 |
| Poxviruses | |||||||
| VV | F11 | Yes | BS-C-1, Vero, PtK2 | ND | ND | ND | 77–80 |
| Orthomyxoviruses | |||||||
| IAV | ND | Yes | MDCK, Vero, A549 | ND | Yes | ND | 81, 82 |
| Paramyxoviruses | |||||||
| PIV5 | ND | Yes | MDCK, Vero, A549 | ND | Yes | ND | 81 |
| MeV | ND | Yes | Astrocytoma cells | ND | ND | ND | 83 |
| Nidoviruses | |||||||
| PRRSV | ND | Yes | MARC-145 | Yes | Yes | ND | 84, 85 |
| Flaviviruses | |||||||
| WNV | NS1 (transfected cells) | Yes | Vero, SH-SY5Y, U-87MG | ND | ND | ND | 86 |
| Arenaviruses | |||||||
| LCMV | ND | Yes | HeLa | ND | ND | ND | 88 |
| Picornaviruses | |||||||
| CVB3 | ND | Yes | GMK | Yes | Yes | ND | 89 |
| Reoviruses | |||||||
| RDV | Pns10 (transduced cells) | Yes | NC-24 and Sf9 insect cells | ND | Yes | ND | 90, 91 |
| RBSDV | P7-1 (transduced cells) | Yes | Sf9 insect cells | ND | ND | ND | 92, 93 |
More detailed information can be found in the text. TEM, transmission electron microscopy; SEM, scanning electron microscopy.
ND, not determined.
RETROVIRUSES
Intercellular retrovirus spread using TNTs was first shown for human immunodeficiency virus (HIV). Transfer of virus was found to occur between macrophages, where virus capsids were observed in endocytic compartments in the TNTs, and also between CD4+ T cells where virus particles may surf on the outer surface of the intercellular membranous conduits (15, 42, 43). Recently, coinfection of HIV and Mycobacterium tuberculosis was found to increase TNT-mediated spread of HIV in macrophages. The interleukin-10 (IL-10)-dominated anti-inflammatory microenvironment associated with tuberculosis (TB) increases TNT formation in macrophages via IL-10/STAT3 signaling, thereby enhancing intercellular HIV spread (44). TNTs in HIV-infected macrophages may also connect to B cells, which results in transfer of the viral protein Nef from infected macrophages to B cells, thereby interfering with antibody isotype class switching in the latter (45). TNT formation in HIV-infected macrophages depends on the viral Nef protein and its ability to interact with the nucleotide exchange factor Vav, thereby affecting Rho GTPase signaling and actin polymerization (45).
Interestingly, the HIV-associated TNTs observed in T cells displayed very different properties compared to those observed in HIV-infected macrophages. In T cells, TNTs contain actin but no microtubules and do not allow the transfer of membrane-associated proteins, small cytoplasmic dyes, or intercellular calcium signaling (15). TNTs between macrophages do contain microtubules, allow the transfer of organelles, and mediate gap junctional communication (43, 46). As another important difference between both cell types, in T-cells, HIV uses existing endogenous TNTs to transfer infectious virus to connected cells, while the virus triggers increased formation of TNTs in macrophages via the viral accessory protein Nef (15, 42, 45, 47) (HIV Nef-induced TNT formation in macrophages is schematically illustrated in Fig. 1). This discrepancy in properties and functionality between the two cell types was explained in 2016 when it was shown that HIV Nef-dependent induction of TNTs in macrophages requires the expression of M-Sec (48). T cells, in contrast to macrophages, do not express M-Sec, and thus the number of TNTs does not increase upon Nef expression in these cells. T-cell lines expressing M-Sec, on the other hand, do show increased TNT formation in response to Nef expression (48). The precise mechanism by which Nef activates M-Sec to induce the formation of TNTs is still largely unknown, although it has been shown that Nef associates with the Rac1/Cdc42 effector p21-activated kinase 2 (PAK2) and several components of the exocyst complex and that Nef expression leads to upregulation of MyoX, which is required for TNT induction (26, 49, 50). Whether and how MyoX and M-Sec act cooperatively during Nef-induced TNT formation remains to be determined.
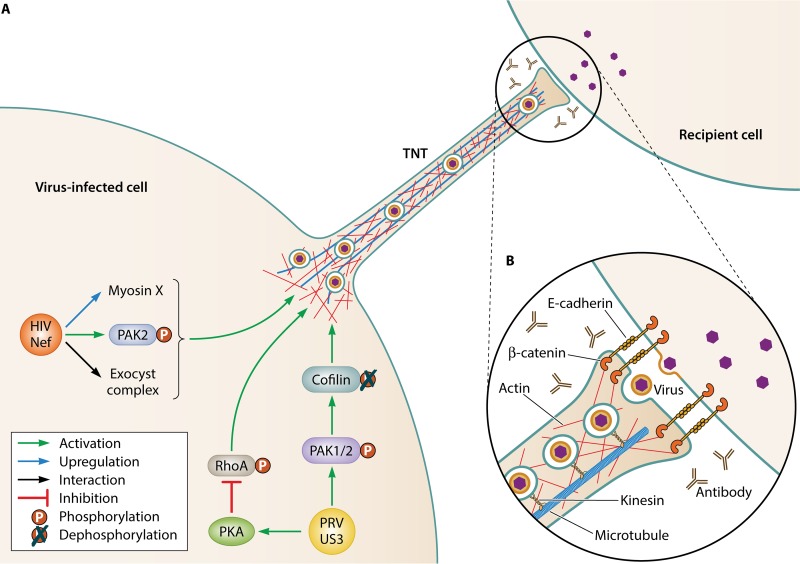
FIG 1.
(A) Schematic representation of two of the best characterized models of virus-induced TNT formation: Nef-induced TNT formation in HIV-infected macrophages and US3-induced TNT formation in PRV-infected epithelial cells. Both types of TNTs have been shown to carry virus particles in vesicles (see, for example, references 17 and 43). Some of the molecular players that have been shown to be involved in these two types of TNT formation are indicated. More detailed information on their role is provided in the present study. (B) An inset image shows a schematic representation of what is known about the contact area between a PRV US3-induced TNT and a recipient cell based on electron microscopy and confocal microscopy analyses (based on reference 17).
Human T-cell leukemia virus type 1 (HTLV-1), another retrovirus, also triggers the formation of TNTs via its p8 protein. HTLV-1 p8-induced TNTs contain mature virus particles that concentrate in the contact area with connected cells. In contrast to HIV Nef, the p8 protein of HTLV-1 is able to trigger formation of TNTs in T cells (51). It was claimed that this is due to the ability of p8 to induce the expression of M-Sec (48). However, another report described that expression of the Tax protein of HTLV-1 also induces the expression of M-Sec, without affecting TNT formation (52). Thus, the involvement of M-Sec in HTLV-1-induced TNT formation has not yet been completely resolved. The p8-induced TNTs facilitate intercellular spread of HTLV-1 between T cells (51).
Other reports also document retrovirus-mediated induction and/or use of intercellular connections, which may constitute bona fide TNTs and/or TNT-related structures. For example, murine leukemia virus (MLV) and HIV both use intercellular filopodial bridges, dubbed viral cytonemes, to spread infection in Cos-1, XC, and HEK293 cell lines (53). These filopodial bridges originate from uninfected cells and contact infected cells. Their formation depends on the interaction of the virus receptor (mCATs1 for MLV and CD4/CXCR4 for HIV) on the surfaces of uninfected cells and the viral ligand Env on the infected cell. Virus particles reach the uninfected cell body by surfing on the outside of the viral cytoneme using actin-based retrograde flow of the receptor. Expression of Env alone is sufficient to induce the formation of these viral cytonemes (53).
