Abstract
TBX3, a member of the ancient and evolutionary conserved T-box transcription factor family, is a critical developmental regulator of several structures including the heart, mammary glands, limbs and lungs. Indeed, mutations in the human TBX3 lead to ulnar mammary syndrome which is characterized by several clinical malformations including hypoplasia of the mammary and apocrine glands, defects of the upper limb, areola, dental structures, heart and genitalia. In contrast, TBX3 has no known function in adult tissues but is frequently overexpressed in a wide range of epithelial and mesenchymal derived cancers. This overexpression greatly impacts several hallmarks of cancer including bypass of senescence, apoptosis and anoikis, promotion of proliferation, tumour formation, angiogenesis, invasion and metastatic capabilities as well as cancer stem cell expansion. The debilitating consequences of having too little or too much TBX3 suggest that its expression levels need to be tightly regulated. While we have a reasonable understanding of the mutations that result in low levels of functional TBX3 during development, very little is known about the factors responsible for the overexpression of TBX3 in cancer. Furthermore, given the plethora of oncogenic processes that TBX3 impacts, it must be regulating several target genes but to date only a few have been identified and characterised. Interestingly, while there is compelling evidence to support oncogenic roles for TBX3, a few studies have indicated that it may also have tumour suppressor functions in certain contexts. Together, the diverse functional elasticity of TBX3 in development and cancer is thought to involve, in part, the protein partners that it interacts with and this area of research has recently received some attention. This review provides an insight into the significance of TBX3 in development and cancer and identifies research gaps that need to be explored to shed more light on this transcription factor.
Keywords: Transcription factor, T-box factors, TBX3, Heart development, Limb development, Mammary gland development, Lung development, Ulnar mammary syndrome, Obesity, Rheumatoid arthritis, Cancer, Signalling, Target genes, Co-factors, Stem cells
1. Introduction
The T-box 3 gene (TBX3) is a member of the ancient T-box gene family which is conserved across a wide spectrum of species. A mouse mutation that results in a short tail identified in 1927 led to the discovery and ultimate cloning of Brachyury, the prototype of the family, in 1990 and, as shown in Table 1, there are currently 17 paralogues in human, mouse and rat that are grouped into five subfamilies, namely Brachyury (T), T-brain (Tbr1), TBX1, TBX2, and TBX6 (Papaioannou, 2014). Tbx3 is a member of the Tbx2 subfamily which includes Tbx2, Tbx4 and Tbx5. Members of this subfamily originated from the duplication of a primordial gene by an unequal crossing over event which initially gave rise to the Tbx2/Tbx3 and Tbx4/Tbx5 cognate gene pairs and their subsequent duplication led to the four independent genes. Functional studies have shown that T-box family members are transcription factors with a highly conserved DNA binding domain known as the T-box. They can activate and/or repress their target genes through binding a partially palindromic sequence (T(G/C)ACACCT AGGTGTGAAATT) known as the T-element, or half sites within this sequence as well as protein co-factor binding sites (Wilson and Conlon, 2002). Their importance has been well established in the field of developmental biology where they play essential roles from as early as cell-fate determination all the way through to organogenesis (Packham and Brook, 2003; Papaioannou, 2001). Not surprisingly, numerous human congenital developmental syndromes are associated with mutated T-box genes and there is significant evidence implicating T-box factors as major contributors of cancer processes as either oncoproteins and/or tumour suppressors (Wansleben et al., 2014).
Table 1.
TBX3 paralogues identified to date in human (green), mouse (blue) and rat (orange) tabulated according to their respective subfamilies (data appropriated from the ensembl genome browser). aa = amino acids, Da = Daltons.
| T-box gene subfamily | Gene name | Chromosome | Ensemble transcript ID | UniProt Code | Protein length (aa) | Mass (Da) |
|---|---|---|---|---|---|---|
| T | T | 6 | ENST00000296946.6 | 015178 | 453 | 47.44 |
| 17 | ENSMUST00000074667.8 | Q78ZW9 | 436 | 47.44 | ||
| 1 | ENSRNOT00000033685.5 | F1LXU5 | 377 | 43.211 | ||
| Tbx19 (Tpit) | 1 | ENST00000367821.8 | O60806 | 448 | 48.24 | |
| 1 | ENSMUST00000027859.il | Q99ME7 | 446 | 48.037 | ||
| 13 | ENSRNOT00000063870.1 | D3Z977 | 212 | 23.695 | ||
| Tbx1 | Tbx1 | 22 | ENST00000649276.1 | A0A3B3IS18 | 504 | 53.505 |
| 16 | ENSMUST00000009241.6 | F6ZP09 | 479 | 51.677 | ||
| 11 | ENSRNOT00000002597.5 | D4A2E9 | 480 | 51.774 | ||
| Tbx10 | 11 | ENST00000335385.3 | 075333 | 385 | 42.341 | |
| 19 | ENSMUST00000041871.8 | Q810F8 | 385 | 42.407 | ||
| 1 | ENSRNOT00000024129.4 | D3ZAQ3 | 344 | 38.267 | ||
| Tbx15 | 1 | ENST00000369429.5 | Q96SF7 | 602 | 65.757 | |
| 3 | ENSMUST00000029462.9 | 070306 | 602 | 65.802 | ||
| 2 | ENSRNOT00000067358.2 | D3ZJ07 | 602 | 65.789 | ||
| Tbx18 | 6 | ENST00000369663.10 | 095935 | 607 | 64.753 | |
| 9 | ENSMUST00000034991.7 | G3X919 | 613 | 65.434 | ||
| 8 | ENSRNOT00000014657.4 | D4A1V6 | 612 | 65.595 | ||
| Tbx20 | 7 | ENSRNOT00000064783.2 | D3ZUF4 | 298 | 33.274 | |
| 9 | ENSMUST00000052946.il | Q9ES03 | 445 | 49.096 | ||
| 8 | ENSRNOT00000082744.1 | A0A0G2KAH3 | 446 | 49.163 | ||
| Tbx22 | X | ENST00000373294.8 | Q9Y458 | 520 | 57.910 | |
| X | ENSMUST00000168174.8 | E9Q5R8 | 531 | 59.869 | ||
| X | ENSRNOT00000003190.5 | D3ZMK6 | 518 | 58.557 | ||
| Tbx2 | Tbx2 | 17 | NST00000240328.4 | A0A024QZ86 | 712 | 75.066 |
| 11 | ENSMUST00000000095.6 | Q60707 | 711 | 75.081 | ||
| 10 | ENSRNOT00000004698.7 | F1M0C0 | 364 | 40.690 | ||
| Tbx3 | 12 | ENST00000257566.7 | 015119 | 743 | 79.389 | |
| 5 | ENSMUST00000018748.8 | P70324 | 741 | 79.16 | ||
| 12 | ENSRNOT00000084018.1 | A0A0G2K8D7 | 723 | 77.124 | ||
| Tbx4 | 17 | ENST00000240335.1 | P57082 | 545 | 60.204 | |
| 11 | ENSMUST00000108047.7 | P70325 | 552 | 61.101 | ||
| 10 | ENSRNOT00000004736.3 | D4A0A2 | 554 | 61.387 | ||
| Tbx5 | 12 | ENST00000310346.8 | Q99593 | 518 | 57.711 | |
| 5 | ENSMUST00000018407.9 | Q5CZX7 | 518 | 57.832 | ||
| 12 | ENSRNOT00000001893.5 | G3V657 | 517 | 57.745 | ||
| Tbx6 | Tbx6 | 16 | ENST00000279386.6 | 095947 | 436 | 47.045 |
| 7 | ENSMUST00000094037.4 | P70327 | 436 | 47.006 | ||
| 1 | ENSRNOT00000068543.1 | D3ZJK7 | 436 | 47.216 | ||
| Mga | 15 | ENST00000219905.il | Q8IWI9 | 3065 | 336.159 | |
| 2 | ENSMUST00000046717.12 | A2AWL7 | 3003 | 328.802 | ||
| 3 | ENSRNOT00000008528.8 | D3ZJB5 | 3005 | 329.343 | ||
| Tbr1 | Tbr1 | 2 | ENST00000389554.8 | Q16650 | 682 | 74.053 |
| 2 | ENSMUST00000048934.14 | Q64336 | 681 | 73.940 | ||
| 3 | ENSRNOT00000065340.3 | D4A6N8 | 680 | 73.632 | ||
| Eomes (Tbr2) | 3 | ENST00000295743.8 | 095936 | 686 | 72.732 | |
| 9 | ENSMUST00000035020.14 | 054839 | 707 | 74.801 | ||
| 8 | ENSRNOT00000013530.5 | D3ZY52 | 699 | 74.203 | ||
| Tbx21 (Tbet) | 17 | ENST00000177694.2 | Q9UL17 | 535 | 58.328 | |
| 11 | ENSMUST00000001484.2 | Q9JKD8 | 530 | 57.852 | ||
| 10 | ENSRNOT00000012538.5 | D3ZCM2 | 528 | 57.674 |
1.1. TBX3 gene location and structure
The human TBX3 gene maps to the reverse strand of chromosome 12 at position 12q23–24.1 and consists of 7 exons within a 4.7 kb region which spans from 114670255 bp to 114684175 bp (ENSEMBL assembly release GRCh38.p12) (Fig. 1). It encodes a 723 amino acid protein with part of exon 1, exons 2 and 3, and part of exon 4, encoding the conserved T-box domain, exon 1 and 2 encode one of two repression domains (R2), exon 6 encodes the activation domain and part of exon 6 and 7 encode the second repression domain (R1) (Bamshad et al., 1997; Carlson et al., 2001; He et al., 1999).
Fig. 1.
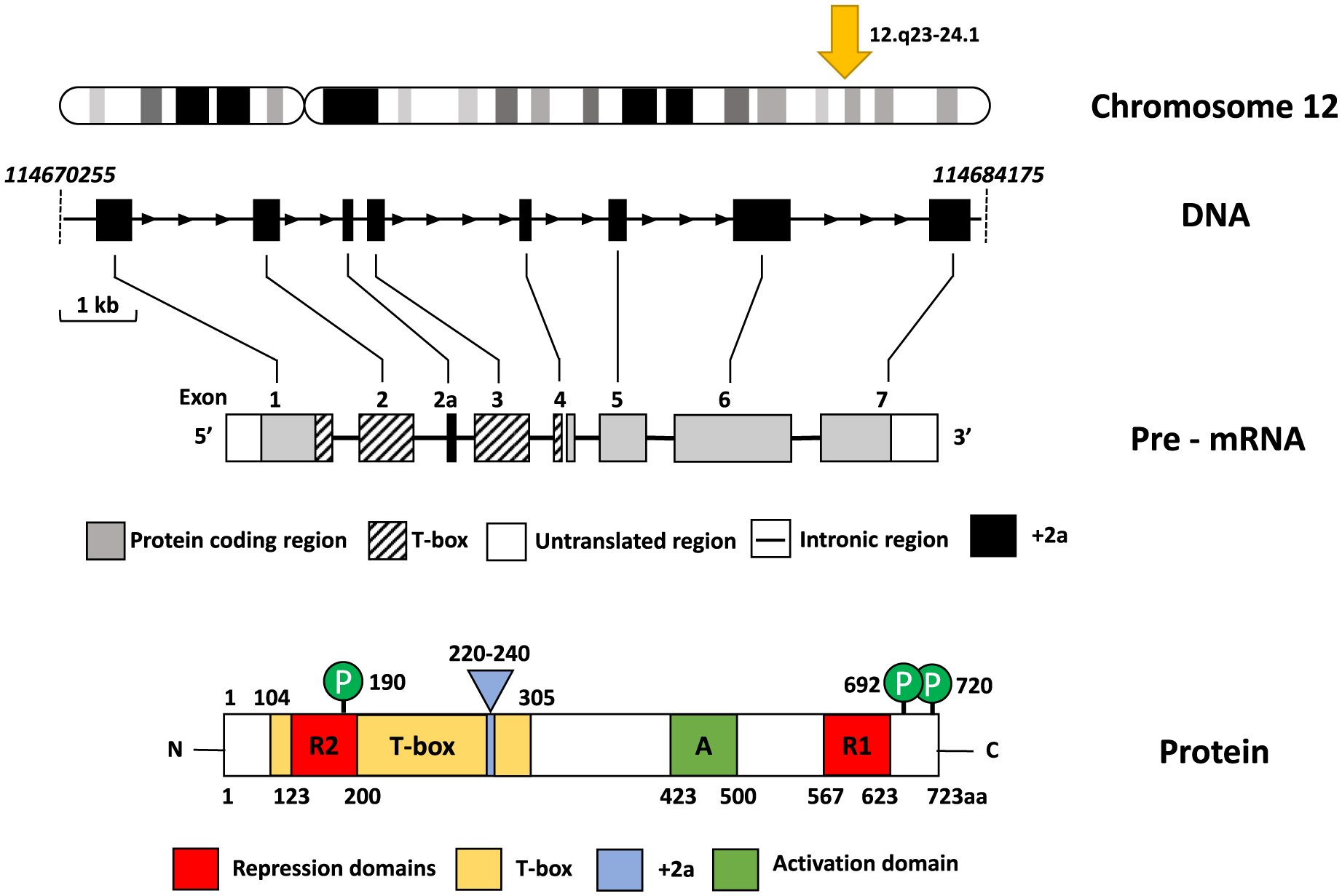
Schematic representation of the human TBX3 gene, pre-mRNA and protein structure. The location of TBX3 on chromosome 12 is depicted with the yellow arrow. The 4.7 kb DNA region is shown with coding regions (exons 1–7) represented by black boxes and the horizontal arrows indicate the direction of transcription. Representative size of region is depicted by thin bracketed horizontal line segment beneath the gene. The exons are linked to the pre-mRNA region depicting relative size, position of exons and the +2a splice variant of TBX3. The diagram depicting the TBX3 protein shows the DNA binding domain (T-box, yellow boxes), two repression domains (R1 and R2, red boxes), activation domain (A, green box) and the +2a splice variant (blue box). The amino acid residue number is displayed below each box and green circles above the protein diagram correspond to phosphorylation sites (adapted from Willmer et al, 2017).
1.2. TBX3 mRNA structure and splicing
Alternative processing and splicing gives rise to at least 4 distinct TBX3 isoforms with TBX3 and TBX3+2a being the predominant isoforms. TBX3+2a results from alternative splicing of the second intron which leads to the addition of the +2a exon and consequently this isoform has an additional 20 amino acids within the T-box DNA binding domain (Fig. 1) (Bamshad et al., 1997; Fan et al., 2004). Considering the location of the extra 20 amino acids in the TBX3+2a isoform, it is tempting to speculate that it may regulate a different set of downstream targets to TBX3. Indeed, an initial study by Fan et al. (2004) indicated that whereas Tbx3 inhibits senescence in mouse embryonic fibroblasts (MEFs), Tbx3+2a accelerated the process. Subsequent studies have, however, shown that the two TBX3 isoforms have similar roles, at least functionally if not always mechanistically. For example, as will be seen later, they can both bind and repress several common target genes during embryonic development and cancer and they can both inhibit the process of mRNA splicing by directly binding RNAs containing the core motif of a T-element (Hoogaars et al., 2008; Krstic et al., 2016, 2019; Rodriguez et al., 2008; Zhao et al., 2014). It therefore seems likely that the function of the TBX3 and Tbx3+2a isoforms may vary slightly across different cell types.
2. TBX3 protein
2.1. TBX3 functional domains
TBX3 is a transcription factor characterised by a DNA-binding domain (DBD) also called the T-box, a nuclear localization signal (NLS), two repression domains (R2 and R1) and an activation domain (A) (Fig. 1). The NLS, T-box and R2 domains are 100% conserved between human and mouse and their R1 and activation domains share 98.4% and 77.5% homology respectively. The T-box is situated in the amino terminus (position 105–287; REFSEQ: accession NM 005996.3) and consists of 182 amino acids. It recognizes highly related DNA sequences, called T-elements, although it can also recognise variations within the consensus T-element sequences. The NLS is a 6 basic amino acid cluster ‘RREKRK’, which spans amino acid residues 292–297. While the R2 consists of 77 amino acids and is located within the T-box (123–200), the dominant repressor domain R1 consists of 56 amino acids and is in the C terminus (567–623). The activation domain consists of 77 amino acids and is located between amino acids 423–500 of the TBX3 protein (Carlson et al., 2001).
2.2. DNA binding properties of TBX3
Coll et al. (2002) resolved the 3-dimensional structure of the TBX3 DBD interacting with its palindromic consensus target DNA (5′TAATT TCACACCTAGGTGTGAAAT3′). The crystallographic data showed that TBX3 recognises the core 10 base pair sequence (5′TTTCACACCT3′) referred to as the half T-element. Furthermore, the authors show that the TBX3 DBD interacts with the GC base pairs 3 and 5 through direct hydrogen bonds, with TA base pairs 8 and 9 through hydrophobic interactions, and with base pairs 1, 2 and 4 through an indirect mechanism. Hoogaars et al. (2008), confirmed that both TBX3 isoforms bind the half T-element efficiently and that the +2a region does not alter the DNA binding ability of the TBX3 DBD. Coll et al. (2002) further showed that Tbx3 binds its consensus sequence as two monomers, with each one recognising one of the half T-elements in the palindromic target sequence. It was however predicted that TBX3, like all other T-box members identified to date, will bind its biological downstream effectors as a single monomer through the half T-element of the palindrome target sequence (Coll et al., 2002; Wilson and Conlon, 2002).
