Abstract
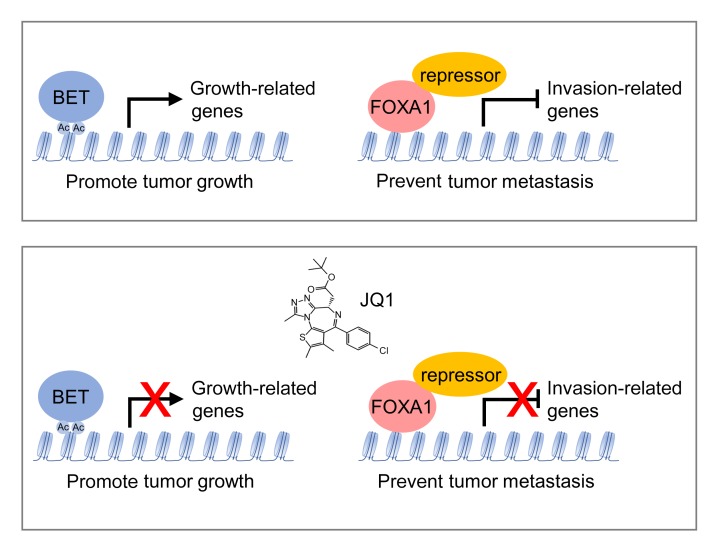
Recent findings have shown that inhibitors targeting bromodomain and extraterminal domain (BET) proteins, such as the small molecule JQ1, are potent growth inhibitors of many cancers and hold promise for cancer therapy. However, some reports have also revealed that JQ1 can activate additional oncogenic pathways and may affect epithelial-to-mesenchymal transition (EMT). Therefore, it is important to address the potential unexpected effect of JQ1 treatment, such as cell invasion and metastasis. Here, we showed that in prostate cancer, JQ1 inhibited cancer cell growth but promoted invasion and metastasis in a BET protein–independent manner. Multiple invasion pathways including EMT, bone morphogenetic protein (BMP) signaling, chemokine signaling, and focal adhesion were activated by JQ1 to promote invasion. Notably, JQ1 induced upregulation of invasion genes through inhibition of Forkhead box protein A1 (FOXA1), an invasion suppressor in prostate cancer. JQ1 directly interacted with FOXA1 and inactivated FOXA1 binding to its interacting repressors TLE3, HDAC7, and NFIC, thereby blocking FOXA1-repressive function and activating the invasion genes. Our findings indicate that JQ1 has an unexpected effect of promoting invasion in prostate cancer. Thus, the ill effect of JQ1 or its derived therapeutic agents cannot be ignored during cancer treatment, especially in FOXA1-related cancers.
Keywords: Oncology
Keywords: Cancer, Prostate cancer
Introduction
Prostate cancer is the most commonly diagnosed cancer in men and the second leading cause of cancer-related deaths in most Western countries. Given the crucial roles of the androgen receptor (AR) in prostate cancer progression, androgen deprivation therapy (ADT) by surgical or chemical castration remains the major treatment regimen (1). However, the response is transient, and most patients develop resistance to ADT and progress to castration-resistant prostate cancer (CRPC), with high mortality (2). AR antagonists such as bicalutamide and enzalutamide were then developed to treat patients with CRPC (3). Unfortunately, some unwanted effects have been associated with treatment using these AR antagonists, including increased cancer metastasis and neuroendocrine differentiation (4–6), eventually leading to treatment failure. Thus, identification of novel treatment approaches and therapeutic targets is imperative for the treatment of patients with prostate cancer.
Epigenetics-based therapies are increasingly promising for the treatment of cancer. Bromodomain and extraterminal domain (BET) proteins are a family of epigenetic regulators that include the 3 ubiquitously expressed bromodomain-containing proteins BRD2, BRD3, and BRD4 and the testis-specific BRDT. Through their 2 acetylated lysine-binding bromodomains and unique ET domain, BET proteins read acetylated histones and interact with histone modifiers as well as transcription factors to regulate gene transcription in different physiological conditions and diseases (7–9). Given their essential roles in oncogene transcription and their upregulation in multiple cancers, the targeting of BET proteins has been considered a novel cancer treatment strategy (10). Small-molecule inhibitors such as JQ1 that target BET proteins (I-BETs) were found to reversibly bind to bromodomains, disrupt the association of BET proteins with acetylated lysine in histones and transcription factors, repress oncogene expression, and eventually lead to cessation of cancer cell growth (8, 11, 12). In fact, I-BETs have emerged as an exciting new epigenetic therapeutic strategy for multiple cancers, especially advanced aggressive cancer types such as CRPC, triple-negative breast cancer (TNBC), and nuclear protein in testis (NUT) midline carcinoma (11–13). However, several recent reports suggested that JQ1 may also have unexpected effects. JQ1 affects both BET protein–dependent and –independent transcription regulation and regulates distinct pathways upon continued treatment (14). JQ1 also induces variable oncogenic pathway responses in ovarian cancer cells (15). BET proteins exhibit transcriptional and functional opposition in epithelial-to-mesenchymal transition (EMT) (16), raising the possibility that JQ1 may promote EMT and metastasis.
Here, we show that the BET protein inhibitor JQ1 suppressed prostate cancer cell proliferation but promoted invasion and metastasis independently of BET proteins. JQ1 directly interacted with Forkhead box protein A1 (FOXA1) and promoted invasion through inhibition of FOXA1 to activate multiple invasion pathways, including BMP signaling and EMT. Our study strongly indicates that more attention should be focused on the potential unexpected effects of JQ1 or JQ1-derived inhibitors. It also implies that combination treatment strategies could be used to overcome the potential metastasis-promoting effect of JQ1.
Results
JQ1 promotes prostate cancer cell invasion.