In infected dendritic cells (DCs) cocultured with T cells, immature budding HIV particles localize to the tips of filopodia of the DCs (54). These virus-capped filopodia scan the surroundings of the DCs and can make up to 800 contacts per hour with neighboring T cells. HIV capping of filopodia depends on the viral Gag polyprotein and the frequency of filopodium formation is enhanced by the viral Nef protein, suggesting functional and/or biochemical similarity to HIV Nef-induced TNTs (54, 55). The presence of virus-capped filopodia correlates with increased spread of HIV in DC T-cell cocultures (54).
HERPESVIRUSES
Around the time of the discovery of TNTs, we described the formation of long cell protrusions in epithelial and fibroblast cells infected with the porcine alphaherpesvirus pseudorabies virus (PRV) (56). These protrusions contact uninfected cells to form intercellular membranous structures, contain F-actin and microtubules, and do not contact the substrate and therefore represent TNTs (17). Expression of the viral serine/threonine protein kinase US3 is necessary and sufficient for their formation (56, 57) (PRV US3-induced TNT formation is schematically illustrated in Fig. 1). In PRV-infected cells, US3-induced TNTs contain mature enveloped virus particles in vesicles that move toward uninfected cells and contribute to viral spread, even in the presence of virus-neutralizing antibodies (17, 56). Electron microscopy showed that US3-induced TNTs are closed ended and that TNT-mediated virus spread appears to occur via egress of enveloped virus particles at the contact area between TNT and recipient cell, followed by viral entry into the recipient cell (17). In line with this, PRV lacking the fusion-driving envelope protein gB is unable to spread via TNTs (R. J. J. Jansens and H. W. Favoreel, unpublished data). Interestingly, the US3 protein is largely dispensable for virus replication in cell cultures, while US3-null viruses are substantially attenuated in vivo, indicating that US3 serves as an important virulence factor (58–60). In contrast to most TNTs described in literature which are only stable for relatively short time periods (typically minutes to a few hours), TNTs induced by the US3 protein are very stable and have lifetimes up to 24 h (17, 56). This property is likely caused by the presence of stabilized microtubules (containing posttranslational tubulin modifications) in the US3-induced TNTs, and a marked enrichment of adhesion molecules (e.g., E-cadherin and β-catenin) at the contact area between TNT and recipient cell, ensuring persistent TNT-mediated contact between donor and recipient cell (17).
The formation of alphaherpesvirus-induced TNTs depends on the kinase activity of US3 (61). Although the underlying signaling network is not entirely clear, there are indications that US3-induced TNT formation occurs via activation of the cellular Cdc42/Rac1 Rho signaling axis and suppression of the opposing RhoA signaling axis. On the one hand, US3 of PRV directly phosphorylates and thereby activates group A PAKs, which are downstream effectors of Cdc42/Rac1 signaling, and PAK inhibition suppresses US3-induced TNT formation (62). US3-mediated activation of group A PAKs triggers the actin-severing protein cofilin via a poorly understood mechanism, which further contributes to the cytoskeletal rearrangements caused by US3 (63). On the other hand, expression of US3 also activates cellular protein kinase A, leading to phosphorylation of RhoA and thereby interfering with RhoA activity (64). The induction of TNTs by the US3 protein thus appears to depend on simultaneous activation of the Cdc42/Rac1 branches and inhibition of the RhoA branch of actin-controlling Rho GTPase signaling.
The US3 protein is conserved among the alphaherpesvirus subfamily of the herpesviruses and, probably as a consequence, many other alphaherpesviruses induce the formation of intercellular structures. Indeed, induction of TNT-like structures has been described upon transfection of cells with the US3 genes of bovine herpesvirus 1 (BoHV1), herpes simplex virus type 2 (HSV2), and bovine herpesvirus 5 (BoHV5) (65–67) and upon infection of cells with HSV1 or BoHV1 (68–71). In contrast to PRV US3-induced TNTs, TNTs formed in BoHV1-infected KOP and EBTR cells were described to be significantly less stable and highly sensitive to chemical fixation. This could indicate that BoHV1 differs from PRV with regard to stabilization of TNTs (71).
Although gammaherpesviruses do not encode a US3 homologue, Epstein-Barr virus (EBV) and murine gammaherpesvirus 68 (MHV-68) also have a dramatic impact on the actin host cytoskeleton, including the formation of TNT-like protrusions that are devoid of tubulin (72, 73). TNT formation was observed in different cell lines that coexpress either MHV-68 gp48 and ORF58 proteins or their distantly related EBV homologues BDLF2 and BMRF2. MHV-68 gp48 has been implicated in intercellular virus spread both in vitro and in vivo and is thought to drive the observed actin rearrangements, whereas ORF58 is thought to assist mainly via recruitment of gp48 to the plasma membrane (73, 74). Despite the homologies in gammaherpesvirus proteins that trigger TNT-like formation, the underlying mechanisms may be different since for EBV, cell protrusion formation was blocked by expression of a dominant active RhoA, whereas MHV-68-induced protrusion formation was blocked by dominant negative RhoA (72, 73).
OTHER VIRUSES
Several members of the alphavirus genus induce actin- and tubulin-containing TNTs, including Sindbis virus (SINV), Semliki Forest virus (SFV), and Chikungunya virus (CHIKV) (75). Alphavirus-induced TNTs depend on expression of viral structural proteins and were observed upon infection of Vero cells, as well as primary human umbilical vein endothelial cells (HUVECs). Although these TNTs allow antibody-resistant infection of connected cells even if the latter are virus receptor depleted, spread of infection still requires fusion-competent virus, indicating that virus transfer may not occur via direct cytoplasmic connectivity between donor and recipient cell (75).
Human metapneumovirus (HMPV) causes dramatic changes in the cytoskeleton of infected human bronchial airway cells, resulting in the formation of complex branched filamentous networks, as well as long intercellular extensions that likely represent TNTs (76). These intercellular connections contain actin and tubulin, and their formation depends on actin polymerization and involves Rho GTPase activity, particularly Cdc42/Rac1 signaling. Transfection of cells with an expression vector encoding the viral P protein was sufficient to trigger formation of TNT-like structures. HMPV-induced TNT-like structures contain viral RNA and promote viral spread in the presence of virus-neutralizing antibodies which, in contrast to cell-free infection, does not require the presence of heparan sulfate on target cells (76).
During the late stage of an infection of epithelial BS-C-1 or Vero cells with vaccinia virus (VV), the prototypic poxvirus, long intercellular connections are formed (77, 78). These cell projections are formed by extending lamellipodia that progressively condense to form a fine projection. These cell projections can exist without contacting other cells and contribute to cell migration, but when they do make contact with other cells, TNT-like structures appear to be formed. The viral F11 protein is necessary and sufficient to trigger the formation of these structures and acts through binding to and inactivation of RhoA (79, 80).
TNT-like structures in MDCK, Vero, and A549 cells infected with the orthomyxovirus influenza A virus (IAV) or the paramyxovirus parainfluenza virus 5 (PIV5) and in IAV-infected primary human bronchial epithelial cells have been reported to contain the viral ribonucleoprotein (vRNP) and the associated viral polymerase complex, which make up the core infectious viral machinery (81, 82). In MDCK cells, IAV and PIV5 trigger formation of TNT-like structures and this formation depends on actin polymerization and the activity of group A PAKs (81). TNT-like structures in IAV- and PIV5-infected cells contribute to intercellular viral spread, even when inhibiting spread of cell-free virus via the addition of virus-neutralizing antibodies or neuraminidase inhibitors in the extracellular milieu (81, 82). Astrocytes infected with measles virus (MeV), another paramyxovirus, form large syncytia by extensive cell fusion. It has been shown that infected cells form TNT-like protrusions that intimately connect to surrounding cells and that these initial contacts then fuse to form the basis of the syncytium, thereby promoting virus spread (83).