To date, only TBX3 target sites with sequences closely related to half-sites of the original palindrome have been identified. Indeed, while there is still a paucity of information available on TBX3 target genes, studies have shown that TBX3 can regulate diverse cellular processes through its ability to transcriptionally repress or activate biologically relevant factors through mechanisms involving single half T-elements. For example, TBX3 directly represses transcription of the tumour suppressors, p19ARF (p14ARF in humans) through CACCTCTGGTGCCA in primary breast tumours (Lingbeek et al., 2002), p21WAF1/CIP1 through a GTGTGA close to the initiator in chondrosarcoma (Willmer et al., 2016a, 2016b), E-cadherin through CAGGTGT in melanoma (Rodriguez et al., 2008) and TBX2 through GACACCT in breast cancer and melanoma cells (Li et al., 2014). Furthermore, Weidgang et al. (2013) showed that in mouse embryonic stem cells (mESCs), Tbx3 directly bound highly conserved T-elements to activate the promoters of Eomes, T and Sox17 which are essential for mesoderm differentiation. Lu et al. (2011) also reported that TBX3 directly binds a conserved T-element at −700 bp of the Gata6 promoter to activate it in mESCs in order to promote extra embryonic endodermal differentiation. Interestingly, TBX3 represses PTEN through a region of its promoter which lacks putative T-elements, but which forms an important regulatory unit for PTEN transcriptional activators. This raises the possibility that TBX3 may also repress some of its target genes through interfering with transcriptional activators (Burgucu et al., 2012).
2.3. Phosphorylation of TBX3
While there are several predicted post-translational modification sites for TBX3 including 10 ubiquitination, 1 acetylation, 2 methylation and 29 phosphorylation sites only the SP190, SP692 and S720 phosphorylation sites have been fully characterised. The kinases involved are cyclin A-CDK2 at either SP190 or SP354, p38 MAP kinase at SP692 and AKT Serine/Threonine Kinase 3 (AKT3) at S720 (Peres et al., 2015; Willmer et al., 2016; Yano et al., 2011). The SP190 motif within the DBD is highly conserved across T-box factors and species suggesting that it must have an important regulatory role. Indeed, while the kinase responsible for phosphorylating TBX3 at this site has not been identified, a SP190 pseudo phosphorylated TBX3 protein has reduced ability to bind and transcriptionally repress p21WAF1/CIP1 and consequently to promote proliferation (Willmer et al., 2016a, 2016b). Phosphorylation within the N-terminal (1–371) half of the TBX3 protein by the cyclin ACDK2 complex is important for stabilizing TBX3 during the S phase of the cell cycle and allowing for its functional role in driving S phase progression (Willmer et al., 2015). SP motifs are the minimum consensus sequences for cyclin A-CDK2 and since SP190 and SP354 are the only SP motifs within this region either one or both must be involved in this phosphorylation. Furthermore, phosphorylation of TBX3 by the p38 MAP kinase at SP692 in embryonic kidney cells enhances its ability to transcriptionally repress its well-known target, E-cadherin, to promote migration (Yano et al., 2011). Importantly, in melanoma, phosphorylation of TBX3 at S720 by AKT3 promotes its protein stability, nuclear localisation, transcriptional repression of E-cadherin, and its role in cell migration and invasion (Peres et al., 2015).
2.4. Interacting proteins
Increasing evidence suggests that the function of TBX3 as either a transcriptional repressor or transcriptional activator is, in part, modulated by protein co-factors. For example, during embryogenesis it can interact with other transcription factors such as Nkx2–5, Msx and Sox4 to assist it binding to its target genes to regulate heart development (Bakker et al., 2008; Boogerd et al., 2008, 2011, Christoffels et al., 2000, 2004; Hoogaars et al., 2008; Stennard and Harvey, 2005). In the cancer context, TBX3 can interact with histone deacetylases (HDACs) to repress target genes. Indeed, it interacts with HDACs 1, 2, 3 and 5 to repress the tumour suppressor p14ARF in breast cancer and with HDAC5 to repress E-cadherin to promote metastasis in hepatocellular carcinoma (Dong et al., 2018a, 2018b; Yarosh et al., 2008). Lastly, Kumar et al. (2014a, 2014b) showed that TBX3 interacts with CAPERα to repress the long non-coding RNA, UCA1, resulting in the bypass of senescence through loss of UCA1-mediated stabilisation of p16INK4A mRNA.
3. Expression and function of TBX3 during development
TBX3 plays multiple roles during embryonic development as evidenced by the abnormalities reported for homozygous and heterozygous mice as well as the phenotype of individuals with the ulnar mammary syndrome (UMS) which results from mutations in human TBX3.
During mouse embryonic development, Tbx3 is expressed in the inner cell mass of the blastocyst, in the extraembryonic mesoderm during gastrulation, and in the developing heart, limbs, musculoskeletal, mammary glands, nervous system, skin, eye, liver, pancreas, lungs, pituitary glands and genitalia (Chapman et al., 1996; Tümpel et al., 2002; Davenport et al., 2003; Moorman et al., 2004; Cho et al., 2006; Lin et al., 2007; Bakker et al., 2008; Mesbah et al., 2008; Pontecorvi et al., 2008; Lüdtke et al., 2009, 2016; Begum and Papaioannou, 2011; Colasanto et al., 2016; Emechebe et al., 2016; Ichijo et al., 2017; López et al., 2018; Quarta et al., 2019; Karolak et al., 2019). Importantly, Tbx3 null embryos show defects in, among other structures, the heart, mammary glands and limbs and they die in utero by embryonic day E16.5, most likely due to yolk sac and heart defects (Davenport et al., 2003). These observations, underscored by studies described below, have illustrated that Tbx3 plays crucial roles in the development of the heart, mammary glands, limbs and lungs.
3.1. Heart development
During the onset of cardiogenesis, the linear heart tube undergoes looping and forms a chamber myocardium, which consists of ventricular and atrial chambers, and a non-chamber myocardium, which consists of the inflow and outflow tract (IFT and OFT), the atrioventricular canal (AVC) and the inner curvatures (Christoffels et al., 2004). During the process of looping, a chamber myocardiumspecific gene program, which includes expression of Nppa, Cx40, Cx43, and Chisel, initiates proliferation and differentiation in specific regions of the heart tube to form the chamber myocardium (Christoffels et al., 2000; Delorme et al., 1997; Van Kempen et al., 1996). In contrast, regions that form the non-chamber myocardium do not express this specific gene program and largely retain the phenotype of the early myocardium. Cells from the non-chamber myocardium form the cardiac conduction system (CCS), which controls the co-ordinated contraction of the heart (Christoffels et al., 2004; Greulich et al., 2011).
During early heart development, Tbx3 is exclusively expressed in the non-chamber myocardium of the AVC and the OFT (Christoffels et al., 2004). At E10.5, Tbx3 expression is found in the AVC, atrioventricular bundle (AVB), sinoatrial node (SAN) and OFT. In the formed heart at E16.5, Tbx3 fully delineates the CCS and is expressed in the SAN, AVB, atrioventricular node (AVN) and proximal bundle branches (BBs) (Fig. 2A). Tbx3 expression is particularly important for the formation of the CCS, AVB, and the ventricular septum of the heart and Tbx3-deficient embryos develop ventricular septal defects, delay in heart looping and outflow tract malformations (Bakker et al., 2008; Mesbah et al., 2008; Ribeiro et al., 2007; Washkowitz et al., 2012). It is believed that Tbx3 contributes to the developing CCS by firstly modulating cell division which results in constrictions between chambers, and secondly by directly repressing chamber myocardium genes by cooperatively binding their promoters along with other transcription factors (Christoffels et al., 2004; Hoogaars et al., 2004; Washkowitz et al., 2012). Indeed, Tbx3 binds cooperatively with Msx1 and Msx2 in the repression of Cx43 and with Nkx2.5 to repress Nppa in the non-chamber myocardium to block chamber formation (Fig. 2A) (Boogerd et al., 2008; Hoogaars et al., 2004). Furthermore, Tbx3 mutant hearts show elevated expression of Cx40, Cx43, and Nppa in the non-chamber AVC and ectopic expression of Tbx3 leads to the upregulation of CCS genes, such as Lbh and Hcn4, and the development of functional conduction tissue (Fig. 2A) (Hoogaars et al., 2007). These results show that Tbx3 exerts an important function by repressing the chamber-specific genetic program in regions from which functional tissues of the non-chamber myocardium are formed.
Fig. 2.
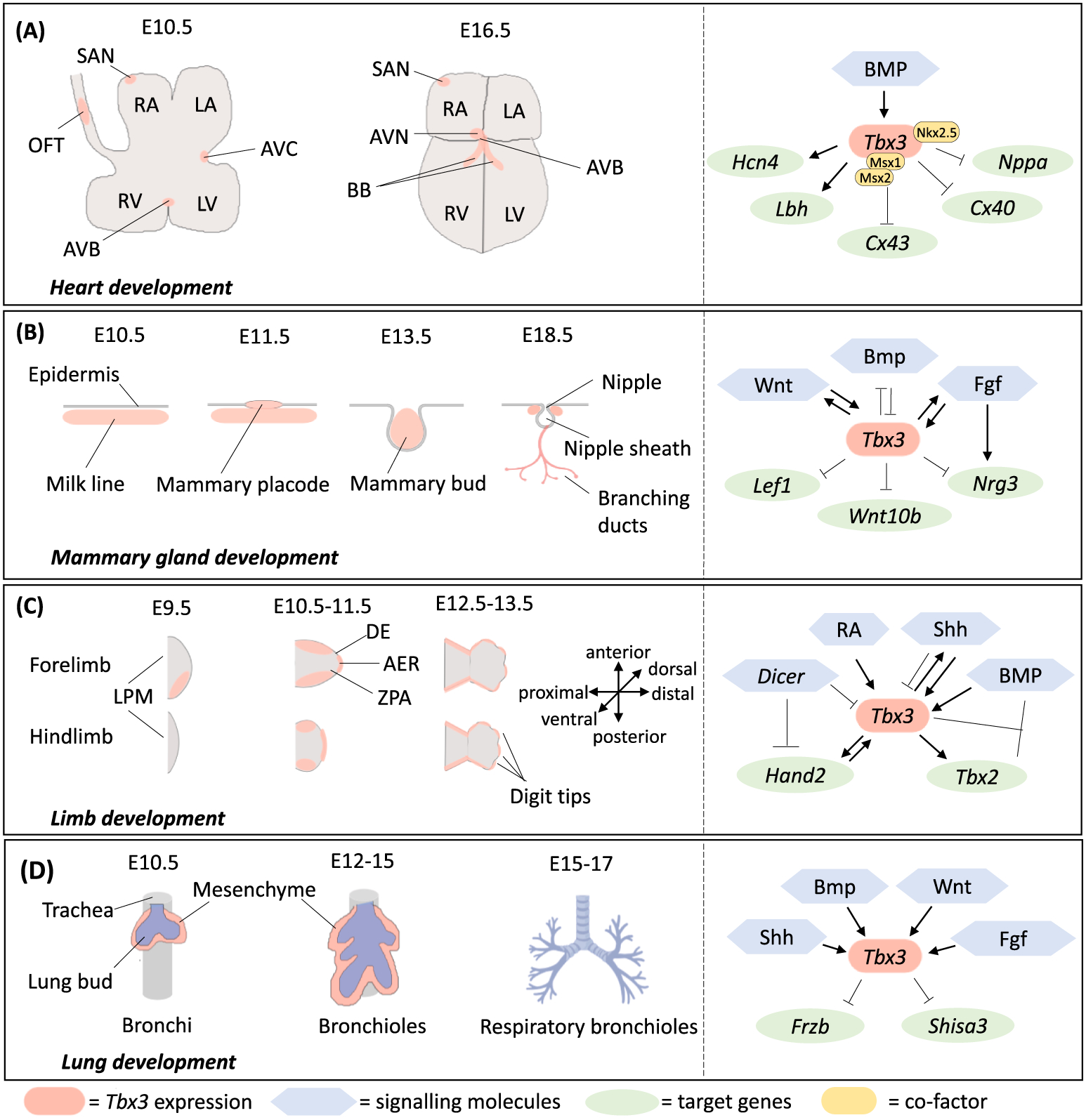
Left panels: Expression of Tbx3 (red) during the development of the mouse (A) heart, (B) mammary gland, (C) limb and (D) lung. (A) At E10.5, Tbx3 is expressed in the SAN, OFT, AVB and AVC, whereas at E16.5 the topography of Tbx3 expression delineates the CCS with expression in the SAN, AVN, AVB and BB. (B) Tbx3 first appears in the mesenchymal milk line at E10.5 and is then expressed in the mammary placodes at E11.5. Tbx3 expression continues during mammary bud formation at E13.5 and the formation of the branching ductal system at E18.5. Furthermore, Tbx3 is expressed in the mesenchyme surrounding the nipples.(C) At E10.5, Tbx3 is expressed in the posterior and anterior margins of the fore and hindlimb buds, as well as the AER. By E12.5, Tbx3 expression is limited to the tips of the digits. (D) Tbx3 is expressed in the lung mesenchyme from E10.5 (embryonic stage) to E14.5 (late pseudoglandular stage). Some of the diagrams in this figure are adapted from Washkowitz et al. (2012) and permission was granted by the corresponding author Prof Virginia Papaioannou. Right panels (A)–(D): Signalling molecules and targets that modulate Tbx3 activity during the relevant developmental processes indicated on the left.
Tbx3, alongside with Tbx18 and Shox2, is also important for the development of the functional SAN (Espinoza-Lewis et al., 2009; Hoogaars et al., 2007; Wiese et al., 2009). The SAN is the pacemaker of the heart and initiates the heartbeat and controls the rate and the rhythm of contraction throughout life (Hoogaars et al., 2007; Protze et al., 2017). Lineage tracing showed that the SAN originates from Tbx3 positive cells in the early heart tube (Mohan et al., 2018). Importantly, Tbx3 was shown to regulate the pacemaker gene program and phenotype by suppressing the atrial differentiation gene program in the SAN. Ectopic expression of Tbx3 in the atria of mouse models resulted in the development of functional ectopic pacemakers and induced Tbx3 expression reprogrammed terminally differentiated cardiomyocytes into pacemaker cells (Bakker et al., 2012; Hoogaars et al., 2007). Recently, SAN-like pacemaker cells were generated from human pluripotent stem cells without genetic manipulation and these cells were positive for Tbx3, Tbx18 and Shox2 and were shown to be able to function as a biological pacemaker in vivo (Protze et al., 2017). Taken together, Tbx3 plays an important role in the pacemaker function of the heart.
To date, very little is known about signalling pathways that regulate Tbx3 expression during heart formation. However, there is increasing evidence that the Bone Morphogenetic Protein (BMP) pathway is an important upstream modulator of Tbx3 expression (Fig. 2A) (Yamada et al., 2000; Yang et al., 2006). BMPs are members of the transforming growth factor β (TGF-β) gene family and play a critical role in the formation of the non-chamber myocardium (Shi et al., 2000). Yamada et al. (2000) showed that Tbx3 expression patterns overlap with those of Bmp2 during chick embryonic heart development and that ectopic expression of Bmp2 induces Tbx3 expression in non-cardiogenic tissue capable of developing into cardiac tissue. Moreover, Yang et al. (2006) showed that Tbx3 expression is downregulated when the Type I Bmp receptor is ablated and that Tbx3 is a direct target of Bmp Smads in vivo.
3.2. Mammary gland development
In mice, the development of the mammary glands (Fig. 2B) begins at E10.5 with the formation of the milk line between the forelimb and hindlimb of each flank, which forms an ectodermal ridge characterized by Wnt10b expression (Veltmaat et al., 2004). At E11.5 the mammary placodes begin to form and by E13.5, five pairs of mammary placodes have developed along the milk line and start to expand into mammary buds and by E18.5 the branching ductal system has formed. Studies have shown that Fibroblast growth factor (Fgf) signalling is critical for the induction and maintenance of mammary placodes 1, 2, 3 and 5 and for the expression of the earliest known breast differentiation marker, Lymphoid enhancer factor 1 (Lef1), a downstream mediator of Wnt signalling (Mailleux et al., 2002). Wnt signalling plays a critical role in mammary bud formation as illustrated by Lef1 null mice developing a reduced number of mammary buds and when the pathway is inhibited by Dickkopf-1, bud formation is completely lost at E11.5 (Andl et al., 2002; van Genderen et al., 1994).
