The development of vaccines and therapeutics to combat viral hemorrhagic fevers remains a top priority within the Implementation Plan of the U.S. Department of Health and Human Services Public Health Emergency Medical Countermeasures Enterprise. The Can strain, derived from the pathogenic XJ strain of JUNV, has been demonstrated to be both safe and protective against AHF. While the vaccine strain is approved for use in regions of endemicity within Argentina, the mechanisms of Can attenuation have not been elucidated. A better understanding of the viral genetic determinants of attenuation will improve our understanding of the mechanisms contributing to disease pathogenesis and provide critical information for the rational design of live attenuated vaccine candidates for other viral hemorrhagic fevers.
KEYWORDS: Junin virus, arenavirus, glycoproteins, hemorrhagic fever, vaccines
ABSTRACT
Argentine hemorrhagic fever is a potentially lethal disease that is caused by Junin virus (JUNV). There are currently around 5 million individuals at risk of infection within regions of endemicity in Argentina. The live attenuated vaccine strain Candid #1 (Can) is approved for use in regions of endemicity and has substantially decreased the number of annual Argentine hemorrhagic fever (AHF) cases. The glycoprotein (GPC) gene is primarily responsible for attenuation of the Can strain, and we have shown that the absence of an N-linked glycosylation motif in the subunit G1 of the glycoprotein complex of Can, which is otherwise present in the wild-type pathogenic JUNV, causes GPC retention in the endoplasmic reticulum (ER). Here, we show that Can GPC aggregates in the ER of infected cells, forming incorrect cross-chain disulfide bonds, which results in impaired GPC processing into G1 and G2. The GPC fails to cleave into its G1 and G2 subunits and is targeted for degradation within lysosomes. Cells infected with the wild-type Romero (Rom) strain do not produce aggregates that are observed in Can infection, and the stress on the ER remains minimal. While the mutation of the N-linked glycosylation motif (T168A) is primarily responsible for the formation of aggregates, other mutations within G1 that occurred earlier in the passage history of the Can strain also contribute to aggregation of the GPC within the ER.
IMPORTANCE The development of vaccines and therapeutics to combat viral hemorrhagic fevers remains a top priority within the Implementation Plan of the U.S. Department of Health and Human Services Public Health Emergency Medical Countermeasures Enterprise. The Can strain, derived from the pathogenic XJ strain of JUNV, has been demonstrated to be both safe and protective against AHF. While the vaccine strain is approved for use in regions of endemicity within Argentina, the mechanisms of Can attenuation have not been elucidated. A better understanding of the viral genetic determinants of attenuation will improve our understanding of the mechanisms contributing to disease pathogenesis and provide critical information for the rational design of live attenuated vaccine candidates for other viral hemorrhagic fevers.
INTRODUCTION
The New World arenavirus Junin virus (JUNV) is the causative agent of Argentine hemorrhagic fever (AHF), a potentially lethal disease endemic to the central Pampas regions of Argentina. JUNV has a bisegmented negative-sense RNA genome consisting of two segments, a small (S) segment and large (L) segment. Each genome segment encodes two viral proteins in opposite orientations. The S RNA segment (∼3.4 kb) encodes both the nucleoprotein (NP) and the glycoprotein precursor (GPC). The NP protein is the most abundant viral protein in infected cells and main component of the virus ribonucleoprotein (vRNP) complex responsible for directing the biosynthetic processes of replication and transcription of the viral genome (1). The Old World arenavirus lymphocytic choriomeningitis virus (LCMV) NP has been shown to inhibit the induction of type I interferon expression through an interaction with interferon regulatory factor 3 (IRF3) and IκB kinase epsilon (IKKɛ) (2, 3). The GPC protein is cotranslationally cleaved by cellular signal peptidases to release the stable signal peptide (SSP) and posttranslationally processed by the proprotein-convertase (PC) subtilisin kexin isozyme-1/site-1 protease (SKI-1/S1P) into the N-terminal G1 and membrane-anchored G2 subunits. Mature arenavirus GP forms a trimer of SSP/G1/G2 heterotrimers that represents the functional unit of virus attachment and entry. The N-terminal G1 binds to cellular receptors, whereas the transmembrane G2 mediates viral fusion (4, 5). The L RNA segment (∼7 kb) encodes the RNA-dependent RNA polymerase (LP) and the matrix (Z) protein. The Z protein contains a central zinc-binding RING domain and carries out a number of functions during the virus life cycle, including the recruitment of GPC and the RNP into the budding virion, inhibition of the viral replication complex through interaction with LP, and antagonism of the host immune response (6).
After delivery of the RNP into the cytoplasm of the host cell, the viral polymerase initiates transcription of the virus genome. Studies conducted with the prototypic arenavirus LCMV revealed that RNA synthesis significantly declined during peak virus titer in cell culture (7), supporting the idea that the Z protein is not required for the biosynthetic activity of the vRNP but rather exhibits a dose-dependent inhibitory effect on viral RNA synthesis (7). GPC is translated into the ER and undergoes a series of glycosylation and processing steps during its trafficking to the cell surface. Myristoylation of both Z protein and the SSP is required for normal GPC-Z interaction at the cell surface (8).