MARC-145 cells infected with the nidovirus porcine reproductive and respiratory syndrome virus (PRRSV) establish numerous TNT-like structures (84, 85). These structures contribute to PRRSV intercellular spread, even in the presence of neutralizing antibodies, and PRRSV structural proteins coprecipitate with F-actin and the motor protein myosin IIA that are present in the nanotubes (84). Interestingly, microtubule-containing TNT-like structures between infected and uninfected cells also resulted in transfer of mitochondria from uninfected to infected cells, thereby rescuing the latter from PRRSV-induced apoptosis/necrosis (85).
For the flavivirus West Nile virus (WNV), it has been shown that expression of the nonstructural viral NS1 protein in different transfected epithelial, neuroblastoma and astrocytoma cell lines (Vero, SH-SY5Y and U-87MG) results in the formation of actin- and NS1-containing TNTs. Similar structures were also observed in WNV-infected Vero cells and contained not only NS1 but also the viral envelope glycoprotein E and have been suggested to contribute to intercellular virus spread (86).
In HeLa cells persistently infected with the MX strain of the arenavirus lymphocytic choriomeningitis virus (LCMV), there are also indications that viral intercellular spread may involve different types of intercellular connections, including TNT-like structures (87, 88).
Coxsackievirus B3 (CVB3), like other picornaviruses, typically requires cell lysis for spread. However, in infected GMK cells, CVB3 also triggers the formation of cell protrusions containing viral capsids, and these structures allow intercellular spread in the presence of neutralizing antibodies. CVB3-induced cell protrusions branch out and appear to embrace neighboring cells (89). Since they appear to adhere to the substrate on which the cells are grown, it is unclear how these structures relate to TNTs.
Finally, some plant reoviruses that are transmitted by insects have also been reported to trigger the formation of TNT-like structures in their insect host cells. Indeed, the viral membrane-associated Pns10 protein of the phytoreovirus rice dwarf virus (RDV) triggers formation of actin-based filopodium-like structures that contact neighboring cells and facilitate intercellular spread in the presence or absence of virus-neutralizing antibodies (90, 91). Tubule formation occurs in RDV-infected and Pns10-transduced insect cells and depends on actin polymerization and myosin activity (91). Insect cells infected with the Fijivirus rice black-streaked dwarf virus (RBSDV) or transduced with the P7-1 nonstructural protein of RBSDV also form filopodium-like structures that may facilitate intercellular spread (92, 93).
OTHER TYPES OF ACTIN-BASED INTERCELLULAR VIRUS SPREAD
In addition to TNTs and long cell projections that closely resemble TNTs, some viruses induce and/or exploit shorter, filopodium-like actin-based structures that may also enhance intercellular virus spread but do not directly contact neighboring cells. The discovery of such virus-induced filopodium-like structures in cells infected with the orthopoxvirus vaccinia virus (VV) in fact led to some of the first convincing evidence that viruses can exploit the host cytoskeleton to enhance intercellular spread.
Over 40 years ago, using transmission electron microscopy, Gerald Stokes reported that VV particles appeared to be released from the tip of microvillus-like short cell projections in infected BSC-1 cells (94). These filopodium-like structures occurred late in infection and contained actin and actin-binding proteins (95, 96). Much later, it was found that these structures in fact constitute “actin tails” that propel progeny VV virions from infected cells to uninfected cells, thereby enhancing virus spread (97). The actin tails are formed underneath VV progeny particles that are sitting on the cell surface upon fusion of their outer envelope with the plasma membrane during virus egress (98, 99). Their formation is initiated via a still incompletely understood outside-in-signaling process involving the SCR4 domain of the viral B5 envelope protein and cellular Src and Abl kinase family members, resulting in tyrosine (Y112) phosphorylation of the cytoplasmic domain of the viral A36 transmembrane protein (100–103). Phosphorylated A36 serves as a signaling hub, recruiting several cellular proteins that trigger actin polymerization, including Nck, WIP, N-WASP, Arp2/3, and Grb2 (100, 102, 104–107).
A36, together with its complex partner A33, is already expressed in the early stages of VV infection of a host cell. The expression of these viral proteins therefore marks the cells as infected. Fascinatingly, this expression prevents superinfection of the cell by other VV particles and allows for a repulsion-mediated mode of virus spread (108). When a VV particle lands on an already infected cell, the A33/A36 protein complex on the plasma membrane of the infected cell initiates the formation of an actin tail underneath the superinfecting virus particle, propelling it away from the infected cell to another cell. Virions can be repelled from infected cells repeatedly, basically “jumping” from one infected cell to another, until an uninfected cell is reached, thereby accelerating virus spread (108).
A36 is highly conserved in orthopoxviruses, and it is therefore not surprising that actin tails similar to those described for VV have been reported for other orthopoxvirus family members, including monkeypox, variola, and ectromelia (109, 110).
Like poxviruses, African swine fever virus (ASFV) is a large DNA virus that replicates in the cytoplasm. ASFV also triggers the formation of virus-tipped filopodium-like cell protrusions in infected cells. Curiously, unlike for poxviruses, ASFV virions are located inside the tip of these cell projections rather than on the outside (111).
Filopodium-associated spread has also been described for some RNA viruses. The paramyxovirus respiratory syncytial virus (RSV) triggers Arp2-dependent filopodium formation in A549 human alveolar epithelial cells, which is associated with enhanced virus spread and migration of infected cells. Expression of the F protein of RSV alone was found to be sufficient to trigger filopodium formation (112).
Filoviruses Marburg virus and Ebola virus use filopodia to leave infected macrophages and Huh-7 human liver cells (113–115), although it is unclear whether these viruses actively trigger filopodium formation. In infected cells, progeny virus capsids that reach the cell periphery via actin-dependent transport enter filopodia, followed by migration in and budding along these filopodia, allowing virus egress close to neighboring cells (113–115).
CONCLUSION AND PERSPECTIVES
It is clear that several viruses trigger the formation of TNTs and TNT-like structures to facilitate intercellular virus spread. However, the exact nature and characteristics of virus-induced TNT-like structures are often poorly resolved. For example, in most cases, it is unclear whether these structures are open ended, allowing direct cytoplasmic transfer of virus or virus-related information to contacted cells, or closed ended. Experiments involving transfer of fluorescent dyes or depolarization signals are often used as evidence for open-ended TNTs, although such transfer can sometimes also be explained by the presence of gap junctions in the contact area between the TNT and the recipient cell. Direct observations of open-ended TNTs remain rather rare in literature (2, 16). In addition, the factors mediating fusion in open-ended TNTs remain unknown. It has been suggested that SNARE and/or viral fusion proteins or the specific curvature of TNT tips could be involved in driving fusion but these hypotheses have not been tested experimentally (116). Although for several viruses it has been shown that TNT-mediated virus spread protects the virus from neutralization by antibodies that are present in the extracellular milieu, this does not automatically imply the involvement of open-ended intercellular structures. Indeed, for PRV, virus-induced TNTs facilitate antibody-resistant intercellular spread, but electron microscopy indicated that these TNTs are closed ended and that virus spread involves virus exit from the TNT and subsequent entry in the contacted cell (17, 56, 62). Apparently, the tight contact area between the tip of the TNT and the contacted cell protects virus particles that are present in this tight gap from neutralization by antibodies (Fig. 1). In the case of PRV, the observation that the contact area is enriched in adhesion molecules (e.g., E-cadherin and β-catenin) probably contributes to a tight, antibody-impermeable region (17). It will be interesting to determine whether other virus-induced TNTs also display increased densities of adhesion molecules at the contact area with the recipient cell and what the underlying mechanism is that drives such locally increased expression of adhesion molecules.
