Abstract
Despite the effective clinical use of steroids for the treatment of Diamond Blackfan anemia (DBA), the mechanisms through which glucocorticoids regulate human erythropoiesis remain poorly understood. We report that the sensitivity of erythroid differentiation to dexamethasone is dependent on the developmental origin of human CD34+ progenitor cells, specifically increasing the expansion of CD34+ progenitors from peripheral blood (PB) but not cord blood (CB). Dexamethasone treatment of erythroid-differentiated PB, but not CB, CD34+ progenitors resulted in the expansion of a newly defined CD34+CD36+CD71hiCD105med immature colony-forming unit–erythroid (CFU-E) population. Furthermore, proteomics analyses revealed the induction of distinct proteins in dexamethasone-treated PB and CB erythroid progenitors. Dexamethasone treatment of PB progenitors resulted in the specific upregulation of p57Kip2, a Cip/Kip cyclin–dependent kinase inhibitor, and we identified this induction as critical; shRNA-mediated downregulation of p57Kip2, but not the related p27Kip1, significantly attenuated the impact of dexamethasone on erythroid differentiation and inhibited the expansion of the immature CFU-E subset. Notably, in the context of DBA, we found that steroid resistance was associated with dysregulated p57Kip2 expression. Altogether, these data identify a unique glucocorticoid-responsive human erythroid progenitor and provide new insights into glucocorticoid-based therapeutic strategies for the treatment of patients with DBA.
Keywords: Development, Hematology
Keywords: Bone marrow differentiation, Cell cycle, Hematopoietic stem cells
Introduction
Diamond Blackfan anemia (DBA) is an inherited bone marrow (BM) failure syndrome with an incidence of 5 to 10 cases per million live births and is characterized by red cell aplasia, a range of physical anomalies, developmental bone defects, and cancer predisposition (1, 2). More than 70% of the patients diagnosed with DBA have defects in ribosome biogenesis due to mutations in genes encoding ribosomal proteins. In addition, mutations in the GATA1 transcription factor, a key regulator of erythroid development, and TSR2, a pre-rRNA processing protein, have recently been identified in several families with DBA (3, 4). The genetic landscape of DBA is heterogeneous, but genotype and phenotype correlations have been noted in association with mutations in the RPL5 and RPL11 ribosomal proteins in patients (5).
The standard of care for patients with DBA after the first year of life is glucocorticoids. Notably, a majority of treated patients have an increase in red cell production and have a reduced dependency on blood transfusions (6–8). However, the therapeutic dose is extremely variable among patients, and many become refractory to treatment over time. Once patients are glucocorticoid refractory they become dependent on chronic RBC transfusions unless they enter remission or undergo curative hematopoietic stem cell transplantation (7). The actions of glucocorticoids have been well studied in many disease contexts. These molecules interact with the glucocorticoid receptor (GR), leading to nuclear translocation of the resulting complex, which binds DNA at glucocorticoid response elements (GREs) and ultimately activates gene transcription (9). However, the specific mechanisms of action of glucocorticoids in the erythroid system in both healthy individuals and patients with DBA remain to be fully elucidated. Several studies have demonstrated that glucocorticoids act at the erythroid progenitor level, but the precise stages of erythroid differentiation at which they exert their effects have not been identified (10–13). This is due in part to the considerable heterogeneity of erythroid progenitor populations and the different markers and model systems that are used for studying glucocorticoid effects on erythropoiesis. Indeed, early burst-forming unit–erythroid (BFU-E) and colony-forming unit–erythroid (CFU-E) progenitors are often characterized on the basis of their functional ability to form erythroid colonies in colony-forming assays (CFAs), and immunophenotypic evaluations (14–18) are still in progress.
A further difficulty in evaluating the impact of glucocorticoids on BFU-E and CFU-E progenitors is due to important differences in murine versus human erythroid differentiation. In mice, the vast majority of research has focused on fetal liver progenitors, with a detailed characterization of early and late populations of fetal liver BFU-Es (19–21). Although several studies performed on murine fetal liver cells have suggested that dexamethasone acts at the BFU-E stage (11, 13, 19), a more recent study showed that dexamethasone enhances the maintenance of proliferative murine CFU-Es by upregulating p57Kip2, a member of the Cip/Kip cyclin–dependent kinase (CDK) inhibitor (CKI) protein family (10). Under these conditions, the proliferative CFU-E population is maintained, and there is a delayed differentiation to the less proliferative proerythroblast (pro-EB) stage. However, it is not clear whether these data in fetal murine erythroid progenitors translate to humans, as regards both the potential ex vivo heterogeneity of BFU-E and CFU-E populations and, more important, the diversity of erythroid progenitor subpopulations in human BM. These limitations have made it challenging to elucidate the mechanisms through which glucocorticoids act at different stages of erythroid progenitor development, under both physiological and pathological conditions.
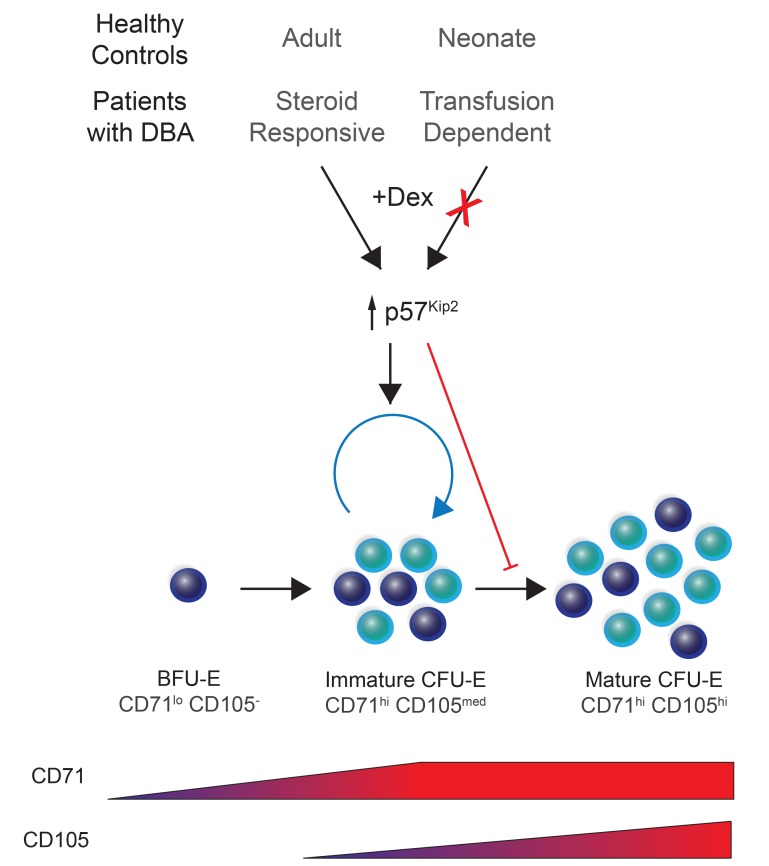
In the present study, we identified significant differences in the potential of dexamethasone to affect physiological as well as disordered human erythropoiesis. In the context of physiological differentiation, we found that, although it affected terminal erythropoiesis independently of the source, dexamethasone had an effect on the expansion of CD34+ hematopoietic stem and progenitor cells (HSPCs) isolated from adult peripheral blood (PB), but not from cord blood (CB). Dexamethasone did not affect BFU-Es, but using a new set of cell-surface markers, we identified a unique, PB progenitor–generated subset of transitional CD34+CD36+CD71hiCD105med CFU-Es that were dexamethasone responsive. Mass spectrometry–based quantitative proteomics analyses revealed substantial differences in the effects of dexamethasone on PB and CB progenitors, with upregulation of nuclear receptor subfamily 4 group A member 1 (NR4A1), a negative cell-cycle regulator (22), in the former cell type. Furthermore, we found that the p57Kip2 Cip/Kip CKI was specifically upregulated by dexamethasone in PB-derived CFU-Es and that its downregulation significantly attenuated the effects of this glucocorticoid. Even more notably, we found that p57Kip2 was not upregulated by dexamethasone in CFU-Es isolated from steroid-resistant patients with DBA. These findings open new avenues for the development of specific therapeutic strategies for these patients.
Results
Dexamethasone increases the proliferation of adult human CFU-Es.
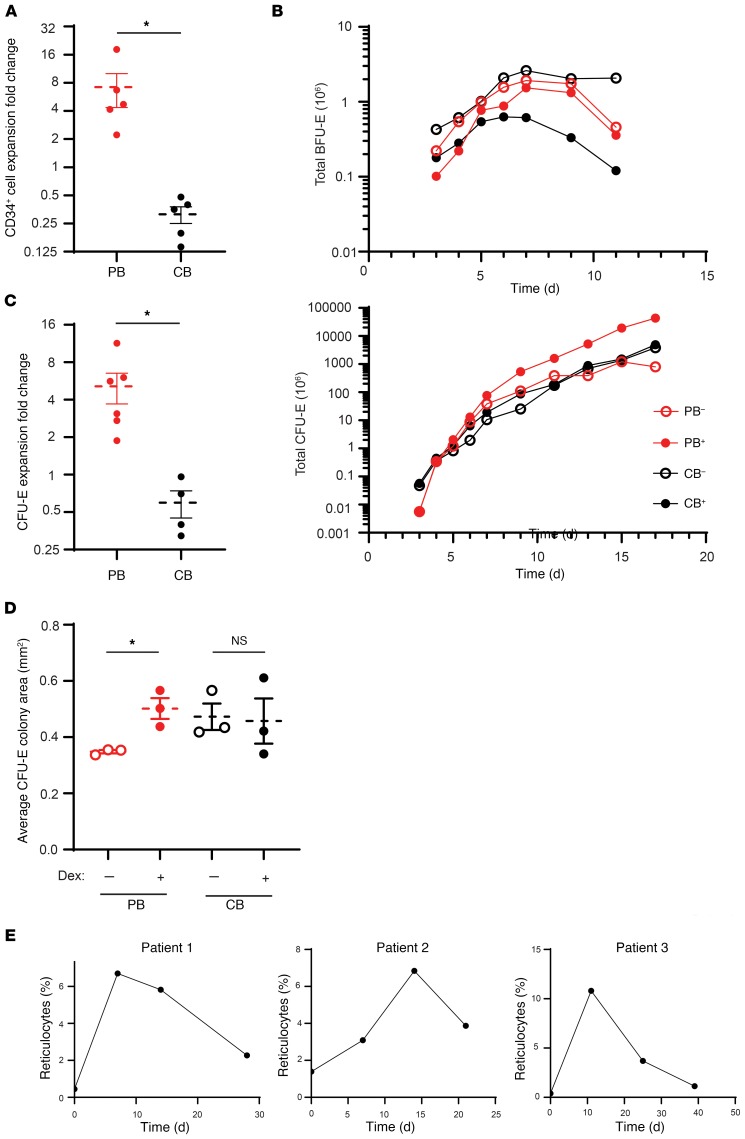
It has previously been shown that dexamethasone increases the proliferation of erythroid progenitors derived from both healthy and DBA HSPCs (23–25). However, the differentiation stage at which dexamethasone exerts its effects is still unclear. Using a serum-free expansion medium that allowed for the effective erythroid differentiation of HSPCs under steady-state conditions (without dexamethasone), we studied the effects of dexamethasone on human erythropoiesis (18, 26). Notably, we found that dexamethasone (100 nM) increased the expansion of CD34+ cells isolated from adult PB by 7-fold as compared with control conditions (Figure 1A). Surprisingly, though, we did not detect a dexamethasone-mediated increase in the expansion of CB-derived CD34+ cells. In fact, total numbers of precursors differentiating from CB CD34+ cells were approximately 4-fold lower in the presence versus absence of dexamethasone (Figure 1A), strongly suggesting marked developmental differences in the responsiveness of human PB and CB progenitors to this glucocorticoid.