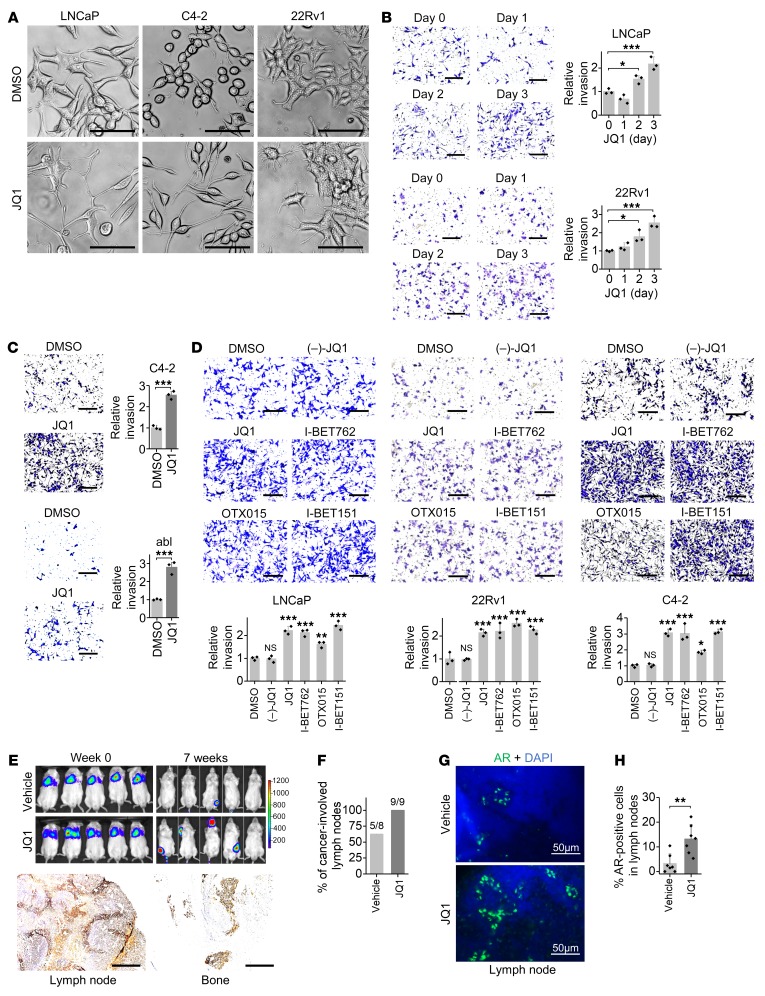
To examine the effect of JQ1 on prostate cancer cell growth and invasion, we first determined the functional concentration of JQ1. We found that JQ1 inhibited prostate cancer cell growth in a dose-dependent manner, with an IC50 of approximately 200 nM on LNCaP, C4-2, and 22Rv1 cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI126327DS1). Cells treated with 200 nM JQ1 showed significant growth inhibition (Supplemental Figure 1B). Interestingly, we observed obvious JQ1-induced changes in cell morphology. JQ1-treated cells appeared to be astrocyte like, with fusiform or fibroblastic phenotypes characteristic of mesenchymal cells with elongated and more leading-edge protrusions (Figure 1A). This morphological change raises the possibility that JQ1 treatment may enhance cell invasion. Indeed, we observed a time- and dose-dependent increase in cell invasion in multiple prostate cancer cells following treatment with JQ1 (Figure 1, B and C, and Supplemental Figure 1C). To further evaluate the effect of JQ1 on invasion, we examined additional JQ1-derived inhibitors. After assessing their functional concentration that could suppress c-Myc expression and reduce cell proliferation (Supplemental Figure 1, D and E), we found that all JQ1-derived inhibitors, including I-BET762, I-BET151, and OTX015, promoted cell invasion (Figure 1D). In accordance with increased protrusions and invasion, JQ1 significantly enhanced the formation of focal adhesions that are crucial for cell migration and invasion (Supplemental Figure 1F). Moreover, we measured JQ1-affected metastasis in mice that received a tail-vein injection of luciferase-labeled prostate cancer cells. JQ1 altered neither luciferase expression driven by a CMV promoter on a lentiviral vector in cells, nor luciferase activity (Supplemental Figure 1, G and H). Importantly, we found that JQ1 injection resulted in enhanced homing of luciferase-labeled cells into typical prostate cancer–metastasized organs such as lymph nodes and bone as well as other sporadic sites (Figure 1E and Supplemental Figure 1I), indicating that JQ1 promoted prostate cancer cell metastasis. Moreover, JQ1 treatment in this metastatic mouse model led to poor survival in comparison with the vehicle-treated control (Supplemental Figure 1J). We used a prostate cancer transgenic mouse model (probasin-Cre–driven Pten-null mice) to determine whether JQ1 increases metastasis in this mouse model. We measured prostate cancer cell metastasis to the draining lumbar lymph nodes through immunohistochemical detection of AR-positive prostatic cells in lymph nodes. We found that AR protein levels were not increased by JQ1 (Supplemental Figure 1K), analogous to previous reports (11, 17). However, the number of AR-positive cells in the draining lumbar lymph nodes was increased after JQ1 treatment (Figure 1, F–H), indicating enhanced prostate cancer cell metastasis to the lymph nodes. Taken together, these results indicate that JQ1 promotes invasion and metastasis of prostate cancer.
Figure 1. JQ1 promotes invasion of prostate cancer cells.
(A) Representative images of cell morphology 2 days after 200 nM JQ1 treatment. Scale bars: 50 μm. (B) Cell invasion was measured on the indicated days after 200 nM JQ1 treatment. Representative images of invasion are shown. Scale bars: 200 μm. n = 3 per group. *P < 0.05 and ***P < 0.001, by 1-way ANOVA. (C) Cell invasion was measured 3 days after 200 nM JQ1 treatment. Representative images of invasion are shown. Scale bars: 200 μm. n = 3 per group. ***P < 0.001, by Student’s t test. abl, LNCaP-abl cells. (D) Cell invasion was measured 3 days after treatment with 200 nM of the indicated inhibitor. Representative images of invasion are shown. Scale bars: 200 μm. n = 3 per group. P > 0.05 (NS), *P < 0.05, **P < 0.01, and ***P < 0.001 versus DMSO, by 1-way ANOVA. (E) 22Rv1-Luc cells were injected into SCID mice via the tail vein. JQ1 (10 mg/kg) was given daily by i.p. injection, and images were taken 7 weeks later. Metastatic sites with luciferase signal in different tissues were stained with an AR antibody. Representative images of AR staining are shown. Scale bars: 400 μm. (F) Probasin-Cre Pten-null mice of approximately 18 weeks of age were given 10 mg/kg JQ1 for 7 weeks. Draining lumbar lymph nodes were collected for AR immunohistochemical staining. The percentage of cancer-involved lymph nodes (AR staining–positive lymph nodes/total collected lymph nodes) is shown. (G) Representative AR staining in lymph nodes. Scale bars: 50 μm. (H) Quantitation of the percentage of AR-positive cells in lymph nodes. n = 8–9 per group. **P < 0.01, by Student’s t test.
JQ1 activates invasion pathways.
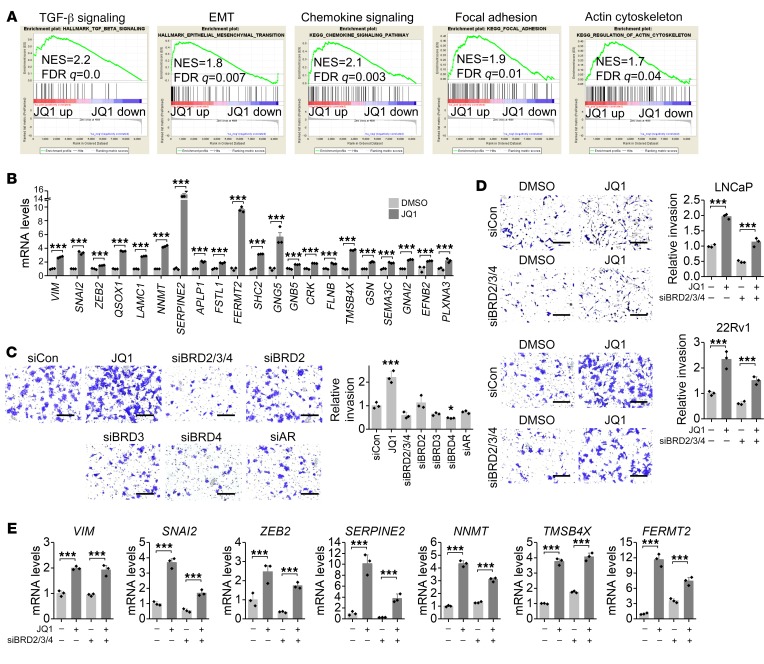
To dissect the molecular events of JQ1-promoted invasion, we performed transcriptome profiling by RNA-Seq of JQ1-treated cells (Supplemental Figure 2A). Gene Ontology (GO) analysis revealed that a JQ1-upregulated gene profile was associated with biological processes such as cell migration and endocytosis, whereas a downregulated gene profile was associated with the cell cycle and transcription (Supplemental Figure 2B). We performed gene set enrichment analysis (GSEA) to analyze hallmarks and pathways that were enriched upon treatment with JQ1 (Supplemental Figure 2, C and D). In the top enrichment list, we found that JQ1 activated multiple invasion pathways such as TGF-β family signaling, EMT, chemokine signaling, focal adhesion, and actin cytoskeleton regulation (Figure 2A). Many key invasion genes in these pathways were confirmed to be upregulated by JQ1 treatment (Figure 2B).