Previously, GPC has been shown to be the primary viral determinant of Can attenuation (9). A single amino acid change in the transmembrane region of Can G2 (F427I), introduced during FRhL-2 cell passaging, attenuated the neurovirulence of JUNV in mice (10), but the G2 F427I mutation was not present in already-attenuated JUNV XJ37 and XJ44 strains which precede the passages in FRhL-2 cells (11). Therefore, other determinants in Can GPC contribute to the attenuated phenotype of Can observed in guinea pig and nonhuman primate (NHP) models of AHF (12). We have shown that a single amino acid substitution within an N-linked glycosylation motif in G1 (T168A) results in the absence of an N-linked glycan on Can G1 that is present on the pathogenic Romero (Rom) strain of JUNV. The absence of this glycan contributes to the retention of Can GPC in the endoplasmic reticulum (ER) and accumulation of nonprocessed GPC within Can-infected cells, whereas the Rom GPC is cleaved and expressed in higher concentrations at the cell surface (13).
Here, we provide evidence that impaired processing of Can GPC activates the unfolded protein response (UPR) in Can-infected cells through upregulation of binding immunoglobulin protein (BiP). We also present evidence of GPC aggregation through the formation of interchain disulfide bonds that promotes its degradation within lysosomal compartments. We also observed decreased levels of total light-chain 3 (LC3) protein, suggesting that autophagy is involved in degradation of Can GPC. The degradation of the GPC may contribute to the enhanced immunogenicity of the protective antigen GPC through a more active presentation of GPC antigens as products of degradation.
RESULTS
Effect of Can-like G1 substitutions on JUNV GPC protein expression levels.
During mouse brain passages of the pathogenic XJ strain to generate Can, a single amino acid change (T168A) occurred between the 13th mouse brain passage (XJ13) and the 44th mouse brain passage (XJ44) that eliminated an N-linked glycosylation motif within G1 (14). Two additional amino acid differences (M109Q and E116A) within G1 and a single amino acid change within the cytoplasmic domain of G2 (R484G) exist between the pathogenic Rom strain and XJ13 (Fig. 1A). During cell culture passaging in FRhL-2 cells, another four mutations (E186G, S206P, F427I, and T446S) accumulated to yield Can GPC. The single transmembrane substitution F427I has been shown to mediate the attenuation of JUNV neurovirulence in mice (10). However, recombinant JUNV (rJUNV) carrying the G2 F427I mutation caused a self-limiting febrile illness in our guinea pig model of AHF (9), suggesting that other GPC-specific mutations contribute to Can attenuation in the guinea pig model of AHF disease. We have shown that XJ13- and XJ44-specific mutations in G1 resulted in impaired processing of GPC compared to the wild-type Rom GPC (13). We also observed reduced surface and total expression of Can GPC compared to Rom GPC in cell transfection assays (13). We therefore used plasmid-supplied GPC in transfected HEK293 cells to determine the effects of XJ13- and XJ44-specific mutations on total cellular GPC expression. We found that cleaved G2 expression as determined by Western blotting was lower than that of Rom in XJ13, XJ44, Can, and Rom-T168A samples at both time points. Additionally, total GPC expression was lower in XJ44, Can, and Rom-T168A than in Rom, XJ13, and Can-A168T GPC at 48 h posttransfection (P < 0.05). We also noticed the presence of GPC dimers and trimers in each sample across both the 24- and 48-h time points. The quantities of dimers and trimers are directly comparable to the quantity of full-length GPC in each sample as opposed to the quantities of cleaved protein (represented by the cleaved G2 band), suggesting that they are forming very early in trafficking, before G1/G2 cleavage occurs (Fig. 1B). To rule out differences in mRNA expression as the cause of the differences in protein between samples, we used quantitative real-time PCR to compare GPC RNA levels among HEK293 cells transfected with GPC expression plasmids for the 24-h time point. We observed similar RNA expression levels for cells transfected with the different GPC expression plasmids (Fig. 1C).
FIG 1.
Comparison of total detectable GPC in HEK293 cells and chaperone upregulation in infected Vero cells. (A) Schematic representation of amino acid substitutions between Rom GPC and the GPCs from key passages in the creation of the Can vaccine. Differences in XJ13 and Rom are indicated in blue, differences between XJ13 and XJ44 are indicated in yellow, and Can-specific amino acids are indicated in red. The green marks represent N-linked glycosylation motifs. (B) HEK293 cells were seeded into 12-well plates (1 × 105 cells/well) and allowed to incubate for 24 h at 37°C prior to transfection. The cells were transfected with plasmids expressing either Rom, XJ13, XJ44, Can, Rom-T168A, or Can-A168T GPC and allowed to incubate for an additional 24 to 48 h. Cells were lysed using Triton X-100 lysis buffer at 24 or 48 h posttransfection, and proteins were detected under nonreducing conditions with an anti-JUNV G2 antibody. Cellular proteins were detected using commercially available antibodies. Total GPC stained in each lane was quantified using ImageJ for each time point, and the results are shown as a bar graph. Whiskers represent the standard error among triplicate results for each value. (C) Duplicates of samples from panel B were treated with TRIzol at 24 h posttransfection, and RNA was isolated from cell lysates. The RNA was quantified using real-time PCR, and the GPC total RNA was normalized to GAPDH RNA. The experiment was repeated in triplicate. Whiskers are the standard error values among sample replicates, and ANOVA was performed to measure significance between groups (see Results). (D) Vero cells were prepared and infected with either rRom or rCan virus (MOI, 1.0). Lysates were collected at the indicated time points (8 hpi, 24 hpi, and 36 hpi) and stained for intracellular PDI and viral NP. PDI bands in each lane were quantified using ImageJ for each time point, and the results are shown as a bar graph. Whiskers represent the standard error among triplicate results for each value. ANOVA was performed to measure significance between groups (see Results).