Tbx3 first appears at E10.5 in the mesenchymal milk line and at E11.5 it is one of the earliest markers of mammary gland epithelium in the placodes. Tbx3 continues to be expressed at E13.5 in the mammary buds and by E18.5 Tbx3 is expressed in the mesenchyme surrounding the nipples (Chapman et al., 1996; Davenport et al., 2003) (Fig. 2B). The functional significance of Tbx3 expression during mammary gland development has been demonstrated in several studies. Loss of Tbx3 in homozygous mutant mice results in failure of placode induction and heterozygous mutant mice have decreased ductal tree development and failed nipple formation (Davenport et al., 2003; Jerome-Majewska et al., 2005; Rowley et al., 2004). Interestingly, failure of placode induction in homozygous mutant mice, places Tbx3 activity upstream of both Fgf and Wnt signalling (Fig. 2B) (Rowley, et al., 2004). This is evidenced by the loss of Wnt10b and Lef1 expression, as well as Fgf signalling when Tbx3 is absent (Davenport, et al., 2003). Interestingly, both Wnt and Fgf signalling, have also been described to feed into the regulatory network of Tbx3 during mammary gland development (Fig. 2B). Indeed, when Wnt or Fgf signalling is inhibited by CK1–7 or SU5402 respectively in early bud formation, Tbx3 expression is completely abolished. Taken together, these results indicate that Wnt, Fgf and Tbx3 are involved in feedforward and feedback loops to regulate the expression of each other (Eblaghie et al., 2004). Cho et al. (2006) also provided evidence that a reciprocal negative regulation between Bmp4 and Tbx3 expression is crucial for mammary gland positioning (Fig. 2B). Furthermore, Nrg3 transmits signals downstream of Tbx3 and Fgf signalling from somite to the overlying ectoderm to promote their local aggregation in the mammary placode and subsequent placode formation (Howard et al., 2005; Howard and Ashworth, 2006).
3.3. Limb and digit development
Limb development (Fig. 2C) is initiated by the emergence of small buds from the lateral body wall, consisting of a lateral plate mesoderm (LPM) and an overlying ectodermal layer. The outgrowth and patterning of the limb buds is dependent on three key signalling centres: the apical ectodermal ridge (AER), the dorsal ectoderm (DE) and the zone of polarizing activity (ZPA). These induce and co-ordinate specific outgrowth of the limb bud along the dorsal-ventral, anterior-posterior, and proximal-distal axes (Capdevila and Belmonte, 2001). Activities of the AER, ZPA and DE depend on complex signalling pathways, with the major contributors being the Fgf and Sonic Hedgehog (Shh) pathways (Fig. 2C). In the limb mesenchyme, Fgf10 induces Fgf8 in the overlying ectoderm and the formation of the AER and Fgf8 induces Shh expression to establish the ZPA. Together, Fgf and Shh signalling promote digit development and control digit number and patterning (Martin, 1998; Ohuchi et al., 2000).
Tbx3 is first expressed at the posterior margin and thereafter in the mesenchyme of the anterior and posterior margins of the early limb buds and at the AER (Fig. 2C). By E13.5, expression of Tbx3 in the AER is restricted to the tips of the digits (Chapman et al., 1996; Gibson-Brown et al., 1996). Importantly, Tbx3 homozygous mutant embryos display forelimb abnormalities, severe reduction in hindlimb bud development and reduced AER formation (Davenport, et al., 2003). Furthermore, a recent study by Emechebe et al. (2016) revealed that Tbx3 positively regulates Shh signalling to control digit number (Fig. 2C). The authors generated Tbx3 fl/fl;Cre mutant mice in which Tbx3 expression was stopped at different stages of mouse limb development and observed different abnormalities depending on when Tbx3 expression was halted. Whereas loss of Tbx3 expression in early development disrupted Shh signalling and resulted in failure of limb initiation and limb abnormalities, later deletion of Tbx3 in the posterior limb mesenchyme resulted in digit loss. It is important to note that Tbx3 also controls digit number via a Shh-independent, cilium-based Hedgehog pathway and loss of Tbx3 in the anterior limb results in preaxial polydactyly. In addition, Tbx3 expression has been reported to be downstream of the retinoic acid (RA) signalling pathway which plays a critical role in early limb development. Indeed, loss of components of the RA pathway in mutant mice leads to various forelimb abnormalities ranging from small limbs with digit anomalies to absent limbs (Lohnes et al., 1994; Sandell et al., 2007). Furthermore, an RA–receptor complex directly activates the Tbx3 promoter and RA deficient embryos show decreased expression of Tbx3 in the limb (Ballim et al., 2012). Hand2 and Tbx3 also form an important regulatory network in limb development. For example, anterior and posterior polarization of the limb bud mesenchyme requires the expression of Tbx3 (and Gli3) which is regulated by Hand2 and Hand2 is downregulated in the limbs of Tbx3 mutant mice (Davenport, et al., 2003; Osterwalder et al., 2014; Sheeba and Logan, 2017). Furthermore, Tbx3 and Hand2 are both regulated by the microRNA-processing enzyme Dicer to ensure proper limb bud positioning (Zhang et al., 2011a, 2011b). In addition, experiments in the chick have shown that Tbx3 plays an important role in posterior digit specification, acting together with Tbx2 and the interdigital BMP signalling cascade (Suzuki et al., 2004).
3.4. Lung development
The formation of the lungs is initiated in the ventral wall of the foregut endoderm at E9.0. Primary lung buds and tracheal primordium start to develop at E9.5 and at E10.5 secondary lung buds develop as outgrowths from the primary buds. From E11.5 onwards, the epithelium undergoes branching morphogenesis and eventually forms a respiratory (bronchial) tree (Cardoso and Lu, 2006). Lung development is mediated by members of the Bmp, Wnt, Fgf, and Shh signalling families and in the lung mesenchyme during E10.5 and E14.5 they converge on Tbx3 and Tbx2 to maintain mesenchymal proliferation and lung branching morphogenesis (Fig. 2D) (Herriges and Morrisey, 2014; Li et al., 2004; Lüdtke et al., 2016). For example, Wnt signalling depends on active Bmp signalling and the loss of Wnt2/2b leads to failure of trachea and branching lung formation and inactivation of the Bmp receptors Bmpr1a and Bmpr1b leads to tracheal agenesis and ectopic primary bronchi (Domyan et al., 2011; Goss et al., 2009). Fgf signalling is regulated by, among other pathways, Bmp4 and Shh and when Fgf signalling is disrupted, branching is abrogated (Ohuchi et al., 2000; Pepicelli et al., 1998; Sekine et al., 1999; Weaver et al., 2000). Moreover, Shh null mice have hypoplastic lungs due to incorrect branching morphogenesis (Litingtung et al., 1998; Pepicelli et al., 1998). Finally, Tbx2 and Tbx3 regulate mesenchymal proliferation by maintaining pro-proliferative Wnt signalling through direct repression of the Wnt antagonists Frzb and Shisa3 (Lüdtke et al., 2016).
4. TBX3 in stem cell biology
Embryonic stem cells (ESCs) and adult stem cells, are undifferentiated cells which when they divide have the potential to either remain a stem cell or to differentiate into other specialised cells (Mo, et al., 2014; Yin and Zhang, 2015). ESCs are pluripotent cells derived from the inner cell mass (ICM) of the blastocyst and give rise to a plethora of mature cell types that make up the body. Adult stem cells are multipotent progenitor cells found in numerous adult tissues and, as part of the body repair system, they can develop into more than one cell type but they are more limited than ESCs (Barbosa et al., 2012; Becker et al., 1963; Gilbert et al., 2012; Kim and Hirth, 2009). BMP/TGF-β, Notch, Wnt/β-catenin, FGF, LIF/STAT, Hedgehog and Hippo are some of the signalling pathways which function in combination with transcription factors/co-factors, including Tbx3, octamer-binding transcription factor 4 (Oct4), SRY box2 (Sox2), kruppel-like factor 5 (KLF5) and homeobox protein Nanog, to regulate pluripotency and self-renewal of ESCs (Andersson et al., 2011; Huang et al., 2015; Ng and Surani, 2011; Niwa et al., 2009; Zhao et al., 2011). Importantly, several lines of evidence suggest that TBX3 enhances and maintains stem cell pluripotency in vitro by preventing differentiation and enhancing self-renewal (Niwa et al., 2009; Russell et al., 2015; Saunders et al., 2013). For example, LIF maintains the pluripotency of mESCs through regulating the Jak/Stat3 and PI(3)K/Akt signalling pathways which activate Klf4/Sox2 and Tbx3/Nanog respectively to maintain expression of Oct3/4 (Niwa et al., 2009). In the absence of LIF, the upregulation of Tbx3 in mouse pluripotent stem cells is sufficient to maintain adequate expression levels of Oct3/4 to keep pluripotency and low levels of Tbx3 results in reduced pluripotency in mESCs (Niwa et al., 2009; Russell et al., 2015). Furthermore, while Tbx3 levels are high in undifferentiated mESCs, its levels are downregulated in mESCs undergoing retinoic acid induced differentiation (Ivanova et al., 2006). Recently Tbx3 was also shown to be highly expressed in the interfollicular epidermal stem cells and it was shown to promote proliferation of these cells and to be required for abdominal skin expansion in mice during pregnancy and regeneration during wound repair (Ichijo et al., 2017).
It is important to note that in mESCs, Tbx3 appears to play a dual role in self-renewal and differentiation. Indeed, Tbx3 was demonstrated to be important for self-renewal and extraembryonic endoderm specification in mESCs (Lu et al., 2011; Semrau et al., 2017). In addition, the chromatin remodelling Baf45 complex maintains the pluripotency and differentiation potential of mESCs and its subunit Dpf2 was recently found to directly activate Tbx3 expression (Zhang et al., 2019). Importantly, the deletion of Dpf2 led to the repression of Tbx3 and a reduction of mesodermal differentiation and when Tbx3 was restored, mesodermal differentiation was recovered. The authors further show that Eed, a subunit of PRC2, can bind an intragenic Tbx3 enhancer to prevent Dpf2 dependent Tbx3 expression in mesodermal differentiation. During differentiation of mESCs into neural cells, miR-137 is upregulated and it was found to bind the 3′ UTR of Tbx3 which resulted in the repression of Tbx3 levels, the inhibition of self-renewal and increased differentiation of mESCs in vitro (Jiang et al., 2013). In early mouse adipocyte precursor cells, miRNA-93 also exhibited the ability to repress Tbx3 to prevent self-renewal (Cioffi et al., 2015).
Induced pluripotent stem cells (iPSCs) are ESC-like cells that can generate scalable quantities of relevant tissue and are of major interest for their application in personalized regenerative medicine, drug screening, and for our understanding of the cell signalling networks that regulate embryonic development and disease. In vitro studies have shown that expressing Tbx3, KLF4, SOX2, OCT4, Nanog, LIN-28A and C-MYC in somatic cells can reprogram them to form iPSCs (Okita and Yamanaka, 2011; Lee, et al., 2013). Importantly, Han et al., (2010) showed that iPS cells generated with Oct4, Sox2, Klf4 and Tbx3 are superior in both germ-cell contribution to the gonads and germline transmission frequency. They further showed using genome-wide chromatin immunoprecipitation sequencing analysis of Tbx3-binding sites in ESCs that Tbx3 regulates pluripotency-associated and reprogramming factors. In addition, co-expression of Tbx3 and Nr5α2 with Oct4, Sox2, Klf4 and c-Myc enhanced the generation of porcine iPSCs which resembled mESCs (Wang et al., 2013). Ke et al. (2018) also showed that LIF enhanced the levels of p-AKT as well as Tbx3 in marmoset iPSCs and an inhibitor of PI3K drastically reduced this regulation. Consistent with this data, naïve cynomolgus monkey (Cm) iPSCs was shown to express Oct3/4, DPPA5, SOX2, TBX3, KLF4, and KLF5 and expression of these genes in Cm ESCs was LIF-dependent (Honda et al., 2017). Interestingly, two studies showed that Tbx3/TBX3 is not entirely critical for the tenacity or generation of iPSCs (Klingenstein et al., 2016; Russell et al., 2015). Indeed, these studies compared the pluripotency potential of MEFs isolated from Tbx3+/+ and Tbx3 null (Tbx3−/−) mice as well as human foreskin fibroblasts and keratinocytes in which TBX3 was inducibly knocked down. They showed that Tbx3−/− MEFs and TBX3 knockdown cells could still be reprogrammed to iPSCs. Together these results indicate that while TBX3 is able to promote the efficacy of iPSC reprogramming, it is not essential for the reprogramming kinetics and maintenance of the pluripotency phenotype. This may be due to alternative pluripotency networks such as DPPA3 being able to substitute for TBX3.
5. TBX3 in human disease
TBX3 has been implicated in human diseases including ulnar mammary syndrome, rheumatoid arthritis, obesity and cancer (Frank et al., 2013; Quarta et al., 2019; Sardar et al., 2019; Willmer et al., 2017).
5.1. TBX3 in ulnar mammary syndrome
In humans, heterozygous mutations of TBX3 that result in haploinsufficiency lead to ulnar mammary syndrome (UMS, OMIM 181450) (Bamshad et al., 1997). UMS is an autosomal dominant developmental disorder, characterized by a number of clinical features including mammary and apocrine gland hypoplasia, upper limb defects, malformations of areola, dental structures, heart and genitalia (Chen and Chen, 2017). Interestingly, not all tissues and organs that express TBX3 are affected in UMS patients. This suggests that specific expression levels of TBX3 may be crucial for its functions in various tissues and/or that other T-box transcription factors, such as TBX2, could substitute for TBX3 in tissues and organs unaffected by UMS. Eighteen UMS causing mutations in the TBX3 gene have been reported which include 5 nonsense, 8 frameshift (due to deletion, duplication and insertion), 3 missense and 2 splice site mutations (Table 2). While these mutations can occur throughout TBX3, those which occur within or upstream of the T-domain (DNA binding) are associated with the most severe phenotype (Meneghini et al., 2006). Missense mutations within the T-domain that alter its structure are responsible for abolishing the DNA binding and transcription activity of TBX3 (Lingbeek, et al., 2002). Furthermore, in vitro studies in which the RD1 was deleted resulted in decreased transcriptional activity of Tbx3 (Carlson et al., 2001). More recent observations suggest that aberrant transcripts and truncated proteins resulting from mutations in TBX3 contribute to UMS through functions unrelated to its transcriptional activity. Indeed, Kumar et al. (2014) found that TBX3 proteins that model different UMS mutations were unable to perform its pre-mRNA splicing regulatory functions and were capable of interfering with the splicing inhibition function of endogenous wild type TBX3. It is interesting to note that numerous UMS patients have been reported to be obese which is consistent with the recent study by Quarta et al. (2019) that linked haploinsufficiency of Tbx3 in mice to obesity. Taken together, clinical phenotypes arising from mutations in TBX3 reveal the importance of this gene during the development of multiple tissues and organs.
Table 2.
TBX3 mutations in ulnar mammary syndrome.
| Exons | Mutation | Type of mutation |
|---|---|---|
| 1 | c.88insA | Ins/Frameshift |
| c.227delT | Del/Frameshift | |
| 2 | L143P | Missense |
| Y149S | Missense | |
| c.465_466insTATTGATGGACATT | Ins/Frameshift | |
| IVS2+1G > C | Splice site mutation | |
| 3 | c. 723del | Frameshift |
| 4 | K273X | Nonsense |
| 5 | c.991C > T | missense |
| c.992dup | Dup/Frameshift | |
| Q331X | Nonsense | |
| S343X | Nonsense | |
| 6 | c.1301_1302insGAGGAGCG | Ins/Frameshift |
| Q360X | Nonsense | |
| Q475X | Nonsense | |
| c.1586_1587insC | Ins/Frameshift | |
| IVS6 + 2T > A | Splice site mutation | |
| 7 | c.1857delC | Frameshift |
5.2. TBX3 in obesity
Heterogeneous populations of hypothalamic arcuate nucleus (ARC) neurons, such as the agouti-related protein (Agrp)-expressing and proopiomelanocortin/cocaine- and amphetamine-regulated transcript (Pomc/Cart)-neurons, release specific neuropeptides that control energy homeostasis by controlling appetite and energy expenditure. Energy imbalances and obesity have been associated with the deregulation of these hypothalamic neurons. Interestingly, Tbx3 is expressed in these hypothalamic neurons and has been implicated in the differentiation of human embryonic stem cells into hypothalamic Pomc neurons (Eriksson and Mignot, 2009; Linden et al., 2009; Quarta et al., 2019). Importantly, patients with UMS have shown symptoms consistent with ARC neuron dysfunction, including deficiency in growth hormone production leading to impaired puberty and obesity (Linden et al., 2009). The ablation of Tbx3 function in Agrp and Pomc neurons was recently shown to cause obesity in mice by interfering with the identity, differentiation and plasticity of these hypothalamic neural networks (Quarta et al., 2019). The Drosophila melanogaster Tbx3 homologue, omb, is expressed in the central nervous system of the adult fly and it was also reported to prevent obesity because depleting it by RNAi led to the induction and consequent increase in body fat content (Quarta et al., 2019). Tbx3 thus appears to be a key player in driving the functional heterogeneity of hypothalamic neurons responsible for governing body weight and energy metabolism and this role is conserved in mice, drosophila and humans.
