Abstract
High-grade glioma (HGG), and particularly Glioblastoma (GBM), can exhibit pronounced intratumoral heterogeneity that confounds clinical diagnosis and management. While conventional contrast-enhanced MRI lacks the capability to resolve this heterogeneity, advanced MRI techniques offer a spectrum of physiologic and biophysical image features to improve the specificity of imaging diagnoses. Published studies have shown how integrating these advanced techniques can help better define histologically distinct targets for surgical and radiation treatment planning, and help evaluate the regional heterogeneity of tumor recurrence and response assessment following standard adjuvant therapy. Application of texture analysis and machine learning (ML) algorithms has also enabled the emerging field of radiogenomics, which can spatially resolve the regional and genetically distinct subpopulations that coexist within a single GBM tumor. This review focuses on the latest advances in neuro-oncologic imaging and their clinical applications for the assessment of intratumoral heterogeneity.
Introduction
High-grade glioma (HGG), and particularly Glioblastoma (GBM), can exhibit pronounced intratumoral heterogeneity that confounds clinical diagnosis and management. While conventional contrast-enhanced MRI lacks the capability to resolve this heterogeneity, advanced imaging techniques offer a spectrum of physiologic and biophysical image features to improve the specificity of imaging diagnoses. This review covers the broad array of physiologic-based advanced imaging techniques that are nearly universally available across imaging sites and are most likely to be encountered in clinical practice. These include Diffusion Weighted Imaging (DWI), Diffusion Tensor Imaging (DTI), Dynamic Susceptibility Contrast (DSC)-MRI, Dynamic Contrast Enhanced (DCE)-MRI, MR Spectroscopy (MRS), as well as Positron Emission tomography (PET). Published studies have shown how integrating these advanced techniques can help better define histologically distinct targets for surgical and radiation treatment planning, and help evaluate the regional heterogeneity of tumor recurrence and response assessment following standard adjuvant therapy. Application of texture analysis and machine learning (ML) algorithms has also enabled the emerging field of radiogenomics, which can spatially resolve the regional and genetically distinct subpopulations that coexist within a single GBM tumor. This review discusses some of the most clinically relevant challenges in diagnosis and treatment of HGG, and particularly GBM, in the context of intratumoral heterogeneity. We describe the latest advances in neuro-oncologic imaging and their clinical applications to address these challenges.
High-grade gliomas (HGG) and glioblastoma (GBM)
After meningiomas, cerebral gliomas represent the most common primary brain tumor in adults overall, with an incidence of 5–6 per 100,000 persons annually. 1 The vast majority (>75%) of gliomas are high-grade (WHO grade III and IV), with the most common and aggressive form of glioma being Glioblastoma (GBM). In fact, GBM (WHO grade IV) accounts for over half (56.1%) of all gliomas. These tumors result in a dismal prognosis, with median survival of ~15 months, despite best available multimodal therapies.2 The clinical challenges of treatment and diagnosis, coupled with high relative incidence, have made high-grade gliomas (HGG), and GBM in particular, the subject of tremendous interest in neuro-oncologic research. For instance, GBM was the first tumor type studied by the foundational work of the Cancer Genome Atlas (TCGA) initiative, which started over a decade ago, and has since extended to other tumor types throughout the body.3 Advances in genomic and molecular profiling have introduced promising new therapeutic paradigms to combat this deadly disease.4,5 Yet, translating these discoveries to improve patient outcomes will rely critically on methods to resolve the internal heterogeneity of these tumors, which exists at multiple levels (e.g., genetic, molecular, histologic).