We have previously observed that recombinant Can (rCan) produces a much higher detectable quantity of full-length GPC during infection, and BiP is upregulated in rCan-infected cells (13). We therefore suspected that rCan infection could be causing a buildup of precleavage GPC dimers and trimers within the ER, triggering stress. We infected Vero cells with either recombinant Rom (rRom) or rCan virus (multiplicity of infection [MOI], 1.0) and collected lysates at 8, 24, and 36 h postinfection. We stained for the ER chaperone protein disulfide-isomerase (PDI) to determine whether the ER was under stress due to the accumulation of misfolded proteins. The rRom-infected cells expressed a uniform amount of PDI across all time points, whereas there was a significantly increased (P < 0.05) upregulation of PDI in rCan-infected cells over time (Fig. 1D).
ER accumulation of GPC promotes its lysosome-mediated degradation.
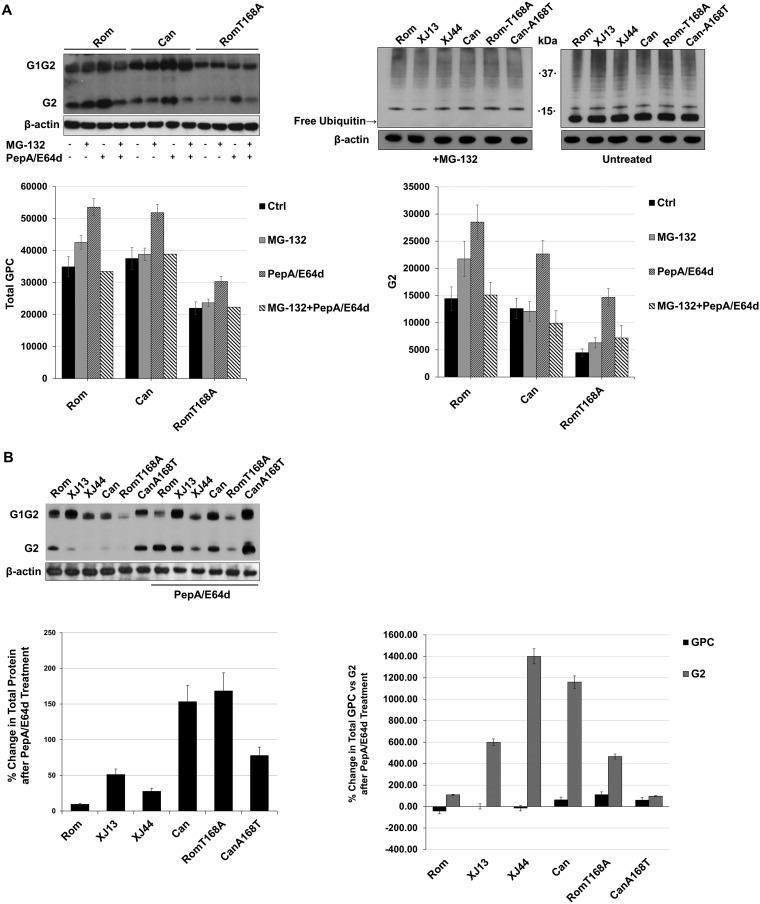
We have shown that the T168A substitution found in XJ44 is the primary substitution responsible for the retention of JUNV GPC within the ER, whereas XJ13-specific substitutions contributed to the retention of GPC to a lesser degree (13). We have documented different degrees of ER retention of XJ13, XJ44, Can, and Rom-T168A GPCs (13), as well as decreased levels in total expression of these GPCs in HEK293 cells, suggesting that posttranslational processing of GPC is affected by one or more of these substitutions, which could result in GPC degradation. Degradation of membrane-associated proteins can occur through two main pathways in mammalian cells, the ubiquitin-proteasome pathway and the lysosomal pathway (15). The small molecule MG-132 has been shown to potently inhibit the 26S proteasome (16), whereas both pepstatin-A (PepA) and E64d are well known to inhibit lysosomal proteases. We examined the effects of MG-132 or PepA/E64d treatment on GPC expression levels in cells transfected with Rom and Can GPC-expressing plasmids. Due to the significant effect that the presence or absence of the N-linked glycan at N166 has previously shown for ER retention (13), we also included the Rom-T168A GPC in the comparison. At 24 h posttransfection, BHK-21 cells were treated with either MG-132 (1 μM) or PepA/E64d (10 μg/ml), and 8 h later, cell lysates were prepared. Western blot analysis revealed that PepA/E64d treatment, but not MG-132 treatment, affected GPC expression levels. Lysosomal protease inhibition caused a greater increase than did proteasomal inhibition in Rom, Can, and Rom-T168A G2 levels (Fig. 2A). Since we could not detect any changes in GPC amounts with MG-132, we measured the free ubiquitin pool in transfected BHK-21 cells after MG-132 treatment to ensure that the drug was depleting free ubiquitin in our transfected cells (Fig. 2A). We were able to confirm that the free ubiquitin pool was depleted, confirming that the proteasome has no effect on GPC levels. We then examined the effect of PepA/E64d treatment on the expression of our full panel of different GPCs in HEK293 cells (Fig. 2B and C). The increase in G2 upon PepA/E64d treatment was visible for all GPCs, confirming that the observation in BHK-21 cells was not cell type specific. Densitometry analysis indicated that PepA/E64d treatment resulted in a 2- to 4-fold increase in Rom and Can-A168T G2 and a much higher increase in XJ13, XJ44, Can, and Rom-T168A G2 (P < 0.05) (Fig. 2C). We did not observe significant differences in full-length GPC expression after PepA/E64d treatment (Fig. 2C).