Figure 1. Dexamethasone enhances the proliferation of human CFU-Es derived from PB CD34+ cells.
(A) Fold change in total expansion of PB-derived (red) and CB-derived (black, n = 5) CD34+ cells upon erythroid differentiation in the presence or absence of dexamethasone for 14 days. Data represent the mean ± SEM. (B) Total numbers of BFU-Es and CFU-Es generated from PB and CB cultures in the absence (PB–, CB–) or presence (PB+, CB+) of dexamethasone. Data are representative of 1 of 3 independent experiments. (C) Fold change in purified CFU-Es derived from PB (n = 6) and CB (n = 4) in the presence of dexamethasone relative to untreated controls after 14 days of expansion. Data represent the mean ± SEM. (D) Average size (area) of colonies generated from purified CFU-Es derived from untreated PB and CB after 5 days of culture in the absence (open circles) or presence (solid circles) of dexamethasone (n = 3). Data represent the mean ± SEM. Dex, dexamethasone. (E) Percentages of reticulocytes in 3 patients with DBA following treatment with prednisone, beginning on day 0. *P < 0.05, by 2-tailed Student’s t test (A, C, and D).
To further elucidate the erythroid differentiation stage at which dexamethasone acts, we used our recently developed experimental strategy for the characterization of BFU-E and CFU-E progenitors, based on surface expression levels of glycophorin A (GPA), IL-3R, CD34, and CD36. The absolute numbers of BFU-Es and CFU-Es generated from 106 PB and CB CD34+ cells were enumerated on the basis of cell-surface marker expression in the absence and presence of dexamethasone (Figure 1B). Beginning on day 7, the number of BFU-Es (defined as GPA–IL3R–CD34+CD36–) cells decreased in cultures starting with PB as well as CB CD34+ progenitors, in the absence and presence of dexamethasone, and the extent of this decrease was enhanced by dexamethasone. In marked contrast, the absolute number of CFU-Es (defined as GPA–IL3R–CD34–CD36+) increased over time, and this increase was more pronounced in dexamethasone-treated adult PB CD34+ cells. Notably, dexamethasone decreased the appearance of terminally differentiated GPA+ cells in both PB- and CB-derived cultures. These data indicate that erythroblasts from both sources were responsive to this glucocorticoid, resulting in a delayed transition to terminal erythroid differentiation (Supplemental Figure 1, A and B). However, cells derived from PB were more affected, possibly because of additional effects at the progenitor levels and at this transitional stage. We also detected this decrease in terminal erythroid differentiation as a function of α4-integrin/band 3 profiles as well as reduced hemoglobinization (Supplemental Figure 1A).
To further validate that the CFU-E was the population primarily responding to dexamethasone, we sorted CFU-Es derived from PB and CB cultures of CD34+ cells and studied their potential for expansion in serum-free expansion media. As shown in Figure 1C, the expansion of CFU-Es purified from adult CD34+ cells increased by 5-fold in the presence of dexamethasone. Once again, dexamethasone had little to no effect on the proliferation of purified CFU-Es derived from CB, supporting the idea that the response to dexamethasone is linked to the specific developmental stage of erythroid progenitors. We performed the same experiments using increasing concentrations of dexamethasone on sorted populations of erythroid progenitors derived from adult PB cultures and observed that the effect on proliferation was maximal at the 100-nM dose (Supplemental Figure 2).
Functional assays using methylcellulose cultures with erythropoietin (Epo) only, a culture condition that supports the growth of CFU-Es but not BFU-Es (requiring both stem cell factor [SCF] and Epo [ref. 18]), revealed that treatment with dexamethasone markedly increased the colony size of PB-derived CFU-Es but had very little or no effect on CB-derived CFU-E colony size (Figure 1D and Supplemental Figure 3), despite the fact that in the absence of dexamethasone the colony size of purified CB-derived CFU-Es was larger than that generated by PB-derived CFU-Es. Taken together, these data imply that only human CFU-Es derived from adult PB respond to dexamethasone by increasing proliferation.
Previous studies have shown that it takes at least 2 weeks in vivo for healthy BM to produce reticulocytes from the BFU-E stage and approximately 7 to 10 days from the CFU-E stage (27–29). We hypothesized that if the CFU-E is the progenitor population that responds to glucocorticoids in vivo, then patients treated with steroids should present with reticulocytosis in less than 2 weeks. We followed patients with DBA over a 1-month period before and after treatment with prednisone. We observed that in each patient, the reticulocyte count increased within 7 to 11 days after initiation of the treatment, strongly suggesting that in vivo, the CFU-E is indeed the glucocorticoid-responsive population (Figure 1E). We further noticed a decline in the reticulocyte response in a similar time frame, in association with a decrease in the dose of prednisone administered to the patient. Our finding that patients with DBA develop reticulocytosis within less than 2 weeks of starting prednisone treatment strongly suggests the presence of a mature, steroid-responsive progenitor, such as a CFU-E.
Dexamethasone targets a subpopulation of adult-derived CFU-Es.
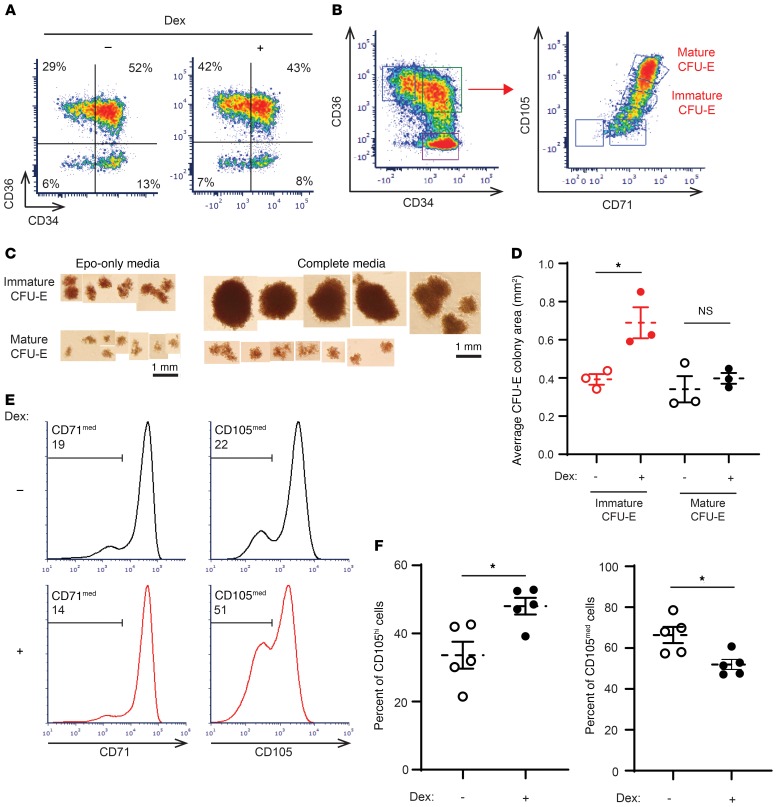
We previously described that, given the surface expression of CD34 and CD36, we could obtain a highly enriched population of BFU-Es (GPA–IL-3R–CD34+CD36–) and CFU-Es (GPA–IL-3R–CD34–CD36+) (18). More recently, we reported a transitional progenitor population defined as GPA–IL-3R–CD34+CD36+, which is more predominant during differentiation of adult PB than that of CB (30). Notably, the kinetics of progression through these differentiation states were also altered with dexamethasone treatment (Figure 2A).
Figure 2. Dexamethasone specifically targets a transitional subpopulation of human CFU-Es derived from PB CD34+ cells.
(A) PB-derived CD34+ cells differentiated in the presence or absence of dexamethasone were evaluated as a function of their CD36 and CD34 expression profiles. Representative plots on day 4 of differentiation are presented. Data shown are representative of 1 of 3 independent experiments. (B) Gating strategy to define mature and immature transitional CFU-E progenitor populations based on the CD71/CD105 profiles of the CD34+CD36+ subset. Data are representative of 1 of 3 independent experiments. (C) Representative images of colonies formed by immature and mature CFU-Es as defined in B, in the presence of Epo alone or Epo, SCF, IL-3, IL-6, G-CSF, and GM-CSF (n = 3). Scale bars: 1 mm. (D) Colony size (area) generated by immature (red) and mature (black) CFU-Es, defined as in B and formed in the presence of Epo alone, in the absence (open circles) or presence (solid circles) of dexamethasone (n = 3). Data represent the mean ± SEM. (E) Representative histograms show CD71 and CD105 expression in PB-derived CD34+ cells differentiated in the absence (black) or presence (red) of dexamethasone (unsorted, day 4). Percentages of CD71med and CD105med are shown in the upper left quadrant of each plot. Data are representative of 1 of 3 independent experiments. (F) Quantification of CD105med and CD105hi cells following differentiation of PB-derived CD34+ cells in the absence (control, open circles) or presence (solid circles) of dexamethasone (day 4, n = 5). Data represent the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test (D and F).
