Abstract
Protein arginine methyltransferase 5 (PRMT5) catalyzes symmetric dimethylation (SDM) of arginine, a posttranslational modification involved in oncogenesis and embryonic development. However, the role and mechanisms by which PRMT5 modulates Th cell polarization and autoimmune disease have not yet been elucidated. Here, we found that PRMT5 promoted SREBP1 SDM and the induction of cholesterol biosynthetic pathway enzymes that produce retinoid-related orphan receptor (ROR) agonists that activate RORγt. Specific loss of PRMT5 in the CD4+ Th cell compartment suppressed Th17 differentiation and protected mice from developing experimental autoimmune encephalomyelitis (EAE). We also found that PRMT5 controlled thymic and peripheral homeostasis in the CD4+ Th cell life cycle and invariant NK (iNK) T cell development and CD8+ T cell maintenance. This work demonstrates that PRMT5 expression in recently activated T cells is necessary for the cholesterol biosynthesis metabolic gene expression program that generates RORγt agonistic activity and promotes Th17 differentiation and EAE. These results point to Th PRMT5 and its downstream cholesterol biosynthesis pathway as promising therapeutic targets in Th17-mediated diseases.
Keywords: Autoimmunity, Immunology
Keywords: Adaptive immunity, Autoimmune diseases, T cells
Introduction
Arginine methylation is a posttranslational modification of histones and other proteins that regulates important physiological processes, including stem cell biology, differentiation, and development. Conversely, arginine methylation dysregulation occurs in diseases such as cancer and autoimmunity (1, 2). Arginine methylation is catalyzed by a group of 10 protein arginine methyltransferase (PRMT) enzymes (3). Type I, II, and III PRMTs catalyze asymmetric dimethylation (ADM), symmetric dimethylation (SDM), and monomethylation of arginine (3), respectively. PRMT5 and 9 belong to the type II PRMTs; both can catalyze SDM. However, PRMT5 is considered responsible for the majority of cellular SDM (4). PRMT5 has been shown to methylate a number of targets, ranging from histones H4 (at R3) and H3 (at R8) to NF-κB and spliceosome proteins (5). The short and long isoforms of the PRMT5 protein are produced from the PRMT5 gene via alternative splicing. Both isoforms are protein coding and can influence PRMT5 cellular localization and function (6). PRMT5 overexpression is common in human cancer cells and has been shown to promote proliferation and survival of cancer and stem cells (5). Proliferation downstream of T cell receptor (TcR) or B cell receptor (BcR) activation is also common in immune cells such as B cells and T cells, and PRMT5 has been shown to play an important role in lymphocyte biology (7).
We recently reported that PRMT5 is induced after T cell activation and that its induction is controlled by NF-κB/MYC/mTOR signaling (8, 9). PRMT5’s SDM mark also has been shown to be dynamically regulated in T cells (10), suggesting that it contributes to the T cell activation process. We have evidence that both PRMT5 inhibitors and shRNA-mediated PRMT5 knockdown impair T cell proliferation after activation (8). Genetic deletion of the long PRMT5 isoform in all T cells recapitulated the proliferation defect (11). However, we do not yet know how the deletion of both PRMT5 isoforms would influence T cell proliferation. In addition, we do not know the impact of PRMT5 on naive T cell polarization toward the Th1/Th2/Th17/Treg phenotypes. Exploring these unknowns is important because Th cell polarization is relevant to protection from or development of disease. For example, inflammatory Th1 and Th17 responses drive chronic tissue damage in autoimmune diseases such as multiple sclerosis (MS) (12).
Metabolic reprogramming upon T cell activation is a phenomenon that is increasingly recognized as an essential part of regulating Th cell function and polarization. Activated T cells grow and proliferate very rapidly, requiring the induction of a biosynthetic phenotype. Thus, quiescent naive or resting memory T cells that rely on oxidative phosphorylation and/or fatty acid oxidation for energy generation rapidly shift upon activation to biosynthetic metabolic pathways, including glycolysis and cholesterol biosynthesis (13, 14). Inflammatory Th1 and Th17 cells require this glycolytic and biosynthetic reprogramming. Moreover, the induction of cholesterol biosynthesis is particularly important for cells differentiating into the Th17 lineage (15). However, the contribution of PRMT5 to metabolic shifts that promote the Th17 program is unknown.
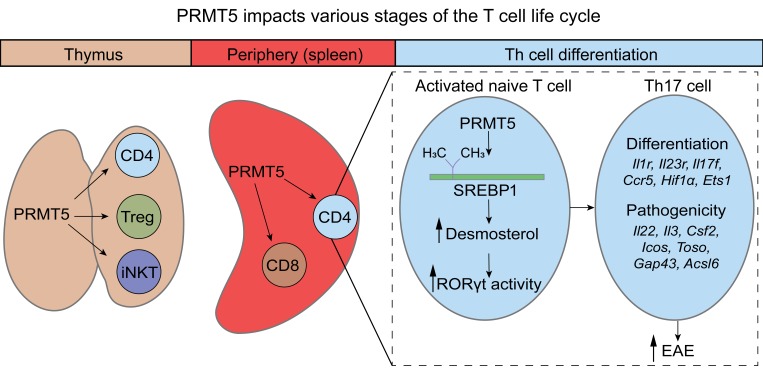
We used the first PRMT5-KO mouse models that delete all protein-coding isoforms, to our knowledge, in order to investigate the T cell–intrinsic role of the Prmt5 gene during T cell development, T cell homeostasis, naive Th cell differentiation, and T cell–mediated autoimmune disease. We focused on the mechanism by which PRMT5 controls T cell cholesterol metabolism and modulates Th17 differentiation and experimental autoimmune encephalomyelitis (EAE) disease. The primary application of this work is to further therapeutic strategies for Th17-mediated disease such as MS autoimmunity, but our findings have implications for PRMT5 inhibitor therapies for other diseases such as cancer.
Results
Development of constitutive pan-T cell–specific and inducible CD4+ Th cell–specific mouse models of PRMT5 deficiency.
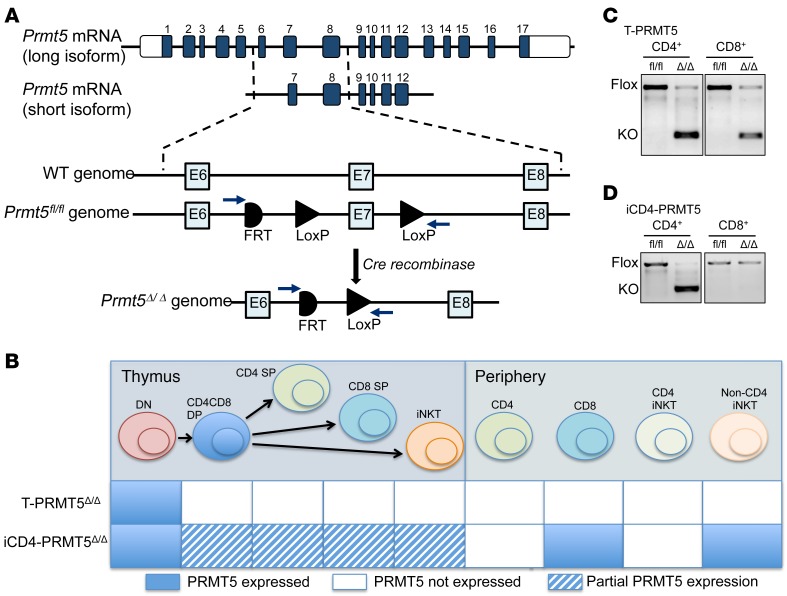
PRMT5 is essential for embryonic development (16, 17) and hematopoietic cell development (18). Therefore, evaluation of PRMT5’s function in T cells requires conditional KO models that allow for a T cell subset–specific and time–controlled PRMT5 deletion. To develop conditional PRMT5-KO mice in which both PRMT5 protein–coding isoforms (Figure 1A) are specifically deleted in T cells, we used the Prmt5tm2c(EUCOMM)wtsi mutation that was engineered to harbor 2 loxP sites flanking exon 7. To inactivate PRMT5 in all T cells (pan-T) or, alternatively, solely in the CD4+ Th compartment, the PRMT5fl/fl mice were crossed to CD4-Cre (19) or CD4-Cre-ERT2 (20) mice, respectively. The CD4-Cre transgene is constitutively expressed in all CD4-expressing cells, including thymic double-positive (DP) T cells. As a result, PRMT5 is inactivated in all CD3+ T cells, providing a mouse model in which all peripheral T cells lack PRMT5 (Figure 1B, hereafter referred to as T-PRMT5Δ/Δ mice). In contrast, the tamoxifen-inducible CD4-Cre-ERT2 transgene induces PRMT5 deletion specifically in peripheral CD4+ T cells only upon tamoxifen treatment (Figure 1B, hereafter these mice will be referred to as iCD4-PRMT5Δ/Δ mice). Such strategy limits deletion on DP thymocytes; only those present during the tamoxifen treatment window may be affected (20). As expected, the short PCR product corresponding to PRMT5-KO was amplified from both CD4+ Th and CD8+ T cells in T-PRMT5Δ/Δ mice (Figure 1C), but only amplified in CD4+ Th cells from tamoxifen-treated iCD4-PRMT5Δ/Δ mice (Figure 1D).
Figure 1. Constitutive and inducible CD4+ Prmt5-KO models.
(A) Schematic of Prmt5 long and short isoform transcripts and Prmt5 genomic locus targeting strategy. Exon 7, an exon common to all protein-coding Prmt5 isoforms, was flanked by loxP sites to provide deletion of both isoforms in Cre-expressing cells. (B) Schematic of expected PRMT5 deletion in thymus T cell precursors and peripheral T cells in 2 different transgenic models, namely T-PRMT5Δ/Δ (constitutive) and iCD4-PRMT5Δ/Δ (inducible). (C and D) PCR amplification of genomic DNA isolated from CD4+ and CD8+ T cells from (C) T-PRMT5Δ/Δ or (D) iCD4-PRMT5Δ/Δ mice, as well as control PRMT5fl/fl mice. PRMT5-KO band: 283 bp; full-length floxed PRMT5 band: 1100 bp. DN, double negative; DP, double positive; SP, single positive.
PRMT5 is necessary for normal CD4+ Th cell and regulatory T cell development.