5.3. TBX3 in rheumatoid arthritis
Rheumatoid arthritis (RA) is characterized by chronic inflammation, which primarily affects the synovial joints leading to tissue damage and physical disability and genome wide association studies have casually linked TBX3 to RA susceptibility (Freudenberg et al., 2011; Julià et al., 2008; Plenge et al., 2007). Furthermore, Tbx3 was identified as a candidate gene for RA in collagen-induced arthritis (CIA) mouse models (Sardar et al., 2019). Compared to control mice, mice with allelic variants in the Eae39r locus (which harbours the Tbx3 and Tbx5 genes) developed more severe CIA which correlated with increased Tbx3 serum levels but decreased TBX3 intracellular levels. Tbx3 was shown to repress B lymphocyte proliferation and it was thus proposed that decrease intracellular levels of Tbx3 results in their increased proliferation and activation. This is likely to cause an activated humoral immune response which is associated with chronic inflammation of the synovium leading to RA. Tbx3 may thus be an important player in regulating the immune system and a candidate biomarker for the diagnosis of RA severity. This is consistent with the limb defects seen in UMS which suggests the involvement of TBX3 in bone development pathways which are closely associated with immune pathways (D’Amelio and Sassi, 2016; Frank et al., 2013).
5.4. TBX3 in cancer
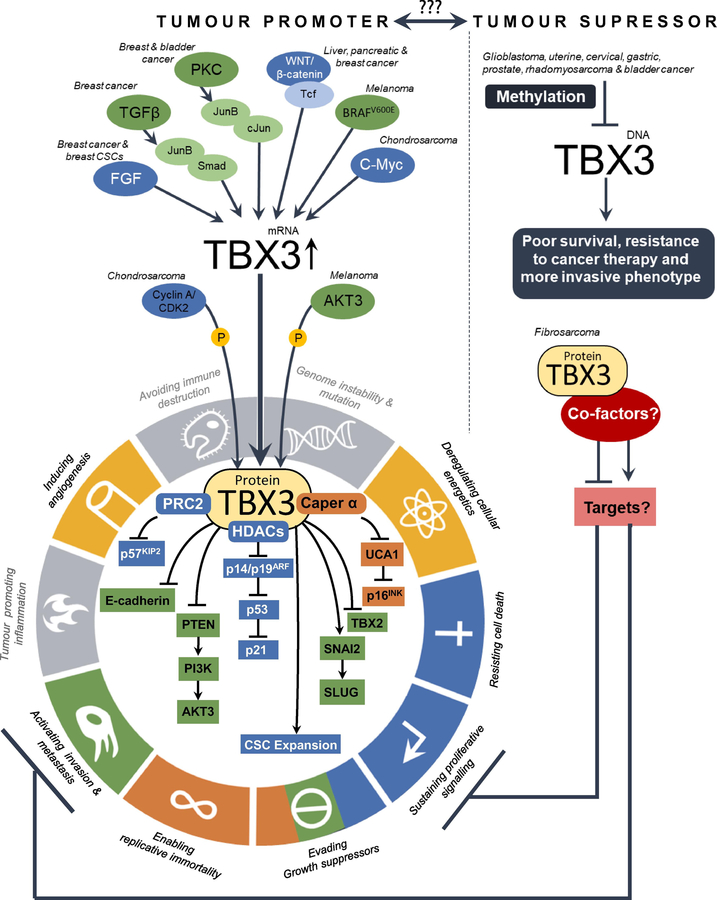
TBX3 is overexpressed in a wide range of carcinomas (breast, pancreatic, melanoma, liver, lung, gastric, ovarian, bladder and head and neck cancers) and sarcomas (chondrosarcoma, fibrosarcoma, liposarcoma, rhabdomyosarcoma and synovial sarcoma) and there is compelling evidence that it contributes to several hallmarks of cancer (Fig. 3). Indeed, uncontrolled cell proliferation and the bypass of senescence and apoptosis are early events in oncogenesis, and TBX3 has been shown to impact these processes as well as to promote tumour formation, angiogenesis and metastasis (Dong et al., 2018b, 2018a; Feng et al., 2018; Krstic et al., 2019; Wang, 2018; Willmer et al., 2017).
Fig. 3.
Summary of the regulation and roles of TBX3 in cancer. TBX3 is overexpressed in numerous cancers where it promotes several hallmarks of cancer as identified by Hanahan and Weinberg (2011) including (1) sustaining proliferative signalling; (2) evading growth suppressors; (3) resisting cell death; (4) enabling replicative immortality; (5) inducing angiogenesis; (6) activating invasion and metastasis and (7) deregulating cellular energetics. The key signalling molecules responsible for this overexpression and the co-factors and downstream targets that mediate the oncogenic functions of TBX3 are depicted in the figure adapted from Hanahan and Weinberg (2011) with colour coding that matches the appropriate hallmarks of cancer. Right panel: TBX3 also exhibits tumour suppressor activity. As indicated in this panel, it is silenced by methylation in certain cancers and it negatively impacts some hallmarks of cancer in fibrosarcoma and rhabdomyosarcoma. The factors that upregulate TBX3 in fibrosarcoma as well as the co-factors and target genes that mediate the tumour suppressor functions of TBX3 are yet to be elucidated.
5.4.1. The role of TBX3 in promoting proliferation and bypassing senescence, apoptosis and anoikis
A fundamental trait of cancer cells is uncontrolled proliferation and normal cells have several checkpoints that serve as barriers to prevent this from happening. For example, cell cycle arrests, senescence (irreversible cell cycle arrest), and programmed cell death pathways, including apoptosis and anoikis, prevent inappropriate cell division and/ or survival and the bypass of these processes can result in cancer (Hanahan and Weinberg, 2011). At a molecular level, these checkpoints are triggered and maintained by negative regulators of the cell cycle such as p14ARF/p19ARF, p53, p21WAF1/CIP1, p16INK4a, the retinoblastoma protein (Rb) and PTEN (Barnum and O’Connell, 2014). For example, in response to diverse oncogenic stresses, p14ARF and p16INK4a are upregulated. This results in p14ARF sequestering the p53 antagonist, MDM2, which leads to the upregulation of p53 and consequently activation of p53 target genes including p21WAF1/CIP1, an important inhibitor of cell cycle progression and an inducer of senescence and apoptosis (Berkovich et al., 2003; Brugarolas et al., 1995; Inoue et al., 1999; Pomerantz et al., 1998). p16INK4a, like other members of the Ink4 family, functions by blocking the kinase active sites of cyclin-dependent kinases (CDKs) 4 and 6 thereby preventing their interaction with their cognate cyclins and thus preventing CDK-cyclin mediated phosphorylation of Rb (DeGregori, 2004). Hypo-phosphorylated Rb interacts with and sequesters the E2F family of transcription factors which results in a G1 cell cycle arrest and the maintenance of the senescence phenotype (DeGregori, 2004). TBX3 contributes to tumour progression, in part, by inhibiting the p14ARF/p53/p21WAF1/CIP1 and p16INK4a/pRb tumour suppressor pathways to bypass key cell cycle checkpoints, cellular senescence, apoptosis and anoikis.
Several groups have reported that TBX3 can promote cell proliferation by directly repressing p14ARF/p19ARF, p21WAF1/CIP1, p57KIP2 or PTEN. Indeed, Tbx3 expression promoted the proliferative ability of normal and tumorigenic mammary epithelial cells (MECs) by transcriptionally repressing p19ARF which was accompanied by the downregulation of p21WAF1/CIP1 (Platonova et al., 2007). It is important to note that p53-null MECs exhibited a similar growth response to TBX3 suggesting that the negative impact of TBX3 on p21WAF1/CIP1, occurs independently of p53. Similarly, Suzuki et al., (2008) demonstrated that Tbx3 expression in hepatic progenitor cells negatively impacts p19ARF levels resulting in significantly increased proliferative potential. In chondrosarcoma cells, TBX3 is upregulated transcriptionally by c-Myc and post-translationally by cyclin A/CDK2 and it is required for transition through S-phase (Willmer et al., 2015). Furthermore, TBX3 promotes chondrosarcoma cell proliferation by directly binding to and repressing the p21WAF1/CIP1 promoter at a T-element at −121 bp (Willmer et al., 2016a, 2016b). More recently, TBX3 was shown to promote proliferation of papillary thyroid carcinoma cells through repressing p57KIP2 (Li et al., 2018a, 2018b). This resulted from TBX3 binding and recruiting the PRC2 and HDACs 1 and 2 to the region of the CDKN1C promoter that regulates p57KIP2 expression. TBX3 may also promote proliferation by repressing PTEN, an inhibitor of PI3K/AKT-mediated cell growth, proliferation and survival (Leslie and Downes, 2004). Indeed, TBX3 levels were significantly upregulated in 33 head and neck squamous cell carcinoma (HNSCC) patients and this correlated with reduced expression of PTEN. Furthermore, in the same study the authors show that TBX3 represses both basal and induced PTEN levels in HeLa and HEK cells (Burgucu et al., 2012).
Carlson et al. (2001) demonstrated that MEFs stably overexpressing wild type Tbx3, but not a Tbx3 protein in which the dominant RD1 is mutated, were able to form colonies and proliferate for more than 50 passages. This suggests that Tbx3 can promote unlimited cell division and bypass senescence and that the RD1 plays an important role in these abilities. Furthermore, Fan et al. (2004) showed that Tbx3, and not its isoform Tbx3+2a, could immortalise MEFs and Brummelkamp et al. (2002) identified Tbx3 as a key anti-senescence factor in a genetic screen of conditionally immortalised mouse striatal cells. The mechanism responsible was shown to involve the ability of Tbx3 to directly repress p19ARF and mutations within the Tbx3 DBD dramatically abrogated this ability. Yarosh et al. (2008) subsequently showed that TBX3 interacts with HDACs 1, 2, 3 and 5 to repress p14ARF through a T-box binding site in its initiator. TBX3 can also promote proliferation and prevent senescence by co-operating with CAPERα to repress UCA1 and consequently the p16INK4a/Rb pathway. UCA1, a long non-coding RNA, stabilises p16INK4a mRNA by sequestering the p16INK4a antagonist HnRNP A1 and in this way promotes senescence. Importantly, knockdown of either CAPERα or TBX3 increased senescence-associated β-galactosidase activity in human foreskin fibroblasts which was accompanied by an increase in p21WAF1/CIP1, p16INK4a and pRb (Kumar et al., 2014b).
Apoptosis is a physiologically ubiquitous cellular program that eliminates damaged or abnormal cells and cancer cells acquire mechanisms to evade apoptosis to confer upon them a survival advantage and resistance to anti-cancer agents (Hanahan and Weinberg, 2011). Tbx3 is upregulated in rat bladder carcinoma cells and depleting Tbx3 in these cells dramatically reduced cell growth and cell adhesion while promoting apoptosis (Ito et al., 2005). On the other hand, the ectopic overexpression of TBX3 or TBX3+2a in human mesangial cells inhibited apoptosis (Wensing and Campos, 2014). Furthermore, the co-expression of Tbx3, with Myc or H-RasVal17 can transform MEFs and bypass Myc-induced apoptosis through the repression of p19ARF/p53/ p21WAF1/CIP1 (Carlson et al., 2002). Interestingly, a Tbx3 N-terminal truncated protein had no effect on Myc-induced apoptosis suggesting that the C-terminus of Tbx3 harbours a motif(s), probably RD1 and/or the activation domain, that may be required for inhibiting apoptosis (Carlson et al., 2002). Importantly, Renard et al. (2007) demonstrated that TBX3 is a direct transcriptional target of β-catenin/Tcf and that β-catenin mediated upregulation of TBX3 confers resistance to doxorubicin-induced apoptosis in U2OS osteosarcoma and HCT116 colorectal carcinoma cells. Similarly, Zhang et al. (2011a, 2011b) showed that human DLD-1 colorectal cancer cells treated with an aqueous extract of the herb, Fructus Ligustri Lucidi, inhibited TBX3 expression which resulted in the upregulation of p14ARF and p53 and subsequently sensitization of the cells to doxorubicin-induced apoptosis. TBX3 can also confer resistance to anoikis, another form of programmed cell death that occurs when cells lose contact with the ECM or neighbouring cells and it serves as a barrier to metastasis (Gilmore, 2005). Indeed, TBX3 overexpression in HNSCC cells increased their resistance to anoikis thus enabling them to survive without appropriate ECM interaction (Humtsoe et al., 2012). Importantly, when TBX3 was depleted in HNSCC cells, they exhibited a significantly reduced ability to adhere to culture plates, had dramatically lower numbers of live cells and they exhibited a two-fold increase in fragmented nuclei and a significant increase in activated caspase 3 suggestive of apoptosis (Humtsoe et al., 2012). It is worth noting that the bypass of anoikis has also been linked to cancer drug resistance (Ko, et al., 2009). It will therefore be interesting to investigate if the ability of TBX3 to confer resistance to anoikis may be another mechanism by which it confers cancer drug resistance.
In contrast to the above findings, it is interesting to note that in pancreatic ductal adenocarcinomas (PDAC), melanoma and breast carcinomas, TBX3 has no effect, or negatively regulates proliferation, in favour of promoting cell migration, a phenotypic trade-off which is common in cancer (Gallaher et al., 2019). Indeed, ectopic overexpression of TBX3 in human PDAC cell lines had no effect on proliferation but enhanced the migratory and invasive ability of the cells (Perkhofer et al., 2016). Furthermore, non-tumourigenic radial growth phase (RGP) melanoma cells genetically engineered to overexpress TBX3 had significantly reduced proliferative ability but increased migratory ability and the opposite was observed when TBX3 was depleted in advanced melanoma cells (Peres et al., 2010; Peres and Prince, 2013). Similarly, the upregulation of TBX3 in breast and melanoma cells stimulated with RA or TGFβ1 led to diminished proliferative rates but increased migration (Ballim et al., 2012; Li et al., 2013). The ability of TBX3 to inhibit proliferation correlated with decreased levels of its homologue, TBX2, a powerful pro-proliferative factor in melanoma and breast cancer. The mechanism by which TBX3 mediated the anti-proliferative effect downstream of TGFβ1 was shown to be through it directly repressing a T-element in the TBX2 promoter (Li et al., 2014). Together this suggests that TBX3 inhibits breast cancer and melanoma proliferation through repressing TBX2 and while the mechanisms that enable it to promote or inhibit proliferation in different cellular contexts are largely unknown, there is strong evidence that it may be co-factor dependent.
5.4.2. The role of TBX3 in tumour formation, angiogenesis and metastasis
Malignant cells form tumours, generate a tumour-associated neo vasculature (angiogenesis) which supplies the tumour with nutrients and oxygen and removes metabolic wastes, and they break away from the primary tumour and metastasise and invade distant organs (Hanahan and Weinberg, 2011). Several studies have suggested that TBX3 contributes to these advanced oncogenic processes in colon cancer, hepatocarcinoma, breast cancer, melanoma, PDAC and chondrosarcoma. Indeed, knocking down TBX3 in colon and liver carcinoma cell lines reduced anchorage-independent growth in vitro, and expressing a dominant negative mutant Tbx3-Y149S in these cell lines, diminished their ability to form tumours in mice (Renard et al., 2007). In hepatocarcinoma patient samples, the expression of TBX3 positively correlated with histological grade, tumour size and cancer cell metastasis (Li et al., 2018a, 2018b). Ectopic overexpression of TBX3 enhanced the migratory and invasive ability of human PDAC cell lines and promoted angiogenesis in vitro and in vivo which correlated with increased expression of angiogenesis-associated genes such as FGF2 and VEGF-A (Perkhofer et al., 2016). The ectopic expression of TBX3 in chondrosarcoma cells also enhanced their ability to form tumours in mice and knockdown of TBX3 in liposarcoma, rhabdomyosarcoma and chondrosarcoma, resulted in diminished substrate-dependent and -independent cell proliferation and migration (Willmer et al., 2016a, 2016b).
Five different mutations were identified in TBX3 in breast tumour samples and there is evidence to suggest that TBX3 is a potential driver gene in breast cancer. High TBX3 mRNA levels were found in breast cancer cells and estrogen receptor (ER)-positive breast tumour samples which correlated positively with a metastatic prognosis (Chen, et al., 2009; Fillmore et al., 2010; Stephens et al., 2012). Furthermore, knocking down TBX3 in ER-positive MCF-7 breast cancer cells resulted in the inhibition of anchorage independent growth and migration (Peres et al., 2010). In addition, TBX3 was shown to mediate breast cancer cell migration downstream of the PKC and TGF-β signalling pathways (Li et al., 2014, 2013; Mowla et al., 2011). Moreover, TBX3 was identified as a potential regulator of the transition from ductal carcinoma in situ (DCIS) to invasive breast cancer. Transient and stable overexpression of TBX3 or TBX3+2a enhanced the survival, colony forming and invasive abilities of DCIS-like and non-invasive breast cancer cells (Krstic et al., 2016, 2019). The mechanism involved was demonstrated to occur through the two TBX3 isoforms directly upregulating SNAI2, which encodes SLUG, and thereby inducing EMT. The authors provide compelling evidence that in breast cancer patient samples, there is a strong correlation between elevated levels of TBX3 and SLUG and that this is associated with poor prognosis.