Having identified 2 phenotypically different CFU-E populations (CD34+CD36+ and CD34–CD36+), we sought to determine whether there were additional cell-surface markers that would provide further insights into the heterogeneity of these CFU-E populations. We focused on CD71, the transferrin receptor, and CD105 (endoglin), both of which demonstrated large differences in their RNA expression during erythroid differentiation at the progenitor stages (Supplemental Figure 4A).
On the basis of the expression patterns of CD71 and CD105 on CD34+CD36+ cells, we identified a continuum of cells with 2 distinct populations: CD71hiCD105med and CD71hiCD105hi (Figure 2B). We then sorted these different cell populations and performed colony-forming assays in the presence of either Epo alone, to produce colonies with the traditional definition of a CFU-E, or in complete medium with SCF, Epo, IL-3, IL-6, granulocyte CSF (G-CSF), and granulocyte-macrophage–CSF (GM-CSF). Interestingly, although both sorted populations from CD34+CD36+ cells generated colonies in the presence of Epo only (Figure 2C), only CD71hi CD105med cells showed marked responsiveness to SCF in complete medium, whereas SCF had little or no effect on CD71hiCD105hi cells (Figure 2C). CD71hiCD105hi cells from CD34+CD36+ cell populations responded similarly to CD3–CD36+ cell populations, with a minimal response to SCF. Given these findings, we propose that CD71hiCD105med cells should be termed immature CFU-Es and CD71hiCD105hi cells mature CFU-Es. Importantly, immature CFU-Es functionally responded to dexamethasone by increasing their colony size in a methylcellulose culture system (Figure 2D). In marked contrast, mature CFU-Es responded marginally to dexamethasone in the same functional colony-forming assays, and this increase was not statistically significant. When treated with dexamethasone, PB-derived CD34+ cells preferentially maintained this immature CFU-E population. Indeed, although both untreated and treated cells expressed comparable levels of CD71 on their surface, approximately 50% of the PB cells treated with dexamethasone were still CD105med compared with untreated controls, approximately 30% of which were CD105med by day 4 (Figure 2, E and F, left). Consequentially, the population of CD105hi cells was decreased in PB cells treated with dexamethasone (Figure 2F, right). Taken together, these data demonstrate that in human erythropoiesis, dexamethasone treatment preferentially maintains the immature CFU-E progenitor population for an extended period to increase the proliferative capacity of this cell population.
Proteomics studies highlight previously unidentified erythroid dexamethasone targets.
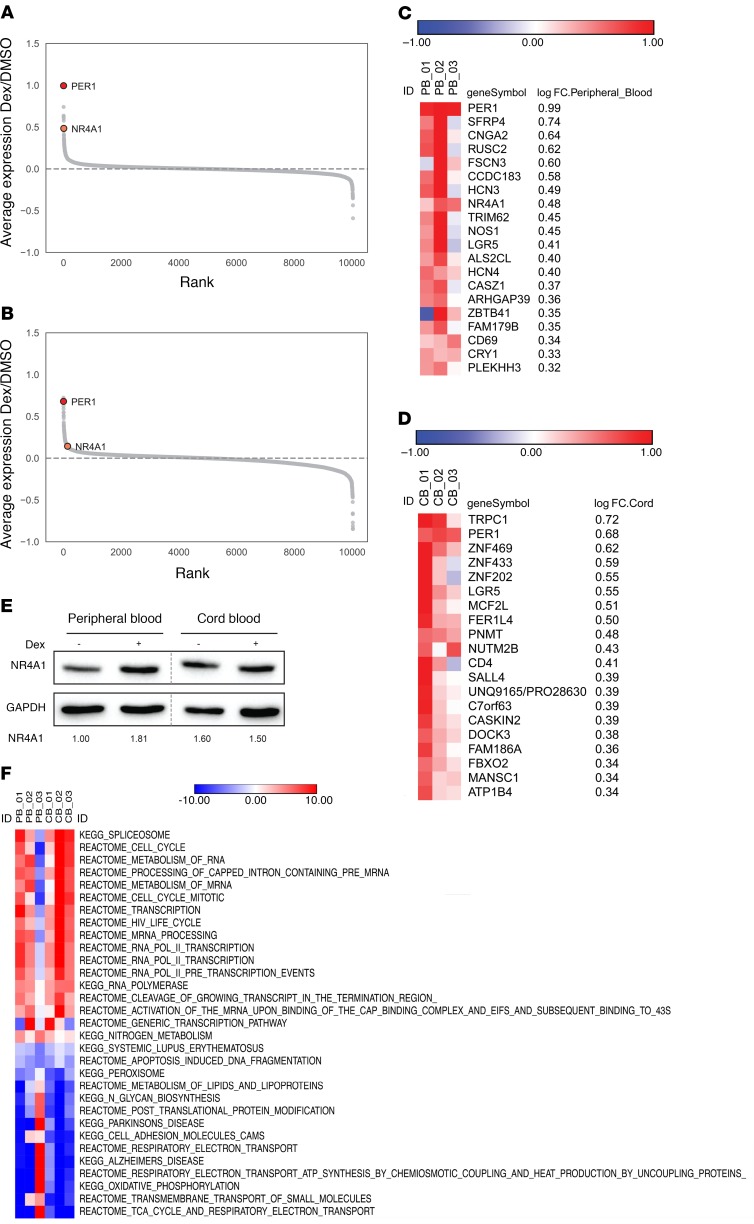
To begin exploring the mechanisms regulating the differential impact of dexamethasone on PB- and CB-derived progenitors, we elected to use a global, comparative proteomics approach. PB- and CB-derived CD34+ cells were cultured in vitro for 5 days to allow for the acquisition of sufficient numbers of flow-sorted purified CFU-Es (~20 × 106 cells) and then subjected to proteomics evaluation. We treated biological triplicates of PB- and CB-derived CFU-Es in the absence or presence of dexamethasone (100 nM) for 24 hours. These cells were then processed for proteomics analysis by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Proteomics quantified 10,045 proteins in PB samples and 10,028 proteins in CB samples (Supplemental Table 1). Proteomic evaluation of PB and CB samples was performed in 2 separate tandem mass tag–6 (TMT-6) plexes. This allowed for precise measurements of changes in protein abundance due to drug treatment in the PB-plex and the CB-plex, but less precise comparisons between PB and CB plexes. Therefore, the observed differences between PB and CB samples were subsequently validated in follow-up experiments.
Importantly, both PB- and CB-derived CFU-Es responded to dexamethasone as demonstrated by the upregulation of the gene for period 1 (PER1) (Figure 3, A and B), a major component of the mammalian circadian clock that has long been known to be regulated by glucocorticoids (31, 32). Notably, however, the majority of dexamethasone-upregulated proteins in PB- and CB-derived CFU-Es differed (Figure 3, C and D), consistent with the distinct phenotypic responses of these CFU-Es. One of the proteins whose abundance was upregulated by dexamethasone in PB-derived CFU-Es, but not CB-derived CFU-Es, was NR4A1 (Figure 3, C and D), a negative cell-cycle regulator. NR4A1 is an interesting target, given its role as a regulator of the cell cycle (22) and of T cell differentiation (33). NR4A1 also binds to HIF1α (34), an important regulator of erythroid differentiation (35). Identification of NR4A1 as a target of dexamethasone was confirmed by Western blot analysis (Figure 3E). Interestingly, the transcription profiles of NR4A1 as well as of 2 other cell-cycle inhibitors, p27Kip1 and p57Kip2, showed a substantial decrease between the CD34+ and BFU-E stages (Supplemental Figure 4B). Although we did not detect p57Kip2 in the proteomics screen, it is notable that only p57Kip2 was expressed at markedly higher levels in PB than in CB CD34+ progenitors (Supplemental Figure 4B). We also performed single-sample gene set enrichment analysis (ssGSEA) to identify the pathways most differentially affected by dexamethasone treatment in both PB and CB samples (Figure 3F). Notably, we observed an increase in proteins involved in the cell cycle and RNA processing as well as a decrease in proteins implicated in oxidative phosphorylation and biosynthetic pathways. Together, these data point to potential differences in the roles of cell-cycle inhibitors in CD34+ progenitors as a function of their developmental origin.
Figure 3. Proteomics studies highlight NR4A1 as a dexamethasone target in erythroid-differentiated PB progenitors.
(A) Ranked average log fold-change plots of differences in protein expression induced by dexamethasone in erythroid-differentiated PB progenitors. (B) Ranked average log fold-change plots of differences in protein expression induced by dexamethasone in erythroid-differentiated CB progenitors. (C) Top-20 proteins upregulated by dexamethasone in erythroid-differentiated PB progenitors based on the log fold change (log FC). (D) Top-20 proteins upregulated by dexamethasone in erythroid-differentiated CB progenitors based on the log fold change. (E) NR4A1 expression levels in purified PB- and CB-derived unsorted progenitors were evaluated by Western blotting on day 4 of expansion. Expression of NR4A1 relative to GAPDH is quantified below each lane. Data are representative of 1 of 3 independent experiments. (F) ssGSEA of proteins differentially regulated by dexamethasone. The top-10 upregulated and downregulated pathways between all samples are listed, and redundant pathways were eliminated.
Dexamethasone increases p57Kip2 expression in human CFU-Es.