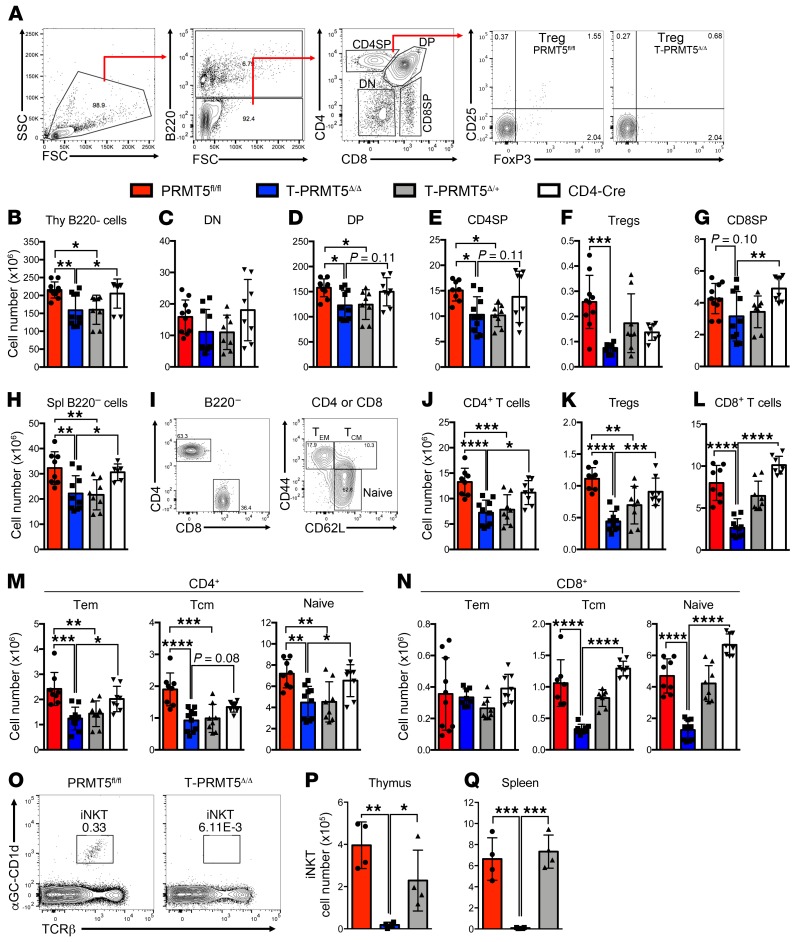
We have previously shown that PRMT5 is essential for T cell proliferation (8). Because thymocyte proliferation is a crucial event during T cell development, we chose to evaluate the impact of PRMT5 deficiency on the thymic immune compartment by flow cytometry (Figure 2, A–G, and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI131254DS1). B220– thymocyte numbers were significantly reduced in T-PRMT5Δ/Δ mice compared with PRMT5fl/fl and CD4-Cre controls (Figure 2B). The CD4-Cre driver is first expressed at the DP stage. As expected, although total CD4/CD8 double-negative (DN) thymocyte numbers were not significantly affected (Figure 2C), a significant loss in the CD4/CD8 DP compartment (Figure 2D) was present. This defect was more prominent in the CD4 single-positive (SP) (Figure 2E) and regulatory T cell (Treg) (Figure 2F) populations. Interestingly, the loss of CD4 SP cells was also observable in heterozygous PRMT5-KO cells (Figure 2E), indicative of PRMT5 haploinsufficiency during thymic CD4+ T cell development. In contrast, the CD8 SP compartment was not significantly affected between PRMT5fl/fl and T-PRMT5Δ/Δ, but CD8 SP cells were significantly reduced in T-PRMT5Δ/Δ compared with CD4-Cre controls (Figure 2G). Consistent with the role of PRMT5 in proliferation, the observed thymic defects appear to be due to reduced thymocyte expansion in that no significant impact on DN, DP, or SP frequencies was observed, with the exception of reduced Treg frequency (Figure 2A and Supplemental Figure 1).
Figure 2. Impaired thymocyte development and peripheral T cell compartment in T-PRMT5Δ/Δ mice.
(A) Gating strategy for analysis of thymocyte populations and representative thymic Treg percentage plots. Live/dead gating was also performed after FSC/SSC but is not displayed. (B–G) Thymocytes were analyzed by flow cytometry and (B) B220–, (C) DN, (D) DP, (E) CD4 SP, (F) Treg, and (G) CD8 SP thymocyte numbers were calculated. (H–N) Splenocytes were analyzed by flow cytometry (gating strategy in I and H) B220– splenocyte, (J) CD4+, (K) Treg, (L) CD8+, (M) CD4+ Tem, Tcm, and naive populations, and (N) CD8+ Tem, Tcm, and naive populations were calculated. (O) iNK T cell flow cytometric analysis gating strategy and total (P) thymic and (Q) splenic iNKT cell numbers. Data are representative of 4 independent experiments. Shown n = 4–5 independent mice of matched age. One-way ANOVA, followed by Sidak’s (B–H and J–N) or Tukey’s (P and Q) multiple-comparisons test: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Bar graphs display mean ± SD. DN, double negative; DP, double positive; SP, single positive; Tem, effector memory T cells; Tcm, central memory T cells.
To evaluate the impact of PRMT5 deletion on peripheral immune populations, we analyzed spleen and lymph nodes (LNs) (Figure 2, H–N, and Supplemental Figure 1). Although total splenocyte and LN cell numbers were unaffected in T-PRMT5Δ/Δ mice, flow cytometry analyses of T cell populations (Figure 2I) showed reduced numbers of B220– cells, CD4+ Th cells, and Tregs in the spleen (Figure 2, H–K) and LNs (Webb, unpublished results), following the same pattern observed in the thymus. These peripheral CD4+ T cell defects could stem from thymic defects alone or, alternatively, combined thymic/peripheral homeostasis defects. In addition, a robust loss of CD8+ T cells was evident in T-PRMT5Δ/Δ mice spleens (Figure 2L) and LNs (Webb, unpublished results). The decrease in CD8+ T cell numbers was much more obvious in the spleen than the thymus, suggesting that CD8+ T cell maintenance requires PRMT5. Among CD4+ Th cells, all naive (CD62L+CD44–), effector memory (Tem, CD62L–CD44+ effector), and central memory (Tcm, CD62L+CD44+) CD4+ T cells were reduced (Figure 2M). Within the CD8+ T cell compartment, both the naive and Tcm CD8+ T cells were drastically reduced, whereas no changes in Tem cells were observed (Figure 2N). Furthermore, in our model where both the long and short isoforms of PRMT5 were deficient, we observed complete loss of invariant NK (iNK) T cell populations in the thymus and spleen of T-PRMT5Δ/Δ mice (Figure 2, O–Q). This corroborates the report of iNK T cell loss in mice with a T cell deficiency in the long isoform of PRMT5 (11). Overall, these data show that PRMT5 is required for thymic CD4+ Th, Treg, and iNK T cell development and for normal CD4+, CD8+, and iNKT peripheral T cell compartments, and that any defects thereof are T cell intrinsic.
Acute PRMT5 knockout in CD4+ T cells does not affect thymic T cell development or peripheral immune cell compartments.
The observation that CD8+ T cell numbers were normal at the thymic level but drastically reduced peripherally suggests that CD8+ T cells are highly dependent on PRMT5 in the periphery. The peripheral CD4+ Th cell loss could result from thymic development defects and/or peripheral homeostasis defects. To address this, we took advantage of the iCD4-PRMT5Δ/Δ model, which allows for temporal control of a directed PRMT5 deletion in peripheral CD4+ Th cells. To rule out any effects on thymic development, we evaluated thymocytes and peripheral immune cell compartments in adult iCD4-PRMT5Δ/Δ mice (Supplemental Figure 2A). As expected, thymic compartments were unaffected in iCD4-PRMT5Δ/Δ mice after 1 week of tamoxifen treatment (Supplemental Figure 2, B–M), which indicates normal T cell development in these mice. We also observed no significant effects of acute PRMT5 deficiency on the proportion or total number of peripheral CD4+, CD8+, and iNK T cells (Supplemental Figure 2, N–Y). Therefore, this model allows investigation of questions concerning the influence of PRMT5 deficiency in an otherwise normal CD4+ T cell compartment. We have focused on this acute deletion iCD4-PRMT5Δ/Δ model throughout this paper, unless otherwise noted. However, we also explored the impact of more extended (5-week) tamoxifen-induced Cre activity (Supplemental Figure 3A). Once again, extended treatment did not substantially affect thymic CD4 SP, but reduced CD4+ T cell numbers in the spleen (Supplemental Figure 3, B–I) and LNs (Webb, unpublished results), confirming PRMT5 promotion of peripheral CD4+ T cell homeostasis. Among peripheral CD4+ T cells, both naive and Tem subsets were lost (Supplemental Figure 3J). This model is selective for CD4+ T cells because we confirmed that CD8+ T cell populations were unaffected by extended tamoxifen treatment (Supplemental Figure 3, F and I). In summary, these data demonstrate that PRMT5 expression promotes thymic T cell development and peripheral CD4+ and CD8+ T cell maintenance.
PRMT5 drives CD4+ Th cell proliferation.
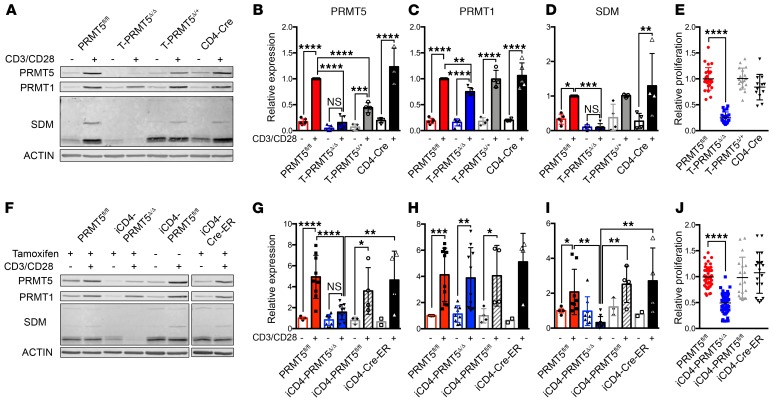
To evaluate the impact of PRMT5 deficiency on peripheral Th cell function, we isolated peripheral CD4+ Th cells from T-PRMT5Δ/Δ mice and analyzed them 48 hours after TcR engagement. We confirmed the near complete loss of PRMT5 protein expression and its SDM mark (detected with the SYM10 antibody) in T-PRMT5Δ/Δ Th cells compared with controls (Figure 3, A, B, and D). The type I methyltransferase PRMT1 was also slightly reduced (Figure 3, A and C), which confirmed our previously observed positive modulation of PRMT1 by PRMT5 (8). Functionally, such PRMT5 loss resulted in a robust (≥80%) suppression of proliferation in CD4+ Th cells (Figure 3E) and total CD3+ T cells (Supplemental Figure 4). To determine whether the proliferative defect was also present in T cells deleted of PRMT5 after thymic development, we analyzed Th cells from iCD4-PRMT5Δ/Δ mice. We found that PRMT5 protein induction after TcR stimulation in the iCD4-PRMT5Δ/Δ model was suppressed 60%–70% (Figure 3, F and G), albeit to a lesser extent than in T-PRMT5Δ/Δ mice. These results are consistent with the lower efficiency of the iCD4-Cre-ER driver (20). Likewise, approximately 80%–90% of PRMT5’s symmetric dimethylation mark SYM10 was lost (Figure 3, F and I), whereas PRMT1 maintained normal expression (Figure 3, F and H). We also found suppressed Th cell proliferation in the iCD4-PRMT5Δ/Δ model (Figure 3J). These data substantively demonstrate a driver role for PRMT5 in TcR-induced CD4+ Th and CD8+ T cell proliferation.
Figure 3. Prmt5 deficiency suppresses T cell proliferation.
Whole spleen CD4+ T cells from (A–E) T-PRMT5Δ/Δ or (F–J) iCD4-PRMT5Δ/Δ mice were isolated and activated with anti-CD3/CD28. Cells were collected for immunoblotting directly ex vivo and after 48 hours of anti-CD3/CD28 activation and (A and F) analyzed by immunoblot. Lysates were run on the same gel but are noncontiguous in F. Bands detected by (B and G) anti-PRMT5, (C and H) anti-PRMT1, and (D and I) PRMT5’s symmetric dimethylation mark (anti-SYM10 antibody) were quantified using ImageStudio software. Data are pooled from at least 3 representative independent experiments (n = 5–7 independent mice). (E and J) Proliferation of CD4+ T cells was analyzed by 3H-thymidine incorporation and expressed as a relative proliferation ratio to the resting PRMT5fl/fl control condition. Data include at least 3 independent experiments (n = 5–12 mice/group). Two-way ANOVA followed by Sidak’s multiple-comparisons test (B–D and G–I) or 1-way ANOVA followed by Dunnett’s multiple-comparisons test (E and J). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Graphs display mean ± SD.