Figure 2. JQ1 activates invasion pathways.
(A) GSEA showed activated invasion pathways by JQ1. (B) JQ1 promoted the expression of invasion genes in the activated invasion pathways. n = 3 per group. ***P < 0.001, by Student’s t test. (C) Cell invasion was measured 3 days after siRNA transfection. Representative images of invasion are shown. Scale bars: 200 μm. n = 3 per group. *P < 0.05 and ***P < 0.001, versus control siRNA (siCon), by 1-way ANOVA. (D) JQ1 promoted invasion in the absence of BET proteins. Invasion was measured 3 days after JQ1 treatment and siRNA transfection. Representative images of invasion are shown. Scale bars: 200 μm. n = 3 per group. ***P < 0.001, by 2-way ANOVA. (E) JQ1-activated invasion genes in the absence of BET proteins. mRNA levels were measured 3 days after JQ1 treatment and siRNA transfection. n = 3 per group. ***P < 0.001, by 2-way ANOVA.
The typical role of JQ1 in prostate cancer is to block BET proteins and AR signaling. Interestingly, JQ1-promoted invasion was unrelated to blocking of BET proteins or the AR, because knockdown of single BET, all BET proteins, or the AR did not promote invasion (Figure 2C and Supplemental Figure 3A). In fact, when all BET proteins were suppressed, we detected a decrease in cell invasion ability, yet JQ1 still promoted invasion in the absence of all BET proteins (Figure 2D). Likewise, we found that knockdown of all BET proteins did not phenocopy JQ1 in activation of invasion and EMT genes, whereas JQ1 activated most invasion and EMT genes in the absence of all BET proteins (Figure 2E and Supplemental Figure 3B). Consistently, we observed that knockdown of BRD4, the key BET protein, regulated distinct pathways compared with JQ1 treatment (Supplemental Figure 4A), and JQ1 still activated similar invasion pathways including TGF-β family signaling and EMT, regardless of BRD4 expression levels (Supplemental Figure 4B). Taken together, these results suggest that JQ1 promotes prostate cancer invasion in a BET protein–independent manner.
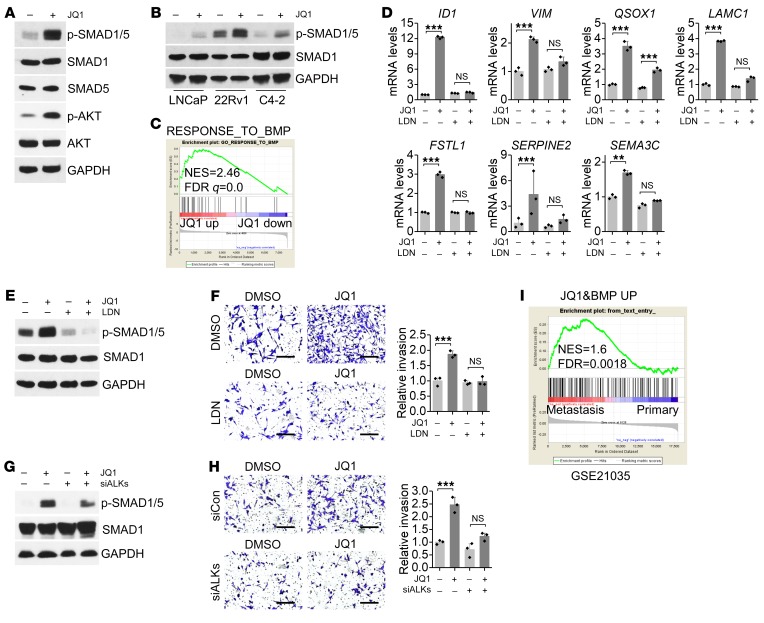
To determine whether JQ1-activated invasion pathways contribute to enhanced invasion, we investigated TGF-β family signaling, as it plays crucial roles in promoting EMT and cancer cell invasion. We found that SMAD1 and SMAD5 (SMAD1/5) phosphorylation was significantly increased by JQ1 treatment (Figure 3A), suggesting that TGF-β family member BMP signaling was activated. Similar results showing that JQ1 promoted SMAD1/5 phosphorylation were obtained in multiple prostate cancer cells (Figure 3B). Furthermore, we observed a time-dependent increase in phosphorylated SMAD1/5 (p-SMAD1/5) as well as p-AKT, a kinase that can be activated by BMP, following treatment with JQ1 (Figure 3A and Supplemental Figure 5A). In addition, GSEA analysis showed that the response to the BMP (GO: 0071772) signature was positively enriched in JQ1-treated cells (Figure 3C). Moreover, we found that multiple JQ1-derived inhibitors activated BMP signaling and promoted expression of the mesenchymal marker vimentin (VIM) (Supplemental Figure 5, B and C), which is consistent with the role of BMP signaling in promoting EMT in cancer cells.
Figure 3. JQ1 activates BMP signaling.
(A) Protein levels were measured in JQ1-treated LNCaP cells. (B) p-SMAD1/5 levels were measured in JQ1-treated cells. (C) GSEA showed that the response to the BMP signature was activated by JQ1. (D) The BMP signaling inhibitor LDN-212854 (LDN) impaired JQ1-induced upregulation of invasion genes. n = 3 per group. P > 0.05 (NS), **P < 0.01, and ***P < 0.001, by 2-way ANOVA. (E) Levels of the indicated proteins after LDN-212854 and JQ1 treatment. (F) Cells were treated with 200 nM JQ1 and 2 μM LDN-212854 for 3 days, and invasion was measured. Representative images of invasion are shown. Scale bars: 200 μm. n = 3 per group. P > 0.05 (NS), ***P < 0.001, by 2-way ANOVA. (G) Levels of the indicated proteins after transfection with siALKs (siRNAs targeting ALK1, ALK2, and ALK3) and JQ1 treatment. (H) Cells were transfected with siALKs and treated with JQ1 for 3 days, and then cell invasion was measured. Representative images of invasion are shown. Scale bars: 200 μm. n = 3 per group. P > 0.05 (NS), ***P < 0.001, by 2-way ANOVA. (I) GSEA showed that the JQ1-activated BMP target gene signature was enriched in human metastatic prostate cancer tissues (GSE21035). A JQ1-activated BMP target gene signature was generated through a combination of JQ1-upregulated genes and BMP positively regulated genes (GSE96914). down, downregulated; up, upregulated.
We further investigated the role of BMP signaling in JQ1-promoted invasion. Through the BMP signaling inhibitor LDN-212854, we found that blocking BMP signaling significantly impaired the upregulation of JQ1-activated invasion genes and the EMT marker VIM (Figure 3D), suggesting that BMP signaling might play an important role in JQ1-promoted invasion. As expected, blocking BMP signaling, either through LDN-212854 or a siRNA against the BMP receptors ALK1, ALK2, and ALK3, significantly impaired JQ1-promoted invasion (Figure 3, E–H, and Supplemental Figure 5D). Next, we asked whether the JQ1-induced BMP signaling detected in cultured cells has any relevance to prostate cancer in humans. Notably, we identified 173 JQ1-induced BMP target genes whose signature was enriched in metastatic prostate cancer tissues as compared with primary cancer tissues (Figure 3I), supporting the idea that BMP signaling plays a role in prostate cancer metastasis. Taken together, our results indicate that JQ1-activated BMP signaling plays an important role in JQ1-promoted invasion.
JQ1 inhibits FOXA1 to promote prostate cancer cell invasion.