Another important but currently unresolved question is whether TNT formation and elongation occur randomly or may show directionality, e.g., preferentially growing in the direction of other (noninfected) cells. Future research may show whether cells express and/or secrete molecules that attract or repel growing TNTs, possibly similar to what is known with regard to axon guidance during neuronal development (117). Interestingly, in the case of HIV Nef-induced TNT-like structures in macrophages that drive Nef transport to B cells, it has been reported that the TNT-like conduits navigate toward B cells through a CXCR4-dependent mechanism (45).
Several viral proteins that trigger formation of TNT-like structures have been described, including Nef of HIV, p8 of HTLV-1, US3 of PRV and other alphaherpesviruses, NS1 of West Nile virus, F11 of VV, and P protein of human metapneumovirus. Elucidating the signaling networks affected by these viral proteins may point to central and common signaling nodes that are involved in the formation of several TNT-like structures. This will shed light on the still poorly understood mechanisms of TNT formation in general and could lead to the identification of much-needed markers to more easily discriminate TNTs from other actin-containing cell projections and to differentiate TNT subtypes. In addition, elucidating the molecular mechanisms of virus-induced formation of TNT-like structures may identify potential targets for drug development aimed at suppressing spread of different types of viruses. From what is currently known, TNT formation by many different viruses involves modulation of actin-controlling Rho GTPase signaling, particularly activation of Cdc42/Rac1 signaling and/or inhibition of RhoA signaling. Of note in this context, actin remodeling by PRV US3 and HIV Nef involves the Cdc42/Rac1 effector PAK protein kinases and PAK activity also contributes to IAV-induced formation of TNT-like structures, pointing toward PAKs as potential common signaling molecules (50, 62, 81).
Whereas most studies on virus-induced TNTs to date have focused on two-dimensional cultures of cell lines and primary cells, an important challenge will be to translate this information to models that more closely mimic natural infection, such as, for example, tissues derived from infected animals and individuals and ex vivo material that is subsequently infected in vitro. TNT-like structures have been observed in vivo and ex vivo between immune cells in inflamed tissue and lymph nodes, between cancer cells, and crossing the basement membrane in different tissues (reviewed in reference 118). Regarding the latter, using ex vivo porcine respiratory mucosa explants, we showed that during infection of respiratory epithelial cells with PRV, the actin-modulating effects of the TNT-inducing US3 protein kinase contribute to passage of virus across the underlying basement membrane toward deeper tissues (119). The development and availability of increasingly sensitive methods of imaging, including multiphoton microscopy, intravital microscopy, and super-resolution microscopy, will be instrumental in further dissecting the role of TNTs in health and different types of disease, including virus infections, and if and how these structures can be targeted for therapeutic purposes. Interestingly, since TNTs have been reported to enhance the cytolytic effects of an oncolytic herpesvirus in mesothelioma cells in vitro by transfer of thymidine kinase to bystander cells (120), such therapeutic purposes may also include strategies to allow or even enhance formation of cell projections.
To date, virus-induced TNTs have been mainly investigated with regard to their potential to transport infectious virus from one cell to another. However, the ability of TNTs to drive intercellular transport of several types of molecular information (e.g., microRNA and other types of RNA, proteins, lipids, and even organelles) may also have a substantial impact on several aspects of the host response to infection and therefore virus pathogenesis. Examples include the TNT-mediated transport of the Nef protein of HIV to B cells, which interferes with isotype class switching, and the transport of mitochondria from healthy cells to PRRSV-infected cells, which protects the latter from virus-induced cells death (45, 84, 85). A more complete characterization of the molecular information that is transported via virus-induced TNTs and its subsequent effect on the recipient (and donor) cells will be crucial to better understand the role and importance of TNTs in virus pathogenesis.
ACKNOWLEDGMENTS
H.W.F., R.J.J.J., and A.T. are supported by grants from FWO-Vlaanderen (G019617N and G060119N) and the Special Research Fund of Ghent University (GOA grant 01G01317).
REFERENCES
- 1.van Niel G, D’Angelo G, Raposo G. 2018. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 2.Rustom A, Saffrich R, Markovic I, Walther P, Gerdes H-H. 2004. Nanotubular highways for intercellular organelle transport. Science 303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 3.Biran A, Perelmutter M, Gal H, Burton DGA, Ovadya Y, Vadai E, Geiger T, Krizhanovsky V. 2015. Senescent cells communicate via intercellular protein transfer. Genes Dev 29:791–802. doi: 10.1101/gad.259341.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watkins SC, Salter RD. 2005. Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity 23:309–318. doi: 10.1016/j.immuni.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Thayanithy V, Dickson EL, Steer C, Subramanian S, Lou E. 2014. Tumor-stromal cross talk: direct cell-to-cell transfer of oncogenic microRNAs via tunneling nanotubes. Transl Res 164:359–365. doi: 10.1016/j.trsl.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Cui J, Sun X, Zhang Y. 2011. Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ 18:732–742. doi: 10.1038/cdd.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koyanagi M, Brandes RP, Haendeler J, Zeiher AM, Dimmeler S. 2005. Cell-to-cell connection of endothelial progenitor cells with cardiac myocytes by nanotubes. Circ Res 96:1039–1041. doi: 10.1161/01.RES.0000168650.23479.0c. [DOI] [PubMed] [Google Scholar]
- 8.Önfelt B, Nedvetzki S, Benninger RKP, Purbhoo MA, Sowinski S, Hume AN, Seabra MC, Neil MAA, French PMW, Davis DM. 2006. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol 177:8476–8483. doi: 10.4049/jimmunol.177.12.8476. [DOI] [PubMed] [Google Scholar]
- 9.Austefjord MW, Gerdes HH, Wang X. 2014. Tunneling nanotubes: diversity in morphology and structure. Commun Integr Biol 7:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurke S, Barroso JFV, Hodneland E, Bukoreshtliev NV, Schlicker O, Gerdes HH. 2008. Tunneling nanotube (TNT)-like structures facilitate a constitutive, actomyosin-dependent exchange of endocytic organelles between normal rat kidney cells. Exp Cell Res 314:3669–3683. doi: 10.1016/j.yexcr.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Gousset K, Marzo L, Commere P-H, Zurzolo C. 2013. Myo10 is a key regulator of TNT formation in neuronal cells. J Cell Sci 126:4424–4435. doi: 10.1242/jcs.129239. [DOI] [PubMed] [Google Scholar]
- 12.Gross SP. 2004. Hither and yon: a review of bidirectional microtubule-based transport. Phys Biol 1:R1–R11. doi: 10.1088/1478-3967/1/2/R01. [DOI] [PubMed] [Google Scholar]