PRMT5 regulates Th cell differentiation.
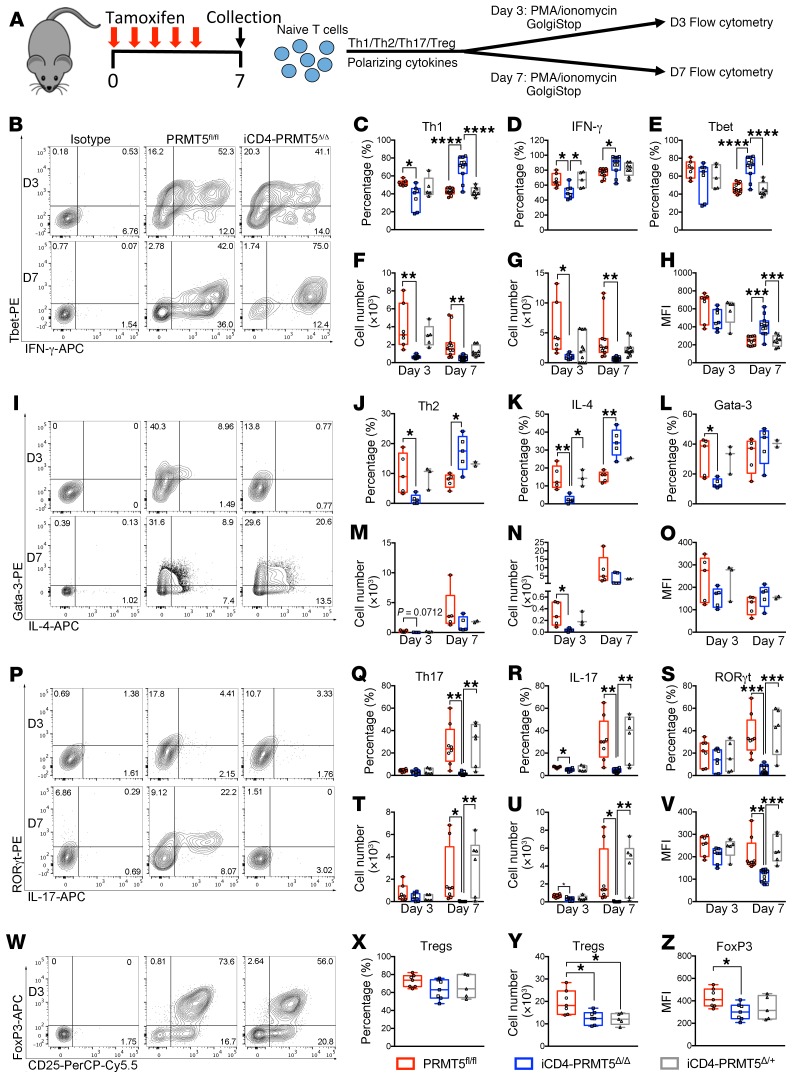
Naive Th cell activation leads to T cell differentiation, which can result in distinct Th cell phenotypes depending on the environmental milieu. Cell-intrinsic parameters also influence a T cell’s poise toward specific Th cell phenotypes. Inflammatory Th1 and Th17 responses are highly pathogenic and drive chronic tissue damage in MS, an autoimmune disease of the CNS, whereas Th2 cells and Tregs are beneficial in this autoimmune disease (12). As such, we evaluated the impact of PRMT5 on Th1/Th2/Th17/iTreg cell differentiation (Figure 4A) in the iCD4-PRMT5Δ/Δ model in which T cells undergo normal thymic development and are present in normal numbers in the periphery before acute PRMT5 deletion (Supplemental Figure 2). Approximately 50% of normal T cell proliferation remains upon TcR engagement in the iCD4-PRMT5Δ/Δ model, ensuring a T cell pool for analysis. Activated CD44+ cells negative for dead cell stain were gated for analysis. Following a slight defect in IFN-γ–secreting Th1 cells on day 3, we observed an increase in the proportion of IFN-γ+T-bet+, IFN-γ+, and T-bet+ T cells among live, activated CD44+ cells (Figure 4, B–E). T-bet mean fluorescence intensity (MFI) also was increased on day 7 (Figure 4H). These data are consistent with PRMT5 acting to suppress signature transcription factor and cytokine expression in committed late-stage Th1 cells. However, the total number of differentiated Th1 cells was reduced overall in iCD4-PRMT5Δ/Δ mice (Figure 4, F and G), which was likely due to reduced proliferation. During Th2 differentiation, we also observed an increased percentage of GATA-3+IL-4+ and IL-4+ in committed day 7 Th2 cells, whereas the percentages of GATA-3 and MFI were maintained (Figure 4, I–L, and O). No significant changes were observed in the total number of day 7 Th2 cells (Figure 4, M and N). Strikingly, Th17 differentiation was severely blunted, with almost complete abrogation of the proportion and number of committed (day 7) RORγt+IL-17+, IL-17+, and RORγt+ Th17 cells and RORγt MFI (Figure 4, P–V). Finally, the proportions of Foxp3+CD25+ Treg cells were maintained in iCD4-PRMT5Δ/Δ mice, but their total number was reduced and a decrease in Foxp3 MFI was also observed (Figure 4, W–Z). Similar results were obtained in the T-PRMT5Δ/Δ model in which PRMT5 was deleted at the thymic level (Supplemental Figure 5). Overall, these data indicate that PRMT5 modulates Th cell polarization and is essential for Th17 differentiation.
Figure 4. Prmt5 deficiency in iCD4-PRMT5Δ/Δ T cells abrogates Th17 cell differentiation.
(A) Experimental design for Th cell differentiation in iCD4-PRMT5Δ/Δ mice. Naive CD4+ T cells isolated from iCD4-PRMT5Δ/Δ mice were polarized into (B–H) Th1, (I–O) Th2, (P–V) Th17, or (W–Z) Tregs and assessed by flow cytometry. Cells shown are gated on live (LiveDead Dye–) CD44+ cells. Th1 cells were assessed by T-bet+IFN-γ+ cell (C) percentage and (F) number, IFN-γ+ cell (D) percentage and (G) number, T-bet+ (E) cell percentage, and (H) mean fluorescence intensity (MFI) by flow cytometry. Th2 cells were assessed by GATA-3+IL-4+ cell (J) percentage and (M) number, IL-4+ cell (K) percentage and (N) number, GATA-3+ (L) cell percentage, and (O) MFI by flow cytometry. Th17 cells were assessed by RORγt+IL-17+ cell (Q) percentage and (T) number, IL-17+ cell (R) percentage and (U) number, RORγt+ (S) cell percentage, and (V) MFI by flow cytometry. Tregs were assessed by Foxp3+CD25+ (X) cell percentage and (Y) number, and (Z) Foxp3 MFI. Data pooled from 3 independent experiments including n = 6–12/group. For Th2 cells, data from 2 independent experiments, n = 2–5/group. One-way ANOVA, followed by Tukey’s multiple-comparisons test was used. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Graphs are box-and-whisker plots (box extends from 25th to 75th percentiles, all points shown, whiskers extend from min to max, line represents median).
PRMT5 modulates Th17 differentiation via cholesterol biosynthesis.
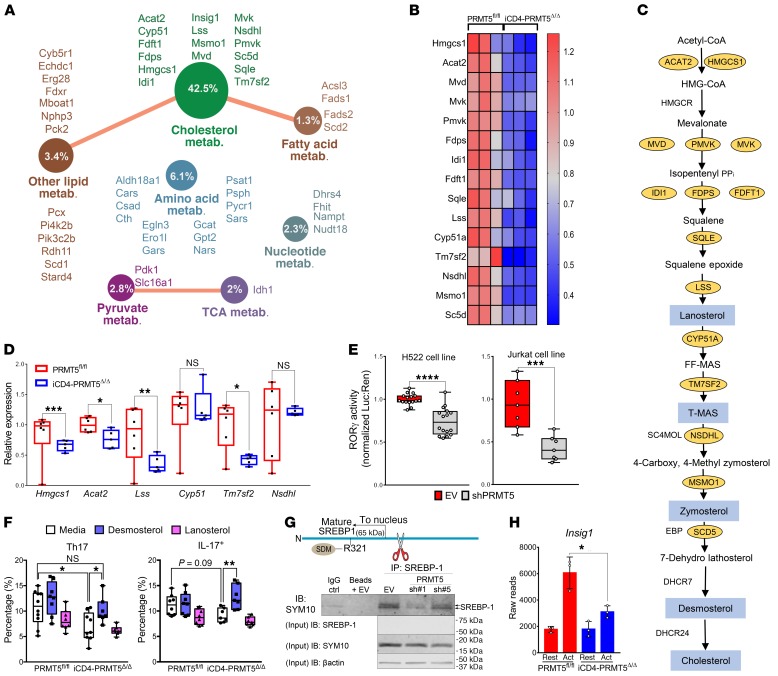
Our data show that PRMT5 deficiency in T cells robustly suppresses Th cell proliferation and Th17 differentiation, suggesting that PRMT5 promotes transcriptional programs essential for Th17 differentiation. To define these programs, we performed RNA sequencing (RNA-Seq) in resting and activated CD4+ Th cells from iCD4-PRMT5Δ/Δ and control PRMT5fl/fl mice. T cells were activated in nonpolarizing (Th0) conditions to capture Th17 milieu–independent PRMT5-driven signatures that poise activated naive T cells toward the Th17 fate. Lack of PRMT5 resulted in differential expression of 545 genes (FDR ≤ 0.05), with 342 exhibiting increased and 203 decreased gene expression (Supplemental Tables 1 and 2), indicating that PRMT5 has both activating and repressing effects in T cells. Gene ontology (GO) pathway analysis of PRMT5-regulated genes (Supplemental Table 3) revealed that PRMT5 controls multiple metabolic pathways, including cholesterol, fatty acid and other lipids, amino acids, pyruvate, TCA cycle, and nucleotide metabolism, among others (Figure 5A). Nonetheless, the most striking effect was on cholesterol metabolism, with 42% of genes in the GO cholesterol metabolic pathway reduced in PRMT5-KO cells (Figure 5A). The expression of 75% (15 out of 20) of the enzymes in the cholesterol biosynthesis pathway was suppressed in PRMT5-deficient Th cells (Figure 5, B and C). The significant loss of several of these enzymes, namely Tm7sf2, Hmgcs1, Lss, and Acat2, was validated by real-time PCR in cells undergoing Th17 differentiation (Figure 5D). Lipid metabolism and cholesterol biosynthesis not only support T cell growth and division (14, 21–23), but also are crucial for Th17 cell differentiation (24–26). Several cholesterol biosynthesis pathway intermediates, including lanosterol, zymosterol, and desmosterol, are strong RORγt agonists essential for Th17 cell differentiation (24) and are highlighted in blue in the pathway (Figure 5C). This led us to hypothesize that PRMT5 promotes Th17 differentiation by promoting production of RORγt agonists. To test whether PRMT5 promotes RORγt agonistic activity, luciferase assays were performed using an RORγt activity GAL-4 reporter system. PRMT5 shRNA knockdown selectively suppressed RORγt activity in both human lung adenocarcinoma (H522) and T cell (Jurkat) lines (Figure 5E). If PRMT5 mediates the Th17 differentiation defect through suppression of cholesterol precursor biosynthesis, restoring such precursors in PRMT5-KO T cells should restore this defect. To test this, we supplemented cholesterol intermediate lanosterol or desmosterol during Th17 differentiation in iCD4-PRMT5Δ/Δ T cells. We found that desmosterol enhanced Th17 differentiation in PRMT5fl/fl T cells and restored normal levels of Th17 cell differentiation in iCD4-PRMT5Δ/Δ T cells (Figure 5F). In contrast, lanosterol, which is further upstream in the pathway of cholesterol biosynthesis, had no effect (Figure 5F). Lipid and sterol biosynthesis are regulated via SREBP transcription factors. Cleavage of SREBP1 produces the mature SREBP1 product that translocates to the nucleus and transactivates expression of genes that promote the biosynthesis of cholesterol and other lipids. SREBP1 is phosphorylated by glycogen synthase kinase 3 (GSK3) at residue S430, and the phosphorylated SREBP1 is targeted for proteasomal degradation (27). This is inhibited by PRMT5-mediated SDM of arginine 321 (R321) within the mature form, which prevents SREBP1 phosphorylation and proteasomal degradation (28). As a result, PRMT5 enhances the stability and transcriptional activity of SREBP1 (28). Immunoprecipitation of SREBP1 followed by SYM10 SDM immunoblotting of activated Jurkat T cells transduced with an empty lentiviral vector detected 2 symmetrically dimethylated bands, sizes consistent with the size of the active mature form of SREBP1. The doublet of mature SREBP1 has been previously reported by Geng et al. (29). This doublet was reduced in Jurkat T cells transduced with PRMT5 shRNA (Figure 5G), consistent with PRMT5-dependent SREBP1 methylation in T cells. Controls had no band (beads + EV) or a faint band (IgG + beads), which was likely due to trace bead carryover in the supernatant. In addition, transcription of the direct SREBP target gene Insig-1 was lost in PRMT5-deficient activated CD4+ Th cells (Figure 5H), which is consistent with PRMT5 controlling lipid metabolism via enhanced SREBP activity after T cell activation.