Clinical standard for imaging diagnosis and treatment planning in high-grade gliomas
Conventional contrast-enhanced MRI serves as the clinical standard for image-guided diagnosis and treatment planning, guiding essentially every aspect of glioma care. This includes surgical neuronavigation for biopsy and resection, as well as dosimetric radiation treatment planning. Contrast-enhanced MRI also guides response assessment after standard adjuvant chemo-radiation therapy. Conventional contrast-enhanced MRI consists of two fundamental types of MRI sequences: T1W contrast-enhanced (T1+C) and T2-Weighted and Fluid Attenuated Inversion Recovery (T2W/FLAIR) imaging. T1-weighted post-contrast imaging (T1+C): T1+C images are obtained following intravenous injection of Gadolinium-based contrast agent (GBCA), with areas of brightness or increased signal on imaging corresponding with tissue regions in which GBCA has extravasated due to vascular leakage and blood-brain-barrier (BBB) disruption. This leakage generally relates to Vascular Endothelial Growth Factor (VEGF) expression and/or neoangiogenesis. Based on current convention, the volume of T1+C enhancement serves as the clinical standard for defining HGG tumor burden, which guides surgical targeting for biopsy and resection, as well as dosimetric radiation treatment planning. Serial MRI and changes in T1+C enhancing volumes also define response assessment following standard adjuvant therapy, such that new or enlarging T1+C lesion volumes indicate the burden of recurrent tumor, as a sign of treatment failure. T2-Weighted and Fluid Attenuated Inversion Recovery (T2W/FLAIR) imaging: In the context of tumor imaging, T2W/FLAIR imaging provides information that generally relates to tissue water content. The T2W signal or brightness is typically qualitatively assessed relative to reference tissues such as normal white matter and gray matter. Particularly notable in low grade gliomas, bulk tumor typically demonstrates T2W signal that is brighter than white matter, and may be similar to slightly brighter than gray matter. In high-grade tumors that demonstrate BBB disruption, there often exists an even brighter, more peripheral “peritumoral” region of T2W signal, surrounding the contrast enhancing volume, comprised predominantly of non-tumoral vasogenic edema. This presumably results from hydrostatic efflux through highly permeable vessel walls, with fluid accumulation within the extravascular extracellular space. This fluid often tracts along white matter and spares cortical gray matter, resulting in a classic “finger-like” pattern on imaging (Figure 1).
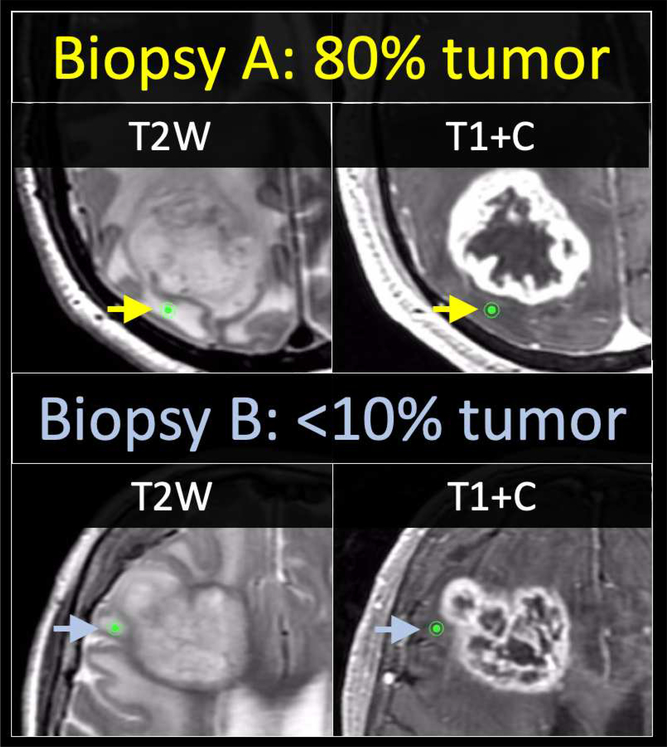
Figure 1: Two separate treatment naive GBM patients undergoing image-guided biopsy within the non-enhancing vasogenic edema.
