LETTER
Computationally optimized broadly reactive antigens (COBRA) designed for different influenza virus subtypes (H1N1, H3N2, and H5N1) elicit a subtype-specific broad antibody (Ab) response in naive as well as in preimmune influenza virus animal models (1–9).
In particular, an H1 hemagglutinin (HA) COBRA candidate, named P1, has been designed by a multiple-sequence alignment of HA sequences belonging to H1 swine and human strains (1). Importantly, immunization with P1 elicits a broad neutralizing Ab response against H1 human and swine viruses (1, 4, 10).
Recently, we generated a panel of P1-specific mouse monoclonal Abs (MAbs) with the aim to dissect the Ab response to P1. As previously described, these MAbs feature different functional activities, spanning from narrowly to broadly reactive against H1N1 human viruses (11).
In this study, we investigated the breadth of hemagglutination inhibition (HAI) featured by representative P1-elicited MAbs along with those generated following immunization with wild-type historic H1N1 human vaccine strains in order to dissect the breadth of HAI activity of the P1-elicited response against influenza swine viruses.
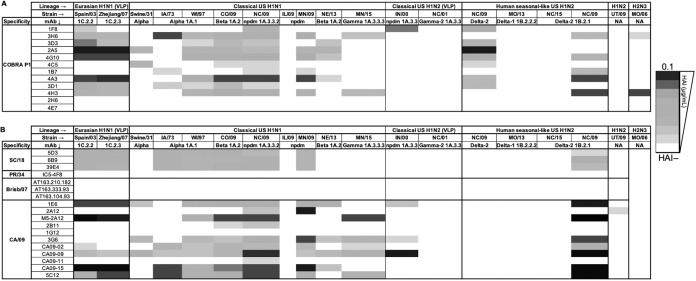
As shown in Fig. 1A, P1-specific MAbs featured a differentiating breadth of HAI activity, spanning from narrowly to broadly reactive against H1N1, H1N2, and H2N3 swine viruses. Interestingly, MAbs endowed with broad HAI activity against a panel of human H1N1 viruses featured a narrower HAI profile against swine viruses. Comparatively, those endowed with a narrower profile against human viruses featured a broader profile against swine viruses belonging to the Eurasian, classical, and human seasonal-like lineages (11).
FIG 1.
HAI and neutralizing activity breadth of P1- (A), seasonal-, and pandemic-specific (B) MAbs against influenza A H1N1, H1N2, and H2N3 swine viruses.
Unsurprisingly, due to its swine origin, CA/09-specific MAbs, previously classified to have a narrow profile of neutralization activity against pandemic and pandemic-like viruses (11), collectively exhibited a broad HAI activity against H1N1 and H1N2 swine viruses and none against the A/Swine/Missouri/4296424/2006 H2N3 virus (Fig. 1B).
Interestingly, a P1-specific MAb (4C5), previously demonstrated to have no HAI activity against any of the human H1N1 strains (11), showed detectable HAI activity against H1N1 and H1N2 swine viruses, suggesting that its epitope is particular to swine viruses and not to human seasonal, pandemic, and pandemic-like HA proteins.
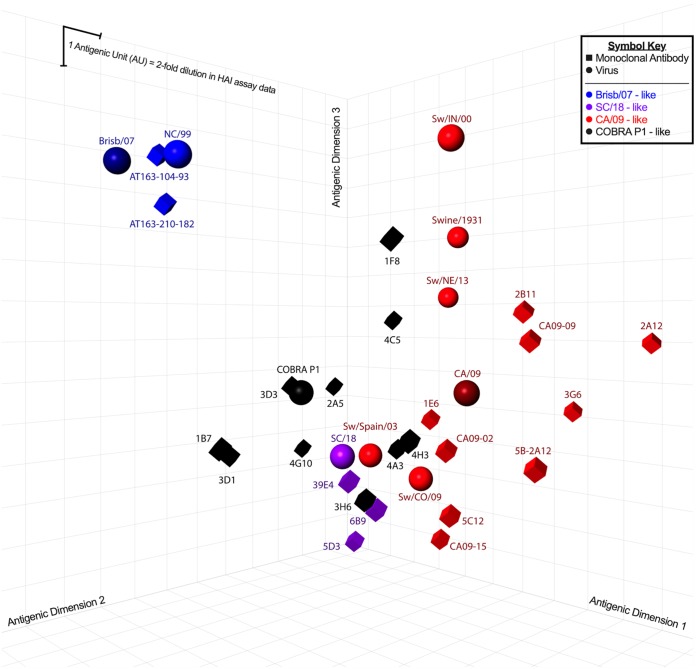
The antigenic cartography segregates MAbs based on their HAI profile against human and swine viruses, with the P1 and pandemic-specific MAbs clustering together as opposed to the seasonal (Brisb/07)-specific MAbs (Fig. 2) (11, 12). However, further investigation aimed at determining the amino acid contact residues of these MAbs will improve the resolution of the recognized epitopes, clarify distinctions between human- and swine-specific H1 epitopes, and elucidate the mechanism of breadth conferred by COBRA immunogens.