FIG 2.
Comparison of cellular degradation pathway activity during JUNV GPC expression. (A) BHK-21 cells were seeded into 12-well plates (1 × 105 cells/well) and incubated at 37°C for 24 h prior to transfecting the cells with plasmids expressing either Rom, Can, or Rom-T168A. After an additional 28 h, the indicated cells were treated with either MG-132 (1 μM) or PepA/E64d (10 μg/ml each) and allowed to incubate for another 8 h. The cells were lysed with Laemmli’s SDS buffer, and GPC was detected with an anti-JUNV G2 antibody. Cellular proteins were detected with commercially available antibodies. Densitometries for both total GPC and G2 were obtained using ImageJ, and the results are shown in graphs to compare either total GPC or G2 among treatments. Whiskers represent the standard error among triplicate results for each value. (B) HEK293 cells were prepared identically to the cells in panel A and transfected with plasmids expressing either Rom, XJ13, XJ44, Can, Rom-T168A, or Can-A168T GPC. Treatments and transfections were repeated using conditions identical to those in panel A, and the protein was detected using anti-JUNV G2 antibodies. Ubiquitin was stained in MG-132-treated cells to confirm that the inhibitor was depleting free ubiquitin to validate the results in panel A. Densitometries were obtained for G2 using ImageJ on a scale of 0 to 100, where 100 represents complete saturation. Fold change in G2 was calculated for untreated samples compared to samples treated with PepA/E64D. Densitometry values are provided for total GPC, full-length GPC, and G2. Whiskers represent the standard error among triplicate results for each value. ANOVA was performed to determine statistical significance between groups (see Results).
Growth properties in cultured cells of recombinant Rom viruses expressing GPCs that contain XJ13-, XJ44-, and Can-specific amino acid substitutions in GPC.
In infected cells, the Z protein regulates the activity of the vRNP, interacts with GPC to promote assembly of infectious mature viral particles, and acts also as the driving force of virus budding. Because of its multiple regulatory roles in the virus life cycle, Z could affect the processing and trafficking of both Rom and Can GPC, a process that would not be captured in our transfection assays with GPC expression plasmids. We therefore used our Pol-I/Pol-II plasmid-driven rescue system to generate infectious recombinant Rom (rRom) viruses expressing either XJ13, XJ44, Can, or Rom-T168A GPC in place of the wild-type Rom GPC to examine differential trafficking of each GPC in the context of a natural infection (Fig. 3A). All of the viruses exhibited similar peak growth kinetics in both Vero cells and A549 cells (Fig. 3B) despite exhibiting differences in plaque phenotypes (Fig. 3C). However, we did note a slightly accelerated growth in early (24 to 48 hours postinfection [hpi]) infection of Rom-infected cells compared to the other clones.
FIG 3.
Growth kinetics of recombinant JUNVs expressing all or part of Can GPC. (A) Schematic representation of viruses rescued. Gray represents Can-like regions. Specific mutations for these regions can be cross-referenced using Fig. 1A. (B) Vero and A549 cell monolayers were infected with working stocks of the viruses (MOI, 0.1) illustrated in panel A. Supernatant was collected every 24 h and subjected to plaque assay to quantify infectious progeny. Plaque assays were performed using Vero cell monolayers cultured in 6-well plates. Serial dilutions of supernatant were used to infect the monolayers and incubated with a 1% agarose overlay for 5 days. Cells were fixed in formalin and stained with crystal violet. The samples were generated in triplicate, and the whiskers represent the standard error among replicates at each point. (C) Plaque phenotypes for each virus. Images were taken from plaque assay titration of the purified working stock in the same manner as described in panel B.
Can GPC aggregates during infection, and T168A promotes degradation via autophagy.
Since our rRom viruses expressing XJ13, XJ44, Can, or Rom-T168A GPC produced similar levels of infectious particles at all times examined, we aimed to determine which substitutions could be contributing to the differences in protein production. Mutant forms of certain ER-targeted proteins are retained in the ER and can form interchain disulfide bridges (17). We therefore compared the GPC expression of each virus under both reducing (+β-mercaptoethanol [+BME]) and nonreducing (−BME) conditions to determine whether GPC aggregates could be detected for any of the viruses. For this, we infected (MOI, 5) Vero cells with our working virus stocks, and at 48 hpi, we prepared cell lysates using Western blotting to compare patterns of GPC expression under +BME and −BME conditions (Fig. 4A). Rom GPC did not exhibit evidence of aggregation, whereas a significant fraction of all the other GPCs contained interchain disulfide bonds that created dimers, trimers, tetramers, and higher-order aggregates (Fig. 4A). These results indicated that both the T168A- and XJ13-specific mutations contribute to GPC the aggregation. Additionally, the absence of aggregates in the Rom GPC sample removes the possibility that these aggregates are an artifact of disulfide bond formation during SDS denaturation. BiP, the master regulator of the UPR, as well as its downstream effector proteins lysosome-associated membrane protein 1 (LAMP-1) and C/EBP homologous protein (CHOP), were significantly upregulated (P < 0.05 for each protein) in cells infected with viruses carrying a non-Rom GPC (Fig. 4A). The upregulation of these UPR-associated proteins correlates with the presence of aggregation.