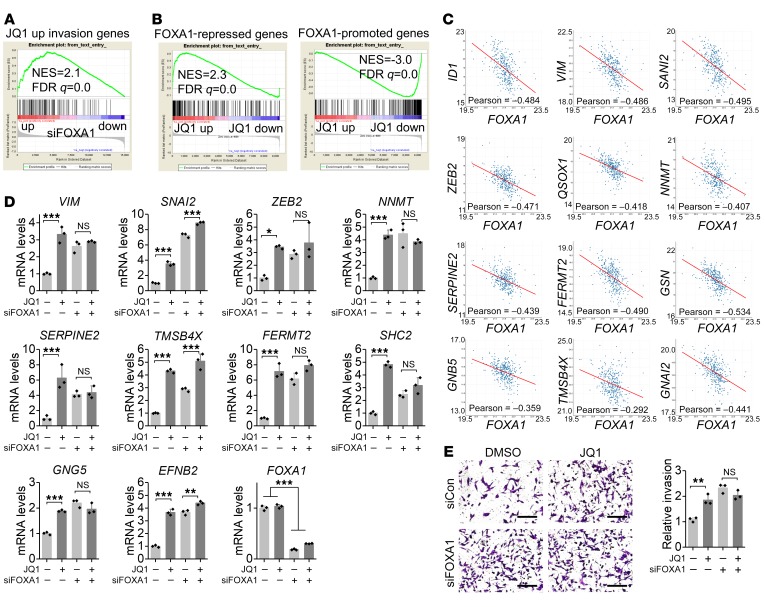
To determine how JQ1 activates invasion pathways and genes, we searched for transcription factors that have binding sites on the promoter of JQ1-activated invasion genes using the oPOSSUM program (http://opossum.cisreg.ca/oPOSSUM3/) (18). We predicted a list of transcription factors that have potential binding sites on the promoters of 114 JQ1-activated invasion genes (Supplemental Figure 6A). Taking into account the expression levels of these transcription factors in cells (Supplemental Figure 6B), GSEA highlighted FOXA1 as a promising candidate. Subsequently, we found that JQ1-activated invasion genes were negatively regulated by FOXA1 (Figure 4A). Importantly, in addition to these invasion genes, GSEA showed that the regulated gene profile of JQ1 was also negatively regulated by FOXA1 (Figure 4B), indicating that JQ1 indeed inhibits FOXA1 activity. Interestingly, we found that many BMP ligands and receptors that contribute to BMP signaling activation were upregulated by JQ1 (Supplemental Figure 6C). FOXA1 also ranked as the top candidate among transcription factors responsible for JQ1 activation of BMP ligands and receptors (Supplemental Figure 6, D and E). Furthermore, we found that FOXA1 indeed negatively regulated BMP ligands and receptors as well as the BMP signaling marker inhibitor of DNA binding 1 (ID1) (Supplemental Figure 6F), further supporting the idea that JQ1 inhibits FOXA1 in prostate cancer cells.
Figure 4. JQ1 represses FOXA1 to promote invasion.
(A) GSEA showed that JQ1-activated invasion genes were repressed by FOXA1 (GSE58309). (B) GSEA showed that the FOXA1 signature was negatively enriched following JQ1 treatment. The FOXA1 gene signature was generated by GSE58309. (C) FOXA1 was negatively correlated with JQ1-activated invasion genes in patients’ prostate cancer tissues from the The Cancer Genome Atlas (TCGA) data set. (D) FOXA1 knockdown impaired JQ1-induced upregulation of invasion genes. mRNA levels were measured 3 days after JQ1 treatment and siRNA transfection. n = 3 per group. P > 0.05 (NS), *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-way ANOVA. (E) FOXA1 knockdown promoted cell invasion, which was measured 3 days after JQ1 treatment and siRNA transfection. Representative images of invasion are shown. Scale bars: 200 μm. n = 3 per group. P > 0.05 (NS), **P < 0.01, by 2-way ANOVA.
In addition, we compared JQ1-induced reduction of FOXA1 activity and BET protein activity. Activity was indicated by the expression of specific target genes regulated by FOXA1 or BRD2, BRD3, or BRD4 (Supplemental Figure 7A). We found that the dosages of JQ1 for inhibition of FOXA1- and BRD2/3/4-specific target gene expression were similar (Supplemental Figure 7B), reinforcing the notion that JQ1 inhibits FOXA1 as well as BET activity. Moreover, JQ1 inhibited FOXA1 activity and promoted invasion in a similar time-dependent manner (Supplemental Figure 7C and Figure 1B). Consistent with the repressive role of FOXA1 in JQ1-activated invasion genes, we found a negative correlation between FOXA1 and JQ1-activated invasion genes in human prostate cancer tissues (Figure 4C). Moreover, in the absence of FOXA1, genes important for invasion were upregulated, and JQ1 failed to further activate the expression of these invasion genes in most cases (Figure 4D), reinforcing the idea that JQ1 inhibits FOXA1 activity to activate invasion genes. Likewise, we found that FOXA1 knockdown promoted invasion and that JQ1 failed to further promote invasion when FOXA1 was repressed (Figure 4E). Together, these results indicate that JQ1 inhibits FOXA1 to activate invasion genes and promote invasion.
JQ1 interacts with FOXA1 and inhibits binding of FOXA1 to repressors.
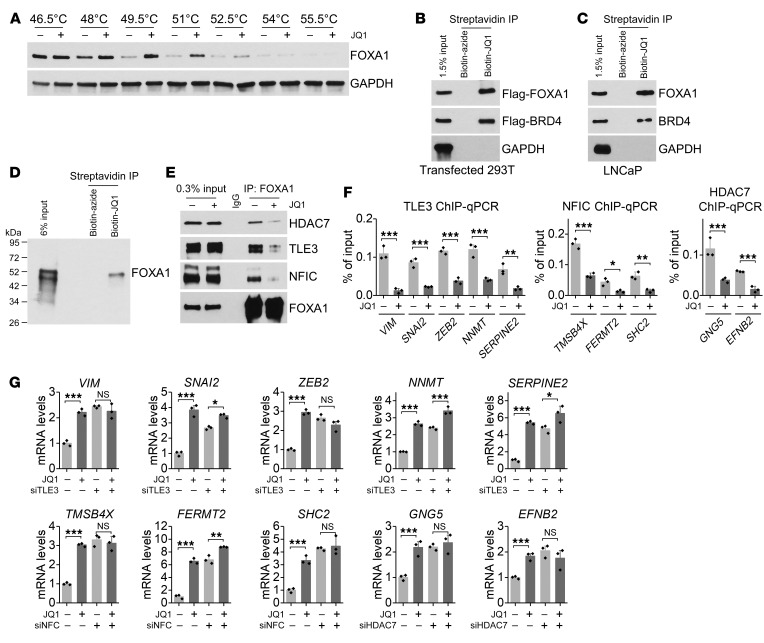
Subsequently, we investigated how JQ1 inhibits FOXA1 to activate invasion genes. FOXA1 protein levels were largely unaffected by JQ1 treatment (Supplemental Figure 8A). Therefore, we speculated that JQ1 might interact with FOXA1 to block FOXA1 invasion suppressor function. The result of cellular thermal shift assays (CETSAs) supported the observation that JQ1 bound to FOXA1 and led to significant thermal stabilization of FOXA1 (Figure 5A). Indeed, through biotinylated JQ1 pull-down assays, we found that JQ1 interacted with both overexpressed Flag-tagged FOXA1 and endogenous FOXA1 in cell lysates as well as with purified FOXA1 protein (Figure 5, B–D). These results indicate that JQ1 interacts directly with FOXA1.