- 13.McCoy-Simandle K, Hanna SJ, Cox D. 2016. Exosomes and nanotubes: control of immune cell communication. Int J Biochem Cell Biol 71:44–54. doi: 10.1016/j.biocel.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupont M, Souriant S, Lugo-Villarino G, Maridonneau-Parini I, Vérollet C. 2018. Tunneling nanotubes: intimate communication between myeloid cells. Front Immunol 9:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Köhler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, Onfelt B, Sattentau Q, Davis DM. 2008. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol 10:211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- 16.Sartori-Rupp A, Cordero Cervantes D, Pepe A, Gousset K, Delage E, Corroyer-Dulmont S, Schmitt C, Krijnse-Locker J, Zurzolo C. 2019. Correlative cryo-electron microscopy reveals the structure of TNTs in neuronal cells. Nat Commun 10:342. doi: 10.1038/s41467-018-08178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansens RJJ, Van den Broeck W, De Pelsmaeker S, Lamote JAS, Van Waesberghe C, Couck L, Favoreel HW. 2017. Pseudorabies virus US3-induced tunneling nanotubes contain stabilized microtubules, interact with neighboring cells via cadherins, and allow intercellular molecular communication. J Virol 91:e00749-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lokar M, Iglič A, Veranič P. 2010. Protruding membrane nanotubes: attachment of tubular protrusions to adjacent cells by several anchoring junctions. Protoplasma 246:81–87. doi: 10.1007/s00709-010-0143-7. [DOI] [PubMed] [Google Scholar]
- 19.Önfelt B, Nedvetzki S, Yanagi K, Davis DM. 2004. Cutting edge: membrane nanotubes connect immune cells. J Immunol 173:1511–1513. doi: 10.4049/jimmunol.173.3.1511. [DOI] [PubMed] [Google Scholar]
- 20.Hase K, Kimura S, Takatsu H, Ohmae M, Kawano S, Kitamura H, Ito M, Watarai H, Hazelett CC, Yeaman C, Ohno H. 2009. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat Cell Biol 11:1427–1432. doi: 10.1038/ncb1990. [DOI] [PubMed] [Google Scholar]
- 21.Schiller C, Diakopoulos KN, Rohwedder I, Kremmer E, von Toerne C, Ueffing M, Weidle UH, Ohno H, Weiss EH. 2013. LST1 promotes the assembly of a molecular machinery responsible for tunneling nanotube formation. J Cell Sci 126:767–777. doi: 10.1242/jcs.114033. [DOI] [PubMed] [Google Scholar]
- 22.Kimura S, Hase K, Ohno H. 2012. Tunneling nanotubes: emerging view of their molecular components and formation mechanisms. Exp Cell Res 318:1699–1706. doi: 10.1016/j.yexcr.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Andresen V, Wang X, Ghimire S, Omsland M, Gjertsen BT, Gerdes HH. 2013. Tunneling nanotube (TNT) formation is independent of p53 expression. Cell Death Differ 20:1124. doi: 10.1038/cdd.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohil AB, Robertson BW, Cheney RE. 2006. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci U S A 103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerber ML, Cheney RE. 2011. Myosin-X: a MyTH-FERM myosin at the tips of filopodia. J Cell Sci 124:3733–3741. doi: 10.1242/jcs.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhl J, Gujarathi S, Waheed AA, Gordon A, Freed EO, Gousset K. 2019. Myosin-X is essential to the intercellular spread of HIV-1 Nef through tunneling nanotubes. J Cell Commun Signal 13:209–224. doi: 10.1007/s12079-018-0493-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinnery HR, Pearlman E, McMenamin PG. 2008. Cutting edge: membrane nanotubes in vivo: a feature of MHC class II+ cells in the mouse cornea. J Immunol 180:5779–5783. doi: 10.4049/jimmunol.180.9.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Gerdes H-H. 2015. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ 22:1181–1191. doi: 10.1038/cdd.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lou E, Fujisawa S, Morozov A, Barlas A, Romin Y, Dogan Y, Gholami S, Moreira AL, Manova-Todorova K, Moore M. 2012. Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS One 7:e33093. doi: 10.1371/journal.pone.0033093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna SJ, McCoy-Simandle K, Leung E, Genna A, Condeelis J, Cox D. 2019. Tunneling nanotubes, a novel mode of tumor cell-macrophage communication in tumor cell invasion. J Cell Sci 132:jcs223321. doi: 10.1242/jcs.223321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polak R, De Rooij B, Pieters R, Den Boer ML. 2015. B-cell precursor acute lymphoblastic leukemia cells use tunneling nanotubes to orchestrate their microenvironment. Blood 126:2404–2414. doi: 10.1182/blood-2015-03-634238. [DOI] [PubMed] [Google Scholar]
- 32.Desir S, O’Hare P, Vogel RI, Sperduto W, Sarkari A, Dickson EL, Wong P, Nelson AC, Fong Y, Steer CJ, Subramanian S, Lou E. 2018. Chemotherapy-induced tunneling nanotubes mediate intercellular drug efflux in pancreatic cancer. Sci Rep 8:9484. doi: 10.1038/s41598-018-27649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, Huang L, Ratliff M, Karimian Jazi K, Kurz FT, Schmenger T, Lemke D, Gömmel M, Pauli M, Liao Y, Häring P, Pusch S, Herl V, Steinhäuser C, Krunic D, Jarahian M, Miletic H, Berghoff AS, Griesbeck O, Kalamakis G, Garaschuk O, Preusser M, Weiss S, Liu H, Heiland S, Platten M, Huber PE, Kuner T, von Deimling A, Wick W, Winkler F. 2015. Brain tumour cells interconnect to a functional and resistant network. Nature 528:93–98. doi: 10.1038/nature16071. [DOI] [PubMed] [Google Scholar]
- 34.Pasquier J, Magal P, Boulangé-Lecomte C, Webb G, Le Foll F. 2011. Consequences of cell-to-cell P-glycoprotein transfer on acquired multidrug resistance in breast cancer: a cell population dynamics model. Biol Direct 6:5. doi: 10.1186/1745-6150-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasquier J, Galas L, Boulangé-Lecomte C, Rioult D, Bultelle F, Magal P, Webb G, Le Foll F. 2012. Different modalities of intercellular membrane exchanges mediate cell-to-cell P-glycoprotein transfers in MCF-7 breast cancer cells. J Biol Chem 287:7374–7387. doi: 10.1074/jbc.M111.312157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hekmatshoar Y, Nakhle J, Galloni M, Vignais ML. 2018. The role of metabolism and tunneling nanotube-mediated intercellular mitochondria exchange in cancer drug resistance. Biochem J 475:2305–2328. doi: 10.1042/BCJ20170712. [DOI] [PubMed] [Google Scholar]