FIG 4.
Aggregation of GPC and upregulation of ER stress-related markers during infection. (A) Expression of total viral protein and stress markers in Vero cells. Vero cells were seeded into 12-well plates (1 × 105 cells/well) and incubated at 37°C for 24 h. The cells were infected (MOI, 5) with each of the viruses from Fig. 4 and allowed to incubate for 48 h before lysing the cells in Laemmli’s SDS buffer. Viral protein was detected using custom anti-JUNV G2 and NP antibodies. The cellular proteins BiP, LAMP-1, LC3, and CHOP were stained using commercially available antibodies. Bands for BiP, LAMP-1, and CHOP were quantified using ImageJ. Whiskers represent the standard error among triplicate results for each value. (B) Western blot images of LC3 were analyzed in ImageJ to quantify LC3-I and LC3-II. Ratios of LC3-II to LC3-I were compared relative to uninfected Vero cells to detect potential increases in autophagy. Whiskers represent the standard error among triplicate results for each value. ANOVA was performed to determine statistical significance between groups (see Results).
In the presence of aggregated protein and the activation of the UPR, lysosomal degradation is thought to be the primary pathway for degradation due to the inability of protein aggregates to fit through the ER-associated degradation (ERAD) pore (17). The process of autophagy can be utilized to package ER vesicles and target them for degradation. We therefore quantified both LC3-I and LC3-II in Vero cell lysates at 48 h postinfection. Only the viruses expressing Can or Rom-T168A GPC exhibited an LC3-II/total LC3 ratio significantly higher than that of the rRom virus (P < 0.05), and the increase was minimal for the Rom-T168A sample (Fig. 4B). Although aggregation and BiP upregulation occurred in cells infected with rRom expressing XJ13 and XJ44, these cells did not exhibit a significant increase in autophagy over Rom-infected cells. Collectively, the data indicate that the Can GPC forms aggregates due to contributions from mutations specific to both XJ13 and XJ44 GPC. The mutation of the N-linked glycosylation motif in G1 alone is sufficient to result in a formation of complex aggregates and in degradation of the protein.
DISCUSSION
Although the Can vaccine is currently approved for use in Argentina within AHF regions of endemicity (18), the mechanisms of Can attenuation remain mostly unknown. We have shown that a rRom virus where Can GPC substituted for Rom GPC (rRom/GPC-Can) is completely attenuated in a guinea pig model of human AHF (9). The GPC of Rom contains eight amino acid differences with respect to Can, and the single substitution F4271 that occurred during FRhL-2 cell passage of XJ attenuates the neurovirulence of JUNV in mice (10). This mutation lies within the transmembrane domain of G2 and has been proposed to destabilize the conformation of GPC, making the conformational change to its fusogenic state more favorable at neutral pH (19). However, intraperitoneal challenge of guinea pigs with rRom virus expressing the F427I Can-like substitution in GPC resulted in only partial attenuation (9). Moreover, the F427I mutation was introduced during cell culture passage following the 44th mouse brain passage of XJ, when the virus had already achieved attenuation in guinea pigs (18, 20). This suggests that other amino acid substitutions within GPC contribute to Can attenuation. The T168A substitution within the G1 lies within a predicted N-linked glycosylation motif (21) and has been shown to prevent glycosylation at G1 N166 in Can, which is present in the pathogenic Rom strain (13). The absence of this N-linked glycan was associated with the retention of GPC within the ER, impaired GPC processing, and a lower surface expression of GPC (13). The T168A substitution was introduced into XJ between the 13th and 44th mouse brain passages, supporting its role in the attenuation of the virus in guinea pigs. It is noteworthy that XJ13 GPC displayed some degree of ER retention and decreased surface expression compared to Rom, but trafficking of XJ13 GPC was less affected than that of Rom GPC carrying only the T168A substitution (13).
The results from the transfection assays with plasmids expressing JUNV GPCs from key historical passages in the creation of Can revealed that the efficiency of GPC processing decreased substantially in comparison to the wild-type Rom strain GPC (13). The impaired processing of GPC by S1P appears to be caused primarily by the T168A substitution. The same substitution also had a significant impact on total protein expression (Fig. 1), and our results suggest that the degradation of GPC lacking the N166 glycan was likely the primary cause for reduced expression levels of Can GPC with respect to Rom GPC. Similar levels of GPC RNA in cells transfected with plasmids expressing the different GPC forms indicated that differences in protein-level expression did not reflect differences at the transcriptional level (Fig. 1C).