Figure 5. JQ1 interacts with FOXA1 to block associated repressor activity.
(A) A CETSA was performed using LNCaP cells. (B) Biotin-JQ1 pull-down assay using lysates of 293T cells that overexpressed Flag-FOXA1 and Flag-BRD4. Biotin-azide was used as a control. (C) Biotin-JQ1 pull-down assay using LNCaP cell lysates. (D) Biotin-JQ1 pull-down assay using FOXA1 recombinant protein. (E) LNCaP cells were treated with JQ1 for 8 hours. IP was performed to measure FOXA1 binding to the indicated proteins. (F) ChIP-qPCR assay of FOXA1-interacting repressors at FOXA1-binding sites on the promoter of JQ1-activated invasion genes. n = 3 per group. *P < 0.05, **P < 0.01, and ***P < 0.001, by Student’s t test. (G) FOXA1-interacting repressors regulated JQ1-activated invasion genes. n = 3 per group. P > 0.05 (NS), *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-way ANOVA.
Next, we performed ChIP assays to determine whether binding of JQ1 affects the binding of FOXA1 to its target genes. As shown in Supplemental Figure 8B, we did not observe an apparent reduction of FOXA1 binding to the promoter of its target invasion genes upon JQ1 treatment. Since FOXA1 is known to recruit corepressors and coactivators to regulate gene expression, we speculated that through interaction with FOXA1, JQ1 might disrupt FOXA1 binding to repressors, which would lead to reexpression of FOXA1-repressed invasion genes and promotion of cellular invasion ability. Among FOXA1-interacting proteins, TLE family member 3 (TLE3) (19, 20), histone deacetylase 7 (HDAC7) (21), and nuclear factor I C (NFIC) (22) have been shown to have a FOXA1 corepressor function and are implicated in the regulation of cell invasion. We found that these repressors were involved in the regulation of invasion genes that were repressed by FOXA1 (Supplemental Figure 8, C and D). Knockdown of these repressors promoted expression of FOXA1-repressed invasion genes, whereas silencing of FOXA1 failed to further induce the upregulation of invasion genes in the absence of these repressors (Supplemental Figure 8E), indicating that TLE3, HDAC7, and NFIC play a role in FOXA1-induced repression of these invasion genes. Importantly, JQ1 inhibited FOXA1 binding to the repressors, such as TLE3, HDAC7, and NFIC (Figure 5E), resulting in a reduction in recruitment of these repressors to FOXA1-binding sites on the promoters of the invasion genes (Figure 5F) as assayed by ChIP–quantitative PCR (ChIP-qPCR). Moreover, in the absence of TLE3, HDAC7, and NFIC, JQ1 failed to further increase the expression of invasion genes (Figure 5G and Supplemental Figure 9A). Our results suggest that JQ1 interacts with FOXA1 to inhibit FOXA1 binding to corepressors, thus allowing the expression of FOXA1-repressed genes that are important for invasion and metastasis. In addition, not only was JQ1 capable of reducing binding of repressors to FOXA1, it could also reduce the protein levels of TLE3, HDAC7, and NFIC in multiple prostate cancer cell lines (Supplemental Figure 9B). JQ1 might reduce protein levels through regulation of their protein stability (Supplemental Figure 9, C and D). Taken together, our results indicate that JQ1 inhibits the repressive activity of FOXA1 by interacting with FOXA1 to disrupt its binding to corepressors, thus allowing for the reexpression of genes important for invasion.
Discussion
Recently, inhibitors such as JQ1 that target BET proteins have been shown to be promising for the treatment of many types of cancer (10, 12). In response to JQ1 treatment, c-Myc expression and AR activity are reduced in prostate cancer, leading to growth inhibition (11). However, it has also been shown that JQ1 may have unexpected effects on cancer metastasis, which is lethal for patients with cancer (15, 16). In this study, we show that JQ1 interacted with FOXA and inhibited the repressor function of FOXA1 to enhance the expression of genes important for invasion, thereby promoting prostate cancer metastasis.
JQ1 has been shown to bind to the bromodomains of BET proteins with a KD of approximately 50–190 nM and to inhibit binding of acetylated histone H4 peptide to BRD4 with IC50 values of 77 and 33 nM for the 2 bromodomains (12). It has also been reported that potent biological effects of JQ1 are observed at 50–100 nM (23), and many cancer cells, including prostate cancer cells, respond to JQ1 with IC50 values below 300 nM (11, 13). However, some cancer cells were treated with a much higher dose of JQ1 in some of these reports. Here, we chose 200 nM JQ1 for prostate cancer cell treatment to minimize off-target toxicity. We also administered JQ1 for a longer period — up to 3 days — to mimic prolonged clinical treatment. JQ1 at 200 nM was effective and sufficient to inhibit cell proliferation and reduce c-Myc expression. We found that invasion of multiple prostate cancer cell lines was promoted by JQ1 as well as its derived inhibitors. In addition, we showed that knockdown of BET proteins or the AR did not promote invasion, indicating that JQ1 may promote invasion through other mechanisms. Consistent with our observation, it has been shown that JQ1 induces variable oncogenic pathways independent of the role of BET protein in transcriptional regulation (14, 15).
Cancer metastasis promotion appears to be a concern with drugs that inhibit cancer growth. For example, chemotherapy has been shown to induce cancer metastasis through a tumor metastasis–receptive microenvironment (24, 25). Targeted therapy such as epigenetic HDAC inhibitors has also been found to promote EMT and metastasis in multiple cancers (26). In addition, the prostate cancer–specific drug enzalutamide has been revealed to induce EMT and promote metastasis (27). Through GSEA analysis of the JQ1-regulated gene profile, we found that JQ1 activated multiple invasion pathways in prostate cancer, including EMT, TGF-β family signaling, chemokine signaling, focal adhesion, and actin cytoskeleton regulation. EMT, which plays a crucial role in drug-induced metastasis, is a cellular process defined by the loss of epithelial characteristics of tight cell-cell adhesion and apico-basal polarization and the gain of mesenchymal characteristics of motility and invasion. EMT renders cancer cells more migratory and resistant to drug treatment, eventually leading to enhanced cancer metastasis (28). In this study, we showed that administration of JQ1 to multiple prostate cancer cell lines resulted in EMT, in which BMP signaling was activated. BMPs, which are members of the TGF-β family, are key inducers of EMT that contribute to metastasis of multiple cancer types, including prostate cancer (29, 30). Interestingly, we showed that treatment with the BMP signaling inhibitor LDN-212854 markedly impaired JQ1-induced cell invasion and expression of some EMT pathway genes. Taking into account the possibility that EMT contributes to the JQ1 resistance that was observed in multiple cancer cells (13, 31, 32), it is foreseeable that combination treatment with BMP inhibitors might be applicable for future cancer treatment.
JQ1-promoted invasion genes have potential binding sites for Forkhead box proteins. Among them, we identified FOXA1, a winged-helix transcription factor, as the top predicted transcription factor that is highly expressed in prostate cancer. Moreover, FOXA1 is essential for prostate organogenesis and plays important roles in prostate cancer development (33, 34). Validation results showed that FOXA1 was responsible for inhibiting the expression of JQ1-induced invasion genes. In addition, we found that many JQ1-activated BMP ligand and receptor genes were also repressed by FOXA1. Moreover, the FOXA1 signature was negatively enriched in the JQ1-regulated gene profile, supporting the idea that JQ1 represses FOXA1 activity. Although JQ1 is a BET protein inhibitor, our data clearly showed that it did not work through BET proteins to exert its effect on invasion and metastasis. Instead, it interacted directly with FOXA1 and hindered the ability of FOXA1 to repress the expression of genes critical for invasion and metastasis.