- 37.Lou E, Gholami S, Romin Y, Thayanithy V, Fujisawa S, Desir S, Steer CJ, Subramanian S, Fong Y, Manova-Todorova K, Moore M. 2017. Imaging tunneling membrane tubes elucidates cell communication in tumors. Trends Cancer 3:678–685. doi: 10.1016/j.trecan.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Valdebenito S, Lou E, Baldoni J, Okafo G, Eugenin E. 2018. The novel roles of connexin channels and tunneling nanotubes in cancer pathogenesis. Int J Mol Sci 19:E1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matejka N, Reindl J. 2019. Perspectives of cellular communication through tunneling nanotubes in cancer cells and the connection to radiation effects. Radiat Oncol 14:218. doi: 10.1186/s13014-019-1416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Victoria GS, Zurzolo C. 2017. The spread of prion-like proteins by lysosomes and tunneling nanotubes: implications for neurodegenerative diseases. J Cell Biol 216:2633–2644. doi: 10.1083/jcb.201701047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim B-W, Lee J-S, Ko Y-G. 2019. Mycoplasma exploits mammalian tunneling nanotubes for cell-to-cell dissemination. BMB Rep 52:490–495. doi: 10.5483/BMBRep.2019.52.8.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eugenin EA, Gaskill PJ, Berman JW. 2009. Tunneling nanotubes (TNT) are induced by HIV-infection of macrophages: a potential mechanism for intercellular HIV trafficking. Cell Immunol 254:142–148. doi: 10.1016/j.cellimm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kadiu I, Gendelman HE. 2011. Human immunodeficiency virus type 1 endocytic trafficking through macrophage bridging conduits facilitates spread of infection. J Neuroimmune Pharmacol 6:658–675. doi: 10.1007/s11481-011-9298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Souriant S, Balboa L, Dupont M, Pingris K, Kviatcovsky D, Cougoule C, Lastrucci C, Bah A, Gasser R, Poincloux R, Raynaud-Messina B, Al Saati T, Inwentarz S, Poggi S, Moraña EJ, González-Montaner P, Corti M, Lagane B, Vergne I, Allers C, Kaushal D, Kuroda MJ, Sasiain MC, Neyrolles O, Maridonneau-Parini I, Lugo-Villarino G, Vérollet C. 2019. Tuberculosis exacerbates HIV-1 infection through IL-10/STAT3-dependent tunneling nanotube formation in macrophages. Cell Rep 26:3586–3599. doi: 10.1016/j.celrep.2019.02.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu W, Santini PA, Sullivan JS, He B, Shan M, Ball SC, Dyer WB, Ketas TJ, Chadburn A, Cohen-Gould L, Knowles DM, Chiu A, Sanders RW, Chen K, Cerutti A. 2009. HIV-1 evades virus-specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long-range intercellular conduits. Nat Immunol 10:1008–1017. doi: 10.1038/ni.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okafo G, Prevedel L, Eugenin E. 2017. Tunneling nanotubes (TNT) mediate long-range gap junctional communication: implications for HIV cell to cell spread. Sci Rep 7:16660. doi: 10.1038/s41598-017-16600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kadiu I, Gendelman HE. 2011. Macrophage bridging conduit trafficking of HIV-1 through the endoplasmic reticulum and Golgi network. J Proteome Res 10:3225–3238. doi: 10.1021/pr200262q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto M, Bhuyan F, Hiyoshi M, Noyori O, Nasser H, Miyazaki M, Saito T, Kondoh Y, Osada H, Kimura S, Hase K, Ohno H, Suzu S. 2016. Potential role of the formation of tunneling nanotubes in HIV-1 spread in macrophages. J Immunol 196:1832–1841. doi: 10.4049/jimmunol.1500845. [DOI] [PubMed] [Google Scholar]
- 49.Mukerji J, Olivieri KC, Misra V, Agopian KA, Gabuzda D. 2012. Proteomic analysis of HIV-1 Nef cellular binding partners reveals a role for exocyst complex proteins in mediating enhancement of intercellular nanotube formation. Retrovirology 9:33. doi: 10.1186/1742-4690-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imle A, Abraham L, Tsopoulidis N, Hoflack B, Saksela K, Fackler OT. 2015. Association with PAK2 enables functional interactions of lentiviral nef proteins with the exocyst complex. mBio 6:e01309-15. doi: 10.1128/mBio.01309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Prooyen N, Gold H, Andresen V, Schwartz O, Jones K, Ruscetti F, Lockett S, Gudla P, Venzon D, Franchini G. 2010. Human T-cell leukemia virus type 1 p8 protein increases cellular conduits and virus transmission. Proc Natl Acad Sci U S A 107:20738–20743. doi: 10.1073/pnas.1009635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Omsland M, Pise-Masison C, Fujikawa D, Galli V, Fenizia C, Parks RW, Gjertsen BT, Franchini G, Andresen V. 2018. Inhibition of tunneling nanotube (TNT) formation and human T-cell leukemia virus type 1 (HTLV-1) transmission by cytarabine. Sci Rep 8:11118. doi: 10.1038/s41598-018-29391-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W. 2007. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol 9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aggarwal A, Iemma TL, Shih I, Newsome TP, McAllery S, Cunningham AL, Turville SG. 2012. Mobilization of HIV spread by diaphanous 2 dependent filopodia in infected dendritic cells. PLoS Pathog 8:e1002762. doi: 10.1371/journal.ppat.1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nobile C, Rudnicka D, Hasan M, Aulner N, Porrot F, Machu C, Renaud O, Prevost M-C, Hivroz C, Schwartz O, Sol-Foulon N. 2010. HIV-1 Nef inhibits ruffles, induces filopodia, and modulates migration of infected lymphocytes. J Virol 84:2282–2293. doi: 10.1128/JVI.02230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Favoreel HW, Van Minnebruggen G, Adriaensen D, Nauwynck HJ. 2005. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an alphaherpesvirus are associated with enhanced spread. Proc Natl Acad Sci U S A 102:8990–8995. doi: 10.1073/pnas.0409099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calton CM, Randall JA, Adkins MW, Banfield BW. 2004. The pseudorabies virus serine/threonine kinase Us3 contains mitochondrial, nuclear and membrane localization signals. Virus Genes 29:131–145. doi: 10.1023/B:VIRU.0000032796.27878.7f. [DOI] [PubMed] [Google Scholar]
- 58.Kimman TG, de Wind N, Oei-Lie N, Pol JMA, Berns AJM, Gielkens A. 1992. Contribution of single genes within the unique short region of Aujeszky’s disease virus (Suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J Gen Virol 73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- 59.Kimman TG, Pol JMA, de Wind N, Oei-Lie N, Berns AJM, Gielkens A. 1992. Role of different genes in the virulence and pathogenesis of Aujeszky’s disease virus. Vet Microbiol 33:45–52. doi: 10.1016/0378-1135(92)90034-q. [DOI] [PubMed] [Google Scholar]
- 60.Olsen LM, Ch’ng TH, Card JP, Enquist LW. 2006. Role of pseudorabies virus Us3 protein kinase during neuronal infection. J Virol 80:6387–6398. doi: 10.1128/JVI.00352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van den Broeke C, Deruelle M, Nauwynck HJ, Coller KE, Smith GA, Van Doorsselaere J, Favoreel HW. 2009. The kinase activity of pseudorabies virus US3 is required for modulation of the actin cytoskeleton. Virology 385:155–160. doi: 10.1016/j.virol.2008.11.050. [DOI] [PubMed] [Google Scholar]
- 62.Van den Broeke C, Radu M, Deruelle M, Nauwynck H, Hofmann C, Jaffer ZM, Chernoff J, Favoreel HW. 2009. Alphaherpesvirus US3-mediated reorganization of the actin cytoskeleton is mediated by group A p21-activated kinases. Proc Natl Acad Sci U S A 106:8707–8712. doi: 10.1073/pnas.0900436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacob T, Van den Broeke C, van Troys M, Waterschoot D, Ampe C, Favoreel HW. 2013. Alphaherpesviral US3 kinase induces cofilin dephosphorylation to reorganize the actin cytoskeleton. J Virol 87:4121–4126. doi: 10.1128/JVI.03107-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacob T, Van den Broeke C, Van Waesberghe C, Van Troys L, Favoreel HW. 2015. Pseudorabies virus US3 triggers RhoA phosphorylation to reorganize the actin cytoskeleton. J Gen Virol 96:2328–2335. doi: 10.1099/vir.0.000152. [DOI] [PubMed] [Google Scholar]