Figure 5. PRMT5 promotes Th17 differentiation via cholesterol biosynthesis.
(A) GO analysis of downregulated (FDR ≤ 0.05) genes in 2-day anti-CD3/CD28–activated CD4+ T cells from iCD4-PRMT5Δ/Δ versus PRMT5fl/fl mice (n = 3 each), revealing effect on metabolic genes. Circle indicates percentage of genes within pathway reduced in iCD4-PRMT5Δ/Δ T cells. (B) Heatmap of cholesterol biosynthesis enzymes differentially expressed in T cells from A (FC = 0–1.3). (C) Cholesterol biosynthesis pathway. Yellow ovals: enzymes significantly decreased in iCD4-PRMT5Δ/Δ T cells from B; blue: cholesterol precursors with RORγt agonistic activity. (D) Real-time PCR of cholesterol biosynthesis pathway enzyme expression 2 days after activation of iCD4-PRMT5Δ/Δ or control Th cells in Th17-polarizing conditions. Three independent experiments (n = 5–6). (E) RORγt luciferase activity using the RORγt-GAL4 luciferase/Renilla-normalized reporter, in human H522 and Jurkat T cells transduced with empty vector control (EV) or PRMT5 shRNA (90%–94% knockdown in H522; 70%–75% in Jurkats). Data pooled from 3 independent experiments (n = 2–6). (F) iCD4-PRMT5Δ/Δ naive CD4+ T cells activated in Th17-polarizing conditions with and without cholesterol precursor desmosterol or lanosterol. Flow cytometric analysis of RORγt+IL-17+ or IL-17+ (gated on live CD4+CD44+) Th17 cells was performed 3 days after activation. Data pooled from 2 independent experiments (n = 5). (G) Diagram and IP of mature SREBP1 including PRMT5 methylation site. After cleavage, mature transcript translocates to nucleus for transcriptional activity. Anti-SREBP1 IP, then IB of SYM10 in Jurkats transduced with EV or PRMT5 shRNA as in E. IB represents 2 to 3 independent experiments (n = 3). (H) Differential expression reads for SREBP target Insig-1 in naive, resting, or activated (CD3/CD28, Th17 conditions) CD4+ T cells from iCD4-PRMT5Δ/Δ or control mice (n = 3). Student’s t test (D, E, and H) or 2-way ANOVA, followed by Dunnett’s (within genotypes) or Sidak’s (across genotypes) multiple-comparisons test (F). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Box-and-whisker plots (points = max to min, line = median, box = 25th–75th percentiles). Bar graphs display mean ± SD.
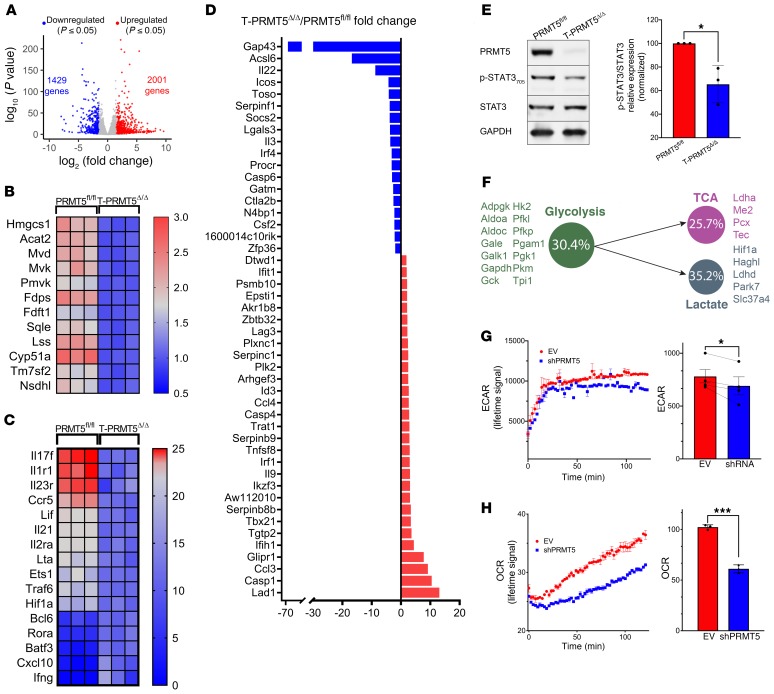
To determine which components of the Th17 signature are controlled by PRMT5, we carried out RNA-Seq of T cells in Th17 differentiation conditions (Figure 6, A–D). We found that 1,429 genes were significantly (FDR ≤ 0.05) decreased (fold change [FC] ≤ 1.5) and 2,001 genes were significantly increased (FC ≥ 1.5) in T-PRMT5Δ/Δ relative to PRMT5fl/fl Th17 cells (Supplemental Tables 4 and 5). The cholesterol biosynthetic pathway was robustly suppressed in PRMT5-deficient Th17 cells (Figure 6B), which is similar to what we observed in Th0 conditions (Figure 5, B and C). In addition, we analyzed the impact of PRMT5 on key Th17 differentiation genes, as defined by Ciofani et al. (30) and Yosef et al. (31). We observed that the ablation of PRMT5 results in robust downregulation of genes crucial for Th17 differentiation, including Il17f, Il1r1, Il23r, and Ccr5. Less robust downregulation was observed for other genes in this pathway, including Lif, Il21, Il2ra, Lta, Ets1, Traf6, and Hif1a (Figure 6C). The IL-23 receptor (IL-23R) is necessary for Th17 cell differentiation and survival, and IL-17 is the signature Th17 cytokine. Hif1a and Ets1 encode transcription factors that promote Th17 differentiation. Rorc was also downregulated. Its decrease did not reach statistical significance, consistent with the notion that PRMT5 regulates RORγt activity rather than expression. In addition, we evaluated the impact of PRMT5 loss on key pathogenic Th17 signature genes, as defined by Lee et al. (32) and Gaublomme et al. (33). Downregulated pathogenic Th17 signature genes include cytokines Il22, Il3, and Csf2; costimulatory gene Icos; IgM receptor Toso/FcmR; the neuromodulin Gap43; and the fatty acid metabolic enzyme Acsl6 (Figure 6D). STAT3 phosphorylation, which is essential for the pathogenicity of Th1 and Th17 cells and occurs downstream of IL-23R signaling (32), was also robustly reduced in Th17 cells from T-PRMT5Δ/Δ mice (Figure 6E). Strikingly, T-bet/T-box21, which is associated with pathogenic Th1 and Th17 cells, was repressed rather than upregulated by PRMT5, as evidenced by its enhanced expression in PRMT5-deficient T cells (Figure 6D). This result was in accordance with the increased percentage of T-bet+ and IFN-γ+ cells observed during Th1 cell differentiation (Figure 4, B–E).
Figure 6. PRMT5 controls key Th17 development, pathogenicity, and metabolic genes and promotes ECAR/OCR during Th17 polarization.
(A) Volcano plot of significantly induced and repressed genes (FDR < 0.05) in Th17 condition–activated naive T cells from T-PRMT5Δ/Δ versus PRMT5fl/fl mice (n = 3 each). (B) Heatmap of cholesterol biosynthesis enzymes differentially expressed in CD4+ Th17 cells from A, showing similar data to that obtained in Th0 conditions (Figure 5). (C) Heatmap of Th17 differentiation signature genes, as defined by Ciofani et al. (30) and Yosef et al. (31), significantly differentially expressed in T-PRMT5Δ/Δ versus PRMT5fl/fl mouse Th17 cells from A. (D) FC in expression of Th17 pathogenic signature genes, as defined by Lee et al. (32) and Gaublomme et al. (33), observed in T-PRMT5Δ/Δ versus PRMT5fl/fl mouse Th17 cells from A. (E) STAT3 phosphorylation Western blot and quantification of p-STAT3 to total STAT3 ratio in T-PRMT5Δ/Δ versus PRMT5fl/fl Th17 cells. Three independent experiments with 2 to 3 mice/experiment. (F) Glycolysis, lactate, and TCA pathway genes were downregulated in T-PRMT5Δ/Δ versus PRMT5fl/fl mouse Th17 cells from A. Circle indicates the percentage of genes within the pathway reduced in T-PRMT5Δ/Δ T cells. (G) Extracellular acidification rate (ECAR) in Jurkat T cell lines stably transduced with control or PRMT5 shRNA (70%–75% knockdown). (H) Oxygen consumption rate (OCR) in Jurkat T cell lines stably transduced with control or PRMT5 shRNA (70%–75% knockdown). For G and H, the left panel corresponds to lifetime ECAR/OCR signal for 1 representative experiment, with each data point corresponding to 3 technical replicates and the right panel corresponds to data pooled from 4 (ECAR) or 3 (OCR) independent experiments; Student’s t test, *P < 0.05, ***P < 0.001. Bar graphs display mean ± SD.
HIF-1α has been shown to be a crucial transcription factor that regulates T cell metabolism and drives Th17 differentiation (34, 35). Hif1a expression was reduced in T cells from PRMT5-KO T cells, suggesting that PRMT5’s modulation of Th17 differentiation may be additionally supported via energy metabolism. To determine whether PRMT5 deletion affected energy metabolic pathways, we performed GO analyses on the Th17 RNA-Seq data. These analyses revealed that at least one-fourth of the genes in the glycolysis, lactate, or TCA pathways were significantly reduced in activated Th17 cells from T-PRMT5Δ/Δ mice (Figure 6F). To determine whether PRMT5 controlled glycolysis to lactate– or TCA-mediated oxidative phosphorylation at the functional level, we performed in vitro metabolic studies in human Jurkat T cells. Knockdown of PRMT5 decreased the extracellular acidification rate (ECAR), which is a measure of lactate generation downstream of glycolysis (Figure 6G). The oxygen consumption rate (OCR), which serves as a measure of TCA oxidative phosphorylation, was also decreased in Jurkat T cells with a PRMT5 knockdown (Figure 6H). Because Jurkat T cells were transformed, it was important to validate these results in primary T cells. So far, we have been able to validate glycolysis effects in primary T cells, as we observed robustly reduced lactate production in Th0, Th1, and Th17 cell supernatants from T-PRMT5Δ/Δ mice (Supplemental Figure 6). Overall, these data show that PRMT5 promotes Th17 cell differentiation by enhancing the biosynthesis of cholesterol intermediates and that it additionally supports energy metabolism.