TBX3 is also overexpressed in advanced melanoma and can drive the transition from non-invasive RGP melanoma to invasive vertical growth phase (VGP) melanoma (Hoek et al., 2004; Peres et al., 2010; Rodriguez et al., 2008). Ectopic expression of TBX3 alone in RGP cells was sufficient to drive them to assume a VGP phenotype and knockdown of TBX3 in advanced melanoma cells inhibited their tumour forming ability and their aggressive phenotype (Peres et al., 2010; Peres and Prince, 2013). A key mechanism by which TBX3 promotes melanoma migration and metastasis was shown to occur through its ability to directly repress E-cadherin (Rodriguez et al., 2008). In the same study, high levels of TBX3 were shown to correlate with low expression of E-cadherin in metastatic melanoma tissue samples and the depletion of TBX3 caused an increase in E-cadherin levels and decreased melanoma invasiveness in vitro. The association between TBX3 and E-cadherin, and its consequences on migration and invasion, were also reported with similar results for squamous cell carcinoma and human hepatocellular carcinoma (Feng et al., 2018; Humtsoe et al., 2012).
The BRAF-MAPK, AKT and PKC pathways have been identified as upstream regulators of the TBX3/E-cadherin axis in melanoma and bladder cancer. BRAFV600E and AKT3 are constitutively activated in approximately 50% and 70% of melanomas respectively and they play critical roles in melanoma formation and invasion (Dhawan et al., 2002; Palmieri et al., 2015; Siroy et al., 2016). The overexpression of TBX3 in a subset of melanomas was shown to result from it being transcriptionally upregulated by BRAFV600E and phosphorylated by AKT3 (Boyd et al., 2013, Peres et al., 2015). AKT3 phosphorylation of TBX3 enhanced its ability to repress E-cadherin and promoted migration (Peres et al., 2015). Levels of miR-137 and TBX3 mRNA correlate inversely in a panel of melanoma cell lines as well as a cohort of primary melanoma patients and miR-137 was shown to be an important component of the TBX3/E-cadherin axis in melanomagenesis (Peres et al., 2017). TBX3 was identified as a direct target of miR-137 in non-malignant RGP cells and re-expression of miR-137 in advanced melanoma cells inhibited their migration by repressing TBX3 and upregulating E-cadherin levels. In human bladder cancer cells, the regulation of E-cadherin by TBX3 occurs in a PLCε/PKC-dependent manner (Du et al., 2014). When PLCε was silenced in bladder cancer cells, TBX3 levels decreased while E-cadherin levels increased, and this correlated with a decrease in invasive capability. Furthermore, this situation was partly reversed when the PKC pathway was stimulated, suggesting that TBX3 is downstream of PLCε/PKC in bladder cancer.
5.4.3. TBX3 in cancer stem cells
Cancer stem cells (CSCs) are a small sub-population of tumour cells which exhibit capabilities such as self-renewal, differentiation and tumorigenicity (Clarke et al., 2006). They are resistant to standard chemotherapies and are thought to be one of the main contributors to cancer development, drug resistance and clinical relapse (Yu et al., 2012). Understanding the role of CSCs within the tumour micro-environment has thus sparked much interest and there is evidence that TBX3 contributes to the expansion of these cells within tumours. The treatment of a panel of ER-positive breast cancer cell lines with 17-β-Estradiol resulted in a significant increase in the number of CSCs and enhanced tumorsphere formation and the downstream effectors were shown to be FGF9 and TBX3 (Fillmore et al., 2010). Additionally, breast tumours that responded best to chemotherapy were shown to express lower levels of TBX3 while tumours that express high levels of TBX3 had the greatest recurrence rates. These results highlight the importance of the FGF/Tbx3 signalling pathway in the expansion of breast CSCs and reveal another mechanism by which TBX3 aids breast cancer progression, recurrence and drug resistance (Fillmore et al., 2010; Dong et al., 2018a, 2018b). CSCs derived from PDACs also express high levels of TBX3 and perpetuate themselves through an autocrine TBX3-ACTIVIN/NODAL signalling loop to sustain stemness (Perkhofer et al., 2016). In these cells, TBX3 co-localized with the pluripotency marker OCT3/4 and was found to be bound to pluripotency genes involved in the ACTIVIN/NODAL pathway. These findings indicate that TBX3 is a key player in regulating pluripotency-related genes in CSCs and that this may be another mechanism by which it contributes to cancer formation and tumour aggressiveness.
5.4.4. The tumour suppressor role of TBX3
During oncogenesis, tumour suppressor genes are frequently silenced by methylation (Jones and Baylin, 2002). Interestingly, TBX3 is methylated in metastatic cervical cancer, DU-145 prostate cancer cells, bladder cancer, urothelial carcinoma, in the AGS gastric cancer cell line, glioblastoma and glioblastoma stem cells, and the methylation of TBX3 was associated with a poor overall survival, resistance to cancer therapy and a more invasive phenotype (Beukers et al., 2015; Eriksson et al., 2015; Etcheverry et al., 2010; Kandimalla et al., 2012; Lee et al., 2014; Lyng et al., 2006; White-Al Habeeb et al., 2014; Yamashita et al., 2006). A comparison of genome wide CpG methylation profiles of three primary glioblastoma cell lines and glioblastoma stem cells with normal brain and neuronal stem cell controls revealed that TBX3 was one of 202 genes that were hypermethylated within their promoters and 5′UTRs in primary glioblastoma cell lines and glioblastoma stem cells (Lee et al., 2014). Gene ontology analyses of this subset of CpG methylated genes implicated them in, amongst other functions, the regulation of metabolism. These findings are interesting because the methylation of TBX3 in glioblastoma is associated with resistance to standard therapy and metabolic pathways have been implicated as important mediators of resistance to anti-cancer agents (Etcheverry et al., 2010; Zaal and Berkers, 2018). It would therefore be important to follow up on whether re-expressing TBX3 in glioblastoma cells alters their metabolic pathways and whether it will lead to the sensitivity of these cells to radiation and chemotherapy.
In a gastric cancer cell line, AGS, de-methylation with 5-aza-2′-deoxycytidine (5-aza-dC) followed by an oligonucleotide array revealed that TBX3 was one of 579 genes which were upregulated 16-fold or more (Yamashita et al., 2006). The authors showed that in this gastric cancer cell line, but not in 5 other gastric cancer cell lines tested, methylation of the CpG island in the 5′ region of the TBX3 gene effectively silenced its expression (Yamashita et al., 2006). Importantly, the authors reveal that 5-aza-dC treatment negatively impacted growth of these cells which may suggest a tumour suppressor role for TBX3 in a subset of gastric cancer cells. However, given the large pool of methylated genes identified, the significance of TBX3 methylation in this context requires further investigation.
Interestingly, TBX3 mRNA and protein levels are overexpressed in fibrosarcoma cells and patient derived tissue samples relative to primary fibroblasts and normal adjacent tissue respectively (Willmer et al., 2016a, 2016b). Investigation of the functional significance of this expression revealed that knockdown of TBX3 promoted substrate dependent and independent proliferation, migration and the formation of tumours in mice with significantly increased volume and weight. In the same study the authors genetically engineered a fibrosarcoma cell line to overexpress TBX3 and they showed that TBX3 conferred tumour suppressor properties on these cells which corroborated their knockdown data (Willmer et al., 2016a, 2016b). Recently, Oh et al. (2019) showed that TBX3 is expressed at very low levels in alveolar and embryonal rhabdomyosarcoma cells and the ectopic overexpression of TBX3 resulted in the inhibition of proliferation and migration of these cells. The authors showed that the mechanism for the inhibition of proliferation involved the direct repression of TBX2 by TBX3. They also demonstrate that PRC2 together with its regulator, JARID2, co-operate with the methyltransferase H3K27me to silence TBX3 in skeletal muscle cells. It would be interesting to investigate whether this is the mechanism by which TBX3 levels are kept low in rhabdomyosarcoma.
5.4.5. TBX3 and its homologue, TBX2, in cancer
TBX2 and TBX3 are highly related members of the TBX2 sub-family. As shown in Fig. 4, they both have a T-box DNA binding domain, 2 repression domains and an activation domain (Paxton et al., 2002; Sinha et al., 2000). The TBX2 and TBX3 DNA binding domains share 95% homology and their repression domains located in the C-terminus share 66.67% homology. However, their second repression domains and their activation domains are found at different positions and share no homology (He et al., 1999). Based on the high degree of homology between their DNA binding domains it was initially expected that they would regulate common target genes and have redundant functions. However, there is now compelling evidence that they also have distinct functions in development and cancer. For example, TBX2 and TBX3 have overlapping expression patterns and co-operate in mammary gland development but there is also evidence that they have distinct spatial expression patterns and functions. Indeed, during the induction of the mammary gland, TBX2 expression is restricted to the mesodermal cells of the milk line and TBX3 is only expressed in the epithelial cells of the emerging mammary placodes (Chapman et al., 1996; Davenport et al., 2003). Importantly, while Tbx3 heterozygous mutations result in failed nipple and ductal tree development, Tbx2 heterozygous mutations have no distinct effect on placode formation but leads to a reduction in ductal tree development (Jerome-Majewska et al., 2005).
Fig. 4.
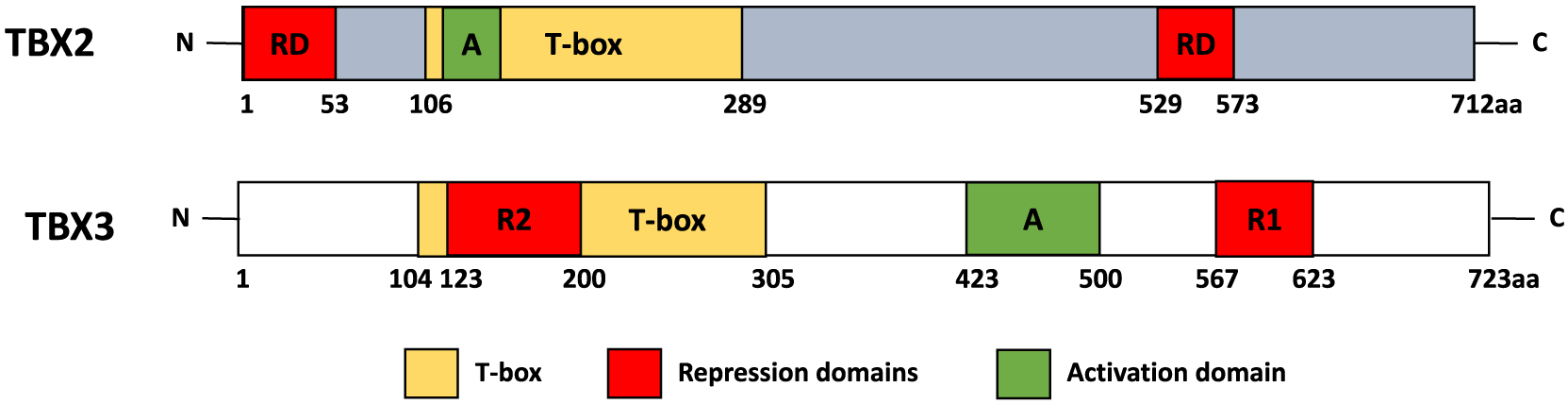
Diagrams depict the structural organisation of the human TBX2 and TBX3 proteins. The DNA binding domains (T-box, yellow boxes), repression domain (R1, R2 and RD, red boxes) and activation domains (A, green boxes) are shown and the amino acid residue number is displayed below each box.
TBX2 and TBX3 are also overexpressed in numerous cancers and they both can contribute to similar oncogenic processes including bypassing senescence and apoptosis, promoting proliferation and EMT and conferring drug resistance (Wansleben et al., 2014). This suggests that they are both able to regulate the same target genes in certain contexts. However, relatively little is known about the genes that they regulate to impact these processes as well as the molecular mechanisms underlying their target gene specificity. There is some evidence that in different contexts, TBX2 and TBX3 are both capable of binding and repressing a variant half T-site that is present close to the p19ARF/ p14ARF and p21WAF1/CIP1 transcriptional start sites to bypass senescence and promote proliferation (Brummelkamp et al., 2002; Jacobs et al., 2000; Lingbeek et al., 2002; Prince et al., 2004; Willmer et al., 2016a, 2016b). This repression of p19ARF/p14ARF and p21WAF1/CIP1 appears to specifically require the homologous DNA binding and C-terminal repression domains of TBX2 and TBX3. It would be interesting to determine whether TBX2 and TBX3 have redundant functions in regulating the p19ARF/p14ARF and p21WAF1/CIP1 promoter in cancers where they are both expressed or if their ability to regulate these target genes is regulated by other factors. In this regard it is worth noting that in breast cancer and melanoma cell lines where TBX2 and TBX3 are both overexpressed, their individual knock down resulted in different phenotypes. While TBX2 functioned as a powerful pro-proliferative factor, TBX3 impacted the later oncogenic processes of tumour formation and cell migration (Peres et al., 2010). This suggests that they must be regulating different target genes when they are both simultaneously expressed. Indeed, whereas TBX2 was shown to be required to maintain proliferation and suppress senescence in melanomas by repressing expression of p21WAF1/CIP1 (Vance et al., 2005), TBX3 was found to enhance melanoma invasiveness by down-regulating expression of E-cadherin (Rodriguez et al., 2008). It is worth noting that in the Rodriguez study, both TBX2 and TBX3 were able to bind the same site in the E-cadherin promotor in vitro, but only TBX3 was able to bind the promoter in vivo and the depletion of TBX3 but not TBX2 led to an increase in endogenous p21WAF1/CIP1 levels. This suggests that while TBX2 and TBX3 can both bind the same sites in vitro, their ability to recognize half T elements in vivo can vary. Interestingly, in two different studies TBX2 and TBX3 were separately implicated in gastric cancer and their levels were shown to correlate inversely with E-cadherin levels (Liu et al., 2019; Miao et al., 2016). This raises the possibility that they are both capable of repressing E-cadherin in some contexts and begs the question as to what regulates this ability in other contexts for example in melanoma. It is possible that their target gene specificity may be regulated by posttranslational modifications by differential signalling cascades or by their associated cofactors. Indeed, the phosphorylation of TBX3 by AKT was shown to enhance its ability to repress E-cadherin in melanoma (Peres et al., 2015). In addition, EGR1 has been identified as a TBX2 co-factor in breast cancer and rhabdomyosarcoma and their interaction has been shown to drive cell proliferation by inhibiting EGR1 dependent gene expression of p21WAF1/CIP1, PTEN, NDRG1 and CST6 (D’Costa et al., 2014; Mohamad et al., 2018; Redmond et al., 2010). It will be interesting to determine if TBX3 is also able to interact with EGR1 in breast cancer and the impact of this on EGR1 target gene regulation. Furthermore, there is evidence that TBX2 and TBX3 are differentially expressed in some cancers which appears to relate, in part, to their ability to repress one another (Mohamad et al., 2018; Rodriguez et al., 2008). TBX3 also mediates the anti-proliferative role of TGF-β by repressing TBX2 at a half T-element site. To better understand the modes of action and functions of TBX2 and TBX3, it is essential that their repertoire of target genes, as well as their interacting proteins and the protein domains involved, are identified.
6. Conclusion
The transcription factor TBX3 is a key player in embryonic development, stem cell maintenance and oncogenesis. During development, mutations resulting in decreased levels of TBX3 lead to ulnar mammary syndrome. This condition affects organs where TBX3 plays a functional role, for example, the heart, mammary gland, limbs and lungs. In contrast, when TBX3 levels are upregulated in postnatal tissue, it contributes to a wide array of epithelial derived cancers and a subset of soft tissue and bone sarcomas by impacting several cancer processes. There is also evidence that in certain cellular contexts TBX3 may function as a brake to prevent tumour progression. A serious gap in TBX3 biology is our understanding of the molecular mechanisms that (i) regulate the overexpression of TBX3 in cancer, (ii) mediate the oncogenic functions of TBX3 and (iii) enables TBX3 to switch between a tumour promoter and tumour suppressor. Investigations into these areas have important implications for identifying versatile ways of targeting TBX3 in anti-cancer therapy.
Acknowledgements
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series—a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. The authors were supported by grants from the South Africa Medical Research Council, the National Research Foundation (NRF), Cancer Association of South Africa (CANSA) and the University of Cape Town. Bianca Del B. Sahm is a recipient of an institutional program for international doctorate fellowship from the CAPES (Coordenação de Aperfeiçoamento de Pessoal de nível Superior) agency. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. We thank Prof Virginia Papaioannou for providing Fig. 2.
The corresponding Gene Wiki entry for this review can be found here: https://en.wikipedia.org/wiki/TBX3.