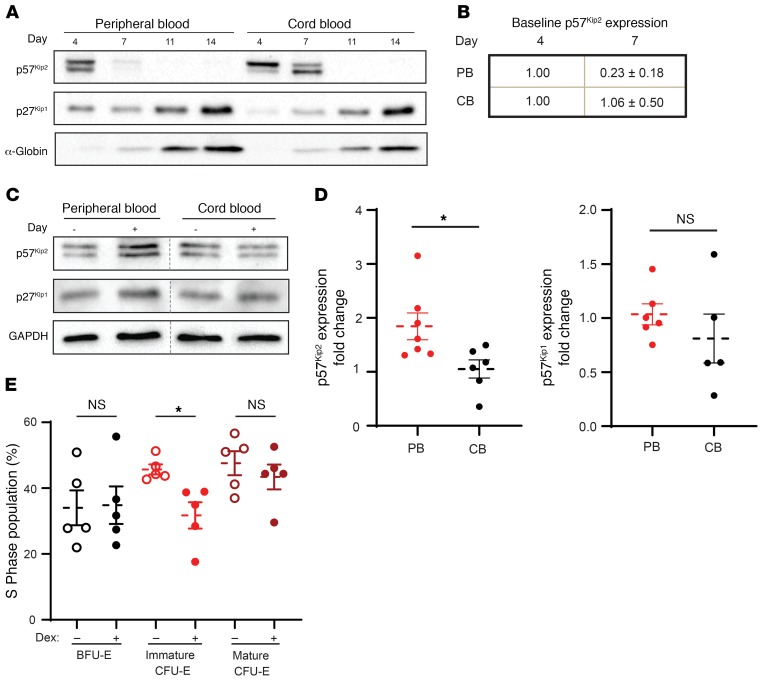
The data presented above, together with an elegant previous study showing that p57Kip2 regulates steroid responsiveness in murine erythroid progenitors (10), suggested that dexamethasone would act through p57Kip2 in PB-derived erythroid progenitors. We first evaluated the expression levels of p57Kip2 protein as a function of erythroid differentiation from PB and CB CD34+ cells (Figure 4A). Although p57Kip2 was expressed in early erythroid progenitors derived from both PB and CB CD34+ cells (unsorted, day 4), the loss of p57Kip2 was substantially more rapid in the CB cells. p57Kip2 levels in PB erythroid progenitors were reduced by 77% by day 7, whereas expression persisted in CB progenitors (Figure 4, A and B). Conversely, p27Kip1, a related Cip/Kip family member, was expressed at minimal levels during early differentiation, but its expression increased dramatically in terminally differentiating erythroblasts (Figure 4A). These distinct expression profiles are in agreement with previous studies showing that p57Kip2 is associated with the quiescence of stem and progenitor cells, whereas p27Kip1 plays a role in the cell-cycle exit that occurs during terminal erythroid differentiation (36, 37). Notably, however, differences in the kinetics of p57Kip2 downregulation in CB and PB erythroid progenitors have not thus far been appreciated.
Figure 4. Dexamethasone increases p57Kip2 expression in Epo-induced PB CD34+ cells.
(A) Expression of p57Kip2 and p27Kip1 was evaluated by Western blotting in CD34+ progenitors during Epo-induced erythroid differentiation. Erythroid differentiation was controlled by evaluating α-globin expression. Data are representative of 5 independent experiments. (B) Quantification of the differences in p57Kip2 protein levels in PB and CB cultures between days 4 and 7 of erythroid differentiation (n = 5; values for day 4 were arbitrarily set at 1.). (C) Expression of p57Kip2, p27Kip1, and GAPDH in PB-derived and CB-derived progenitors was evaluated in sorted CFU-Es following 4 days of Epo-induced differentiation in the absence (–) or presence (+) of dexamethasone. Representative Western blots from 1 of 3 independent experiments are shown. (D) Quantification of the fold changes in p57Kip2 and p27Kip1 expression in purified PB-derived (n = 7, red circles) and CB-derived (n = 6, black circles) CFU-Es in the presence of dexamethasone relative to control conditions (arbitrarily set at 1). Data represent the mean ± SEM. (E) Quantification of the percentages of PB-derived BFU-Es, immature CFU-Es, and mature CFU-Es that were in S phase in the absence (open circles) and presence (solid circles) of dexamethasone. S phase was quantified by Hoechst 33342 staining (n = 5). Data represent the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test (D and E).
Given these findings, it was of interest to assess whether p57Kip2 levels would be differentially affected by dexamethasone in purified PB-derived and CB-derived progenitors. To this end, progenitors derived from both sources were treated with dexamethasone, and the expression of p57Kip2 was quantified by Western blotting. We evaluated total CFU-Es, because the numbers of immature CFU-Es were insufficient in the absence of dexamethasone, particularly in CB cultures, in which the kinetics of differentiation is increased (30). Notably, within CFU-Es, we found that p57Kip2 protein levels were increased by 1.8-fold in PB-derived progenitors but remained unchanged in CB-derived progenitors (P < 0.05; Figure 4, C and D). These data are even more striking in light of the finding that p27Kip1 levels were not significantly altered in progenitors derived from either PB or CB sources (Figure 4, C and D). Altogether, these data strongly suggest a role for p57Kip2 in mediating the dexamethasone-induced changes in PB-derived, but not CB-derived, progenitors.
Given that p57Kip2 is a regulator of the cell cycle, we inhibited the transition from the G1 to the S phase (38) to determine whether dexamethasone-mediated changes in p57Kip2 are associated with differences in the cell-cycle dynamics of PB erythroid progenitors. We found that the percentages of progenitors in the S phase of the cell cycle increased significantly between the BFU-E and CFU-E stages, from a mean of 35% to 48% (P < 0.05; Figure 4E). Notably however, dexamethasone significantly decreased S-phase cells in immature CFU-Es, but not in the BFU-Es or mature CFU-E subsets (Figure 4E and Supplemental Figure 5). In addition, the percentages of cells in the non–S phase of the cell cycle were increased in immature CFU-Es (Supplemental Figure 5B). Thus, in agreement with our data showing that the immature PB-derived CFU-E subset preferentially undergoes a dexamethasone-mediated expansion (Figure 2), only this subset responded to upregulated p57Kip2 levels with a significant decrease in the number of cells in the S phase. Despite this reduction of cells in the S phase, immature CFU-Es continued to divide, with increases in G2/M, before further differentiating into mature CFU-Es and then proerythroblasts and resulting in increased expansion.
p57Kip2 expression is altered in erythroid progenitors from transfusion-dependent patients with DBA.
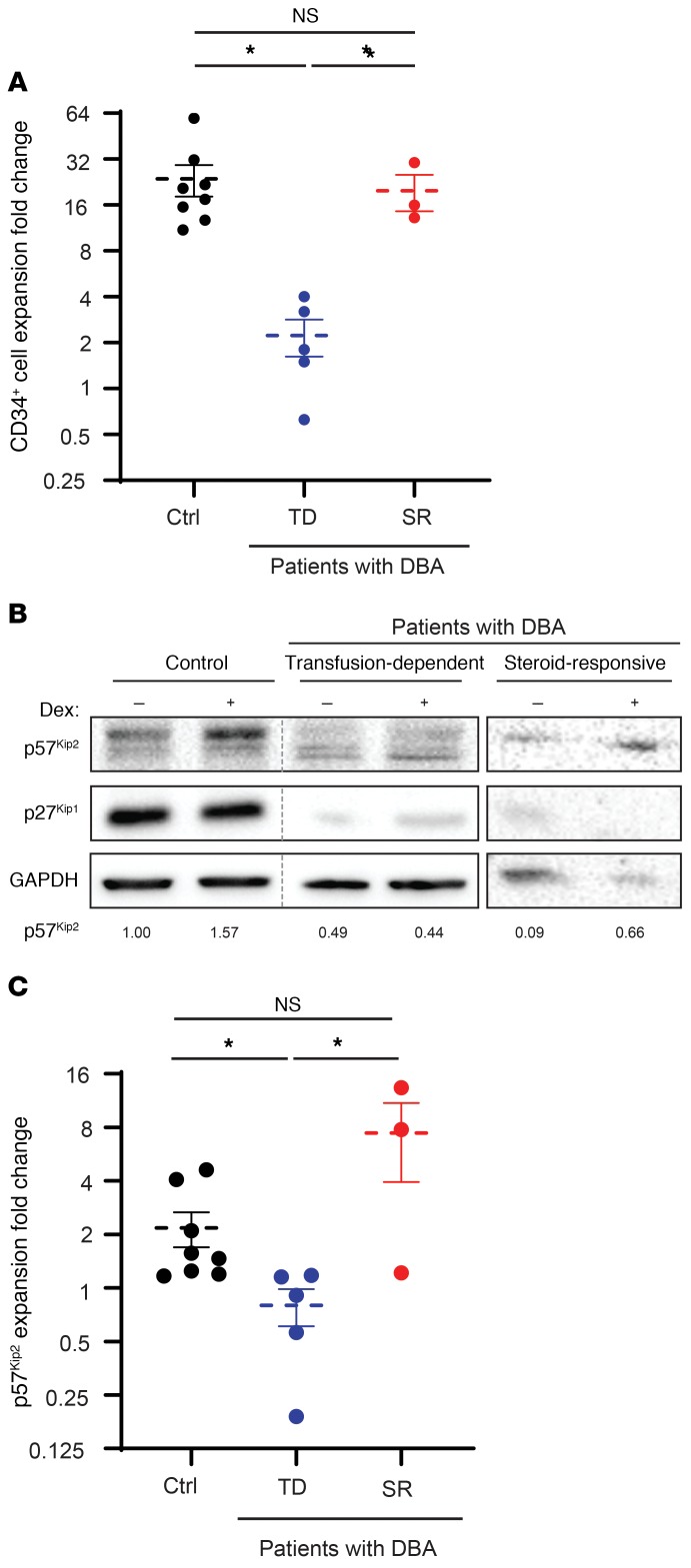
We further hypothesized that the resistance of patients with DBA to glucocorticoids is mediated, at least in part, by p57Kip2. To test this hypothesis, we compared dexamethasone-induced changes in p57Kip2 levels in unsorted cells on day 7 of culture in response to dexamethasone in CD34+ cells from healthy controls and transfusion-dependent patients with DBA. Given the strong reduction in growth of cells from transfusion-dependent patients with DBA, we evaluated p57Kip2 levels in unsorted progenitors on day 7 of expansion (Figure 5A). Importantly, p57Kip2 levels, which were increased in healthy controls and steroid-responsive patients with DBA in response to dexamethasone, were not affected in transfusion-dependent patients with DBA (Figure 5, B and C). As expected from the data presented in Figure 4C, p27Kip1 levels were not affected by dexamethasone, but it is notable that they were dysregulated and pointedly higher or lower in samples from patients with DBA, probably due to early differentiation or defective terminal erythroid differentiation, respectively, in these unsorted cells. Most critically, p57Kip2 levels in progenitors derived from steroid-responsive patients with DBA were upregulated in response to dexamethasone, similar to what was observed in healthy controls (Figure 5, B and C). Notably, the expansion of PB-derived CD34+ cells from healthy controls and steroid-responsive patients with DBA was similar after 7 days of culture, whereas cells from transfusion-dependent patients with DBA showed significantly less expansion (Figure 5A). Taken together, these results suggest a critical role for p57Kip2-associated cell-cycle changes in the steroid responsiveness of both physiological and pathological human erythropoiesis.