PRMT5 is necessary to drive CD4+ Th cell pathogenesis and EAE autoimmunity.
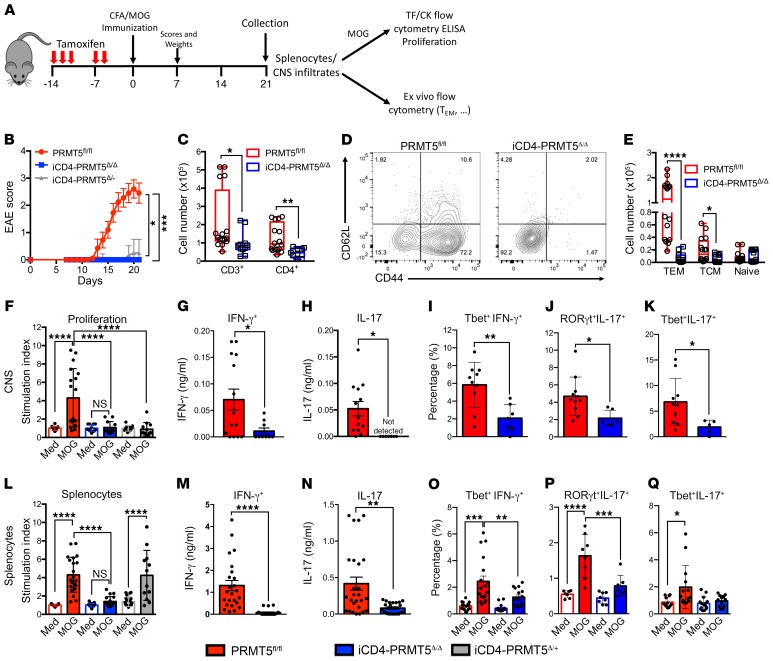
Th17 responses drive several autoimmune and inflammatory diseases, including MS. Given the crucial role of PRMT5 in Th17 differentiation, we hypothesized that loss of PRMT5 in the Th cell compartment would suppress EAE autoimmunity. To test this hypothesis, we used the CD4+ Th cell–specific (Figure 7A) iCD4-PRMT5Δ/Δ PRMT5-KO model, characterized by a normal proportion and number of T cells in the thymus and periphery (Supplemental Figure 2). EAE was abolished in iCD4-PRMT5Δ/Δ mice, whereas mice with a heterozygous deletion of PRMT5 in Th cells experienced delayed mild disease (Figure 7B). Disease development in PRMT5fl/fl mice was associated with significant weight loss, whereas iCD4-PRMT5Δ/+ and iCD4-PRMT5Δ/Δ maintained their weight (Supplemental Figure 7). Total CNS-infiltrating T cell numbers were substantially reduced in iCD4-PRMT5Δ/Δ mice (Figure 7C), even as normal peripheral T cell numbers existed in this conditional KO model (Supplemental Figure 2, T–Y). Within CD4+ Th cells, important losses were evident in the CD44+CD62L– Tem and CD44+CD62L+ Tcm Th cell compartments (Figure 7, D and E). This indicates that Th-specific PRMT5 deficiency in Th cells is sufficient to impair recruitment of memory Th cells, presumably with pathogenic phenotypes, into the CNS. To address this, myelin-specific T cell proliferation and pathogenic Th1 and Th17 responses were evaluated in the CNS. Infiltrating CNS cells from iCD4-PRMT5Δ/Δ mice did not proliferate (Figure 7F) or secrete IFN-γ (Figure 7G) or IL-17 (Figure 7H) in response to myelin oligodendrocyte glycoprotein (MOG). In particular, a substantial loss of MOG-specific T-bet+IFN-γ+ Th1 cells (Figure 7I), RORγt+IL-17+ Th17 cells (Figure 7J), and the particularly pathogenic T-bet+IL-17+ Th17 population (Figure 7K) was observed.
Figure 7. Th cell–specific Prmt5 deficiency prevents induction of EAE autoimmunity.
(A) Schematic of tamoxifen treatment/EAE experimental design and downstream analyses. (B) EAE score in iCD4-PRMT5Δ/Δ and indicated controls after MOG35–55/CFA immunization. All mice were treated with tamoxifen by oral gavage before immunization. (C–E) Flow analysis and quantification of CNS-infiltrating (C) CD3+ and CD3+CD4+ T cells or (D and E) naive, Tem, and Tcm phenotype CD4+ T cells from iCD4-PRMT5Δ/Δ and indicated controls 21 days after MOG35–55/CFA immunization. (F–Q) CNS-infiltrating cells (F–K) or splenocytes (L–Q) from day 14 iCD4-PRMT5Δ/Δ and indicated controls were reactivated with MOG35–55. (F and L) Proliferation was monitored by 3H-thymidine incorporation and expressed as a relative proliferation ratio to the resting PRMT5fl/fl media control condition. MOG35–55-reactivated cells were analyzed by ELISA for (G and M) IFN-γ and (H and N) IL-17 secretion, and flow cytometry for (I and O) T-bet+IFN-γ+ Th1, (J and P) RORγt+IL-17+ Th17, and (K and Q) T-bet+IL-17+ cell populations. Data are pooled from 4 independent experiments, n = 6–10 mice. Kruskal-Wallis with Dunn’s multiple-comparisons test (B and Q); 1-way ANOVA followed by Sidak’s multiple-comparisons test (F, L, O, and P) or Student’s t test (C, E, G–K, M, and N). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Box-and-whisker plots boxes extend from 25th to 75th percentiles, all points shown, whiskers extend from min to max, line represents median. Bar graphs are indicated as mean ± SD for F, I–L, O–Q and mean ± SEM for G, H, M, and N. Tem, effector memory T cells; Tcm, central memory T cells; TF, transcription factor; Ck, cytokine.
Because Th17 differentiation is abrogated in the absence of PRMT5, we expected this defect to be evident in peripheral T cell responses in the spleen. MOG-specific T cell proliferation (Figure 7L), Th1 (Figure 7, M and O), and Th17 (Figure 7, N and P) responses were notably reduced among splenocytes, whereas T-bet+IL-17+ Th17 cell responses did not reach statistical significance (Figure 7Q). Similar abrogation of EAE disease and loss of Th1 and Th17 responses was observed in the T-PRMT5Δ/Δ mouse model where all T cells lacked PRMT5 (Supplemental Figure 8). Overall, these data reveal that Th-specific PRMT5 deficiency leads to marked suppression of MOG-specific, pathogenic Th17 and Th1 cell responses, demonstrating that PRMT5 expression in CD4+ T cells is necessary to drive pathogenic Th cell–mediated EAE autoimmunity.
Discussion
Here we report that the arginine methyltransferase PRMT5 in the Th cell compartment is absolutely required for Th17 differentiation and EAE. We also have provided evidence that reveals the mechanism by which PRMT5 promotes Th17 differentiation. PRMT5 methylates the cholesterol biosynthesis regulator SREBP1 in T cells, setting the stage for Th17 differentiation by promoting cholesterol biosynthesis pathway enzymes that produce RORγt agonists. PRMT5 thereby promotes RORγt activity and drives expression of key Th17 differentiation and pathogenicity genes. PRMT5 also modulates the thymic development and peripheral homeostasis stages of CD4+ Th cells. We also observed broad and marked T cell–intrinsic defects in iNK T and CD8+ T cell development or maintenance, respectively, in the absence of PRMT5. Overall, the PRMT5Δ/Δ mouse models revealed crucial roles of PRMT5 during thymic T cell development, peripheral homeostasis, Th cell polarization, and T cell–mediated autoimmune disease.
The posttranslational modifier PRMT5 is essential for life (36) and has been implicated in stem cell function and development (5, 37, 38). PRMT5 plays crucial roles in carcinogenesis (39, 40) and autoimmune pathogenesis (8). Although multiple mechanisms tightly control PRMT5 expression in nontransformed T cells (9), uncontrolled PRMT5 expression is a hallmark of hematologic and solid oncogenic tumors (41–45). The driver role of PRMT5 in cancer has led to efforts to develop PRMT5-selective inhibitors as a therapy for solid and hematologic cancers that are currently being tested for safety and efficacy (Clinicaltrials.gov, registration ID: NCT03573310, NCT03854227, NCT02783300, NCT03614728). Data presented in this report now suggest that PRMT5 inhibition can potentially be beneficial in the treatment of autoimmune diseases. Homozygous, but not heterozygous, PRMT5 deficiency early in bone marrow hematopoiesis results in severe bone marrow aplasia and eventual death (18). Because of this, safety evaluation studies during clinical trials pay special attention to bone marrow aplasia. We previously reported that PRMT5 inhibitors suppress already-polarized Th1 and Th17 cell function and EAE (8). Those effects could have emanated from drug off-target effects or from loss of PRMT5 activity on cells other than T cells. We have now genetically validated these results and shown that PRMT5 deletion plays crucial roles in T cell development and biology and EAE disease, with robust suppression of Th17 and Th1 responses generally involved in pathogen-protective immunity. In addition, we have provided evidence that PRMT5 expression during the initial exposure of naive T cells to Th polarizing cytokines results in substantive reductions in the total number of committed Th1 and Th17 cells as observed in PRMT5-KO T cells. The impact of PRMT5 on immune responses will be important to consider when PRMT5 inhibitors are tested as therapeutic agents for T cell–mediated diseases. It is important to note that more subtle immune defects were observed in heterozygote Th PRMT5-KO mice, suggesting that careful PRMT5 inhibitor dosing could be the key to avoiding unintended effects. Nonetheless, it will be important to carefully monitor potential hematopoietic and immune effects in PRMT5 inhibitor–based therapeutic approaches.
Our data support the concept that PRMT5 promotes a metabolic switch in cholesterol biosynthesis, leading to expression of multiple enzymes in the cholesterol pathway. PRMT5 can promote gene expression in cancer cells by enhancing constitutive and alternative splicing (46, 47). In addition, PRMT5 can methylate several targets that contribute to cellular metabolic control, including SREBP1 (28). Specifically, PRMT5 can methylate SREBP1 at R321. This modification in turn inhibits SREBP1 phosphorylation and proteasomal degradation (28). Our findings further demonstrate that SREBP1 indeed undergoes PRMT5-dependent methylation in T cells, which supports our proposed model of SREBP1 regulation by PRMT5 in T cells. We surmise that R321 methylation by PRMT5 stabilizes the cleaved nuclear form of SREBP1 in activated T cells, thereby promoting expression of the cholesterol biosynthesis program, a known function of SREBP1 (48). Consistent with this hypothesis, we observed PRMT5-dependent SDM of SREBP1 in T cells and Insig-1 transcription, a direct and obligatory consequence of nuclear SREBP translocation (48, 49). Conclusively identifying the specific R residue methylated by PRMT5 in SREBP1 that mediates SREBP1 stability and Th17 phenotype will be the focus of future studies. Several cholesterol pathway intermediates, including desmosterol, serve as agonists of RORγt and are required for Th17 differentiation (24–26). We found that RORγt activity is dependent on PRMT5 expression. In addition, supplementation with the cholesterol intermediate desmosterol restored Th17 differentiation in iCD4-PRMT5Δ/Δ T cells, demonstrating that PRMT5 is necessary for Th17 cell differentiation by promoting cholesterol biosynthesis. It is important to note that PRMT5 promotes cholesterol biosynthesis enzyme expression even in Th0 (no polarizing cytokines) conditions (Figure 5, A–C). This is consistent with the concept that PRMT5 induction at the Th0 stage poises recently activated naive T cells toward a Th17 fate via production of RORγt agonistic cholesterol pathway intermediates. PRMT5 also promotes T cell proliferation, raising the question of whether the promotion of proliferation contributes to differentiation toward the Th17 lineage. We believe that this is unlikely, as there is strong evidence that conditions that reduce proliferation, such as IL-2 blockade/deletion, do not reduce Th17 differentiation, but instead promote it (50). Besides effects on Th17 differentiation via cholesterol biosynthesis, PRMT5 also promotes expression of Hif1a, which encodes the HIF-1α transcription factor that promotes Th17 differentiation via direct RORγt activation and promotion of the glycolytic pathway (34, 35). We also observed reduced glycolytic metabolism in PRMT5-KO T cells, which could be a consequence of Hif1a modulation.