FOXA1 is regarded as a pioneer factor that binds to condensed chromatin and opens chromatin to facilitate subsequent recruitment of other transcription factors and regulators (35, 36). FOXA1 plays important roles in prostate cancer, because FOXA1 recruits the AR to regulate genes crucial for prostate cancer cell growth (35, 37). Therefore, blocking FOXA1 reduces AR transcription activity to inhibit prostate cancer cell growth. Furthermore, FOXA1 is a maintenance factor for the epithelial cell phenotype and exhibits inhibitory activity on EMT and cancer metastasis in prostate, breast, and pancreatic cancer (38–41). FOXA1 represses the transcription of genes in cell motility, EMT, and neuroendocrine differentiation in prostate cancer though an AR-independent mechanism (42, 43). Consistent with increasing evidence that FOXA1 promotes prostate cancer cell proliferation but prevents metastasis, our findings indicate that through inhibition of FOXA1, JQ1 blocks prostate cancer proliferation but enhances metastasis.
A variety of FOXA1-interacting partners have been shown to confer AR-dependent and -independent roles of FOXA1 in prostate cancer, which may explain the complexity of FOXA1-regulated genes in different cells. The role of FOXA1 in transcriptional repression is likely associated with its binding to repressors or modulators. We found that there was reduced binding of FOXA1-interacting corepressors, including TLE3, NFIC, and HDAC7, to FOXA1. TLE3 is a repressor that was found to associate with WNT/β-catenin–driven EMT (44). FOXA1 was shown to recruit TLE3 to specific genomic target sites to elicit transcriptional repression. NFIC, which is involved in EMT regulation (45), was found to interact with FOXA1 to regulate the expression of prostate-specific genes (22). FOXA1 also interacts with HDAC7, which regulates NCOR-SMRT corepressor complex recruitment (21). We have shown that these repressors contributed to the ability of FOXA1 to suppress the expression of invasion genes. Our studies further provide mechanistic insights into how FOXA1 represses invasion and how JQ1 activates invasion through the inhibition of FOXA1. Our results have shown, for the first time to our knowledge, that JQ1 interacts with FOXA1 and disrupts the ability of FOXA1 to bind to repressors, hampering its repressor activity and allowing the expression of FOXA1-repressed invasion genes. We further showed that JQ1 reduced the stability of TLE3, HDAC7, and NFIC in prostate cancer cells. The destabilization of the repressors by JQ1 could either be due to disturbance of their interactions with FOXA1, which stabilize them in the cellular environment, or through other mechanisms.
Taken together, our results indicate that JQ1 inactivates FOXA1-repressive activity, through direct binding to FOXA1 and disrupting interactions between FOXA1 and its corepressors, to promote prostate cancer invasion. This finding reveals that perturbation of FOXA1 activity by JQ1 may introduce an unexpected effect of JQ1 on cancer, especially in FOXA1-related cancers like prostate, breast, and pancreatic cancer. It raises the possibility that combination treatment strategies, such as those involving BMP inhibition, could be used to overcome the potential metastasis-promoting effect of JQ1.
Methods
Cell lines, culturing conditions, and transfection.
LNCaP, 22Rv1, C4-2, PC3, and 293T cells were purchased from the American Type Culture Collection (ATCC) and maintained at the Tissue and Cell Culture Core Facility at Baylor College of Medicine. Cells were cultured in RPMI 1640 medium (11875093, Thermo Fisher Scientific) supplemented with 10% FBS (F2442, Sigma-Aldrich). The LNCaP-abl cell line was obtained from Zoran Culig (Medical University of Innsbruck, Innsbruck, Austria) and maintained in RPMI 1640 medium supplemented with 10% charcoal-stripped FBS (F6765, Sigma-Aldrich). The authenticity of all cell lines was verified in the previous 6 months. Cell transfection was performed using Lipofectamine 2000 (11668019, Thermo Fisher Scientific) for vectors and Lipofectamine RNAiMAX Reagent (13778075, Thermo Fisher Scientific) for siRNAs, according to the manufacturer’s instructions.
Chemicals.
(-)-JQ1 (1268524-71-5, Cayman Chemical), JQ1 (SML0974, Sigma-Aldrich), I-BET762 (SML1272, Sigma-Aldrich), OTX015 (202590-98-5, Cayman Chemical), I-BET151 (SML0666, Sigma-Aldrich), and LDN-212854 (SML0965, Sigma-Aldrich) were used.
Cell proliferation assay.
Cells were seeded in 96-well plates at 1000–3000 cells per well. JQ1 was added the following day. After 96 hours of incubation, cell viability was assessed by CellTiter-Glo Assay (G9241, Promega). Cell proliferation was measured by CellTiter 96 AQueous One Solution Cell Proliferation Assay (G3582, Promega). The value was measured at 490 nm using a Multiskan FC Microplate Photometer (Thermo Fisher Scientific) and then normalized.
Transwell invasion assay.
Cells were treated with JQ1 for 72 hours. LNCaP (2 × 105), LNCaP-abl (2 × 105), 22Rv1 (0.5 × 105), and C4-2 (1 × 105) cells were seeded with serum-free medium in Transwell chambers precoated with Matrigel (354483, BD Biosciences). Medium with 10% FBS was added to the lower chamber. JQ1 was added to both upper and lower chambers. After 24 hours (22Rv1) or 48 hours (LNCaP, LNCaP-abl, and C4-2), the cells were fixed with methanol. Noninvading cells were gently removed, and invaded cells on the lower side of the chamber were stained with crystal violet, photographed, and counted.
Focal adhesion measurement.
Cells were treated with JQ1 for 72 hours and then seeded on a fibronectin-coated plate for 6 hours. An immunofluorescence assay using a paxillin antibody (ab32084, Abcam) was performed to detect focal adhesion. Cells were fixed with 4% PFA with 0.1% Triton X-100 for 10 minutes and then washed and blocked with PBS with 1% BSA and 0.2% Triton X-100. Cells were incubated with a primary antibody (1:100 dilution in PBS with 1% BSA and 0.2% Triton X-100) overnight at 4°C and were further incubated with a secondary antibody (A11002, Thermo Fisher Scientific, 1:1000 dilution) at room temperature for 1 hour. Focal adhesion numbers per cell were calculated using ImageJ software (NIH).
siRNA transfection.
Cells were transfected with siRNA for 72 hours. The siRNA targeting sequences are shown in Supplemental Table 1.
RNA-Seq and GSEA analysis.
LNCaP cells were treated with 200 nM JQ1 for 72 hours, and total RNA was extracted for RNA-Seq analysis. The sequencing was done by Q2 Solutions. The sequencing data were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE139230). Upregulated and downregulated genes were generated by filtering, with a log2 fold-change cutoff of 0.6. GSEA was performed using the GSEA Java program (https://www.gsea-msigdb.org/gsea/index.jsp). Normalized enrichment score (NES) and FDR q values are shown in the figures.
RNA isolation and qPCR.
Total RNA was extracted from cells using TRIzol Reagent (15596018, Thermo Fisher Scientific). cDNA was synthesized using a Maxima First Strand cDNA Synthesis Kit (FERK1641, Thermo Fisher Scientific). Real-time PCR was performed with FastStart Universal SYBR Green Master Mix (4913850001, Roche) on a StepOnePlus Real-Time PCR system (Applied Biosystems). Relative mRNA levels were normalized to ACTB. All primers were synthesized by Sigma-Aldrich, and the primer sequences are listed in Supplemental Table 2.
Western blot assay.