Figure 5. Aberrant steroid-mediated induction of p57Kip2 in erythroid progenitors from transfusion-dependent patients with DBA.
(A) The expansion of CD34+ cells derived from healthy controls (Ctrl) (black, n = 8), transfusion-dependent (TD) patients with DBA (blue, n = 5), and steroid-responsive (SR) patients with DBA (red, n = 3) is presented following a 7-day stimulation. Data represent the mean ± SEM. (B) Expression of p57Kip2 and p27Kip1 in erythroid progenitors from healthy controls and patients with DBA, either transfusion dependent or steroid responsive, was evaluated by Western blotting on day 7 of differentiation. Expression of p57Kip2 relative to GAPDH is quantified below each lane, with control levels in the healthy donor arbitrarily set at 1. (C) Quantification of the fold change in dexamethasone-induced p57Kip2 expression following expansion of CD34+ cells from healthy controls (n = 8) as compared with transfusion-dependent patients with DBA (n = 5) and steroid-responsive patients with DBA (n = 3). Data represent the mean ± SEM. *P < 0.05, by Kruskal-Wallis test with Dunn’s post hoc analysis with corrections for multiple comparisons (A and C).
Dexamethasone responsiveness is mediated by CDK activity.
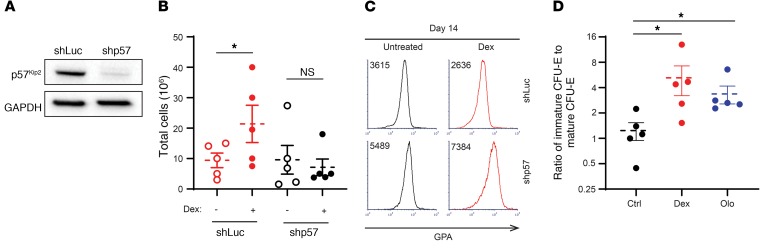
In order to directly assess the role of p57Kip2 in mediating dexamethasone effects on erythroid progenitors, we used a lentivirus-mediated shRNA approach to induce downregulation of p57Kip2 levels. Following transduction, p57Kip2 was downregulated by 80% as compared with cells transduced with a control luciferase–targeting shRNA construct (Figure 6A). Notably, p57Kip2 downregulation abrogated the ability of PB-derived CD34+ cells to respond to dexamethasone, monitored as a function of their expansion (Figure 6B). Moreover, we observed that erythroid differentiation was accelerated, as demonstrated by 1.5- and 3-fold increases in GPA expression levels in control- and dexamethasone-treated progenitors, respectively (Figure 6C). Finally, this effect on erythroid progenitors was specific to the p57Kip2 CKI, as downregulation of p27Kip1 did not alter the expansion of these progenitors (Supplemental Figure 6, A and B). The effect on p27Kip1 was only noticed at later stages and was indicated by a delay in terminal differentiation measured by the surface markers α4-integrin and band3 (Supplemental Figure 6C). Altogether, these data reveal the function of p57Kip2 in regulating the balance between human erythroid progenitor proliferation and differentiation and, furthermore, in controlling glucocorticoid responsiveness under both physiological and pathological conditions.
Figure 6. Downregulation of CDKN1C (p57Kip2) in CD34+ progenitors attenuates the impact of dexamethasone, accelerating erythroid differentiation, whereas olomoucine, a CKI, mimics the effect of dexamethasone.
(A) CD34+ progenitors were transduced with a lentiviral vector harboring an shRNA targeting luciferase (shLuc) or p57Kip2 (shp57). Expression of p57Kip2 was evaluated by Western blotting relative to GAPDH levels on day 7 after transduction. Data are representative of 1 of 3 independent experiments. (B) Expansion of control (shLuc) and p57Kip2-downregulated (shp57) PB CD34+ cells was evaluated in the absence (open circles) or presence (solid circles) of dexamethasone after 14 days of culture (n = 5). Data represent the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test. (C) Representative histograms of GPA expression in control (shLuc) and p57Kip2-downregulated (shp57) progenitors on day 14 of differentiation. MFIs are indicated. Data are representative of 1 of 3 independent experiments. (D) Quantification of the ratio of immature CFU-Es to mature CFU-Es following expansion of PB-derived CD34+ cells in the absence (black circles) or presence of 100 nM dexamethasone (red circles) or 1 μM olomoucine (blue circles) on day 4 of culture. n = 5. Data represent the mean ± SEM. *P < 0.05, by Kruskal-Wallis test with Dunn’s post hoc analysis with corrections for multiple comparisons.
Given our finding that an increase in levels of p57Kip2, a CKI, is critical for dexamethasone responsiveness, we hypothesized that inhibiting cyclin kinases would have the same effect as increasing p57Kip2. Notably, treatment of PB-derived CD34+ cells with olomoucine (1 μM), a small-molecule CDK1 and CDK2 inhibitor (39), resulted in a significant increase in the ratio of immature CFU-Es/mature CFU-Es to levels similar to those induced by dexamethasone (Figure 6D). Together, these data demonstrate the importance of CDK/CKI balance in the expansion of the immature CFU-E population.
Dexamethasone upregulates p57Kip2 expression in erythroid progenitors derived from human BM.
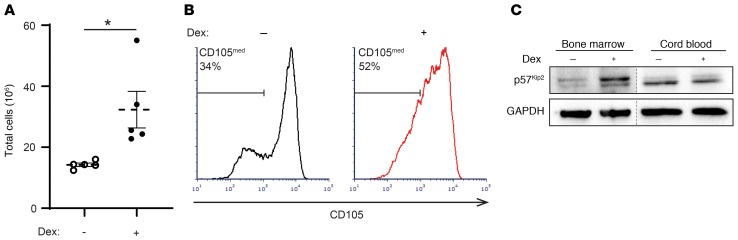
To assess the effects of dexamethasone on populations of cells that are directly targeted by the drug in vivo, we repeated key experiments with erythroid progenitors derived from human BM. In culture with serum-free expansion media, we found that the expansion of BM-derived CD34+ cells increased significantly in the presence of dexamethasone (Figure 7A). When examining the expression of CD105 in BM-derived progenitors, we observed that dexamethasone treatment maintained the population of CD105med cells and correspondingly decreased the population of CD105hi cells (Figure 7B). Furthermore, treatment of BM-derived CFU-Es with dexamethasone led to an increase in p57Kip2 expression that we did not observe in CB-derived CFU-Es treated with dexamethasone (Figure 7C). Overall, these data indicate that PB- and BM-derived erythroid progenitors respond to dexamethasone in a similar manner and that the phenotypes we observed may be relevant in the clinical use of dexamethasone for the treatment of red cell disorders.
Figure 7. Dexamethasone increases p57Kip2 levels in Epo-induced, BM-derived progenitors, expanding the immature CD105med CFU-E subset.
(A) BM CD34+ progenitors were differentiated in the absence (open circles) or presence (solid circles) of dexamethasone, and total cell numbers on day 14 are presented (n = 5 independent experiments). Data represent the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test. (B) Representative histograms of CD105 expression in BM-derived CD34+ cells differentiated in the absence (black) or presence (red) of dexamethasone on day 4 are shown, and the percentages of CD105med cells are indicated. (C) Expression of p57Kip2 and GAPDH in purified BM-derived and CB-derived mature CFU-Es, generated in the absence or presence of dexamethasone, was evaluated by Western blotting (blots from 1 of 3 independent experiments are shown).
Discussion
We believe the present study, which focused on identifying the role of dexamethasone during physiological and pathological human erythropoiesis, has generated several unique insights. Our finding that dexamethasone markedly enhanced the erythroid proliferation of CD34+ cells from adult PB but not from CB was unexpected and very surprising. In this context, it is of interest that the transition trajectories from BFU-E to CFU-E differ considerably following erythroid differentiation of CD34+ cells derived from adult PB and CB (30). Together, these data strongly suggest that dexamethasone has differential effects on transitional erythroid progenitor populations that are regulated by the source of the CD34+ progenitors. Importantly, these findings are relevant to BM populations, as we found that CFU-Es from BM-derived CD34+ cells had dexamethasone-induced responses that were similar to those detected in PB-derived CFU-Es (Figure 7).
Although BFU-Es derived from human either PB or CB were not responsive to dexamethasone, BFU-Es from murine fetal liver have been shown to be dexamethasone responsive (19). Murine CFU-Es have also been shown to be specifically dexamethasone responsive (10), suggesting that dexamethasone-dependent erythroid developmental stages may be species dependent. Collectively, these previous studies (10, 19), together with the present work, point to important differences between murine and human systems and, moreover, demonstrate the importance of the developmental origin of hematopoietic stem and progenitor cells in regulating their responsiveness to glucocorticoids during erythropoiesis. These developmental differences as well as steroid resistance in DBA may be mediated by epigenetic regulators (40–43), and further investigations of the mechanisms may offer additional insights into the heterogeneity of progenitor cell populations. Finally, our data show the importance of the p57Kip2 cell-cycle inhibitor in mediating dexamethasone effects during human erythroid differentiation and reveal the critical nature of the p57Kip2 axis in the dexamethasone responsiveness of patients with DBA.
The marked heterogeneity of human erythroid progenitors has long been recognized (44, 45). Indeed, it has been known for decades that erythroid progenitors give rise to colonies of different sizes and morphology in the methylcellulose culture system, which led to the concept of large, intermediate, and small BFU-E colonies, with the small BFU-E–forming cells further differentiating into CFU-Es (46). The recent progress of single-cell technologies has enabled stringent analyses of these varied cell populations with regard to both steroid sensitivity and cell-fate decisions (12, 47). Although we previously found that human BFU-Es and CFU-Es can be immunophenotypically defined on the basis of IL-3R, GPA, CD34, and CD36 cell-surface markers, this characterization did not allow us to fully resolve the heterogeneity of human erythroid progenitors. In the present study, we found that the CD71 and CD105 markers allowed for further immunophenotyping, discriminating subpopulations of erythroid BFU-Es and CFU-Es. Using this method, we identified a transitional human progenitor CFU-E population, resulting in the designation of immature and mature CFU-Es.