- 65.Brzozowska A, Rychłowski M, Lipińska AD, Bieńkowska-Szewczyk K. 2010. Point mutations in BHV-1 Us3 gene abolish its ability to induce cytoskeletal changes in various cell types. Vet Microbiol 143:8–13. doi: 10.1016/j.vetmic.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 66.Finnen RL, Roy BB, Zhang H, Banfield BW. 2010. Analysis of filamentous process induction and nuclear localization properties of the HSV-2 serine/threonine kinase Us3. Virology 397:23–33. doi: 10.1016/j.virol.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ladelfa MF, Kotsias F, Del Médico Zajac MP, Van den Broeke C, Favoreel H, Romera SA, Calamante G. 2011. Effect of the US3 protein of bovine herpesvirus 5 on the actin cytoskeleton and apoptosis. Vet Microbiol 153:361–366. doi: 10.1016/j.vetmic.2011.05.037. [DOI] [PubMed] [Google Scholar]
- 68.van Leeuwen H, Elliott G, O’Hare P. 2002. Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J Virol 76:3471–3481. doi: 10.1128/jvi.76.7.3471-3481.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.La Boissière S, Izeta A, Malcomber S, O’Hare P. 2004. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J Virol 78:8002–8014. doi: 10.1128/JVI.78.15.8002-8014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dixit R, Tiwari V, Shukla D. 2008. Herpes simplex virus type 1 induces filopodia in differentiated P19 neural cells to facilitate viral spread. Neurosci Lett 440:113–118. doi: 10.1016/j.neulet.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panasiuk M, Rychłowski M, Derewońko N, Bieńkowska-Szewczyk K. 2018. Tunneling nanotubes as a novel route of cell-to-cell spread of herpesviruses. J Virol 92:e00090-18. doi: 10.1128/JVI.00090-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loesing JB, Di Fiore S, Ritter K, Fischer R, Kleines M. 2009. Epstein-Barr virus BDLF2-BMRF2 complex affects cellular morphology. J Gen Virol 90:1440–1449. doi: 10.1099/vir.0.009571-0. [DOI] [PubMed] [Google Scholar]
- 73.Gill MB, Edgar R, May JS, Stevenson PG. 2008. A gamma-herpesvirus glycoprotein complex manipulates actin to promote viral spread. PLoS One 3:e1808. doi: 10.1371/journal.pone.0001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.May JS, Walker J, Colaco S, Stevenson PG. 2005. The murine gammaherpesvirus 68 ORF27 gene product contributes to intercellular viral spread. J Virol 79:5059–5068. doi: 10.1128/JVI.79.8.5059-5068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez MG, Kielian M. 2016. Intercellular extensions are induced by the alphavirus structural proteins and mediate virus transmission. PLoS Pathog 12:e1006061. doi: 10.1371/journal.ppat.1006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.El Najjar F, Cifuentes-Muñoz N, Chen J, Zhu H, Buchholz UJ, Moncman CL, Dutch RE. 2016. Human metapneumovirus induces reorganization of the actin cytoskeleton for direct cell-to-cell spread. PLoS Pathog 12:e1005922. doi: 10.1371/journal.ppat.1005922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanderson CM, Smith GL. 1998. Vaccinia virus induces Ca2+-independent cell-matrix adhesion during the motile phase of infection. J Virol 72:9924–9933. doi: 10.1128/JVI.72.12.9924-9933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xiao M, Xu N, Wang C, Pang D-W, Zhang Z-L. 2017. Dynamic monitoring of membrane nanotubes formation induced by vaccinia virus on a high throughput microfluidic chip. Sci Rep 7:44835. doi: 10.1038/srep44835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Valderrama F, Cordeiro JV, Schleich S, Frischknecht F, Way M. 2006. Vaccinia virus-induced cell motility requires F11L-mediated inhibition of RhoA signaling. Science 311:377–381. doi: 10.1126/science.1122411. [DOI] [PubMed] [Google Scholar]
- 80.Morales I, Carbajal MA, Bohn S, Holzer D, Kato SEM, Greco FAB, Moussatché N, Locker JK. 2008. The vaccinia virus F11L gene product facilitates cell detachment and promotes migration. Traffic 9:1283–1298. doi: 10.1111/j.1600-0854.2008.00762.x. [DOI] [PubMed] [Google Scholar]
- 81.Roberts KL, Manicassamy B, Lamb RA. 2015. Influenza A virus uses intercellular connections to spread to neighboring cells. J Virol 89:1537–1549. doi: 10.1128/JVI.03306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar A, Kim JH, Ranjan P, Metcalfe MG, Cao W, Mishina M, Gangappa S, Guo Z, Boyden ES, Zaki S, York I, García-Sastre A, Shaw M, Sambhara S. 2017. Influenza virus exploits tunneling nanotubes for cell-to-cell spread. Sci Rep 7:40360. doi: 10.1038/srep40360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duprex WP, Rima BK, McQuaid S, Hangartner L, Billeter MA. 1999. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J Virol 73:9568–9575. doi: 10.1128/JVI.73.11.9568-9575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guo R, Katz BB, Tomich JM, Gallagher T, Fang Y. 2016. Porcine reproductive and respiratory syndrome virus utilizes nanotubes for intercellular spread. J Virol 90:5163–5175. doi: 10.1128/JVI.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo R, Davis D, Fang Y. 2018. Intercellular transfer of mitochondria rescues virus-induced cell death but facilitates cell-to-cell spreading of porcine reproductive and respiratory syndrome virus. Virology 517:122–134. doi: 10.1016/j.virol.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 86.Furnon W, Fender P, Confort M, Desloire S, Nangola S, Kitidee K, Leroux C, Ratinier M, Arnaud F, Lecollinet S, Boulanger P, Hong S. 2019. Remodeling of the actin network associated with the nonstructural protein 1 (NS1) of West Nile virus and formation of NS1-containing tunneling nanotubes. Viruses 11:901. doi: 10.3390/v11100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Závada J, Závadová Z, Pastoreková S, Čiampor F, Pastorek J, Zelník V. 1993. Expression of MaTu‐MN protein in human tumor cultures and in clinical specimens. Int J Cancer 54:268–274. doi: 10.1002/ijc.2910540218. [DOI] [PubMed] [Google Scholar]
- 88.Labudová M, Čiampor F, Pastoreková S, Pastorek J. 2018. Cell-to-cell transmission of lymphocytic choriomeningitis virus MX strain during persistent infection and its influence on cell migration. Acta Virol 62:424–434. doi: 10.4149/av_2018_411. [DOI] [PubMed] [Google Scholar]
- 89.Paloheimo O, Ihalainen TO, Tauriainen S, Valilehto O, Kirjavainen S, Niskanen EA, Laakkonen JP, Hyoty H, Vihinen-Ranta M. 2011. Coxsackievirus B3-induced cellular protrusions: structural characteristics and functional competence. J Virol 85:6714–6724. doi: 10.1128/JVI.00247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei T, Kikuchi A, Moriyasu Y, Suzuki N, Shimizu T, Hagiwara K, Chen H, Takahashi M, Ichiki-Uehara T, Omura T. 2006. The spread of Rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J Virol 80:8593–8602. doi: 10.1128/JVI.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei T, Shimizu T, Omura T. 2008. Endomembranes and myosin mediate assembly into tubules of Pns10 of Rice dwarf virus and intercellular spreading of the virus in cultured insect vector cells. Virology 372:349–356. doi: 10.1016/j.virol.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 92.Isogai M, Uyeda I, Lee BC. 1998. Detection and assignment of proteins encoded by Rice black streaked dwarf fijivirus S7, S8, S9, and S10. J Gen Virol 79:1487–1494. doi: 10.1099/0022-1317-79-6-1487. [DOI] [PubMed] [Google Scholar]