We found that PRMT5 expression in peripheral CD4+ T cells is an essential driver for the development of EAE disease. Although pan-T cell T-PRMT5Δ/Δ mice were completely resistant to EAE, this effect could be due to these mice lacking full CD4+, CD8+, and iNK T cell compartments. Using the inducible peripheral CD4+ Th cell–specific PRMT5-KO model (iCD4-PRMT5Δ/Δ) with normal CD4+ T cell numbers, we found that PRMT5 in CD4+ Th cells was still required for EAE development through mechanisms beyond lack of CD4+ Th cells. We observed strongly reduced proliferative, Th17, and Th1 recall responses in these mice, as well as severely reduced numbers of memory T cell infiltration in the CNS. The loss of Th17 responses was consistent with reduced Th17 differentiation. However, the Th1 cell response was also lost during EAE, in apparent contradiction to increased Th1 polarization observed in vitro (Figure 4, B–H). This result could emanate from decreased Th1 and Th17 cell expansion in PRMT5-KO T cells, as observed in vitro. Th1 polarization enhancement in the KO may be compensated in vivo by reduced expansion of Th1 cells during differentiation or reactivation stages. Indeed, PRMT5 inhibitors strongly suppress expansion of nontransformed Th1 memory T cell lines (8). Therefore, the overall impact of in vivo Th cell PRMT5 deficiency appears to stem from loss of pathogenic Th1 and Th17 responses. The connections between cholesterol pathway intermediates and pathogenic Th17 cells corroborate with and explain observations from statin-treated MS patients. Statin drugs inhibit the HMG-CoA reductase rate-limiting step in the cholesterol biosynthesis pathway (51) and substantially reduce disability scores and atrophy in MS patients (52, 53). The mechanism behind these effects has been unclear, but causal modeling analyses have suggested an effect independent of peripheral cholesterol levels (54). Instead, these analyses implicate upstream intermediates of the cholesterol biosynthesis pathway intermediates (54), such as those induced by PRMT5. Overall, the links between PRMT5, cholesterol biosynthesis pathway intermediates, and pathogenic Th17 responses have therapeutic and biomarker implications in MS.
Given the well-established links between PRMT5 and proliferation, it is conceivable that PRMT5 deletion at the DP stage would significantly impair thymocyte development. Indeed, loss of DP, CD4 SP, and Treg cells was observed in the T-PRMT5Δ/Δ model, consistent with CD4-driven Cre-mediated deletion at the DP stage. Thymic defects were not observed in another report of a constitutive CD4-Cre–driven PRMT5-KO mouse (11). This difference may be due to the deletion of all protein-coding isoforms in our KO model, compared with a KO of only the long isoform in the model previously reported from the lab of Hiroshi Takayanagi (11). We shall refer to this model as the Takayanagi model henceforth. Alternatively, the lack of thymic defects in the Takayanagi model may be mediated by the ubiquitous Prmt5 heterozygous–deficient background used in that model (11). The second possibility is highly likely, as we observed thymic defects in Prmt5 homozygous and heterozygous KO mice in comparison with PRMT5fl/fl and CD4-Cre controls (Figure 2). Although DP and CD4 SP numbers were decreased, their relative distribution remained stable, suggesting a defect mediated by impaired expansion. In contrast, CD8 SP numbers were not as strongly reduced, consistent with the higher proliferation that DP and CD4 SP cells versus CD8 SP undergo during thymic development (55). Finally, as in the Takayanagi model, a striking defect was observed in iNK T cells, again consistent with the reported high proliferative needs of iNK T cells during thymic development (56).
Beyond thymus-level defects, we also observed reduced numbers of CD4+, CD8+, and iNK T cells in the periphery, consistent with the observations in the Takayanagi model (11). The peripheral CD4+ T cell loss likely derived from reduced entry into the peripheral compartment from the thymus and inability to homeostatically proliferate in response to IL-7 signaling, as previously proposed (11). The striking CD8 peripheral loss instead appeared to largely stem from inability to survive and expand in the peripheral compartment, as thymic CD8+ T cell numbers were less affected. Finally, T cell proliferation in response to antigenic stimuli was abrogated in the absence of PRMT5. These data demonstrated that robust inhibition of T cell proliferation with PRMT5 inhibitors (8) was not due to off-target effects, but was indeed due to PRMT5 loss of function.
In summary, our recent work described herein identifies a potentially novel T cell–intrinsic role for PRMT5 as a regulator of T cell development, maintenance, and Th17 cell differentiation and defines a metabolic switch mechanism by which PRMT5 controls Th17 differentiation. PRMT5 methylates SREBPs, which in turn promote the expression of enzymes in the cholesterol biosynthesis pathway, increasing RORγt agonist activity and activating the Th17 program. The role of PRMT5 in driving the differentiation of pathogenic Th17 cell responses identifies PRMT5 as a promising therapeutic target in MS and other Th17-mediated inflammatory diseases.
Methods
Mice.
All mice were bred and maintained under protocol number 2013A00000151-R1. Sperm carrying the Prmt5tm2c(EUCOMM)wtsi mutation were acquired from the Wellcome Trust Sanger Institute (Cambridgeshire, United Kingdom). A Prmt5tm2c(EUCOMM)wtsi founder was obtained via in vitro fertilization at The Ohio State University Comprehensive Cancer Center Animal Modeling Core (supported in part by grant P30 CA016058). Homozygous Prmt5tm2c(EUCOMM)wtsi mice were bred with B6(129X1)-Tg(Cd4-Cre ERT2)11Gnri/J or Tg(Cd4-Cre)1Cwi CD4-Cre mice from The Jackson Laboratory. Males and females were used for experiments without significant differences observed between genders.
PCR genotyping.
PCR reaction for genotyping was performed using the following primers: F: 5′-TCACACCCAGTCTCTTAC-3′, R: 5′-ACACACATGGCACATATACAGA-3′. Cycling conditions were 94°C, 2 minutes; (94°C, 30 seconds; 63°C, 1 minute; 72°C, 1 minute) for 30 cycles; 72°C, 2 minutes; 4°C hold in an Eppendorf Mastercycler Gradient Thermocycler.
In vivo tamoxifen treatment.
Tamoxifen (Sigma-Aldrich, catalog no. T5648) was solubilized at 40 mg/mL in corn oil by shaking at 37°C overnight. iCD4-PRMT5Δ/Δ mice were given 300 mg/kg tamoxifen (150 μL per 20 g mouse) daily for 3 to 5 days by oral gavage. Seven days after starting treatment, mice were euthanized or used for experiments. For extended tamoxifen treatment, mice were treated daily for 5 days every other week for a total of 3 sets of tamoxifen treatment. Mice were collected 3 days after the last day of tamoxifen treatment.
T cell in vitro assays.
Total CD4+ T cells were isolated using a CD4+ T cell isolation kit (Miltenyi Biotec, catalog no. 130-104-454 or Stem Cell Technologies, catalog no. 19852) and an autoMACS Pro (Miltenyi Biotec) or EasyEights magnet (Stem Cell Technologies). CD3+ (Stem Cell Technologies, catalog no. 19851), CD8+ (Stem Cell Technologies, catalog no. 19853), and/or naive CD4+ T cells (Stem Cell Technologies, catalog no. 19765) were isolated using Stem Cell EasyEights Magnet. For naive T cell differentiation experiments, naive T cells were differentiated in the presence of Th1 (IL-12, IL-2, anti–IL-4), Th2 (IL-4, IL-2, anti–IL-12, anti–IFN-γ), Th17 (TGF-β, IL-6), or Treg (TGF-β, all-trans retinoic acid, IL-2) polarizing conditions. Isolated T cells were activated with 5 μg/mL coated anti-CD3 and 2 μg/mL soluble anti-CD28. T cells from iCD4-PRMT5Δ/Δ mice were supplemented with 2 μg/mL 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich, catalog no. H7904) during in vitro cell culture. Desmosterol and lanosterol (Avanti Polar Lipids) were directly added to FBS-containing media at 15 μM final concentrations.
Flow cytometry.
Thymocytes, splenocytes, and LN cells were taken directly ex vivo for flow cytometric analyses. MOG35–55-restimulated splenocytes and LN cells were treated with PMA/ionomycin and GolgiStop (BD Biosciences, catalog no. 51-2092KZ) for 4 hours before collection for staining. Cells were treated with Fc region blockade and then incubated with surface stain markers, including B220 (BD Biosciences, catalog no. 553087, clone RA3-6B2), CD3 (BioLegend, catalog no. 100334, clone 145-2C11), CD4 (BioLegend, catalog no. 100531 or eBioscience, catalog no. 12-0042-85, clone RM4-5), CD25 (Invitrogen, catalog no. RM6017 or eBioscience, catalog no. 45-0251-82, clone PC61 5.3), CD44 (eBioscience, catalog no. 48-0441-82 or 25-0441-82, clone IM7), CD62L (BioLegend, catalog no. 104426 or 104411, clone MEL-14), and CD8 (BD Biosciences, catalog no. 560182, clone 53-6.7) for 15 minutes at 4°C. Cells were then fixed with eBioscience fixation/permeabilization buffer (eBioscience, catalog no. 00-5523-00), or BD Biosciences fixation buffer (BD Biosciences, catalog no. 554715). Intracellular staining with FoxP3 (eBioscience, catalog no. 17-5773-82, clone FJK-16s), T-bet (BioLegend, catalog no. 644810 or 644808, clone 4B10), IL-17 (BioLegend, catalog no. 506916, clone TC11-1810.1), IFN-γ (BioLegend, catalog no. 505830, clone XMG1.2), or RORγt (eBioscience, catalog no. 12-698880, clone AFKJS-9) was performed for 30 to 45 minutes at 4°C. Flow cytometry was run on a FACSCalibur with DxP multicolor upgrades (Cytek) and analysis was performed using FlowJo.
3H-thymidine proliferation assay.
Isolated CD4+ T cells were plated at a density of 100,000 to 125,000 cells per well in a 96-well plate. After 48 hours of culture, cells were given 1 μCi of tritiated (3H)-thymidine; 16 hours later, cells were harvested onto a Unifilter-96 plate (PerkinElmer, catalog no. 6005174). Scintillation fluid was added and CPM was measured by a scintillation counter (Beckman Coulter).
Western blot.