Using this optimized immunophenotyping assay, we identified the immature CFU-E population as the steroid-responsive erythroid progenitor subset, as determined by both proliferation and a decrease in the percentages of cells in the S phase of the cell cycle. This cell-cycle phenotype was due to a dexamethasone-mediated increase in p57Kip2, and under these conditions, differentiation was delayed, resulting in a paradoxical expansion of this progenitor subset. We propose that this occurs through the maintenance of the immature CFU-E population, generating larger numbers of mature CFU-Es.
The critical role of p57Kip2 in CFU-Es was shown by shRNA knockdown, which resulted in an opposite phenotype and an acceleration of erythroid differentiation that was associated with a decrease in proliferation. Together with recent work that identifies a role for cell-cycle status in cell-fate decisions (48), the ensemble of these data demonstrate a clear link between cell-cycle progression and differentiation of erythroid progenitors.
Importantly, erythroid progenitors from transfusion-dependent patients with DBA harbored altered levels of p57Kip2, and expression was not sensitive to dexamethasone treatment. Indeed, the absence of changes in p57Kip2 levels in transfusion-dependent patients is a proof of concept for the importance of this cell-cycle regulator in the response to steroids. Notably, though, there was substantial variability in the phenotype of progenitors from transfusion-dependent patients with DBA that was likely mediated by other mechanisms. In the future, it will be important to determine whether steroid responsiveness in patients with DBA is regulated at the level of a specific subset or subsets of erythroid progenitors and, furthermore, to assess the role of p57Kip2 levels in these subsets.
Our proteomics data revealed previously unidentified dexamethasone targets that may offer additional insights into how dexamethasone influences the cell cycle and increases erythroid proliferation. Of the identified targets, many have been shown to be involved in cell-cycle regulation including PER1 and NR4A1, indicating that these proteins may function to negatively regulate the cell cycle in association with p57Kip2 (31, 49). In this regard, it is of interest that NR4A1 mediates TGF-β signaling, which is also aberrant in DBA and could thus be involved in corticosteroid resistance mechanisms (50). Our finding that dexamethasone globally regulates RNA processing and biosynthetic pathways may also open additional avenues for studies on the effects of dexamethasone on erythroid progenitors.
The present findings have direct relevance for the clinical use of corticosteroids for the treatment of hypoproliferative anemias, most notably DBA. Although corticosteroid use stably increases red cell mass and induces hematopoietic remission or marked improvement in many patients with DBA, other patients are not responsive or cannot continue glucocorticoid treatment because of adverse side effects. We anticipate that our insights into the mechanism of action of glucocorticoids on human erythropoiesis, and specifically on the immature CFU-E subset, will promote the development of targeted drugs and treatment strategies that will induce a sustained effect on erythroid progenitors in more patients with fewer side effects. In summary, we have identified a transitional CFU-E subset, between late BFU-E and mature CFU-E stages, that is responsive to dexamethasone. These findings contribute to our understanding of erythroid progenitor biology and will, we believe, lead to the development of new treatment strategies for erythroid disorders.
Methods
Human studies
CD34+ cells were obtained from deidentified control adult PB leukoreduction filters, deidentified CB units, deidentified BM samples, or phlebotomized patients with DBA. In order to limit sample variability, blood samples from multiple control PB or CB donors were pooled.
Patients with DBA were defined as transfusion dependent or steroid responsive on the basis of their clinical need for chronic RBC transfusion or successful management with corticosteroids, respectively.
The 3 patients with DBA presented in Figure 1E were followed after diagnosis and initial treatment with prednisone. The dose of prednisone was decreased after the initial response.
Isolation and culturing of CD34+ cells
Mononuclear cells from PB, CB, or BM were separated using Lymphoprep (STEMCELL Technologies), and CD34+ cells were purified with anti-CD34 microbeads and MACS Columns (Miltenyi Biotec) according to manufacturer’s protocol. CD34+ cells were cultured at a density of 105 cells/mL in serum-free expansion media as previously described (26), with a base media of StemSpan SFEM (STEMCELL Technologies) initially supplemented with 100 ng/mL SCF, 10 ng/mL IL-3, 0.5 U/mL Epo, 4 μL/mL lipid mixture 1 (MilliporeSigma), 2 mM l-glutamine, and 200 μg/mL transferrin. Beginning on day 7, the dose of Epo was increased to 3 U/mL, and on day 11, the dose of transferrin was increased to 1 mg/mL. Dexamethasone (MilliporeSigma) was added to cultures at 100 nM as indicated, as this dose was found to be optimal (Supplemental Figure 2). Olomoucine (MilliporeSigma) was added at 1 μM as indicated.
Flow cytometry and cell sorting
Erythroid progenitors were examined using surface markers as previously described (18). Erythroid progenitors were analyzed on day 4 of differentiation, at which point 105 cells were stained for 15 minutes at room temperature with an antibody cocktail containing anti–IL-3R phycoerythrin-Cy7 (PE-Cy7), anti–GPA PE, anti–CD34 FITC, anti–CD36 allophycocyanin (APC), anti–CD71 Alexa Fluor 700, and anti–CD105 Brilliant Violet 421 (BD Biosciences; 560826, 555570, 555821, 550956, 560566, and 563920, respectively). BFU-Es were defined as GPA–IL-3R–CD34+CD36–, CFU-Es as GPA–IL-3R–CD34–CD36+, and transitional progenitors as GPA–IL-3R–CD34+CD36+. Dead cells were excluded from further analysis with 7–aminoactinomycin D (BD Biosciences) staining. Analysis was performed using a BD Fortessa flow cytometer with FCS Express 6 and FlowJo 10 software. BFU-Es, CFU-Es, and transitional progenitors were sorted for downstream experiments using a BD FACSAria at low pressure with a 100-μm nozzle. Fluorescence minus 1 and isotype controls were used to define the different populations of progenitor cells and resolve the continuum. Functional assays using sorted cells in colony-forming assays were performed to further confirm the identity and purity of the different cell populations.
Colony-forming assays
Sorted erythroid progenitors were seeded onto methylcellulose media H4230 (STEMCELL Technologies) supplemented with Epo alone at 0.5 U/mL or onto complete methylcellulose media H4435 (STEMCELL Technologies) at 200 cells/mL. BFU-E and CFU-E colonies were counted and measured on days 14 and 7 of culturing, respectively. Colony area was determined by modeling each colony as an ellipse and measuring its major axis a and minor axis b to calculate the area using the formula area = πab/4. Dexamethasone (100 nM) was added to cultures as indicated.
Cell-cycle staining
For live cell-cycle staining to examine cell-cycle profile, erythroid progenitors were incubated with 5 mg/mL Hoechst 33342 for 3 hours at 37°C before staining with an antibody cocktail for surface markers.
Western blot analysis
Cells were lysed in RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) with 1:100 protease inhibitor cocktail (MilliporeSigma) on ice for 10 minutes and then centrifuged at maximum speed for 10 minutes. Supernatants were mixed at 1:1 (vol/vol) with 2× Laemmli Sample Buffer (Bio-Rad) with 0.1 M DTT and boiled for 5 minutes. Samples were then separated with SDS-PAGE for 1.5 hours at 150 V and transferred onto nitrocellulose membranes for 1 hour at 95 V. Membranes were then blocked with 4% (wt/vol) milk powder and 1% (wt/vol) BSA in 0.1% Tween-20 (vol/vol) PBST for 3 hours. Membranes were then incubated with the following primary antibodies overnight at 4°C: p57Kip2 (BD Biosciences; 556346), p27Kip1 (BD Biosciences; 610241), NR4A1 (BD Biosciences; 554088), GAPDH (MilliporeSigma; CB1001), and α-globin (Santa Cruz Biotechnology; sc-514378). Membranes were washed 5 times for 5 minutes with PBST and then incubated with HRP-conjugated secondary antibodies (Bio-Rad) for 2 hours at room temperature. Membranes were imaged with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) using a ChemiDoc MP Imaging System (Bio-Rad). Western blot images are representative of multiple experiments and were quantified with ImageJ software (NIH).
Lentivirus transduction
p57Kip2- and p27Kip1-knockdown experiments were each carried out with 2 lentiviral shRNA-knockdown constructs targeting CDKN1C and CDKN1B, respectively (clone IDs: NM_000076.2-1451s21c1 and NM_000076.2-1216s21c1 for CDKN1C and NM_004064.3-841s21c1 and NM_004064.3-643s21c1 for CDKN1B; MilliporeSigma). shRNA-knockdown constructs targeting luciferase were used as controls. After 2 days of culturing in serum-free expansion media, erythroid progenitors were placed in 10% FBS IMDM with 3 U/mL heparin and 8 μg/mL polybrene. Lentivirus particles were then added at a MOI of 30 and spinoculated for 2 hours at 1500 g. Cells were then incubated overnight and placed in serum-free expansion media. After a 24-hour recovery, lentivirus transduction was positively selected with 1 μg/mL puromycin, which was maintained until day 11 of culture.
Proteomics profiling
In-solution digestion.
CD34+ cell pellets were lysed for 30 minutes at 4°C in 8 M urea, 50 mM Tris-HCl, pH 8.0, 75 mM NaCl, 1 mM EDTA, 2 μg/μL aprotinin (MilliporeSigma), 10 μg/μL leupeptin (Roche), and 1 mM PMSF (MilliporeSigma). Lysates were cleared via centrifugation at 20,000 g, and protein concentration was determined using a bicinchoninic acid (BCA) protein assay (Pierce, Thermo Fisher Scientific). Remaining lysis buffer was used to equalize sample concentrations to the lowest measured concentration before proceeding. Protein reduction was performed with 5 mM DTT for 1 hour at room temperature, followed by alkylation with 10 mM iodoacetamide for 45 minutes at room temperature in the dark. Sample volumes were then adjusted with 50 mM Tris-HCl, pH 8.0, to reduce urea concentration to 2 M preceding enzymatic digestion. Proteins were digested first with endoproteinase LysC (Wako Laboratories) for 2 hours at 25°C and then overnight with sequencing-grade trypsin (Promega) at 25°C, both at enzyme/substrate ratios of 1:50. Following digestion, samples were acidified to a concentration of 1% with neat formic acid, and insoluble peptides and urea were removed via centrifugation at 20,000 g. The remaining soluble peptides were desalted using a 100-mg reverse-phase tC18 SepPak cartridge (Waters). Cartridges were conditioned with 1 mL 100% MeCN and 1 mL 50% MeCN/0.1% formic acid (FA) and then equilibrated with 4× 1 mL 0.1% trifluoroacetic acid (TFA). Samples were loaded onto the cartridge and washed 3 times with 1 mL 0.1% TFA and once with 1 mL 1% FA, and then eluted with 2× 600 μL 50% acetonitrile (MeCN) and 0.1% FA. The peptide concentration of desalted samples was again estimated with a BCA assay, such that the proper amount for TMT labeling could be removed, dried in a vacuum centrifuge, and stored at –80°C.