- 93.Liu Y, Jia D, Chen H, Chen Q, Xie L, Wu Z, Wei T. 2011. The P7-1 protein of southern rice black-streaked dwarf virus, a fijivirus, induces the formation of tubular structures in insect cells. Arch Virol 156:1729–1736. doi: 10.1007/s00705-011-1041-9. [DOI] [PubMed] [Google Scholar]
- 94.Stokes GV. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J Virol 18:636–643. doi: 10.1128/JVI.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. 1979. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology 98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 96.Krempien U, Schneider L, Hiller G, Weber K, Katz E, Jungwirth C. 1981. Conditions for pox virus-specific microvilli formation studied during synchronized virus assembly. Virology 113:556–564. doi: 10.1016/0042-6822(81)90183-5. [DOI] [PubMed] [Google Scholar]
- 97.Cudmore S, Cossart P, Griffiths G, Way M. 1995. Actin-based motility of vaccinia virus. Nature 378:636–785. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 98.Cudmore S, Reckmann I, Griffiths G, Way M. 1996. Vaccinia virus: a model system for actin-membrane interactions. J Cell Sci 109:1739–1747. [DOI] [PubMed] [Google Scholar]
- 99.Rietdorf J, Ploubidou A, Reckmann I, Holmström A, Frischknecht F, Zettl M, Zimmermann T, Way M. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat Cell Biol 3:992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- 100.Frischknecht F, Moreau V, Röttger S, Gonfloni S, Reckmann I, Superti-Furga G, Way M. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signaling. Nature 401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 101.Newsome TP, Scaplehorn N, Way M. 2004. Src mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science 306:124–129. doi: 10.1126/science.1101509. [DOI] [PubMed] [Google Scholar]
- 102.Newsome TP, Weisswange I, Frischknecht F, Way M. 2006. Abl collaborates with Src family kinases to stimulate actin-based motility of vaccinia virus. Cell Microbiol 8:233–241. doi: 10.1111/j.1462-5822.2005.00613.x. [DOI] [PubMed] [Google Scholar]
- 103.Reeves PM, Bommarius B, Lebeis S, McNulty S, Christensen J, Swimm A, Chahroudi A, Chavan R, Feinberg MB, Veach D, Bornmann W, Sherman M, Kalman D. 2005. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat Med 11:731–739. doi: 10.1038/nm1265. [DOI] [PubMed] [Google Scholar]
- 104.Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M. 2000. A complex of N-WASP and WIP integrates signaling cascades that lead to actin polymerization. Nat Cell Biol 2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- 105.Snapper SB, Takeshima F, Antón I, Liu CH, Thomas SM, Nguyen D, Dudley D, Fraser H, Purich D, Lopez-Ilasaca M, Klein C, Davidson L, Bronson R, Mulligan RC, Southwick F, Geha R, Goldberg MB, Rosen FS, Hartwig JH, Alt FW. 2001. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol 3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 106.Scaplehorn N, Holmström A, Moreau V, Frischknecht F, Reckmann I, Way M. 2002. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr Biol 12:740–745. doi: 10.1016/s0960-9822(02)00812-6. [DOI] [PubMed] [Google Scholar]
- 107.Donnelly SK, Weisswange I, Zettl M, Way M. 2013. WIP provides an essential link between nck and n-wasp during arp2/3-dependent actin polymerization. Curr Biol 23:999–1006. doi: 10.1016/j.cub.2013.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Doceul V, Hollinshead M, Van Der Linden L, Smith GL. 2010. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science 327:873–876. doi: 10.1126/science.1183173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reeves PM, Smith SK, Olson VA, Thorne SH, Bornmann W, Damon IK, Kalman D. 2011. Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on Abl and Src family tyrosine kinases. J Virol 85:21–31. doi: 10.1128/JVI.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lynn H, Horsington J, Ter LK, Han S, Chew YL, Diefenbach RJ, Way M, Chaudhri G, Karupiah G, Newsome TP. 2012. Loss of cytoskeletal transport during egress critically attenuates ectromelia virus infection in vivo. J Virol 86:7427–7443. doi: 10.1128/JVI.06636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jouvenet N, Windsor M, Rietdorf J, Hawes P, Monaghan P, Way M, Wileman T. 2006. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell Microbiol 8:1803–1811. doi: 10.1111/j.1462-5822.2006.00750.x. [DOI] [PubMed] [Google Scholar]
- 112.Mehedi M, McCarty T, Martin SE, Le Nouën C, Buehler E, Chen Y, Smelkinson M, Ganesan S, Fischer ER, Brock LG, Liang B, Munir S, Collins PL, Buchholz UJ. 2016. Actin-related protein 2 (ARP2) and virus-induced filopodia facilitate human respiratory syncytial virus spread. PLoS Pathog 12:e1006062. doi: 10.1371/journal.ppat.1006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kolesnikova L, Bohil AB, Cheney RE, Becker S. 2007. Budding of Marburgvirus is associated with filopodia. Cell Microbiol 9:939–951. doi: 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 114.Schudt G, Kolesnikova L, Dolnik O, Sodeik B, Becker S. 2013. Live-cell imaging of Marburg virus-infected cells uncovers actin-dependent transport of nucleocapsids over long distances. Proc Natl Acad Sci U S A 110:14402–14407. doi: 10.1073/pnas.1307681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schudt G, Dolnik O, Kolesnikova L, Biedenkopf N, Herwig A, Becker S. 2015. Transport of ebolavirus nucleocapsids is dependent on actin polymerization: live-cell imaging analysis of ebolavirus-infected cells. J Infect Dis 212:S160–S166. doi: 10.1093/infdis/jiv083. [DOI] [PubMed] [Google Scholar]
- 116.Abounit S, Zurzolo C. 2012. Wiring through tunneling nanotubes: from electrical signals to organelle transfer. J Cell Sci 125:1089–1098. doi: 10.1242/jcs.083279. [DOI] [PubMed] [Google Scholar]
- 117.Roig-Puiggros S, Vigouroux RJ, Beckman D, Bocai NI, Chiou B, Davimes J, Gomez G, Grassi S, Hoque A, Karikari TK, Kiffer F, Lopez M, Lunghi G, Mazengenya P, Meier S, Olguín-Albuerne M, Oliveira MM, Paraíso-Luna J, Pradhan J, Radiske A, Ramos-Hryb AB, Ribeiro MC, Schellino R, Selles MC, Singh S, Theotokis P, Chédotal A. 2019. Construction and reconstruction of brain circuits: normal and pathological axon guidance. J Neurochem doi: 10.1111/jnc.14900. [DOI] [PubMed] [Google Scholar]
- 118.Ariazi J, Benowitz A, De Biasi V, Den Boer ML, Cherqui S, Cui H, Douillet N, Eugenin EA, Favre D, Goodman S, Gousset K, Hanein D, Israel DI, Kimura S, Kirkpatrick RB, Kuhn N, Jeong C, Lou E, Mailliard R, Maio S, Okafo G, Osswald M, Pasquier J, Polak R, Pradel G, de Rooij B, Schaeffer P, Skeberdis VA, Smith IF, Tanveer A, Volkmann N, Wu Z, Zurzolo C. 2017. Tunneling nanotubes and gap junctions: their role in long-range intercellular communication during development, health, and disease conditions. Front Mol Neurosci 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lamote JAS, Glorieux S, Nauwynck HJ, Favoreel HW. 2016. The US3 protein of pseudorabies virus drives viral passage across the basement membrane in porcine respiratory mucosa explants. J Virol 90:10945–10950. doi: 10.1128/JVI.01577-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ady J, Thayanithy V, Mojica K, Wong P, Carson J, Rao P, Fong Y, Lou E. 2016. Tunneling nanotubes: an alternate route for propagation of the bystander effect following oncolytic viral infection. Mol Ther Oncolytics 3:16029. doi: 10.1038/mto.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]