Activated primary CD4+ T cells were collected directly ex vivo or activated and collected at indicated time points, and cell pellets were frozen at –80°C. Cell pellets were lysed in RIPA buffer (10 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholate) containing protease and phosphatase inhibitors (Thermo Fisher Scientific, catalog no. 1862209 and 1862495). Five to 10 μg of protein were run on a 14% SDS-PAGE gel and transferred onto PVDF membrane. Blots were blocked with Odyssey blocking buffer (LI-COR, catalog no. 927-50000) and primary antibodies were probed for 3 hours at room temperature. Primary antibodies used include PRMT5 (Abcam, catalog no. ab31751, 1:1000), PRMT1 (Cell Signaling Technologies, catalog no. 2449, 1:500), SYM10 (MilliporeSigma, catalog no. 07-412, 1:300), STAT3 (Santa Cruz Biotechnology, catalog no. sc-8019), p-STAT3 (Cell Signaling Technologies, catalog no. 91315) and SREBP1 (BD Biosciences, catalog no. 557036), and β-actin (Sigma-Aldrich, catalog no. A1978, 1:20,000). IP of SREBP1 was done following the general protocol suggested by Santa Cruz Biotechnology. Samples for IP were lysed from empty vector control and shPRMT5 Jurkat T cells using RIPA buffer (25 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 0.1% Triton X-100, 0.5% sodium deoxycholate). IP antibodies used were SREBP1 and normal mouse IgG (Santa Cruz Biotechnology, catalog no. sc-2025). Protein A/G Plus Agarose beads were used for the pull down (Santa Cruz Biotechnology, catalog no. sc-2003). After final wash steps discussed in the IP protocol, protein samples were eluted from the beads by heating at 60°C for 10 minutes to prevent the release of antibodies from the beads. Samples were electrophoresed as discussed above. Secondary antibodies anti-mouse 680RD and/or anti-rabbit 800CW (LI-COR, catalog no. 926-68072 and 926-32213) were then used at 1:20,000. Blots were imaged on an Odyssey CLx (LI-COR) and quantification of protein was performed using ImageStudio software.
RNA-Seq.
Isolated resting and activated (anti-CD3/CD28, no cytokines, 2 days) CD4+ T cells from PRMT5fl/fl and iCD4-PRMT5Δ/Δ mice (n = 3 pooled mice/sample and n = 3 samples per group) were stabilized in RNAlater (Thermo Fisher Scientific, catalog no. AM7020) until RNA isolation. When all samples were ready for RNA isolation, the cell suspension was diluted in a 1:1 volume with 1× PBS before lysis with TRIzol (Thermo Fisher Scientific, catalog no. 15596018). RNA isolation was performed with the Direct-zol RNA Miniprep (Zymo Research, catalog no. R2052) according to the manufacturer’s instructions. One nanogram of total RNA was used for quality control (QC), library preparation, and RNA-Seq. The quality of total RNA was evaluated using Agilent 2100 Bioanalyzer and RNA Nano chip (Agilent Technologies), and only samples with an RNA integrity number of 7.7 or greater were sequenced.
RNA-Seq was performed by the Genomic Services Laboratory of the Abigail Wexner Research Institute at Nationwide Children’s Hospital, Columbus, Ohio. RNA-Seq libraries were prepared using Illumina’s TruSeq Stranded protocol. In summary, ribosomal RNA (rRNA) was removed from 250 ng of total RNA with Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) (Illumina). The kit depletes samples of cytoplasmic and mitochondrial rRNA using biotinylated, target-specific oligos combined with rRNA removal beads. After rRNA removal, mRNA is fragmented using divalent cations under elevated temperature and converted into ds cDNA. Incorporation of dUTP in place of dTTP during second strand synthesis inhibits the amplification of the second strand. The subsequent addition of a single “A” base allows for ligation of dual unique tagging sequences. Adaptor-ligated cDNA was amplified by limit-cycle PCR. Quality of libraries were determined via Agilent 4200 TapeStation using a High Sensitivity D1000 ScreenTape Assay kit, and quantified by KAPA qPCR (KAPA BioSystems). Approximately 60–80 million paired-end 150-bp sequence reads were generated for each library on the Illumina HiSeq 4000 platform. RNA-Seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) (57) and are accessible through GEO Series accession numbers GSE141196 (includes Th0 and Th17 data), GSE141168 (Th0 data), and GSE141194 (Th17 data).
For analysis, low-quality reads (q < 10) and adaptor sequences were eliminated from raw reads using BBDuk version 37.64 (DOE Joint Genome Institute). Each sample was aligned to the GRCm38.p3 assembly of the Mus musculus reference from NCBI using version 2.6.0c of the RNA-Seq aligner STAR (58). Features were identified from the GFF file that came with the assembly from Gencode (Release M19) and feature coverage counts were calculated using feature counts (59). Differentially expressed features were calculated using DESeq2 (Bioconductor Release 3.9).
Real-time PCR.
To evaluate mRNA expression, 300–500 ng of RNA was reverse transcribed using oligo d(T) or random primers and Superscript III (Thermo Fisher Scientific, catalog no. 18080051) according to the manufacturer’s instructions; TaqMan quantitative real-time PCR was performed using mouse Prmt5 (Mm00550472_m1), mouse Hprt (Mm0044968_m1), mouse Cyp51a (Mm00490968_m1), mouse Tm7sf2 (Mn0123354_g1), mouse Lss (Mm00461312_m1), mouse Nsdhl (Mm00477897_m1), mouse Acat2 (Mm00782408_s1), and mouse Hmgcs1 (Mm01304569_m1), as previously described (Thermo Fisher Scientific). Samples were run on Quant Studio 5 (Thermo Fisher Scientific). An initial denaturation step at 95°C for 10 minutes was followed by 40 cycles of denaturation at 95°C for 15 seconds and primer annealing/extension at 60°C for 60 seconds. Results were analyzed using the comparative Ct method after ensuring comparable amplification efficiencies for test and housekeeping transcripts.
Cell lines.
The human NCI-H522 non–small cell lung cancer adenocarcinoma cell line originated from an NCI-60 cell line panel obtained from Daniel Haber at Massachusetts General Hospital. The HEK-293T human embryonic kidney cell line originated from Richard Van Etten’s laboratory at Tufts Molecular Oncology Research Institute. The human Jurkat T cell line has been maintained in the Tsichlis lab since his work at Fox Chase University and originated from David Wiest. NCI-H522, HEK-293T, and Jurkat cells were grown in Dulbecco’s modified Eagle’s medium (MilliporeSigma, catalog no. D5796) and RPMI-1640 (MilliporeSigma, catalog no. R8758), respectively. The media were supplemented with penicillin/streptomycin, nonessential amino acids, L-glutamine, and 10% FBS. Cells were used within 3 months of resuscitation. Cell lines were periodically checked for mycoplasma.
PRMT5 knockdown.
The human PRMT5 gene was subcloned in the pENTR/D-TOPO cloning vector (Invitrogen, catalog no. 45-0218) using cDNA derived from NCI-H522 shCon after gel purification using the Nucleospin PCR and Gel Clean Up kit (Macherey-Nagel, catalog no. 740609.50). Following this, the 2 clones were recombined with pLx304 DEST (Addgene plasmid 25890) using standard Clonase II LR mix (Thermo Fisher Scientific, catalog no. 11791100) following the standard manufacturer’s protocol. pLx304 DEST EV was used as a control. The empty vector control (Open Biosystems, catalog no. RSH4080) or PRMT5 shRNA (MilliporeSigma, catalog no. SHCLNG-NM_006109, clone ID TRCN0000107085 and clone ID TRCN0000379612) was used. Lentiviral constructs were packaged in HEK-293T cells by transient transfection, in combination with psPax2 and pMΔ2.G. Transfections were carried out using 2× HEPES-buffered saline (Sigma-Aldrich, catalog no. 51558) and CaCl2 protocol. Spinfections were carried out in the presence of 8 μg/mL polybrene (Sigma-Aldrich, catalog no. 107689) rotating at 32 degrees at 2,400 rpm. Depending on the selection marker in the vector, infected cells were selected for resistance to 10 μg/mL puromycin (Gibco, catalog no. A11138) or 10 μg/mL blasticidin (Gibco, catalog no. A1113903). Cells infected with multiple constructs were selected for infection with the first construct before the next infection.
RORγt-GAL4 luciferase assays.
The RORγt-GAL4 fusion protein was a gift from the lab of Dan R. Littman at New York University Langone Health. This protein was used to drive luciferase activity from the PGL4.31[luc2p/GAL4UAS/Hygro] (luc2p) vector (Promega, catalog no. C935A). The pRL-CMV vector (Promega, catalog no. E226A) was used as the internal control Renilla luciferase reporter. Cells were plated according to manufacturer instructions for transfection reagents. Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, catalog no. 11668019) was used for human lung adenocarcinoma H522 cells and TransIT-X2 transfection reagent (Mirus Bio, catalog no. MIR6000) was used for human Jurkat T cells. At 48 hours after plating, the H522 cells were washed twice with ice-cold PBS in the plate, and Jurkat cells were collected into Eppendorf tubes and spun down before PBS washes and luciferase activity measurements using the Dual Luciferase Reporter Assay (Promega, catalog no. E1910). Cells were lysed in 1 part Passive Lysis Buffer (Promega) made from a 1:5 dilution of 5× Passive Lysis Buffer. Samples were allowed to rock at room temperature for 30 minutes before a quick centrifugation step at 12,000 g and 4°C for 2 minutes. Samples were plated in a solid white polystyrene 96-well plate (Corning, catalog no. 353296) at 20 μL/well. LARII and Stop & Glo Reagent were prepared according to manufacturer’s instructions. Plates were read in a GloMax 96 Luminometer (Promega, catalog no. E4861).
Measurement of mitochondrial respiration and glycolysis.
The OCR for mitochondrial oxidation and ECAR for glycolytic activity were determined using mitoXpress assay (Agilent Technologies, catalog no. MX-200-4) and pHXtra assay (Agilent Technologies, catalog no. PH-200-4), respectively. Briefly, Jurkat T cells lines stably transduced with control or PRMT5 shRNA were plated at 600,000 cells/well using fresh media. For glycolytic rates, cells were incubated in absence of CO2 for 2 hours before the assay. Both assays were read during 120 minutes at 37°C in a Cytation 1 imaging multimode reader (BioTek Instruments). Additional glycolysis measurements were obtained via lactate assay, using Cayman’s glycolysis cell-based assay kit (Cayman Chemical, catalog no. 600450). T cells were activated with anti-CD3/CD28 for 3 days in the indicated conditions. Supernatants were collected to measure the amount of L-lactate released, as per the manufacturer’s recommended procedure.
EAE.
To induce EAE, mice were immunized with MOG35–55 (CS Bio) and CFA (Difco, BD Biosciences, catalog no. 231131) emulsion, as previously described (8). Mice were monitored for disease every day and scored blinded. At the indicated time points, mice were euthanized by injection with 20 mg/mL ketamine and 4 mg/mL xylazine (150 μL/20 g mouse) and perfused with PBS. Spleens, brains, and spinal cords were collected from representative mice and processed for in vitro studies. To isolate brain and spinal cord mononuclear cells, brains and spinal cords were processed through a 70-μm strainer and separated by a 70%–30% isotonic Percoll gradient. Spleens were processed through a 70-μm strainer and red blood cells were lysed by incubating for 1 minute in hypotonic solution. Splenocytes and infiltrating CNS cells were used for ex vivo flow cytometry or reactivated with MOG antigen as indicated.
Statistics.