TMT labeling of peptides.
Samples were divided into 2 groups: PB- and CB-derived cells, and each set was separately labeled with TMT plex isobaric mass tagging reagents (Thermo Fisher Scientific) as previously described (51). Each 6-plex contained triplicate samples of dexamethasone- and DMSO-treated cells. Digested peptides were resuspended in 50 mM HEPES, pH 8.5, at a concentration of 2.5 mg/mL. Dried TMT reagent was reconstituted at 20 μg/μL in 100% anhydrous MeCN and added to samples at a 1:1 TMT/peptide mass ratio (100 μg for PB samples and 70 μg for CB samples due to the limited amount of material). Labeling was performed for 1 hour at 25°C with shaking. The TMT reaction was quenched with 5% hydroxylamine to a final concentration of 0.2% and shaken for 15 minutes at 25°C. TMT-labeled samples within each plex were then combined, dried to completion via vacuum centrifugation, reconstituted in 1 mL 0.1% FA, and desalted with a 100-mg SepPak cartridge as described above.
Basic reverse-phase fractionation.
TMT-labeled peptides were fractionated via offline basic reverse-phase (bRP) chromatography as previously described (52). Chromatography was performed with a Zorbax 300 Extend-C18 column (4.6 × 250 mm, 3.5 μm; Agilent Technologies) on an Agilent 1100 HPLC system. Samples were reconstituted in 900 μL bRP solvent A (5 mM ammonium formate, pH 10.0, in 2% vol/vol MeCN) and injected with this solvent at a flow rate of 1 mL/min. Peptides were separated at the same flow rate with a 96-minute gradient, beginning with an initial increase to 16% bRP solvent B (5 mM ammonium formate, pH 10.0, in 90% vol/vol MeCN), followed by a linear 60-minute gradient to 40% and stepwise ramping to 44% and finally 60% bRP solvent B. A total of 96 fractions were collected in a row-wise snaking pattern into a Whatman 2-mL 96-well plate (GE Healthcare) and were then concatenated nonsequentially into a final 24 fractions for proteomics analysis. The fractions were dried via vacuum centrifugation.
LC-MS/MS.
Dried fractions were reconstituted in 3% MeCN/0.1% FA to a peptide concentration of 1 μg/μL and analyzed via coupled Nanoflow LC-MS/MS using a Proxeon Easy-nLC 1000 (Thermo Fisher Scientific) and a Q-Exactive Plus Series Mass Spectrometer (Thermo Fisher Scientific). A sample load of 1 μg for each fraction was separated on a capillary column (36-μm outer diameter × 75-μm inner diameter) containing an integrated emitter tip and heated to 50°C, followed by packing to a length of approximately 30 cm with ReproSil-Pur C18-AQ 1.9 μm beads (Maisch GmbH). Chromatography was performed with a 110-minute gradient consisting of solvent A (3% MeCN/0.1% FA) and solvent B (90% MeCN/0.1% FA). The gradient profile, described as minutes/percentage solvent B, was 0:2, 1:6, 85:30, 94:60, 95:90, 100:90, 101:50, and 110:50, with the first 6 steps being performed at a flow rate of 200 nL/min and the last 2 at a flow rate of 500 nL/min. Ion acquisition on the Q-Exactive Plus was performed in data-dependent mode, acquiring HCD-MS/MS scans at a resolution of 17,500 on the top-12 most abundant precursor ions in each full MS scan (70,000 resolution). The automatic gain control (AGC) target was set to 3 × 106 ions for MS1 and 5 × 104 for MS2, and the maximum ion time was set to 120 ms for MS2. The collision energy was set to 30, peptide matching was set to “preferred,” isotope exclusion was enabled, and dynamic exclusion time was set to 20 seconds.
The original mass spectra and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000084614.
Data analysis.
Data were analyzed using Spectrum Mill, version 6.01.202 (Agilent Technologies). In extracting spectra from the raw format for MS/MS searching, spectra from the same precursor or within a retention time window of ±60 seconds and an m/z range of ±1.4 were merged. Spectra were filtered to include only those with a precursor mass range of 750 to 6000 Da and a sequence tag length greater than 0. MS/MS searching was performed against a human UniProt database (www.uniprot.org). Digestion enzyme conditions were set to “Trypsin allow P” for the search, allowing up to 4 missed cleavages within a matched peptide. Fixed modifications were carbamidomethylation of cysteine and TMT6 on the N-terminus and internal lysine. Variable modifications were oxidized methionine and acetylation of the protein N-terminus. Matching criteria included a 30% minimum matched peak intensity and a precursor and product mass tolerance of ±20 ppm. Peptide-level matches were validated if found to be below the 1.0% FDR threshold and within a precursor charge range of 2 to 6. A second round of validation was then performed for protein-level matches, requiring a minimum protein score of 0. Protein-centric information, including experimental ratios, was then summarized in a table, which was quality filtered for nonhuman contaminants, keratins, and any proteins not identified by at least 2 fully quantified peptides with 2 ratio counts.
Statistics
All statistical evaluations comparing the different experimental groups were performed using GraphPad Prism 8 software for unpaired 2-tailed Student’s t test, 2-way ANOVA with Tukey’s post hoc test, and Kruskal-Wallis test with Dunn’s post hoc analysis with corrections for multiple comparisons. A P value of less than 0.05 was considered statistically significant. Raw data and complete, unedited blots corresponding to the additional independent experiments are presented in Supplemental Figure 7. For proteomics analyses, PB and CB plexes were analyzed separately, each using biologically paired samples to compare dexamethasone treatment with a DMSO control. Data were median normalized and subjected to a 1-sample moderated t test using an internal R-Shiny package based in the limma R library. Correction for multiple testing was performed using the Benjamini-Hochberg FDR method. ssGSEA was performed as previously described (53) using an R script (available at https://github.com/broadinstitute/ssGSEA2.0). ssGSEA was performed separately on PB and CB plexes, using the log2-transformed ratios of dexamethasone/control as input. Parameters were set as follows: sample.norm.type = “rank”, weight = 0.75, statistic = “area.under.RES”, output.score.type = “NES”, nperm = 1e3, min.overlap = 10, correl.type = “z.score”, par = T, spare.cores = 1. The heatmap in Figure 3F was generated using the average of triplicate enrichment scores, ranked to create a list of top-10 and bottom-10, eliminating redundant rows. It should be noted that the TMT plex design used limits direct quantitative comparisons between PB and CB samples, as the stochastic sampling of spectra on the mass spectrometer can lead to technical differences in the proteins identified in different plexes that are not necessarily driven by biological causes. Therefore, the observed differences between PB and CB samples were validated in follow-up experiments.
Study approval
All human studies were approved by the IRBs of Northwell Health and Stanford University. Written, informed consent was obtained from all participants prior to their inclusion in the study.
Author contributions
The co–first authors RJA and HY are listed in alphabetical order; they designed and performed most of the experiments, analyzed data, and wrote the manuscript. NW, JH, BMD, and JP performed experiments. MEO, NDU, and SAC designed, performed, and analyzed the proteomics studies and edited the manuscript. AV, JML, and LDC analyzed data related to patients with DBA. CH, SK, and NT designed and analyzed data and edited the manuscript. NM, AN, and LB designed the project, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank the patients and their families for their contributions. We thank Betsy J. Barnes, Philippe Marambaud, and Mark J. Koury for helpful discussions and critical reading of the manuscript. We also thank the Tissue Donation Program at Northwell Health for providing access to BM samples. This research was supported in part by NIH grants DK32094 (to NM), HL079571 (to JML and AV), HL134812 (to JML, AV, and LB), CA210986 and CA214125 (to SAC), CA210979 (to DRM), and HL144436 (to LB and AN); the Diamond Blackfan Anemia Foundation (to LB); and the Pediatric Cancer Foundation (to JML and LB). AN is the recipient of an American Society of Hematology (ASH) Bridge Grant and a Faculty Scholar grant from the Maternal and Child Health Research Institute at Stanford University. LB is the recipient of a St. Baldrick’s Scholar award.
Version 1. 01/21/2020
In-Press Preview
Version 2. 03/16/2020
Electronic publication
Version 3. 03/17/2020
Corrected Naomi Taylor's name in the author list.
Version 4. 04/01/2020
Print issue publication
Funding Statement
210979
079571
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(4):2097–2110.https://doi.org/10.1172/JCI132284.
Contributor Information
Ryan J. Ashley, Email: rashley1@northwell.edu.
Hongxia Yan, Email: HYan@nybc.org.
Nan Wang, Email: nanwang@stanford.edu.
John Hale, Email: jhale@nybloodcenter.org.
Julien Papoin, Email: Jpapoin@northwell.edu.
Meagan E. Olive, Email: molive@broadinstitute.org.
Steven A. Carr, Email: scarr@broad.mit.edu.
Adrianna Vlachos, Email: avlachos@nshs.edu.
Jeffrey M. Lipton, Email: JLipton@northwell.edu.
Lydie Da Costa, Email: lydie.dacosta@rdb.aphp.fr.
Sandrina Kinet, Email: kinet@igmm.cnrs.fr.
Narla Mohandas, Email: MNarla@NYBloodcenter.org.
Anupama Narla, Email: anunarla@stanford.edu.
Lionel Blanc, Email: Lblanc@northwell.edu.