FIG 2.
Antigenic cartography map of P1-, seasonal-, and pandemic-specific MAbs. Map was drawn based on the minimum HAI concentration from this and previous studies (11).
ACKNOWLEDGMENTS
The MAbs 5D3, 6B9, 39E4, IC5-4F8, RA5-22, 1C5, 5C12, CA09-02, CA09-09, CA09-11, and CA09-15 were obtained from the Biodefense and Emerging Infections (BEI) Resources Repository while AT170.558.146, AT170.119.5, AT163.272.54, AT163.210.182, AT163.333.93, AT163.104.93, AT163.329.189 MAbs were obtained from the Influenza Reagent Resource (IRR).
The A/North Carolina/152702/2015, A/North Carolina/02744/2009, A/Illinois/02860/2009, A/Minnesota/02751/2009, A/Missouri/A01444664/2013, A/Swine/1931, A/Utah/02861/2009, and A/Nebraska/A01444614/2013 viruses were kindly provided by S. Mark Tompkins, Ralph Tripp (University of Georgia), and the Minnesota Veterinary Diagnostic Laboratory (University of Minnesota). Finally, we thank Silvia Carnaccini for helpful discussions.
REFERENCES
- 1.Carter DM, Darby CA, Lefoley BC, Crevar CJ, Alefantis T, Oomen R, Anderson SF, Strugnell T, Cortes-Garcia G, Vogel TU, Parrington M, Kleanthous H, Ross TM. 2016. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol 90:4720–4734. doi: 10.1128/JVI.03152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen JD, Jang H, DiNapoli J, Kleanthous H, Ross TM. 2019. Elicitation of protective antibodies against 20 years of future H3N2 cocirculating influenza virus variants in ferrets preimmune to historical H3N2 influenza viruses. J Virol 93:e00946-18. doi: 10.1128/JVI.00946-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen JD, Ray S, Ross TM. 2018. Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses. PLoS One 13:e0204284. doi: 10.1371/journal.pone.0204284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sautto GA, Kirchenbaum GA, Ecker JW, Bebin-Blackwell AG, Pierce SR, Ross TM. 2018. Elicitation of broadly protective antibodies following infection with influenza viruses expressing H1N1 computationally optimized broadly reactive hemagglutinin antigens. Immunohorizons 2:226–237. doi: 10.4049/immunohorizons.1800044. [DOI] [PubMed] [Google Scholar]
- 5.Sautto GA, Ross TM. 2019. Hemagglutinin consensus-based prophylactic approaches to overcome influenza virus diversity. Vet Ital 55:195–201. doi: 10.12834/VetIt.1944.10352.1. [DOI] [PubMed] [Google Scholar]
- 6.Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, Ross TM. 2012. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis 205:1562–1570. doi: 10.1093/infdis/jis232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter DM, Darby CA, Johnson SK, Carlock MA, Kirchenbaum GA, Allen JD, Vogel TU, Delagrave S, DiNapoli J, Kleanthous H, Ross TM. 2017. Elicitation of protective antibodies against a broad panel of H1N1 viruses in ferrets preimmune to historical H1N1 influenza viruses. J Virol 91:e01283-17. doi: 10.1128/JVI.01283-17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Sautto GA, Kirchenbaum GA, Ross TM. 2018. Towards a universal influenza vaccine: different approaches for one goal. Virol J 15:17. doi: 10.1186/s12985-017-0918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sautto GA, Kirchenbaum GA, Diotti RA, Criscuolo E, Ferrara F. 2019. Next generation vaccines for infectious diseases. J Immunol Res 2019:5890962. doi: 10.1155/2019/5890962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skarlupka AL, Owino SO, Suzuki-Williams LP, Crevar CJ, Carter DM, Ross TM. 2019. Computationally optimized broadly reactive vaccine based upon swine H1N1 influenza hemagglutinin sequences protects against both swine and human isolated viruses. Hum Vaccin Immunother 15:2013–2029. doi: 10.1080/21645515.2019.1653743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sautto GA, Kirchenbaum GA, Abreu RB, Ecker JW, Pierce SR, Kleanthous H, Ross TM. 2020. A computationally optimized broadly reactive antigen subtype-specific influenza vaccine strategy elicits unique potent broadly neutralizing antibodies against hemagglutinin. J Immunol 204:375–385. doi: 10.4049/jimmunol.1900379. [DOI] [PubMed] [Google Scholar]
- 12.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]