Abbreviations:
- 3′ UTR
3′ untranslated region
- AER
apical ectodermal ridge
- AKT3
AKT Serine/Threonine Kinase 3
- AVB
atrioventricular bundle
- AVC
atrioventricular canal
- AVN
atrioventricular node
- Axin2
axis inhibition protein 2
- Baf45
BAF complex
- BB
bundle branches
- BMP
Bone Morphogenetic Protein
- CAPERα
Coactivator of AP1 and Estrogen Receptor
- CCS
cardiac conduction system
- CDKs
cyclin-dependent kinases
- Cm
cynomolgus monkey
- DBD
DNA-binding domain
- DCIS
ductal carcinoma in situ
- DPPA5
developmental pluripotency-associated 5
- ECM
extracellular matrix
- ESCs
Embryonic stem cells
- Fgf
Fibroblast growth factor
- Gata6
GATA-binding protein 6
- Gli3
GLI Family Zinc Finger 3
- Hand2
Heart- and neural crest derivatives-expressed protein 2
- Hcn4
Hyperpolarization Activated Cyclic Nucleotide Gated Potassium Channel 4
- HDACs
histone deacetylases
- HnRNP A1
Heterogeneous Ribonucleoprotein A1
- HNSCC
head and neck squamous cell carcinoma
- IFT
inflow tract
- OFT
outflow tract
- iPSCs
Induced pluripotent stem cells
- Jak
Janus kinase
- KLF5
kruppel-like factor 5
- Lbh
Limb bud and heart development
- Lef1
Lymphoid enhancer factor 1
- LIF
leukemia inhibitory factor
- LPM
lateral plate mesoderm
- MAP
mitogen-activated protein
- MDM2
Mouse double minute 2 homolog
- MECs
mammary epithelial cells
- MEFs
mouse embryonic fibroblasts
- mESC
mouse ESCs
- mESCs
mouse embryonic stem cells
- NLS
nuclear localization signal
- NRE
non-ridge ectoderm
- Oct3/4
octamer-binding transcription factor ¾
- p-AKT
phosphorylated AKT
- PDAC
pancreatic ductal adenocarcinomas
- PI(3)K
Phosphatidylinositol-3 kinases
- PKC
Protein kinase C
- PRC2
polycomb repressive complex 2
- PTEN
phosphatase and TENsin Homolog
- R2 and R1
repression domains
- Rb
retinoblastoma protein
- RGP
radial growth phase
- SAN
sinoatrial node
- Shh
Sonic Hedgehog
- Shisa3
Shisa family member 3
- Sox2
SRY box2
- STAT
signal transducer and activator of transcription
- TBX2
T-box factor 2
- TBX3
T-box factor 3
- TGF-β
transforming growth factor β
- UCA1
Urothelial Cancer Associated 1
- UMS
ulnar mammary syndrome
- VEGF-A
Vascular endothelial growth factor A
- VGP
vertical growth phase
- WNT
Wingless-related integration site
- ZIC4
Zic Family Member 4
- ZPA
zone of polarizing activity
- Frzb
Frizzled related protein
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Andersson ER, Sandberg R, Lendahl U, 2011. Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612. 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- Andl T, Reddy ST, Gaddapara T, Millar SE, 2002. WNT signals are required for the initiation of hair follicle development. Dev. Cell 2, 643–653. 10.1016/S1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Boukens BJ, Mommersteeg MTM, Brons JF, Wakker V, Moorman AFM, Christoffels VM, 2008. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res 102, 1340–1349. 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Boink GJJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WMH, Buermans HP, de Bakker JMT, Seppen J, Tan HL, Moorman AFM, ť Hoen PAC, Christoffels VM, 2012. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc. Res 94, 439–449. 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- Ballim RD, Mendelsohn C, Papaioannou VE, Prince S, 2012. The ulnar-mammary syndrome gene, Tbx3, is a direct target of the retinoic acid signaling pathway, which regulates its expression during mouse limb development. Mol. Biol. Cell 23, 2362–2372. 10.1091/mbc.e11-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Lin RC, Law DJ, Watkins WS, Krakowiak PA, Moore ME,Franceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau BG, Schinzel A, Seidman JG, Seidman CE, Jorde LB, 1997. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat. Genet 16, 311–315. 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- Barbosa HSC, Fernandes TG, Dias TP, Diogo MM, Cabral JMS, 2012. New insights into the mechanisms of embryonic stem cell self-renewal under hypoxia: a multifactorial analysis approach. e38963 PLoS One 7 10.1371/journal.pone.0038963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnum KJ, O’Connell MJ, 2014. Cell Cycle Regulation by Checkpoints, in: Cell CycleControl. pp. 29–40. doi: 10.1007/978-1-4939-0888-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker AJ, McCulloch EA, Till JE, 1963. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 197, 452–454. 10.1038/197452a0. [DOI] [PubMed] [Google Scholar]
- Begum S, Papaioannou VE, 2011. Dynamic expression of Tbx2 and Tbx3 in developing mouse pancreas. Gene Expr. Patterns 11, 476–483. 10.1016/j.gep.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Lamed Y, Ginsberg D, 2003. E2F and Ras synergize in transcriptionally activating p14 ARF expression. Cell Cycle 2, 127–134. 10.4161/cc.2.2.293. [DOI] [PubMed] [Google Scholar]
- Beukers W, Kandimalla R, Masius RG, Vermeij M, Kranse R, Van Leenders GJJLH, Zwarthoff EC, 2015. Stratification based on methylation of TBX2 and TBX3 into three molecular grades predicts progression in patients with pTa-bladder cancer. Mod. Pathol 28, 515–522. 10.1038/modpathol.2014.145. [DOI] [PubMed] [Google Scholar]
- Boogerd K-J, Wong LYE, Christoffels VM, Klarenbeek M, Ruijter JM, Moorman AFM, Barnett P, 2008. Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of Connexin43. Cardiovasc. Res 78, 485–493. 10.1093/cvr/cvn049. [DOI] [PubMed] [Google Scholar]
- Boogerd CJJJ, Wong LYEE, Van Den Boogaard M, Bakker ML, Tessadori F, Bakkers J, ‘t Hoen PAC, Moorman AF, Christoffels VM, Barnett P, 2011. Sox4 mediates Tbx3 transcriptional regulation of the gap junction protein Cx43. Cell. Mol. Life Sci 68, 3949–3961. 10.1007/s00018-011-0693-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SC, Mijatov B, Pupo GM, Tran SL, Gowrishankar K, Shaw HM, Goding CR, Scolyer RA, Mann GJ, Kefford RF, Rizos H, Becker TM, 2013. Oncogenic B-RAFV600E signaling induces the T-Box3 transcriptional repressor to repress E-cadherin and enhance melanoma cell invasion. J. Invest. Dermatol 133, 1269–1277. 10.1038/jid.2012.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ, 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377, 552–557. 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, van Lohuizen M, Bernards R, 2002. TBX-3, the gene mutated in ulnar-mammary syndrome, is a negative regulator of p19 ARF and inhibits senescence. J. Biol. Chem 277, 6567–6572. 10.1074/jbc.M110492200. [DOI] [PubMed] [Google Scholar]
- Burgucu D, Guney K, Sahinturk D, Ozbudak IHIH, Ozel D, Ozbilim G, Yavuzer U, 2012. Tbx3 represses PTEN and is over-expressed in head and neck squamous cell carcinoma. BMC Cancer 12, 481 10.1186/1471-2407-12-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Belmonte JCI, 2001. Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell Dev. Biol 17, 87–132. 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- Cardoso WV, Lu J, 2006. Regulation of early lung morphogenesis: questions, facts and controversies. Development 133, 1611–1624. 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- Carlson H, Ota S, Campbell CE, Hurlin PJ, 2001. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum. Mol. Genet 10, 2403–2413. 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- Carlson H, Ota S, Song Y, Chen Y, Hurlin PJ, 2002. Tbx3 impinges on the p53 pathway to suppress apoptosis, facilitate cell transformation and block myogenic differentiation. Oncogene 21, 3827–3835. 10.1038/sj.onc.1205476. [DOI] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE, 1996. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev. Dyn 206, 379–390. . [DOI] [PubMed] [Google Scholar]
- Chen H, Chen H, 2017. Ulnar-Mammary Syndrome, Atlas of Genetic Diagnosis and Counseling.
- Chen Z, Lü G, Ji T, 2009. Expression of TBX3 mRNA and its role in the pathogenesis and metastasis of breast cancer. Nan Fang Yi Ke Da Xue Xue Bao 29, 87–89. [PubMed] [Google Scholar]
- Cho K-W, Kim J-Y, Song S-J, Farrell E, Eblaghie MC, Kim H-J, Tickle C, Jung H-S, 2006. Molecular interactions between Tbx3 and Bmp4 and a model for dorsoventral positioning of mammary gland development. Proc. Natl. Acad. Sci. U.S.A 103, 16788–16793. 10.1073/pnas.0604645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Habets PEMH, Franco D, Campione M, De Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, Moorman AFM, 2000. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol 223, 266–278. 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Hoogaars WMH, Tessari A, Clout DEW, Moorman AFM, Campione M, 2004. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev. Dyn 229, 763–770. 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- Cioffi M, Vallespinos-Serrano M, Trabulo SM, Fernandez-Marcos PJ, Firment AN, Vazquez BN, Vieira CR, Mulero F, Camara JA, Cronin UP, Perez M, Soriano J, Galvez GB, Castells-Garcia A, Haage V, Raj D, Megias D, Hahn S, Serrano L, Moon A, Aicher A, Heeschen C, 2015. MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3. Cell Rep. 12, 1594–1605. 10.1016/j.celrep.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CHM, Jones DL, Visvader J, Weissman IL, Wahl GM, 2006. Cancer stem cells—perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 66,9339–9344. 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Colasanto MP, Eyal S, Mohassel P, Bamshad M, Bonnemann CG, Zelzer E, Moon AM, Kardon G, 2016. Development of a subset of forelimb muscles and their attachment sites requires the ulnar-mammary syndrome gene Tbx3. Dis. Model. Mech 9, 1257–1269. 10.1242/dmm.025874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M, Seidman JG, Müller CW, 2002. Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure 10, 343–356. 10.1016/S0969-2126(02)00722-0. [DOI] [PubMed] [Google Scholar]
- D’Amelio P, Sassi F, 2016. Osteoimmunology: from mice to humans. Bonekey Rep. 5, 1–6. 10.1038/bonekey.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Costa ZC, Higgins CA, Ong CW, Irwin G, Boyle D, McArt DG, McCloskey K, Buckley NE, Crawford NT, Thiagarajan L, Murray JT, Kennedy RD, Mulligan KA, Harkin DP, Waugh DJJ, Scott CJ, Salto-Tellez M, Williams R, Mullan PB, 2014. TBX2 represses CST6 resulting in uncontrolled legumain activity to sustain breast cancer proliferation: a novel cancer-selective target pathway with therapeutic opportunities. Oncotarget 5 10.18632/oncotarget.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport TG, Jerome-Majewska LA, Papaioannou VE, 2003. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development 130, 2263–2273. 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- DeGregori J, 2004. The Rb network. J. Cell Sci 117, 3411–3413. 10.1242/jcs.01189. [DOI] [PubMed] [Google Scholar]
- Delorme B, Dahl E, Jarry-Guichard T, Briand J, Willecke K, Gros D, Théveniau-Ruissy M, 1997. Expression pattern of connexin gene products at the early developmental stages of the mouse cardiovascular system. Circ. Res 81, 423–437. 10.1161/01.RES.81.3.423. [DOI] [PubMed] [Google Scholar]
- Dhawan P, Singh AB, Ellis DL, Richmond A, 2002. Constitutive activation of Akt/protein kinase B in melanoma leads to up-regulation of nuclear factor-kappaB and tumor progression. Cancer Res. 62, 7335–7342. [PubMed] [Google Scholar]
- Domyan ET, Ferretti E, Throckmorton K, Mishina Y, Nicolis SK, Sun X, 2011. Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development 138, 971–981. 10.1242/dev.053694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Dong Q, Chen Y, Li Y, Zhang B, Zhou F, Lyu X, Chen GG, Lai P, Kung H, He M-L, 2018a. Novel HDAC5-interacting motifs of Tbx3 are essential for the suppression of E-cadherin expression and for the promotion of metastasis in hepatocellular carcinoma. Signal Transduct. Target. Ther 3, 22 10.1038/s41392-018-0025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Lyu X, Faleti OD, He M-L, 2018b. The special stemness functions of Tbx3 in stem cells and cancer development. Semin. Cancer Biol 10.1016/j.semcancer.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Du HF, Ou LP, Yang X, Song XD, Fan YR, Tan B, Luo CL, Wu XH, 2014. A new PKCα/β/TBX3/E-cadherin pathway is involved in PLCε-regulated invasion and migration in human bladder cancer cells. Cell. Signal 26, 580–593. 10.1016/j.cellsig.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Eblaghie MC, Song S-J, Kim J-Y, Akita K, Tickle C, Jung H-S, 2004. Interactions between FGF and Wnt signals and Tbx3 gene expression in mammary gland initiation in mouse embryos. J. Anat 205, 1–13. 10.1111/j.0021-8782.2004.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emechebe U, Kumar P, Rozenberg JM, Moore B, Firment A, Mirshahi T, Moon AM, 2016. T-box3 is a ciliary protein and regulates stability of the Gli3 transcription factor to control digit number. Elife 5, 1–28. 10.7554/eLife.07897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P, Aine M, Veerla S, Liedberg F, Sjödahl G, Höglund M, 2015. Molecular subtypes of urothelial carcinoma are defined by specific gene regulatory systems. BMC Med. Genomics 8 (1), 25 10.1186/s12920-015-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson KS, Mignot E, 2009. T-box 3 is expressed in the adult mouse hypothalamus and medulla. Brain Res. 1302, 233–239. 10.1016/j.brainres.2009.08.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, Chen Y, 2009. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2–5. Dev. Biol 327, 376–385. 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, Saikali S, Hamlat A, Riffaud L, Menei P, Quillien V, 2010. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics 11 (1), 701 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Huang X, Chen C, Gray J, Huang T, 2004. TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res. 64, 5132–5139. 10.1158/0008-5472.CAN-04-0615. [DOI] [PubMed] [Google Scholar]
- Feng X, Yao W, Zhang Z, Yuan F, Liang L, Zhou J, Liu S, Song J, 2018. T-box transcription factor Tbx3 contributes to human hepatocellular carcinoma cell migration and invasion by repressing E-cadherin expression. Oncol. Res. Featur. Preclin. Clin. Cancer Ther 26, 959–966. 10.3727/096504017X15145624664031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C, 2010. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc. Natl. Acad. Sci. U.S.A 107, 21737–21742. 10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DU, Emechebe U, Thomas KR, Moon AM, 2013. Mouse Tbx3 mutants suggest novel molecular mechanisms for ulnar-mammary syndrome. e67841 PLoS One 8 10.1371/journal.pone.0067841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg J, Lee H-S, Han B-G, Shin H. Do, Kang YM, Sung Y-K, Shim S-C, Choi C-B, Lee AT, Gregersen PK, Bae S-C, 2011. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 63, 884–893. 10.1002/art.30235. [DOI] [PubMed] [Google Scholar]
- Gallaher JA, Brown JS, Anderson ARA, 2019. The impact of proliferation-migration tradeoffs on phenotypic evolution in cancer. Sci. Rep 9, 2425 10.1038/s41598-019-39636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Chapman DL, Alexiou M, Garvey N, Lee SM, Papaioannou VE, 1996. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech. Dev 56, 93–101. 10.1016/0925-4773(96)00514-X. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Corbel S, Doyonnas R, Havenstrite K, Magnusson KEG, Blau HM, 2012. A single cell bioengineering approach to elucidate mechanisms of adult stem cell self-renewal. Integr. Biol 4, 360–367. 10.1039/c2ib00148a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AP, 2005. Anoikis. Cell Death Differ. 12, 1473–1477. 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE, 2009. Wnt2/2b and β-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev. Cell 17, 290–298. 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich F, Rudat C, Kispert A, 2011. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res 91, 212–222. 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, Lufkin T, Ng H-H, Tam W-L, Lim B, 2010. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 463, 1096–1100. 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA, 2011. Hallmarks of cancer: the next generation. Cell 144,646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- He MI, Wen L, Campbell CE, Wu JY, Rao Y, 1999. Transcription repression by Xenopus ET and its human ortholog TBX3, a gene involved in ulnar-mammary syndrome. Proc. Natl. Acad. Sci. U.S.A 96, 10212–10217. 10.1073/pnas.96.18.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges M, Morrisey EE, 2014. Lung development: orchestrating the generation and regeneration of a complex organ. Development 141, 502–513. 10.1242/dev.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, Wu T, Niinobe M, Yoshikawa K, Hannigan GE, Halaban R, 2004. Expression Profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 64, 5270–5282. 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- Honda A, Kawano Y, Izu H, Choijookhuu N, Honsho K, Nakamura T, Yabuta Y, Yamamoto T, Takashima Y, Hirose M, Sankai T, Hishikawa Y, Ogura A, Saitou M, 2017. discrimination of stem cell status after subjecting cynomolgus monkey pluripotent stem cells to naïve conversion. Sci. Rep 7, 45285 10.1038/srep45285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WMH, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LYE, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AFM, Verheijck EE, Christoffels VM, 2007. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 21, 1098–1112. 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WMH, Barnett P, Rodriguez M, Clout DE, Moorman AFM, Goding CR, Christoffels VM, 2008. TBX3 and its splice variant TBX3 + exon 2a are functionally similar. Pigment Cell Melanoma Res. 21, 379–387. 10.1111/j.1755-148X.2008.00461.x. [DOI] [PubMed] [Google Scholar]
- Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM, 2004. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res 62, 489–499. 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Howard B, Ashworth A, 2006. Signalling pathways implicated in early mammary gland morphogenesis and breast cancer. PLoS Genet. 2, e112 10.1371/journal.pgen.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B, Panchal H, McCarthy A, Ashworth A, 2005. Identification of the scar-amanga gene implicates Neuregulin3 in mammary gland specification. Genes Dev. 19, 2078–2090. 10.1101/gad.338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Ye S, Zhou X, Liu D, Ying Q-L, 2015. Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network. Cell. Mol. Life Sci 72, 1741–1757. 10.1007/s00018-015-1833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humtsoe JO, Koya E, Pham E, Aramoto T, Zuo J, Ishikawa T, Kramer RH, 2012. Transcriptional profiling identifies upregulated genes following induction of epithelial-mesenchymal transition in squamous carcinoma cells. Exp. Cell Res 318, 379–390. 10.1016/j.yexcr.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Ichijo R, Kobayashi H, Yoneda S, Iizuka Y, Kubo H, Matsumura S, Kitano S, Miyachi H, Honda T, Toyoshima F, 2017. Tbx3-dependent amplifying stem cell progeny drives interfollicular epidermal expansion during pregnancy and regeneration. Nat. Commun 8, 1–12. 10.1038/s41467-017-00433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue R, Asker C, Klangby U, Pisa P, Wiman KG, 1999. Induction of the humanARF protein by serum starvation. Anticancer Res. 19, 2939–2943. [PubMed] [Google Scholar]
- Ito A, Asamoto M, Hokaiwado N, Takahashi S, Shirai T, 2005. Tbx3 expression is related to apoptosis and cell proliferation in rat bladder both hyperplastic epithelial cells and carcinoma cells. Cancer Lett. 219, 105–112. 10.1016/j.canlet.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR, 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538. 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jacobs JJL, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, van Lohuizen M, 2000. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19ARF) and is amplified in a subset of human breast cancers. Nat. Genet 26, 291–299. 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- Jerome-Majewska LA, Jenkins GP, Ernstoff E, Zindy F, Sherr CJ, Papaioannou VE, 2005. Tbx3, the ulnar-mammary syndrome gene, and Tbx2 interact in mammary gland development through a p19 Arf/p53-independent pathway. Dev. Dyn 234, 922–933. 10.1002/dvdy.20575. [DOI] [PubMed] [Google Scholar]
- Jiang K, Ren C, Nair VD, 2013. MicroRNA-137 represses Klf4 and Tbx3 during differentiation of mouse embryonic stem cells. Stem Cell Res. 11, 1299–1313. 10.1016/j.scr.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB, 2002. The fundamental role of epigenetic events in cancer. Nat.Rev. Genet 3 (6), 415 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Julià A, Ballina J, Cañete JD, Balsa A, Tornero-Molina J, Naranjo A, Alperi-López M, Erra AA, Pascual-Salcedo D, Barcelò P, Camps J, Marsal S, Barceló P, Camps J, Marsal S, 2008. Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility. Arthritis Rheum. 58, 2275–2286. 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- Kandimalla R, van Tilborg AAG, Kompier LC, Stumpel DJPM, Stam RW, Bangma CH, Zwarthoff EC, 2012. Genome-wide analysis of CpG island methylation in bladder cancer identified TBX2, TBX3, GATA2, and ZIC4 as pTa-specific prognostic markers. Eur. Urol 61, 1245–1256. 10.1016/j.eururo.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Karolak JA, Vincent M, Deutsch G, Gambin T, Cogné B, Pichon O, Vetrini F, Mefford HC, Dines JN, Golden-Grant K, Dipple K, Freed AS, Leppig KA, Dishop M, Mowat D, Bennetts B, Gifford AJ, Weber MA, Lee AF, Boerkoel CF, Bartell TM, Ward-Melver C, Besnard T, Petit F, Bache I, Tümer Z, Denis-Musquer M, Joubert M, Martinovic J, Bénéteau C, Molin A, Carles D, André G, Bieth E, Chassaing N, Devisme L, Chalabreysse L, Pasquier L, Secq V, Don M, Orsaria M, Missirian C, Mortreux J, Sanlaville D, Pons L, Küry S, Bézieau S, Liet J-MM, Joram N, Bihouée T, Scott DA, Brown CW, Scaglia F, Tsai AC-HH, Grange DK, Phillips JA, Pfotenhauer JP, Jhangiani SN, Gonzaga-Jauregui CG, Chung WK, Schauer GM, Lipson MH, Mercer CL, van Haeringen A, Liu Q, Popek E, Coban Akdemir ZH, Lupski JR, Szafranski P, Isidor B, Le Caignec C, Stankiewicz P, 2019. Complex compound inheritance of lethal lung developmental disorders due to disruption of the TBX-FGF pathway. Am.J. Hum. Genet 104, 213–228. 10.1016/j.ajhg.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M, He Q, Hong D, Li O, Zhu M, Ou W, He Y, Wu Y, 2018. Leukemia inhibitory factor regulates marmoset induced pluripotent stem cell proliferation via a PI3K/Akt-dependent Tbx-3 activation pathway. Int. J. Mol. Med 42, 131–140. 10.3892/ijmm.2018.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Hirth F, 2009. Genetic mechanisms regulating stem cell self-renewal and differentiation in the central nervous system of Drosophila. Cell Adh. Migr 3, 402–411. 10.4161/cam.3.4.8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenstein M, Raab S, Achberger K, Kleger A, Liebau S, Linta L, 2016. TBX3 knockdown decreases reprogramming efficiency of human cells. Stem Cells Int. 2016, 1–7. 10.1155/2016/6759343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko E, Han W, Noh D, 2009. Reduced self-renewal ability and drug resistance by inhibition of notch-4 and ABCG2 in anoikis-resistant MDA-MB-231 breast cancer cells In: Poster Session Abstracts. American Association for Cancer Research, p. 5059. doi: 10.1158/0008-5472.SABCS-5059. [DOI] [Google Scholar]
- Krstic M, Macmillan CD, Leong HS, Clifford AG, Souter LH, Dales DW,Postenka CO, Chambers AF, Tuck AB, 2016. The transcriptional regulator TBX3 promotes progression from non-invasive to invasive breast cancer. BMC Cancer 16, 671 10.1186/s12885-016-2697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krstic M, Kolendowski B, Cecchini MJ, Postenka CO, Hassan HM, Andrews J, MacMillan CD, Williams KC, Leong HS, Brackstone M, Torchia J, Chambers AF, Tuck AB, 2019. TBX3 promotes progression of pre-invasive breast cancer cells by inducing EMT and directly up-regulating SLUG. J. Pathol 248, 191–203. 10.1002/path.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Franklin S, Emechebe U, Hu H, Moore B, Lehman C, Yandell M, Moon AM, 2014a. TBX3 regulates splicing in vivo: a novel molecular mechanism for ulnar-mammary syndrome. PLoS Genet 10 10.1371/journal.pgen.1004247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Emechebe U, Smith R, Franklin S, Moore B, Yandell M, Lessnick SL, Moon AM, 2014b. Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. Elife 3, 1–28. 10.7554/eLife.02805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Rath P, Liu J, Ryu D, Free A, Pei L, Anthony DC, Sharma S, Kirk MD, Laterra JJ, Ryu DH, Choi J-H, Shi H, Miller DC, Litofsky NS, Feng Q, 2014. Abstract 1379: Identification of global DNA methylation signatures in glioblastoma-derived cancer stem cells In: Molecular and Cellular Biology. American Association for Cancer Research; pp. 1379–1379. [Google Scholar]
- Lee K, Wong W, Feng B, 2013. Decoding the pluripotency network: the emergence of new transcription factors. Biomedicines 1, 49–78. 10.3390/biomedicines1010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie NR, Downes CP, 2004. PTEN function: how normal cells control it and tumour cells lose it. Biochem. J 382, 1–11. 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ballim D, Rodriguez M, Cui R, Goding CR, Teng H, Prince S, 2014. The anti-proliferative function of the TGF-β1 signaling pathway involves the repression of the oncogenic TBX2 by its homologue TBX3. J. Biol. Chem 289, 35633–35643. 10.1074/jbc.M114.596411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ruan X, Zhang P, Yu Y, Gao M, Yuan S, Zhao Z, Yang J, Zhao L, 2018a. TBX3 promotes proliferation of papillary thyroid carcinoma cells through facilitating PRC2-mediated p57KIP2 repression. Oncogene 37, 2773–2792. 10.1038/s41388-017-0090-2. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang Y, Duan S, Shi Y, Li S, Zhang X, Ren J, 2018b. Expression of TBX3 in hepatocellular carcinoma and its clinical implication. Med. Sci. Monit 24, 9324–9333. 10.12659/MSM.909378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Weinberg MS, Zerbini L, Prince S, 2013. The oncogenic TBX3 is a downstream target and mediator of the TGF-β1 signaling pathway. Mol. Biol. Cell 24, 3569–3576. 10.1091/mbc.e13-05-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang H, Choi SC, Litingtung Y, Chiang C, 2004. Sonic hedgehog signaling regulates Gli3 processing, mesenchymal proliferation, and differentiation during mouse lung organogenesis. Dev. Biol 270, 214–231. 10.1016/j.ydbio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai C-L, Bu L, Yang L, Martin J, Kemler R, Rosenfeld MG, Chen J, Evans SM, 2007. beta-Catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc. Natl. Acad. Sci. U.S.A 104, 9313–9318. 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H, Williams R, King J, Blair E, Kini U, 2009. Ulnar mammary syndrome and TBX3: expanding the phenotype. Am. J. Med. Genet. Part A 149A, 2809–2812. 10.1002/ajmg.a.33096. [DOI] [PubMed] [Google Scholar]
- Lingbeek ME, Jacobs JJL, Van Lohuizen M, 2002. The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. J. Biol. Chem 277, 26120–26127. 10.1074/jbc.M200403200. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Lei L, Westphal H, Chiang C, 1998. Sonic hedgehog is essential to foregut development. Nat. Genet 20, 58–61. 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- Liu X, Miao Z, Wang Z, Zhao T, Xu Y, Song Y, Huang J, Zhang J, Xu Hao, Wu J, Xu Huimian, 2019. TBX2 overexpression promotes proliferation and invasion through epithelial-mesenchymal transition and ERK signaling pathway. Exp. Ther. Med 17, 723–729. 10.3892/etm.2018.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dollé P, Dierich A, Gorry P, Gansmuller A, Chambon P, 1994. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development 120, 2723–2748. [DOI] [PubMed] [Google Scholar]
- López SH, Avetisyan M, Wright CM, Mesbah K, Kelly RG, Moon AM,Heuckeroth RO, 2018. Loss of Tbx3 in murine neural crest reduces enteric glia and causes cleft palate, but does not influence heart development or bowel transit. Dev. Biol 444, S337–S351. 10.1016/j.ydbio.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Yang A, Jin Y, 2011. Dual functions of T-Box 3 (Tbx3) in the control of self-renewal and extraembryonic endoderm differentiation in mouse embryonic stem cells. J. Biol. Chem 286, 8425–8436. 10.1074/jbc.M110.202150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüdtke TH-W, Christoffels VM, Petry M, Kispert A, 2009. Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology 49, 969–978. 10.1002/hep.22700. [DOI] [PubMed] [Google Scholar]
- Lüdtke TH, Rudat C, Wojahn I, Weiss AC, Kleppa MJ, Kurz J, Farin HF, Moon A, Christoffels VM, Kispert A, 2016. Tbx2 and Tbx3 act downstream of Shh to maintain canonical Wnt signaling during branching morphogenesis of the murine lung. Dev. Cell 39, 239–253. 10.1016/j.devcel.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Lyng H, Brøvig RS, Svendsrud DH, Holm R, Kaalhus O, Knutstad K, Oksefjell H, Sundfør K, Kristensen GB, Stokke T, 2006. Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer. BMC Genomics 7, 268 10.1186/1471-2164-7-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailleux AA, Spencer-Dene B, Dillon C, Ndiaye D, Savona-Baron C, Itoh N, Kato S, Dickson C, Thiery JP, Bellusci S, 2002. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development 129, 53–60. [DOI] [PubMed] [Google Scholar]
- Martin GR, 1998. The roles of FGFs in the early development of vertebrate limbs. GenesDev. 12, 1571–1586. 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- Meneghini V, Odent S, Platonova N, Egeo A, Merlo GR, 2006. Novel TBX3 mutation data in families with Ulnar-Mammary syndrome indicate a genotype–phenotype relationship: mutations that do not disrupt the T-domain are associated with less severe limb defects. Eur. J. Med. Genet 49, 151–158. 10.1016/j.ejmg.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Mesbah K, Harrelson Z, Théveniau-Ruissy M, Papaioannou VE, Kelly RG, 2008. Tbx3 is required for outflow tract development. Circ. Res 103, 743–750. 10.1161/CIRCRESAHA.108.172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z-F, Liu X-Y, Xu H-M, Wang Z-N, Zhao T-T, Song Y-X, Xing Y-N,Huang J-Y, Zhang J-Y, Xu H, Xu Y-Y, 2016. Tbx3 overexpression in human gastric cancer is correlated with advanced tumor stage and nodal status and promotes cancer cell growth and invasion. Virchows Arch. 469, 505–513. 10.1007/s00428-016-2007-9. [DOI] [PubMed] [Google Scholar]
- Mo J-S, Park HW, Guan K-L, 2014. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 15, 642–656. 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamad T, Kazim N, Adhikari A, Davie JK, 2018. EGR1 interacts with TBX2 and functions as a tumor suppressor in rhabdomyosarcoma. Oncotarget 9, 18084–18098. 10.18632/oncotarget.24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan RA, Mommersteeg MTM, Domínguez JN, Choquet C, Wakker V, de Gier-de Vries C, Boink GJJ, Boukens BJ, Miquerol L, Verkerk AO, Christoffels VM, Dom JN, Choquet C, Wakker V, Vries CDG, Boink GJJ, Boukens BJ, Miquerol L, Verkerk AO, Christoffels VM, 2018. Embryonic Tbx3 + cardiomyocytes form the mature cardiac conduction system by progressive fate restriction. Development 145, dev167361. 10.1242/dev.167361. [DOI] [PubMed] [Google Scholar]
- Moorman AFM, Soufan AT, Hagoort J, de Boer PAJ, Christoffels VM, 2004. Development of the building plan of the heart. Ann. N.Y. Acad. Sci 1015, 171–181. 10.1196/annals.1302.014. [DOI] [PubMed] [Google Scholar]
- Mowla S, Pinnock R, Leaner VD, Goding CR, Prince S, 2011. PMA-induced up-regulation of TBX3 is mediated by AP-1 and contributes to breast cancer cell migration. Biochem. J 433, 145–153. 10.1042/BJ20100886. [DOI] [PubMed] [Google Scholar]
- Ng H, Surani MA, 2011. The transcriptional and signalling networks of pluripotency. Nat. Cell Biol 13, 490–496. 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K, 2009. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460, 118–122. 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Oh T-J, Adhikari A, Mohamad T, Althobaiti A, Davie J, 2019. TBX3 represses TBX2 under the control of the PRC2 complex in skeletal muscle and rhabdomyosarcoma. Oncogenesis 8, 27 10.1038/s41389-019-0137-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N, 2000. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun 277, 643–649. 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- Okita K, Yamanaka S, 2011. Induced pluripotent stem cells: opportunities and challenges. Philos. Trans. R. Soc. B Biol. Sci 366, 2198–2207. 10.1098/rstb.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Speziale D, Shoukry M, Mohan R, Ivanek R, Kohler M, Beisel C, Wen X, Scales SJ, Christoffels VM, Visel A, Lopez-Rios J, Zeller R, 2014. HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev. Cell 31, 345–357. 10.1016/j.devcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packham EAA, Brook JD, 2003. T-box genes in human disorders. Hum. Mol. Genet 12, R37–R44. 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- Palmieri G, Ombra M, Colombino M, Casula M, Sini M, Manca A, Paliogiannis P, Ascierto PA, Cossu A, 2015. Multiple molecular pathways in melanomagenesis: characterization of therapeutic targets. Front. Oncol 5, 183 10.3389/fonc.2015.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou VE, 2001. T-box genes in development: from hydra to humans. Int. Rev. Cytol 207, 1–70. 10.1016/S0074-7696(01)07002-4. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE, 2014. The T-box gene family: emerging roles in development, stem cells and cancer. Development 141, 3819–3833. 10.1242/dev.104471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton C, Zhao H, Chin Y, Langner K, Reecy J, 2002. Murine Tbx2 contains domains that activate and repress gene transcription. Gene 283, 117–124. 10.1016/s0378-1119(01)00878-2. [DOI] [PubMed] [Google Scholar]