During the sequence of events leading to the generation of Can, impaired GPC processing efficiency correlated with a decrease in total protein expression. Membrane-associated proteins must pass a series of checkpoints within the ER, and the addition of glycans to these proteins serves as a ticket into the glycoprotein quality control (GQC) system. The presence of these glycans allows for close association of the glycoprotein with ER chaperones to promote the correct folding of the protein. Prior to being exported to the Golgi, the protein must pass a series of checkpoints involving the calnexin-calreticulin cycle and BiP association, and the checkpoints are highly dependent on glycan modifications (17). Can GPC is less efficient than Rom GPC in progressing through the ER folding checkpoints that are required for export to the Golgi. The T168A substitution in Can G1 appears to be the primary contributor to Can GPC retention within the ER, but XJ13-specific amino acids also contribute to a lesser degree to the ER retention of GPC (13). The absence of a glycan in Can that is present in Rom could impact the ability of the Can GPC to interact with either calnexin or calreticulin, thereby preventing the protein from passing a specific GQC checkpoint and marking the protein for ER-associated degradation (ERAD). GPC processing by S1P occurs in the late Golgi (22) and is therefore directly linked to the ability of JUNV GPC to escape the ER.
During GQC, the preferential path of protein degradation is the proteasomal pathway. If incorrectly folded proteins accumulate faster than they can be exported to the proteasome, the proteins are susceptible to aggregation and must be degraded in bulk (17). Bulk degradation through the lysosome appears to be the main degradation pathway for Can GPC (Fig. 2A). While the T168A substitution seems to account for most of the lysosome-mediated degradation of Can GPC, the XJ13-specific substitutions also contribute to this process during infection (Fig. 2).
We observed a marginal increase in Rom GPC expression levels in samples treated with lysosomal protease inhibitors (Fig. 2). At later time points (>24 h) posttransfection, GPC overexpression eventually results in activation of the UPR even in the case of Rom GPC (13). Within the context of virus infection, the JUNV protein expression is highly regulated, and Rom GPC does not reach expression levels that trigger UPR. In contrast, there is a robust BiP upregulation in Can-infected cells (13). The presence of aggregates in rRom viruses expressing XJ13, XJ44, Can, and Rom-T168A GPCs further suggests ER stress-induced lysosomal degradation of these proteins. Both BiP and PDI were upregulated in cells expressing non-Rom GPC but were unable to resolve the ER stress or prevent GPC aggregates from forming. The activation of the UPR during infection has also been described in association with another arenavirus. The GPC of LCMV has been demonstrated to activate the activating transcription factor 6 (ATF6) branch of the UPR during infection. The activation of the ATF6-regulated branch of the UPR was caused by the GPC of LCMV, and the downregulation of GPC expression during the persistent infection resolved the ER stress and returned the ATF6 activity to basal levels (23). The increase in CHOP levels caused by the T168A substitution suggests that either the protein kinase R-like endoplasmic reticulum kinase (PERK) or inositol-requiring enzyme 1 (IRE1) branches of the UPR are activated as well. Regardless, this leads to the cleavage of caspases 3 and 7 (13), which would explain the potent induction of apoptosis during Can infection in Vero cells (24). It is interesting that XJ13 GPCs were present primarily as dimers and trimers, whereas GPCs containing the T168A substitution formed more complex aggregates during infection (Fig. 4A). XJ13-specific amino acid substitutions affect ER processing of GPC, but the impact of the T168A substitution appears to be more severe. However, further investigation of each substitution is necessary in order to determine the exact GQC steps being impacted. When examining the dimer and trimer formation in cells transfected with JUNV GPCs at supraphysiological levels, even Rom GPC begins to form a significant amount of these precleavage aggregates at 48 h posttransfection (Fig. 1B). However, these aggregates are not present during infection with rRom (Fig. 4A). This is likely due to the regulatory effects of the other viral proteins present in the cell, preventing the wild-type protein from accumulating in the ER.
The pathways through which aggregated proteins arrive in lysosomes are not completely understood, but autophagosomes have been implicated (17). In certain instances, aggregated proteins are exported to the cytoplasm (17). Although autophagy is commonly involved in the removal of aggregates sequestered in the ER, there are instances in which autophagy-independent lysosomal degradation pathways are utilized to clear aggregated proteins within the cytoplasm (25). It is interesting that XJ13, XJ44, Can, and Rom-T168A GPCs all form aggregates, but only the Can GPC, and to a lesser extent Rom-T168A GPC, displays an increase in LC3-II conversion over Rom-infected cells (Fig. 4B). This suggests that lysosomal degradation could be occurring through two separate processes concurrently.