Total proteins were extracted from cells following the standard protocol. The protein concentration was measured using the BCA Protein Assay Kit (23228, Thermo Fisher Scientific). Protein samples were separated by SDS-PAGE and transferred onto a nitrocellulose membrane (1620112, Bio-Rad). The membrane was incubated for 30 minutes in blocking buffer (TBST with 5% nonfat dry milk), followed by overnight incubation at 4°C with the primary antibody. Membrane was then incubated with HRP-conjugated secondary antibodies (7074 or 7076, Cell Signaling Technology) for 1 hour. The signals were visualized with SuperSignal West Dura Extended Duration Substrate (34075, Thermo Fisher Scientific). The primary antibodies used in this study were as follows: FOXA1 (ab23738, Abcam), GAPDH (SC-25778 HRP, Santa Cruz Biotechnology), c-Myc (5605, Cell Signaling Technology), SMAD1 (6944, Cell Signaling Technology), SMAD5 (9517, Cell Signaling Technology), p-SMAD1/5 (9516, Cell Signaling Technology), p-AKT (4060, Cell Signaling Technology), AKT (9272, Cell Signaling Technology), TLE3 (11372-1-AP, Proteintech), HDAC7 (A301-384A-T, Bethyl), NFIC (A303-123A-T, Bethyl), luciferase (ab21176, Abcam), and AR (5153, Cell Signaling Technology). See complete unedited blots in the supplemental material.
CETSA.
The CETSA assay was performed using the standard protocol (46). LNCaP cells were treated with 10 μM JQ1 for 1 hour. Cells were suspended in PBS with protease inhibitors, heated at the indicated temperature for 3 minutes, and immediately snap-frozen. Samples were subjected to 2 freeze-thaw cycles and centrifuged. Supernatants were collected and Western blot assays were performed.
Pull-down assay.
Cells were lysed in IP buffer (20 mM Tris, pH7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, protease inhibitor). Biotinylated JQ1 (biotin-JQ1) was generated through a Cu(I)-catalyzed azide alkyne click chemistry reaction (CuAAC) (47). Briefly, 100 μM JQ1-PA (6589, Tocris), 200 μM biotin-azide (762024, Sigma-Aldrich), 4 mM CuSO4, and 5 mM (+)-sodium l-ascorbate (11140, Sigma-Aldrich) were incubated for 1 hour, followed by a 1-hour incubation with streptavidin-coupled Dynabeads (65601, Thermo Fisher Scientific). Biotin-JQ1– or control compound–loaded (biotin-azide–loaded) beads were washed and incubated with cell lysate or purified FOXA1 protein (ab98301, Abcam) for 4 hours at 4°C. Beads were washed in IP buffer, resuspended in loading buffer, and boiled at 95°C for 5 minutes for separation of the protein and beads. Samples were then analyzed by Western blotting.
IP.
Cells were treated with 10 μM JQ1 for 8 hours and harvested. Lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, and 0.5 mM DTT with protease inhibitor) was used to isolate cell nuclei. Cell nuclei were lysed in IP buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, and protease inhibitor). After preclearance with protein G Dynabeads (10003D, Thermo Fisher Scientific) for 1 hour, lysates were incubated with protein G Dynabeads preloaded with FOXA1 antibody (A305-249A, Bethyl) overnight at 4°C. Beads were washed 3 times in IP buffer and resuspended in loading buffer and then boiled at 95°C for 5 minutes to separate the protein and beads. Samples were then analyzed by Western blotting.
Mouse metastasis assay.
Eight-week-old male SCID mice (NOD-CB17 PrkdcSCID/J, stock no. 001303) were purchased from The Jackson Laboratory. 22Rv1-luciferase and C4-2–luciferase cells were generated by lentivirus-induced luciferase overexpression from pCDH-CMV-Luc2-T2A-tGFP-EF1-hygro. 22Rv1-luciferase cells (1.5 × 106) or C4-2–luciferase cells (4 × 106) were injected into mice through the tail vein. The following day, mice were treated daily with 10 mg/kg JQ1 i.p. After 7 weeks, bioluminescence was measured using an in vivo imaging system (IVIS) system after i.p. injection of luciferin (LUCK-1G, Gold Biotechnology).
Mice of prostate cancer mouse models (probasin-Cre–driven Pten-null mice, Pb-Cre Ptenfl/fl) that were fully backcrossed onto a C57/BL6 background were maintained in our laboratory as previously described (48). Male mice of approximately 18 weeks of age were treated daily with 10 mg/kg JQ1 i.p. for 7 weeks. Draining lumbar lymph nodes of mice were collected for immunohistochemical assay to measure AR-positive cells.
Immunohistochemical assay.
Immunohistochemistry was performed as described previously (49). A primary AR antibody (ab133273, Abcam, 1:500 dilution) was incubated overnight at 4°C, and a secondary antibody (711-066-152, Jackson ImmunoResearch, 1:500 dilution) was incubated for 1 hour at room temperature.
ChIP-qPCR assays.
ChIP assays were performed using a Magna ChIP A/G Kit from Sigma-Aldrich according to the manufacturer’s protocol. FOXA1 (ab23738, Abcam), TLE3 (11372-1-AP, Proteintech), NFIC (A303-123A-T, Bethyl), HDAC7 (PA5-29937, Thermo Fisher Scientific), and corresponding control IgG antibodies were used. The qPCR assays were carried out on the chromatin samples as prepared above. The primer sequences are listed in Supplemental Table 3.
Statistics.
The statistical analyses were performed using Origin 2017 software (OriginLab), and data are presented as dot plots, with the mean ± SD indicated. An unpaired, 2 tailed Student’s t test was used for 2-group comparisons. A 1-way ANOVA was used for comparisons among multiple groups with 1 independent variable followed by a post hoc Tukey’s multiple comparisons test. A 2-way ANOVA was used for comparisons among multiple groups with 2 independent variables followed by Sidak’s multiple comparisons test. Kaplan-Meier survival curves were analyzed using a log-rank test. The difference was regarded as significant when the P value was less than 0.05. All experiments were repeated 2–4 times.
Study approval.
All animal experimental procedures were reviewed and approved by the IACUC of Baylor College of Medicine, and all experiments were performed in compliance with the institutional guidelines of Baylor College of Medicine.
Author contributions
LW, SYT, and MJT conceptualized and designed the study. LW performed and analyzed the experiments. LW and MX completed cell assays and co-IP studies. CYK assisted with in vivo studies. LW, SYT, and MJT wrote, discussed, and edited the manuscript.
Supplementary Material
Acknowledgments
We thank Li-Yuan Yu-Lee and Jodie R. Hebert for editorial assistance. We appreciate the technical support of Pei Li and Guannu Xu. We also thank the Tissue Culture Core of the Molecular and Cellular Biology Department, the Cell-Based Assay Screening Service Core, and the Genetically Engineered Mouse Core for their assistance. This work was supported by grants from the Cancer Prevention and Research Institute of Texas (CPRIT) (RP150648) and the NIH (HL114539).
Version 1. 12/24/2019
In-Press Preview
Version 2. 03/03/2020
Electronic publication
Version 3. 04/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(4):1782–1792.https://doi.org/10.1172/JCI126327.
Contributor Information
Leiming Wang, Email: leimingw@bcm.edu.
Mafei Xu, Email: mafeix@bcm.edu.
Chung-Yang Kao, Email: Chung-Yang.Kao@bcm.edu.
Sophia Y. Tsai, Email: stsai@bcm.edu.
Ming-Jer Tsai, Email: mtsai@bcm.edu.