- Pepicelli CV, Lewis PM, McMahon AP, 1998. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr. Biol 8, 1083–1086. 10.1016/S0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- Peres J, Davis E, Mowla S, Bennett DC, Li JA, Wansleben S, Prince S, 2010. The highly homologous T-box transcription factors, TBX2 and TBX3, have distinct roles in the oncogenic process. Genes Cancer 1, 272–282. 10.1177/1947601910365160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres J, Mowla S, Prince S, 2015. The T-box transcription factor, TBX3, is a key substrate of AKT3 in melanomagenesis. Oncotarget 6, 1821–1833. 10.18632/oncotarget.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres J, Kwesi-Maliepaard EM, Rambow F, Larue L, Prince S, 2017. The tumour suppressor, miR-137, inhibits malignant melanoma migration by targetting the TBX3 transcription factor. Cancer Lett. 405, 111–119. 10.1016/j.canlet.2017.07.018. [DOI] [PubMed] [Google Scholar]
- Peres J, Prince S, 2013. The T-box transcription factor, TBX3, is sufficient to promote melanoma formation and invasion. Mol. Cancer 12, 117 10.1186/1476-4598-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkhofer L, Walter K, Costa IG, Carrasco MCRR, Eiseler T, Hafner S, Genze F, Zenke M, Bergmann W, Illing A, Hohwieler M, Köhntop R, Lin Q, Holzmann K-HH, Seufferlein T, Wagner M, Liebau S, Hermann PC, Kleger A, Müller M, 2016. Tbx3 fosters pancreatic cancer growth by increased angiogenesis and activin/ nodal-dependent induction of stemness. Stem Cell Res. 17, 367–378. 10.1016/j.scr.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Platonova N, Scotti M, Babich P, Bertoli G, Mento E, Meneghini V, Egeo A,Zucchi I, Merlo GR, 2007. TBX3, the gene mutated in ulnar-mammary syndrome, promotes growth of mammary epithelial cells via repression of p19ARF, independently of p53. Cell Tissue Res. 328, 301–316. 10.1007/s00441-006-0364-4. [DOI] [PubMed] [Google Scholar]
- Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LRL, Li W, Tan AKS, Bonnard C, Ong RTH, Thalamuthu A, Pettersson S, Liu C, Tian C, Chen WV, Carulli JP, Beckman EM, Altshuler D, Alfredsson L, Criswell LA, Amos CI, Seldin MF, Kastner DL, Klareskog L, Gregersen PK, 2007. TRAF1–C5 as a risk locus for rheumatoid arthritis — a genomewide study. N. Engl. J. Med 357, 1199–1209. 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liégeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho RA, 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 92, 713–723. 10.1016/S0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Pontecorvi M, Goding CR, Richardson WD, Kessaris N, 2008. Expression of Tbx2 and Tbx3 in the developing hypothalamic–pituitary axis. Gene Expr. Patterns 8, 411–417. 10.1016/j.gep.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Prince S, Carreira S, Vance K, Abrahams A, Goding C, 2004. Tbx2 directly represses the expression of the p21 WAF1 cyclin-dependent kinase inhibitor kinase inhibitor. Cancer Res. 64, 1669–1674. [DOI] [PubMed] [Google Scholar]
- Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, Keller GM, 2017. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol 35, 56–68. 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- Quarta C, Fisette A, Xu Y, Colldén G, Legutko B, Tseng Y, Reim A, Wierer M, De Rosa MC, Klaus V, Rausch R, Thaker VV, Graf E, Strom TM, Poher A-L, Gruber T, Le Thuc O, Cebrian-Serrano A, Kabra D, Bellocchio L, Woods SC, Pflugfelder GO, Nogueiras R, Zeltser L, Grunwald Kadow IC, Moon A, García-Cáceres C, Mann M, Treier M, Doege CA, Tschöp MH, 2019. Functional identity of hypothalamic melanocortin neurons depends on Tbx3. Nat. Metab 1, 222–235. 10.1038/s42255-018-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond KL, Crawford NT, Farmer H, D’Costa ZC, O’Brien GJ, Buckley NE, Kennedy RD, Johnston PG, Harkin DP, Mullan PB, 2010. T-box 2 represses NDRG1 through an EGR1-dependent mechanism to drive the proliferation of breast cancer cells. Oncogene 29, 3252–3262. 10.1038/onc.2010.84. [DOI] [PubMed] [Google Scholar]
- Renard C-A, Labalette C, Armengol C, Cougot D, Wei Y, Cairo S, Pineau P,Neuveut C, de Reyniès A, Dejean A, Perret C, Buendia M-A, 2007. Tbx3 Is a downstream target of the Wnt/β-catenin pathway and a critical mediator of β-catenin survival functions in liver cancer. Cancer Res. 67, 901–910. 10.1158/0008-5472.CAN-06-2344. [DOI] [PubMed] [Google Scholar]
- Ribeiro I, Kawakami Y, Büscher D, Raya Á, Rodríguez-León J, Morita M,Rodríguez Esteban C, Izpisúa Belmonte JC, Rodriguez-León J, Morita M, Rodríguez Esteban C, Izpisúa Belmonte JC, 2007. Tbx2 and Tbx3 regulate the dynamics of cell proliferation during heart remodeling. e398 PLoS One 2 10.1371/journal.pone.0000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR, 2008. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 68, 7872–7881. 10.1158/0008-5472.CAN-08-0301. [DOI] [PubMed] [Google Scholar]
- Rowley M, Grothey E, Couch FJ, 2004. The role of Tbx2 and Tbx3 in mammary development and tumorigenesis. J. Mammary Gland Biol. Neoplasia 9, 109–118. 10.1023/B:JOMG.0000037156.64331.3f. [DOI] [PubMed] [Google Scholar]
- Russell R, Ilg M, Lin Q, Wu G, Lechel A, Bergmann W, Eiseler T, Linta L, Kumar P, Klingenstein M, Adachi K, Hohwieler M, Sakk O, Raab S, Moon A, Zenke M, Seufferlein T, Schöler HR, Illing A, Liebau S, Kleger A, 2015. A dynamic role of TBX3 in the pluripotency circuitry. Stem Cell Rep. 5, 1155–1170. 10.1016/j.stemcr.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, Rey J-P, Ma J-X, Staehling-Hampton K, Trainor PA, 2007. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 21, 1113–1124. 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar S, Kerr A, Vaartjes D, Moltved ER, Karosiene E, Gupta R, Andersson Å,2019. The oncoprotein TBX3 is controlling severity in experimental arthritis. Arthritis Res. Ther 21, 16 10.1186/s13075-018-1797-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Faiola F, Wang J, 2013. Concise review: pursuing self-renewal and pluripotency with the stem cell factor Nanog. Stem Cells 31, 1227–1236. 10.1002/stem.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S, 1999. Fgf10 is essential for limb and lung formation. Nat. Genet 21, 138–141. 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Semrau S, Goldmann JE, Soumillon M, Mikkelsen TS, Jaenisch R, vanOudenaarden A, 2017. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat. Commun 8, 1096 10.1038/s41467-017-01076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba CJ, Logan MPO, 2017. The Roles of T-Box Genes in Vertebrate Limb Development In: Current Topics in Developmental Biology. Elsevier Inc., pp. 355–381. doi: 10.1016/bs.ctdb.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Shi Y, Katsev S, Cai C, Evans S, 2000. BMP signaling is required for heart formation in vertebrates. Dev. Biol 224, 226–237. 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- Sinha S, Abraham S, Gronostajski RM, Campbell CE, 2000. Differential DNA binding and transcription modulation by three T-box proteins, T, TBX1 and TBX2. Gene 258, 15–29. 10.1016/S0378-1119(00)00417-0. [DOI] [PubMed] [Google Scholar]
- Siroy AE, Davies MA, Lazar AJ, 2016. The PI3K-AKT Pathway in Melanoma In: Genetics of Melanoma. Springer; New York, New York, NY, pp. 165–180. doi: 10.1007/978-1-4939-3554-3_7. [DOI] [Google Scholar]
- Stennard FA, Harvey RP, 2005. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development 132 (22), 4897–4910. 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerød A, Karesen R, Schlichting E, Naume B, Sauer T, Ottestad L, Lee MTM, Shen C-YY, Tee BTK, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin S-FF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, Van’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SAJRJR, Salomon AV, Børresen-Dale A-LL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR, Spencer Chapman M, Teague J, Easton D, Langerød A, Lee MTM, Shen C-YY, Tee BTK, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin S-FF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van ‘t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SAJRJR, Salomon AV, Børresen-Dale A-LL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR, 2012. The landscape of cancer genes and mutational processes in breast cancer. Nature 486, 400–404. 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Sekiya S, Buscher D, Izpisua Belmonte JC, Taniguchi H, 2008. Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development 135, 1589–1595. 10.1242/dev.016634. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Takeuchi J, Koshiba-Takeuchi K, Ogura T, 2004. Tbx genes specify posterior digit identity through Shh and BMP signaling. Dev. Cell 6, 43–53. 10.1016/S1534-5807(03)00401-5. [DOI] [PubMed] [Google Scholar]
- Tümpel S, Sanz-Ezquerro JJ, Isaac A, Eblaghie MC, Dobson J, Tickle C, 2002. Regulation of Tbx3 expression by anteroposterior signalling in vertebrate limb development. Dev. Biol 250, 251–262. 10.1006/dbio.2002.0762. [DOI] [PubMed] [Google Scholar]
- van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R, Grosschedl R, 1994. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes & development 8 (22), 2691–2703. 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- Van Kempen MJAA, Vermeulen JLMM, Moorman AFMM, Gros D, Paul DL, Lamers WH, 1996. Developmental changes of connexin40 and connexin43 mRNA distribution patterns in the rat heart. Cardiovasc. Res 32, 886–890. 10.1016/0008-6363(96)00131-9. [DOI] [PubMed] [Google Scholar]
- Vance KW, Carreira S, Brosch G, Goding CR, 2005. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 65, 2260–2268. 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- Veltmaat JM, Van Veelen W, Thiery JP, Bellusci S, 2004. Identification of the mammary line in mouse byWnt10b expression. Dev. Dyn 229, 349–356. 10.1002/dvdy.10441. [DOI] [PubMed] [Google Scholar]
- Wang Y, 2018. Expression level of TBX3 gene in renal carcinoma and its clinical significance. Oncol. Lett 15, 4235–4240. 10.3892/ol.2018.7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gu Q, Hao J, Jia Y, Xue B, Jin H, Ma J, Wei R, Hai T, Kong Q, Bou G, Xia P, Zhou Q, Wang L, Liu Z, 2013. Tbx3 and Nr5α2 play important roles in pig pluripotent stem cells. Stem Cell Rev. Rep 9, 700–708. 10.1007/s12015-013-9439-2. [DOI] [PubMed] [Google Scholar]
- Wansleben S, Peres J, Hare S, Goding CR, Prince S, 2014. T-box transcription factors in cancer biology. Biochim. Biophys. Acta - Rev. Cancer 1846, 380–391. 10.1016/j.bbcan.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Washkowitz AJ, Gavrilov S, Begum S, Papaioannou VE, 2012. Diverse functional networks of Tbx3 in development and disease. Wiley Interdiscip. Rev. Syst. Biol. Med 4, 273–283. 10.1002/wsbm.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Dunn NR, Hogan BL, 2000. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development 127, 2695–2704. [DOI] [PubMed] [Google Scholar]
- Weidgang CE, Russell R, Tata PR, Kühl SJ, Illing A, Müller M, Lin Q, Brunner C, Boeckers TM, Bauer K, Kartikasari AE, Kartikasari AE, 2013. TBX3 directs cell-fate decision toward mesendoderm. Stem cell reports 1 (3), 248–265. 10.1016/j.stemcr.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensing LA, Campos AH, 2014. TBX3, a downstream target of TGF-β1, inhibits mesangial cell apoptosis. Exp. Cell Res 328, 340–350. 10.1016/j.yexcr.2014.08.022. [DOI] [PubMed] [Google Scholar]
- White-Al Habeeb NM, Ho LT, Olkhov-Mitsel E, Kron K, Pethe V, Lehman M, Jovanovic L, Fleshner N, van der Kwast T, Nelson CC, Bapat B, White-Al Habeeb NMA, Ho LT, Olkhov-Mitsel E, Kron K, Pethe V, Lehman M, Jovanovic L, Fleshner N, van der Kwast T, Nelson CC, Bapat B, 2014. Integrated analysis of epigenomic and genomic changes by DNA methylation dependent mechanisms provides potential novel biomarkers for prostate cancer. Oncotarget 5 (17), 7858–7869. 10.18632/oncotarget.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, Mommersteeg MTM, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AFM, Kispert A, Christoffels VM, 2009. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res 104, 388–397. 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Willmer T, Cooper A, Peres J, Omar R, Prince S, 2017. The T-Box transcription factor 3 in development and cancer. Biosci. Trends 11, 254–266. 10.5582/bst.2017.01043. [DOI] [PubMed] [Google Scholar]
- Willmer T, Cooper A, Sims D, Govender D, Prince S, 2016. The T-box transcription factor 3 is a promising biomarker and a key regulator of the oncogenic phenotype of a diverse range of sarcoma subtypes. Oncogenesis 5 10.1038/oncsis.2016.11. e199–e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmer T, Hare S, Peres J, Prince S, 2016. The T-box transcription factor TBX3 drives proliferation by direct repression of the p21WAF1 cyclin-dependent kinase inhibitor. Cell Div. 11, 6 10.1186/s13008-016-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmer T, Peres J, Mowla S, Abrahams A, Prince S, 2015. The T-Box factor TBX3 is important in S-phase and is regulated by c-Myc and cyclin A-CDK2. Cell Cycle 14, 3173–3183. 10.1080/15384101.2015.1080398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Conlon FL, 2002. The T-box family. Genome Biol. 3 10.1186/gb-2002-3-6-reviews3008.REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Revelli JP, Eichele G, Barron M, Schwartz RJ, 2000. Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes during early heart development: evidence for BMP2 induction of Tbx2. Dev. Biol 228, 95–105. 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Tsujino Y, Moriguchi K, Tatematsu M, Ushijima T, 2006. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2′-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 97, 64–71. 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL,Fishman GI, Cogen A, Evans S, 2006. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development 133, 1575–1585. 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Yamazaki Y, Adachi M, Okawa K, Fort P, Uji M, Tsukita Shoichiro,Tsukita Sachiko, 2011. Tara up-regulates E-cadherin transcription by binding to the Trio RhoGEF and inhibiting Rac signaling. J. Cell Biol 193, 319–332. 10.1083/jcb.201009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarosh W, Barrientos T, Esmailpour T, Lin L, Carpenter PM, Osann K, Anton-Culver H, Huang T, 2008. TBX3 is overexpressed in breast cancer and represses p14ARF by interacting with histone deacetylases. Cancer Res. 68, 693–699. 10.1158/0008-5472.CAN-07-5012. [DOI] [PubMed] [Google Scholar]
- Yin M-X, Zhang L, 2015. Hippo signaling in epithelial stem cells. Acta Biochim. Biophys. Sin. (Shanghai) 47, 39–45. 10.1093/abbs/gmu111. [DOI] [PubMed] [Google Scholar]
- Yu Z, Pestell TG, Lisanti MP, Pestell RG, 2012. Cancer stem cells. Int. J. Biochem.Cell Biol 44, 2144–2151. 10.1016/j.biocel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal EA, Berkers CR, 2018. The influence of metabolism on drug response in cancer. Front. Oncol 8, 500 10.3389/fonc.2018.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chronis C, Chen X, Zhang H, Spalinskas R, Pardo M, Chen L, Wu G, Zhu Z, Yu Y, Yu L, Choudhary J, Nichols J, Parast MM, Greber B, Sahlén P, Plath K, 2019. The BAF and PRC2 complex subunits Dpf2 and Eed antagonistically converge on Tbx3 to control ESC differentiation. Cell Stem Cell 24, 138–152.e8. 10.1016/j.stem.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, He M, Dong Qi, Xie W, Chen Y, Lin MCM, Leung P, Zhang Y, Kung H, 2011a. Aqueous extracts of fructus ligustri lucidi enhance the sensitivity of human colorectal carcinoma DLD-1 cells to doxorubicin-induced apoptosis via Tbx3 suppression. Integr. Cancer Ther. 10, 85–91. 10.1177/1534735410373921. [DOI] [PubMed] [Google Scholar]
- Zhang Z, O’Rourke JR, McManus MT, Lewandoski M, Harfe BD, Sun X, 2011b. The microRNA-processing enzyme Dicer is dispensable for somite segmentation but essential for limb bud positioning. Dev. Biol 351, 254–265. 10.1016/j.ydbio.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan K-L, 2011. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol 13, 877–883. 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Wu Y, Chen K, 2014. Tbx3 isoforms are involved in pluripotency maintaining through distinct regulation of Nanog transcriptional activity. Biochem. Biophys. Res. Commun 444, 411–414. 10.1016/j.bbrc.2014.01.093. [DOI] [PubMed] [Google Scholar]