Our findings indicate that both the XJ13-specific and XJ44-specific substitutions within Can GPC negatively impact the processing of the protein within the ER and result in the accumulation of GPC aggregates. These aggregates are degraded in bulk within the lysosome. This process could potentially have a significant impact on both the attenuation and the immunogenicity of the Can vaccine strain by potentially limiting early spread of the virus due to decreased surface GPC and inducing an early immune response to infection by causing a more robust antigen presentation.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney 293 (HEK293), baby hamster kidney 21 (BHK-21), and African green monkey kidney epithelial (Vero) cells (American Tissue Culture Collection) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin antibiotic solution (Gibco). The murine RNA polymerase-I (Pol-I)/RNA polymerase-II (Pol-II) system utilized for the rescue and characterization of rJUNV clones has been described in detail previously (12). Using the cDNA from our Pol-II expression plasmids for XJ13, XJ44, Can, and Rom-T168A GPC (13), we amplified the open reading frames of each GPC and cloned them into our mPol-I Rom S segment (mPol-I-Sag) expression plasmid in place of Rom GPC. These plasmids were transfected into BHK-21 cells along with our Rom L segment (mPol-I-Lag), NP expression (pC-NP), and LP expression (pC-LP) plasmids described previously (9) in order to rescue the rJUNV clones. The virus clone stocks were generated by infecting Vero cells (multiplicity of infection [MOI], 0.01) and collecting the supernatants at 4 days postinfection. The cell debris was removed by centrifugation at 10,000 × g for 10 min. Virus growth curves were performed by infecting Vero cell monolayers seeded in 6-well plates with rJUNV (MOI, 0.01) using our passage 1 (P1) working stock. Supernatant was collected every 24 h through 96 h, and the infectious titer was determined using a plaque assay. Plaque assays were conducted in Vero cell monolayers in 6-well tissue culture plates. Serial dilutions of the collected supernatants were performed and used to infect the Vero cell monolayers. The infected monolayers were overlaid with a solution of minimum Eagle’s medium (MEM) and 0.5% agarose and allowed to incubate at 37°C for 5 days. The cells were fixed with formalin for 30 min and stained with crystal violet. All work with the rJUNVs was conducted within the University of Texas Medical Branch (UTMB) biosafety level 4 (BSL-4) facility in accordance with the institutional health and safety guidelines.
Western blotting.
Vero or HEK293 cells were seeded into 12-well tissue culture plates (1 × 105 cells/well) and allowed to incubate for 24 h at 37°C. The cells were then infected with rRom viruses expressing either Rom, XJ13, XJ44, Can, or Rom-T168A GPCs (MOI, 5) and allowed to incubate at 37°C for 48 h. The cells were rinsed with 1× phosphate-buffered saline and then lysed using 1× Laemmli buffer, or with Triton X-100 lysis buffer and later treated with Laemmli buffer with or without 5% β-mercaptoethanol (BME). The lysates were incubated at 95°C for 10 min, and the indicated samples were then treated with BME equal to 10% of the sample volume. The lysates were loaded onto 10% polyacrylamide gels using equal protein loading and subjected to electrophoresis. The protein was transferred onto polyvinylidene difluoride (PVDF) membranes and blocked with 5% nonfat dry milk in PBS-T (10 mM sodium phosphate, 0.15 M NaCl, 0.1% Tween 20) for 1 h at room temperature prior to antibody staining. The protein of interest was stained with primary antibody diluted into blocking buffer (1:1,000) at 4°C overnight, washed 3 times for 15 min in PBS-T, and stained with secondary antibody conjugated with horseradish peroxidase (HRP; Cell Signaling) diluted in PBS-T (1:1,000) for 1 h at room temperature. After a final PBS-T rinse, the membranes were treated with the Amersham enhanced chemiluminescence (ECL) reagent. The primary antibodies used to detect JUNV G2 and NP have been described previously (13). The primary antibodies against BiP, LC3, PDI, and β-actin were obtained from Cell Signaling Technology, Inc. The MG-132 (SelleckChem), pepstatin-A (PepA; Thermo Scientific), and E64d (Thermo Scientific) inhibitors were dissolved into the supernatant for 8 h at concentrations of 1 μM (MG-132) and 10 μg/ml (PepA/E64d). LC3-I and LC3-II were quantified using the ImageJ blot densitometry analysis. All blots were performed in triplicate, and blot densitometries were analyzed using a one-way analysis of variance (ANOVA).
Quantitative real-time PCR.
The total RNA from HEK293 cells transfected with JUNV GPC was isolated using TRIzol (Thermo Scientific) and purified using the Direct-Zol RNA kit (Zymo Research). RNA encoding the GPC open reading frame (ORF) was quantified using the iScript one-step PCR kit with SYBR green (Bio-Rad) using 100 ng total RNA. The following primers were used for detection: GPC direct, TCGGACTGCACACTGAGTTC; GPC reverse, ACTTGTGGGTTGTGCCTTCA; GAPDH direct, CTCTGCTCCTCCTGTTCGAC; and GAPDH reverse, AAATGAGCCCCAGCCTTCTC (GAPDH, glyceraldehyde-3-phosphate dehydrogenase). Melt curve analysis was performed at the end of the PCR cycle to confirm the PCR fragment specificity. The relative amounts of GPC per sample were calculated using the equation ΔCTsample = (CTsample – CTGAPDH/sample) – (CTGPC – CTGAPDH/GPC) (26).
ACKNOWLEDGMENTS
J.T.M. was supported by the Sealy Center for Vaccine Development and additionally by the Clinical and Translational Science Award NRSA (TL1) Training Core (TL1TR001440) from the National Center for Advancing Translational Sciences at the National Institutes of Health. T.K. was supported in part by a JSPS Postdoctoral Fellowship for Research Abroad. Both S.P. and J.C.D.L.T. were supported by Public Health Service grant R01AI093445, and S.P. was supported by pilot/bridge funding from the Department of Pathology, University of Texas Medical Branch.