Statistical analyses were performed using GraphPad Prism unless otherwise stated. One-way or 2-way ANOVA followed by Tukey’s, Dunnett’s, or Sidak’s post hoc multiple-comparisons test was performed as appropriate. Normality of data distribution was ensured via the D’Agostino-Pearson test or Shapiro-Wilk test, and equal variance was ensured via Bartlett’s test, automatically included in GraphPad Prism ANOVA analyses. If the distribution was nonparametric or could not be calculated because of sample size, significance was confirmed via nonparametric Kruskal-Wallis test or log-transformed to normalize and confirmed by 1-way ANOVA. The post hoc Dunnett’s test was solely used when means from multiple groups were compared against a single control group. Otherwise, Tukey’s (all means compared to all means) or Sidak’s (selected means compared) tests were used. The 2-tailed Student’s t test was solely used when the means of 2 independently and equally distributed samples were compared against each other. The Mann-Whitney test was performed for nonparametric EAE score data analysis. Outliers were identified using the ROUT method in GraphPad Prism. For RNA-Seq, raw RNA-Seq gene expression data were normalized and postalignment statistical analyses were performed using DESeq2 and custom analysis scripts written in R. Comparisons of gene expression and associated statistical analysis were made between different conditions of interest using the normalized read counts. All FC values are expressed as test condition/control condition, where values less than 1 are denoted as the negative of its inverse (note that there will be no FC values between –1 and 1, and that the FCs of “1” and “–1” represent the same value). Transcripts were considered significantly differentially expressed using a 10% FDR (DESeq2 adjusted P value ≤ 0.1).
Study approval.
All animal studies were performed after approval was obtained from The Ohio State University’s Institutional Animal Care and Use Committee (IACUC) under protocol 2013A00000151-R1.
Author contributions
MGA, PNT, LMW, SS, and GL contributed to conceptual experimental design. LMW, SS, CE, ZLPQ, GL, SAA, MB, AK, and JNM performed experiments and analyzed data. MGA, LMW, SS, SAA, ZLPQ, and MB interpreted data and wrote the manuscript. LMW and SS are listed as co–first authors. LMW initiated studies of conditional KO mouse characterization, immunological assessments, and EAE studies, several of which were performed collaboratively with SS’s assistance until her graduation. SS generated and analyzed RNA-Seq data, evaluated cholesterol biosynthesis enzyme expression and RORγt activity, and performed the IP experiments leading to the identification of SREBP1 as a PRMT5 methylation target. All authors thoroughly reviewed the manuscript.
Supplementary Material
Acknowledgments
This work was supported by funds from the NIH National Institute of Allergy and Infectious Diseases grants R01AI121405 and 1R21AI127354 (both to MGA), The Ohio State University School of Health and Rehabilitation Sciences start-up funds (to MGA), the Comprehensive Cancer Center Medicinal Chemistry Shared Resource (Core Cancer Center Support Grant P30CA016058), the NIH National Cancer Institute grant 01-CA186729 (to PNT), the Pelotonia Postdoctoral Fellowship (to GL), and the Center for Clinical and Translational Science (CCTS) Award Number Grant UL1TR002733 from the National Center For Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Advancing Translational Sciences or the NIH. We would like to thank the Genomic Services Laboratory of the Abigail Wexner Research Institute at Nationwide Children’s Hospital for their help with RNA-Seq. We thank Amy Wetzel, Shireen Woodiga, Anthony Miller, and Saranga Wijeratne of the Genomic Services Laboratory at the Abigail Wexner Research Institute at Nationwide Children’s Hospital, Columbus, Ohio, for their help with sample QC, library preparation, RNA-Seq, and analysis of data. We also thank Dan Littman and Fabio Santori for their gift of the RORγt-GAL4 fusion protein, Ning Quan and Gene Oltz for their insightful discussions and comments, Menglin Xu for biostatistics support, and Carson Reider at the Neurological Research Institute for expert manuscript editing support.
Version 1. 02/24/2020
Electronic publication
Version 2. 04/01/2020
Print issue publication
Funding Statement
This work was supported by funds from the National Institutes of Health National Institute of Allergy and Infectious Diseases Grants R01AI121405 and 1R21AI127354 (both to M.G.-d.-A.), The Ohio State University School of Health and Rehabilitation Sciences start-up funds (to M.G.-d.-A.), the Comprehensive Cancer Center Medicinal Chemistry Shared Resource (Core Cancer Center Support Grant P30CA016058) and the Center for Clinical and Translational Science (CCTS) Award Number Grant UL1TR002733 from the National Center For Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Advancing Translational Sciences or the National Institutes of Health.
Footnotes
Conflict of interest: MGA is listed as an inventor in a pending patent for PRMT5 inhibitors and a licensing deal with Prelude Therapeutics.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(4):1683–1698.https://doi.org/10.1172/JCI131254.
Contributor Information
Shouvonik Sengupta, Email: shouvonik.sengupta@osumc.edu.
Claudia Edell, Email: edell.7@osu.edu.
Zayda L. Piedra-Quintero, Email: zayda.piedra-quintero@osumc.edu.
Stephanie A. Amici, Email: stephanie.amici@osumc.edu.
Makenzie Bevins, Email: makenzie.bevins@osumc.edu.
Philip N. Tsichlis, Email: philip.tsichlis@osumc.edu.
Mireia Guerau-de-Arellano, Email: mireia.guerau@osumc.edu.
References
- 1.Guccione E, Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol. 2019;20(10):642–657. doi: 10.1038/s41580-019-0155-x. [DOI] [PubMed] [Google Scholar]
- 2.Yang ML, et al. Lupus autoimmunity altered by cellular methylation metabolism. Autoimmunity. 2013;46(1):21–31. doi: 10.3109/08916934.2012.732133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120(pt 24):4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 4.Branscombe TL, et al. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276(35):32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 5.Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72(11):2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohail M, Zhang M, Litchfield D, Wang L, Kung S, Xie J. Differential expression, distinct localization and opposite effect on Golgi structure and cell differentiation by a novel splice variant of human PRMT5. Biochim Biophys Acta. 2015;1853(10 pt A):2444–2452. doi: 10.1016/j.bbamcr.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Litzler LC, et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat Commun. 2019;10(1):22. doi: 10.1038/s41467-018-07884-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb LM, et al. PRMT5-selective inhibitors suppress inflammatory T cell responses and experimental autoimmune encephalomyelitis. J Immunol. 2017;198(4):1439–1451. doi: 10.4049/jimmunol.1601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webb LM, Narvaez Miranda J, Amici SA, Sengupta S, Nagy G, Guerau-de-Arellano M. NF-κB/mTOR/MYC axis drives PRMT5 protein induction after T cell activation via transcriptional and non-transcriptional mechanisms. Front Immunol. 2019;10:524. doi: 10.3389/fimmu.2019.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geoghegan V, Guo A, Trudgian D, Thomas B, Acuto O. Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat Commun. 2015;6:6758. doi: 10.1038/ncomms7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue M, et al. Arginine methylation controls the strength of γc-family cytokine signaling in T cell maintenance. Nat Immunol. 2018;19(11):1265–1276. doi: 10.1038/s41590-018-0222-z. [DOI] [PubMed] [Google Scholar]
- 12.Dendrou CA, Fugger L, Friese MA. Immunopathology of multiple sclerosis. Nat Rev Immunol. 2015;15(9):545–558. doi: 10.1038/nri3871. [DOI] [PubMed] [Google Scholar]
- 13.Buck MD, O’Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med. 2015;212(9):1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bietz A, Zhu H, Xue M, Xu C. Cholesterol metabolism in T cells. Front Immunol. 2017;8:1664. doi: 10.3389/fimmu.2017.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jetten AM, Takeda Y, Slominski A, Kang HS. Retinoic acid-related orphan receptor γ (RORγ): connecting sterol metabolism to regulation of the immune system and autoimmune disease. Curr Opin Toxicol. 2018;8:66–80. doi: 10.1016/j.cotox.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parry RV, Ward SG. Protein arginine methylation: a new handle on T lymphocytes? Trends Immunol. 2010;31(4):164–169. doi: 10.1016/j.it.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci. 2011;36(12):633–641. doi: 10.1016/j.tibs.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J Clin Invest. 2015;125(9):3532–3544. doi: 10.1172/JCI81749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/S1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 20.Aghajani K, Keerthivasan S, Yu Y, Gounari F. Generation of CD4CreER(T2) transgenic mice to study development of peripheral CD4-T-cells. Genesis. 2012;50(12):908–913. doi: 10.1002/dvg.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14(5):489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499(7459):485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016;531(7596):651–655. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, et al. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORγ agonists. Nat Chem Biol. 2015;11(2):141–147. doi: 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, et al. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell. 2015;163(6):1413–1427. doi: 10.1016/j.cell.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santori FR, et al. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab. 2015;21(2):286–298. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundqvist A, et al. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1(6):379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, et al. Arginine methylation of SREBP1a via PRMT5 promotes de novo lipogenesis and tumor growth. Cancer Res. 2016;76(5):1260–1272. doi: 10.1158/0008-5472.CAN-15-1766. [DOI] [PubMed] [Google Scholar]
- 29.Geng F, et al. Inhibition of SOAT1 suppresses glioblastoma growth via blocking SREBP-1-mediated lipogenesis. Clin Cancer Res. 2016;22(21):5337–5348. doi: 10.1158/1078-0432.CCR-15-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151(2):289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yosef N, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496(7446):461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13(10):991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaublomme JT, et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163(6):1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146(5):772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tee WW, et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24(24):2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang T, et al. Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat Commun. 2015;6:7140. doi: 10.1038/ncomms8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5(3):288–303. doi: 10.1158/2159-8290.CD-14-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao X, Zhao S, Liu T, Liu Y, Liu Y, Yang X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J Histochem Cytochem. 2013;61(3):206–217. doi: 10.1369/0022155413475452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26(15):3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan F, et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014;74(6):1752–1765. doi: 10.1158/0008-5472.CAN-13-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powers MA, Fay MM, Factor RE, Welm AL, Ullman KS. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 2011;71(16):5579–5587. doi: 10.1158/0008-5472.CAN-11-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Z, et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J. 2012;446(2):235–241. doi: 10.1042/BJ20120768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin Y, et al. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun Signal. 2019;17(1):30. doi: 10.1186/s12964-019-0344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bezzi M, et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013;27(17):1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun CJ, et al. Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell. 2017;32(4):411–426.e11. doi: 10.1016/j.ccell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janowski BA. The hypocholesterolemic agent LY295427 up-regulates INSIG-1, identifying the INSIG-1 protein as a mediator of cholesterol homeostasis through SREBP. Proc Natl Acad Sci U S A. 2002;99(20):12675–12680. doi: 10.1073/pnas.202471599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26(3):371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Endo A. The discovery and development of HMG-CoA reductase inhibitors. J Lipid Res. 1992;33(11):1569–1582. [PubMed] [Google Scholar]
- 52.Chataway J, et al. Effect of high-dose simvastatin on brain atrophy and disability in secondary progressive multiple sclerosis (MS-STAT): a randomised, placebo-controlled, phase 2 trial. Lancet. 2014;383(9936):2213–2221. doi: 10.1016/S0140-6736(13)62242-4. [DOI] [PubMed] [Google Scholar]
- 53.Pihl-Jensen G, Tsakiri A, Frederiksen JL. Statin treatment in multiple sclerosis: a systematic review and meta-analysis. CNS Drugs. 2015;29(4):277–291. doi: 10.1007/s40263-015-0239-x. [DOI] [PubMed] [Google Scholar]
- 54.Eshaghi A, et al. Applying causal models to explore the mechanism of action of simvastatin in progressive multiple sclerosis. Proc Natl Acad Sci U S A. 2019;116(22):11020–11027. doi: 10.1073/pnas.1818978116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawicka M, et al. From pre-DP, post-DP, SP4, and SP8 thymocyte cell counts to a dynamical model of cortical and medullary selection. Front Immunol. 2014;5:19. doi: 10.3389/fimmu.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.et al. Intrathymic proliferation wave essential for Vα14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci U S A. 2009;106(21):8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.