References
- 1.Nevedomskaya E, Baumgart SJ, Haendler B. Recent advances in prostate cancer treatment and drug discovery. Int J Mol Sci. 2018;19(5):E1359. doi: 10.3390/ijms19051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32(49):5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rathkopf D, Scher HI. Androgen receptor antagonists in castration-resistant prostate cancer. Cancer J. 2013;19(1):43–49. doi: 10.1097/PPO.0b013e318282635a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin TH, Izumi K, Lee SO, Lin WJ, Yeh S, Chang C. Anti-androgen receptor ASC-J9 versus anti-androgens MDV3100 (Enzalutamide) or Casodex (Bicalutamide) leads to opposite effects on prostate cancer metastasis via differential modulation of macrophage infiltration and STAT3-CCL2 signaling. Cell Death Dis. 2013;4:e764. doi: 10.1038/cddis.2013.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin TH, et al. Differential androgen deprivation therapies with anti-androgens casodex/bicalutamide or MDV3100/enzalutamide versus anti-androgen receptor ASC-J9(R) Lead to promotion versus suppression of prostate cancer metastasis. J Biol Chem. 2013;288(27):19359–19369. doi: 10.1074/jbc.M113.477216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, et al. Blocking the feedback loop between neuroendocrine differentiation and macrophages improves the therapeutic effects of enzalutamide (MDV3100) on prostate cancer. Clin Cancer Res. 2018;24(3):708–723. doi: 10.1158/1078-0432.CCR-17-2446. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 8.Shi J, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25(2):210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155(7):1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stathis A, Bertoni F. BET Proteins as targets for anticancer treatment. Cancer Discov. 2018;8(1):24–36. doi: 10.1158/2159-8290.CD-17-0605. [DOI] [PubMed] [Google Scholar]
- 11.Asangani IA, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu S, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529(7586):413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handoko L, et al. JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states. Epigenetics. 2018;13(4):410–431. doi: 10.1080/15592294.2018.1469891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurimchak AM, et al. Resistance to BET bromodomain inhibitors is mediated by kinome reprogramming in ovarian cancer. Cell Rep. 2016;16(5):1273–1286. doi: 10.1016/j.celrep.2016.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrieu GP, Denis GV. BET proteins exhibit transcriptional and functional opposition in the epithelial-to-mesenchymal transition. Mol Cancer Res. 2018;16(4):580–586. doi: 10.1158/1541-7786.MCR-17-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan SC, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015;43(12):5880–5897. doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2012;2(9):987–1002. doi: 10.1534/g3.112.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28(2):291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jangal M, Couture JP, Bianco S, Magnani L, Mohammed H, Gévry N. The transcriptional co-repressor TLE3 suppresses basal signaling on a subset of estrogen receptor α target genes. Nucleic Acids Res. 2014;42(18):11339–11348. doi: 10.1093/nar/gku791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik S, et al. Histone deacetylase 7 and FoxA1 in estrogen-mediated repression of RPRM. Mol Cell Biol. 2010;30(2):399–412. doi: 10.1128/MCB.00907-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabowska MM, et al. NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression. Mol Endocrinol. 2014;28(6):949–964. doi: 10.1210/me.2013-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190(7):3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karagiannis GS, et al. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci Transl Med. 2017;9(397):eaan0026. doi: 10.1126/scitranslmed.aan0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karagiannis GS, Condeelis JS, Oktay MH. Chemotherapy-induced metastasis: mechanisms and translational opportunities. Clin Exp Metastasis. 2018;35(4):269–284. doi: 10.1007/s10585-017-9870-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong D, Ahmad A, Bao B, Li Y, Banerjee S, Sarkar FH. Histone deacetylase inhibitors induce epithelial-to-mesenchymal transition in prostate cancer cells. PLoS One. 2012;7(9):e45045. doi: 10.1371/journal.pone.0045045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao L, et al. Disrupting androgen receptor signaling induces snail-mediated epithelial-mesenchymal plasticity in prostate cancer. Cancer Res. 2017;77(11):3101–3112. doi: 10.1158/0008-5472.CAN-16-2169. [DOI] [PubMed] [Google Scholar]
- 28.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darby S, Cross SS, Brown NJ, Hamdy FC, Robson CN. BMP-6 over-expression in prostate cancer is associated with increased Id-1 protein and a more invasive phenotype. J Pathol. 2008;214(3):394–404. doi: 10.1002/path.2292. [DOI] [PubMed] [Google Scholar]
- 30.Haudenschild DR, Palmer SM, Moseley TA, You Z, Reddi AH. Bone morphogenetic protein (BMP)-6 signaling and BMP antagonist noggin in prostate cancer. Cancer Res. 2004;64(22):8276–8284. doi: 10.1158/0008-5472.CAN-04-2251. [DOI] [PubMed] [Google Scholar]
- 31.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525(7570):538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang Y, Cai X, Fan D. Roles of epithelial-mesenchymal transition in cancer drug resistance. Curr Cancer Drug Targets. 2013;13(9):915–929. doi: 10.2174/15680096113136660097. [DOI] [PubMed] [Google Scholar]
- 33.DeGraff DJ, et al. FOXA1 deletion in luminal epithelium causes prostatic hyperplasia and alteration of differentiated phenotype. Lab Invest. 2014;94(7):726–739. doi: 10.1038/labinvest.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Tindall DJ, Huang H. Modulation of androgen receptor by FOXA1 and FOXO1 factors in prostate cancer. Int J Biol Sci. 2014;10(6):614–619. doi: 10.7150/ijbs.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahu B, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30(19):3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Augello MA, Hickey TE, Knudsen KE. FOXA1: master of steroid receptor function in cancer. EMBO J. 2011;30(19):3885–3894. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson JL, et al. Elevated levels of FOXA1 facilitate androgen receptor chromatin binding resulting in a CRPC-like phenotype. Oncogene. 2014;33(50):5666–5674. doi: 10.1038/onc.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70(5):2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang JZ, et al. Down-regulation of TRPS1 stimulates epithelial-mesenchymal transition and metastasis through repression of FOXA1. J Pathol. 2016;239(2):186–196. doi: 10.1002/path.4716. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2017;36(8):1157–1166. doi: 10.1038/onc.2016.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jägle S, et al. SNAIL1-mediated downregulation of FOXA proteins facilitates the inactivation of transcriptional enhancer elements at key epithelial genes in colorectal cancer cells. PLoS Genet. 2017;13(11):e1007109. doi: 10.1371/journal.pgen.1007109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, et al. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene. 2017;36(28):4072–4080. doi: 10.1038/onc.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin HJ, Zhao JC, Ogden I, Bergan RC, Yu J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013;73(12):3725–3736. doi: 10.1158/0008-5472.CAN-12-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, et al. RNF6 promotes colorectal cancer by activating the Wnt/β-catenin pathway via ubiquitination of TLE3. Cancer Res. 2018;78(8):1958–1971. doi: 10.1158/0008-5472.CAN-17-2683. [DOI] [PubMed] [Google Scholar]
- 45.Lee HK, Lee DS, Park JC. Nuclear factor I-C regulates E-cadherin via control of KLF4 in breast cancer. BMC Cancer. 2015;15:113. doi: 10.1186/s12885-015-1118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jafari R, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9(9):2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 47.Tyler DS, et al. Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science. 2017;356(6345):1397–1401. doi: 10.1126/science.aal2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin J, et al. COUP-TFII inhibits TGF-β-induced growth barrier to promote prostate tumorigenesis. Nature. 2013;493(7431):236–240. doi: 10.1038/nature11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, et al. MPC1, a key gene in cancer metabolism, is regulated by COUPTFII in human prostate cancer. Oncotarget. 2016;7(12):14673–14683. doi: 10.18632/oncotarget.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.