REFERENCES
- 1.Buchmeier MJ, De la Torre JC, Peters CJ. 2013. Arenaviridae, p 1283–1303. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Pythoud C, Rodrigo WW, Pasqual G, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. 2012. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKε. J Virol 86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martínez-Sobrido L, Zúñiga EI, Rosario D, García-Sastre A, de la Torre JC. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A 98:12701–12705. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmeier MJ. 2002. Arenaviruses: protein structure and function. Curr Top Microbiol Immunol 262:159–173. doi: 10.1007/978-3-642-56029-3_7. [DOI] [PubMed] [Google Scholar]
- 6.Fehling SK, Lennartz F, Strecker T. 2012. Multifunctional nature of the arenavirus RING finger protein Z. Viruses 4:2973–3011. doi: 10.3390/v4112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornu TI, de la Torre JC. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol 75:9415–9426. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capul AA, Perez M, Burke E, Kunz S, Buchmeier MJ, de la Torre JC. 2007. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. J Virol 81:9451–9460. doi: 10.1128/JVI.00499-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seregin AV, Yun NE, Miller M, Aronson J, Smith JK, Walker AG, Smith JN, Huang C, Manning JT, de la Torre JC, Paessler S. 2015. The glycoprotein precursor gene of Junin virus determines the virulence of the Romero strain and the attenuation of the Candid #1 strain in a representative animal model of Argentine hemorrhagic fever. J Virol 89:5949–5956. doi: 10.1128/JVI.00104-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albariño CG, Bird BH, Chakrabarti AK, Dodd KA, Flint M, Bergeron E, White DM, Nichol ST. 2011. The major determinant of attenuation in mice of the Candid1 vaccine for Argentine hemorrhagic fever is located in the G2 glycoprotein transmembrane domain. J Virol 85:10404–10408. doi: 10.1128/JVI.00856-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yun NE, Linde NS, Dziuba N, Zacks MA, Smith JN, Smith JK, Aronson JF, Chumakova OV, Lander HM, Peters CJ, Paessler S. 2008. Pathogenesis of XJ and Romero strains of Junin virus in two strains of guinea pigs. Am J Trop Med Hyg 79:275–282. doi: 10.4269/ajtmh.2008.79.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emonet SF, Seregin AV, Yun NE, Poussard AL, Walker AG, de la Torre JC, Paessler S. 2011. Rescue from cloned cDNAs and in vivo characterization of recombinant pathogenic Romero and live-attenuated Candid #1 strains of Junin virus, the causative agent of Argentine hemorrhagic fever disease. J Virol 85:1473–1483. doi: 10.1128/JVI.02102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning J, Seregin A, Yun N, Koma T, Huang C, Barral J, De La Torre J, Paessler S. 2017. Absence of an N-linked glycosylation motif in the glycoprotein of the live-attenuated Argentine hemorrhagic fever vaccine, Candid #1, results in its improper processing and reduced surface expression. Front Cell Infect Microbiol 7:20. doi: 10.3389/fcimb.2017.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goñi SE, Iserte JA, Stephan BI, Borio CS, Ghiringhelli PD, Lozano ME. 2010. Molecular analysis of the virulence attenuation process in Junin virus vaccine genealogy. Virus Genes 40:320–328. doi: 10.1007/s11262-010-0450-2. [DOI] [PubMed] [Google Scholar]
- 15.Ciechanover A. 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol 6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 16.Tsubuki S, Saito Y, Tomioka M, Ito H, Kawashima S. 1996. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J Biochem 119:572–576. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- 17.Ferris SP, Kodali VK, Kaufman RJ. 2014. Glycoprotein folding and quality-control mechanisms in protein-folding diseases. Dis Model Mech 7:331–341. doi: 10.1242/dmm.014589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambrosio A, Saavedra M, Mariani M, Gamboa G, Maiza A. 2011. Argentine hemorrhagic fever vaccines. Hum Vaccin 7:694–700. doi: 10.4161/hv.7.6.15198. [DOI] [PubMed] [Google Scholar]
- 19.Droniou-Bonzom ME, Reignier T, Oldenburg JE, Cox AU, Exline CM, Rathbun JY, Cannon PM. 2011. Substitutions in the glycoprotein (GP) of the Candid#1 vaccine strain of Junin virus increase dependence on human transferrin receptor 1 for entry and destabilize the metastable conformation of GP. J Virol 85:13457–13462. doi: 10.1128/JVI.05616-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephan BI, Lozano ME, Goni SE. 2013. Watching every step of the way: Junin virus attenuation markers in the vaccine lineage. Curr Genomics 14:415–424. doi: 10.2174/138920291407131220153526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albariño CG, Bergeron E, Erickson BR, Khristova ML, Rollin PE, Nichol ST. 2009. Efficient reverse genetics generation of infectious Junin viruses differing in glycoprotein processing. J Virol 83:5606–5614. doi: 10.1128/JVI.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damonte EB, Mersich SE, Candurra NA. 1994. Intracellular processing and transport of Junin virus glycoproteins influences virion infectivity. Virus Res 34:317–326. doi: 10.1016/0168-1702(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 23.Pasqual G, Burri DJ, Pasquato A, de la Torre JC, Kunz S. 2011. Role of the host cell’s unfolded protein response in arenavirus infection. J Virol 85:1662–1670. doi: 10.1128/JVI.01782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolokoltsova OA, Grant AM, Huang C, Smith JK, Poussard AL, Tian B, Brasier AR, Peters CJ, Tseng CT, de la Torre JC, Paessler S. 2014. RIG-I enhanced interferon independent apoptosis upon Junin virus infection. PLoS One 9:e99610. doi: 10.1371/journal.pone.0099610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juenemann K, Reits EA. 2012. Alternative macroautophagic pathways. Int J Cell Biol 2012:189794. doi: 10.1155/2012/189794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]