References
- 1.Danilova N, Gazda HT. Ribosomopathies: how a common root can cause a tree of pathologies. Dis Model Mech. 2015;8(9):1013–1026. doi: 10.1242/dmm.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vlachos A, Blanc L, Lipton JM. Diamond Blackfan anemia: a model for the translational approach to understanding human disease. Expert Rev Hematol. 2014;7(3):359–372. doi: 10.1586/17474086.2014.897923. [DOI] [PubMed] [Google Scholar]
- 3.Khajuria RK, et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 2018;173(1):90–103.e19. doi: 10.1016/j.cell.2018.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ludwig LS, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20(7):748–753. doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quarello P, et al. Diamond-Blackfan anemia: genotype-phenotype correlations in Italian patients with RPL5 and RPL11 mutations. Haematologica. 2010;95(2):206–213. doi: 10.3324/haematol.2009.011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Costa L, Moniz H, Simansour M, Tchernia G, Mohandas N, Leblanc T. Diamond-Blackfan anemia, ribosome and erythropoiesis. Transfus Clin Biol. 2010;17(3):112–119. doi: 10.1016/j.tracli.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlachos A, Muir E. How I treat Diamond-Blackfan anemia. Blood. 2010;116(19):3715–3723. doi: 10.1182/blood-2010-02-251090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan HS, Saunders EF, Freedman MH. Diamond-Blackfan syndrome. II. In vitro corticosteroid effect on erythropoiesis. Pediatr Res. 1982;16(6):477–478. doi: 10.1203/00006450-198206000-00015. [DOI] [PubMed] [Google Scholar]
- 9.Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci. 2013;34(9):518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang Y, et al. Global increase in replication fork speed during a p57KIP2-regulated erythroid cell fate switch. Sci Adv. 2017;3(5):e1700298. doi: 10.1126/sciadv.1700298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HY, et al. PPAR-α and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature. 2015;522(7557):474–477. doi: 10.1038/nature14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, et al. Rate of progression through a continuum of transit-amplifying progenitor cell states regulates blood cell production. Dev Cell. 2019;49(1):118–129.e7. doi: 10.1016/j.devcel.2019.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, et al. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature. 2013;499(7456):92–96. doi: 10.1038/nature12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory CJ, Eaves AC. Human marrow cells capable of erythropoietic differentiation in vitro: definition of three erythroid colony responses. Blood. 1977;49(6):855–864. [PubMed] [Google Scholar]
- 15.Gregory CJ, Eaves AC. Three stages of erythropoietic progenitor cell differentiation distinguished by a number of physical and biologic properties. Blood. 1978;51(3):527–537. [PubMed] [Google Scholar]
- 16.McLeod DL, Shreeve MM, Axelrad AA. Improved plasma culture system for production of erythrocytic colonies in vitro: quantitative assay method for CFU-E. Blood. 1974;44(4):517–534. doi: 10.1182/blood.V44.4.517.517. [DOI] [PubMed] [Google Scholar]
- 17.Stephenson JR, Axelrad AA, McLeod DL, Shreeve MM. Induction of colonies of hemoglobin-synthesizing cells by erythropoietin in vitro. Proc Natl Acad Sci U S A. 1971;68(7):1542–1546. doi: 10.1073/pnas.68.7.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, et al. Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E. Blood. 2014;124(24):3636–3645. doi: 10.1182/blood-2014-07-588806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flygare J, Rayon Estrada V, Shin C, Gupta S, Lodish HF. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood. 2011;117(12):3435–3444. doi: 10.1182/blood-2010-07-295550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lodish H, Flygare J, Chou S. From stem cell to erythroblast: regulation of red cell production at multiple levels by multiple hormones. IUBMB Life. 2010;62(7):492–496. doi: 10.1002/iub.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, et al. TGF-β inhibitors stimulate red blood cell production by enhancing self-renewal of BFU-E erythroid progenitors. Blood. 2016;128(23):2637–2641. doi: 10.1182/blood-2016-05-718320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freire PR, Conneely OM. NR4A1 and NR4A3 restrict HSC proliferation via reciprocal regulation of C/EBPα and inflammatory signaling. Blood. 2018;131(10):1081–1093. doi: 10.1182/blood-2017-07-795757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golde DW, Bersch N, Cline MJ. Potentiation of erythropoiesis in vitro by dexamethasone. J Clin Invest. 1976;57(1):57–62. doi: 10.1172/JCI108269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohene-Abuakwa Y, Orfali KA, Marius C, Ball SE. Two-phase culture in Diamond Blackfan anemia: localization of erythroid defect. Blood. 2005;105(2):838–846. doi: 10.1182/blood-2004-03-1016. [DOI] [PubMed] [Google Scholar]
- 25.Iskander D, et al. Elucidation of the EP defect in Diamond-Blackfan anemia by characterization and prospective isolation of human EPs. Blood. 2015;125(16):2553–2557. doi: 10.1182/blood-2014-10-608042. [DOI] [PubMed] [Google Scholar]
- 26.Dulmovits BM, et al. Pomalidomide reverses γ-globin silencing through the transcriptional reprogramming of adult hematopoietic progenitors. Blood. 2016;127(11):1481–1492. doi: 10.1182/blood-2015-09-667923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nathan DG, Gardner FH. Erythroid cell maturation and hemoglobin synthesis in megaloblastic anemia. J Clin Invest. 1962;41:1086–1093. doi: 10.1172/JCI104559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies SV, Cavill I, Bentley N, Fegan CD, Poynton CH, Whittaker JA. Evaluation of erythropoiesis after bone marrow transplantation: quantitative reticulocyte counting. Br J Haematol. 1992;81(1):12–17. doi: 10.1111/j.1365-2141.1992.tb08163.x. [DOI] [PubMed] [Google Scholar]
- 29.d’Onofrio G, Chirillo R, Zini G, Caenaro G, Tommasi M, Micciulli G. Simultaneous measurement of reticulocyte and red blood cell indices in healthy subjects and patients with microcytic and macrocytic anemia. Blood. 1995;85(3):818–823. doi: 10.1182/blood.V85.3.818.bloodjournal853818. [DOI] [PubMed] [Google Scholar]
- 30.Yan H, et al. Developmental differences between neonatal and adult human erythropoiesis. Am J Hematol. 2018;93(4):494–503. doi: 10.1002/ajh.25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012;32(18):3756–3767. doi: 10.1128/MCB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy Z, Marta A, Butz H, Liko I, Racz K, Patocs A. Modulation of the circadian clock by glucocorticoid receptor isoforms in the H295R cell line. Steroids. 2016;116:20–27. doi: 10.1016/j.steroids.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Fassett MS, Jiang W, D’Alise AM, Mathis D, Benoist C. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proc Natl Acad Sci U S A. 2012;109(10):3891–3896. doi: 10.1073/pnas.1200090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo YG, Yeo MG, Kim DK, Park H, Lee MO. Novel function of orphan nuclear receptor Nur77 in stabilizing hypoxia-inducible factor-1alpha. J Biol Chem. 2004;279(51):53365–53373. doi: 10.1074/jbc.M408554200. [DOI] [PubMed] [Google Scholar]
- 35.Zhang FL, Shen GM, Liu XL, Wang F, Zhao YZ, Zhang JW. Hypoxia-inducible factor 1-mediated human GATA1 induction promotes erythroid differentiation under hypoxic conditions. J Cell Mol Med. 2012;16(8):1889–1899. doi: 10.1111/j.1582-4934.2011.01484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto A, et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2011;9(3):262–271. doi: 10.1016/j.stem.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Hsieh FF, et al. Cell cycle exit during terminal erythroid differentiation is associated with accumulation of p27(Kip1) and inactivation of cdk2 kinase. Blood. 2000;96(8):2746–2754. [PubMed] [Google Scholar]
- 38.Pateras IS, Apostolopoulou K, Niforou K, Kotsinas A, Gorgoulis VG. p57KIP2: “Kip”ing the cell under control. Mol Cancer Res. 2009;7(12):1902–1919. doi: 10.1158/1541-7786.MCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 39.Abraham RT, et al. Cellular effects of olomoucine, an inhibitor of cyclin-dependent kinases. Biol Cell. 1995;83(2-3):105–120. doi: 10.1016/0248-4900(96)81298-6. [DOI] [PubMed] [Google Scholar]
- 40.Ludwig LS, et al. Transcriptional states and chromatin accessibility underlying human erythropoiesis. Cell Rep. 2019;27(11):3228–3240.e7. doi: 10.1016/j.celrep.2019.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz VP, et al. A unique epigenomic landscape defines human erythropoiesis. Cell Rep. 2019;28(11):2996–3009.e7. doi: 10.1016/j.celrep.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heuston EF, et al. Establishment of regulatory elements during erythro-megakaryopoiesis identifies hematopoietic lineage-commitment points. Epigenetics Chromatin. 2018;11(1):22. doi: 10.1186/s13072-018-0195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farrar JE, et al. Altered epigenetic maturation in early erythroid cells from Diamond Blackfan anemia patients treated with transfusions, corticosteroids, or in remission. Blood. 2018;132(Suppl 1):752. doi: 10.1182/blood-2018-99-116994. [DOI] [Google Scholar]
- 44.D’Arena G, Musto P, Cascavilla N, Di Giorgio G, Zendoli F, Carotenuto M. Human umbilical cord blood: immunophenotypic heterogeneity of CD34+ hematopoietic progenitor cells. Haematologica. 1996;81(5):404–409. [PubMed] [Google Scholar]
- 45.Fauser AA, Messner HA. Fetal hemoglobin in mixed hemopoietic colonies (CFU-GEMM), erythroid bursts (BFU-E) and erythroid colonies (CFU-E): assessment by radioimmune assay and immunofluorescence. Blood. 1979;54(6):1384–1394. [PubMed] [Google Scholar]
- 46.Nathan DG, et al. Human erythroid burst-forming unit: T-cell requirement for proliferation in vitro. J Exp Med. 1978;147(2):324–339. doi: 10.1084/jem.147.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tusi BK, et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature. 2018;555(7694):54–60. doi: 10.1038/nature25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155(1):135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palumbo-Zerr K, et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat Med. 2015;21(2):150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 50.Ge J, et al. Dysregulation of the transforming growth factor β pathway in induced pluripotent stem cells generated from patients with Diamond Blackfan anemia. PLoS ONE. 2015;10(8):e0134878. doi: 10.1371/journal.pone.0134878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zecha J, et al. TMT Labeling for the masses: a robust and cost-efficient, in-solution labeling approach. Mol Cell Proteomics. 2019;18(7):1468–1478. doi: 10.1074/mcp.TIR119.001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mertins P, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc. 2018;13(7):1632–1661. doi: 10.1038/s41596-018-0006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.