The location of biopsy A (top row, green dot) in patient A is shown to originate from within the T2 hyperintense region outside of the T1+C enhancing volume. This biopsy revealed 80% tumor burden at the time of histologic analysis. The location of biopsy B (bottom row, green dot) in patient B is also seen to originate from within the T2 hyperintense region outside of T1+C enhancement. While the imaging appearance appears identical to the previous patient case, biopsy B showed predominance of non-tumoral edematous brain, with a minimal amount (<10%) of tumor. The location of biopsy B (bottom row, green dot) in patient B is also seen to originate from within the T2 hyperintense region outside of T1+C enhancement. While the imaging appearance appears identical to the previous patient case, biopsy B showed predominance of non-tumoral edematous brain, with a minimal amount (<10%) of tumor.
Persisting challenges in imaging-based diagnosis and treatment planning
The intratumoral heterogeneity of HGGs accounts for many persisting clinical challenges in diagnosis and treatment planning. For instance, while T1+C enhancement (i.e., BBB disruption) has classically served as a surrogate of tumor burden, mounting evidence suggests that MRI enhancement generally underestimates true tumor burden, particularly during first-line treatments, because of heterogeneity in the imaging phenotype of these tumors. Namely, there are substantial portions of each tumor that can express preserved BBB and therefore lack enhancement.6–12 These non-enhancing tumor populations may be undertreated by surgery and radiation, which contributes to recurrent disease and poor clinical outcomes. Even in completely non-enhancing gliomas, the lack of MRI enhancing regions precludes the use of MRI enhancement for conventional surgical targeting of high-grade components. This creates challenges because low-grade and high-grade components of tumor can demonstrate an identical appearance on T2W/FLAIR imaging.13–15 Studies using post-contrast T2W/FLAIR imaging suggest increased conspicuity of areas of BBB disruption following contrast administration, suggesting improved delineation of tumor extent. 16,17 However, further studies employing spatially localized histologic correlations are likely needed to validate these assertions. This also does not address the lack of specificity of BBB disruption for distinguishing different histologic entities (e.g., tumor recurrence from post-treatment effects).
Non-contrast enhanced T2/FLAIR abnormalities surrounding areas of T1+C enhancement have also been used to define extent of disease. Yet, despite the predominance of non-tumoral vasogenic edema within this peritumoral region, there can also exist a variable degree of infiltrative and/or bulk tumor that can demonstrate an identical T2 signal appearance to that of non-tumoral edema (Figure 1). Because visual qualitative assessment lacks the specificity to resolve these tumor populations, dosimetric radiation planning must assume that the entire peritumoral region is at risk for containing undetected, non-enhancing tumor. As a result, this region typically receives uniform sub-maximal radiation doses, which would unnecessarily expose normal non-tumoral brain to radiation, while potentially undertreating the non-enhancing infiltrative/bulk tumor populations. Developing better methods to detect non-enhancing tumor, and to distinguish these regions from non-tumoral vasogenic edema, would improve radiation dosimetric strategies to minimize exposure risk to normal brain, while optimizing therapeutic doses to non-enhancing tumor. In regards to image-based response assessment, there are also challenges to using T1+C MRI enhancement as a marker of recurrent tumor burden. Namely, MRI enhancement fails to distinguish non-tumoral post-treatment related inflammation - namely pseudoprogression (PsP) and radiation necrosis(RN) - from tumor recurrence.18,19 While non-tumoral related changes represent a positive response to treatment and a good prognosis, their identical appearance to tumor regrowth can misguide treatment decisions.18,19 Intralesional heterogeneity and admixture between tumor and post-treatment changes can also impact surgical targeting when histologic confirmation of recurrent disease is needed.20,21
Goals of integrating advanced imaging for improved diagnosis and treatment planning
The integration of advanced imaging techniques can help address a variety of unmet clinical challenges in neuro-oncology. These include: 1) Increasing the specificity of biopsy targeting for differentiating high-grade vs. low-grade glioma regions - particularly in non-enhancing gliomas lacking conventional MRI targets for high-grade tumor; 2) Increasing the specificity for differentiating regions of HGG vs. non-tumoral edema within the non-enhancing T2W/FLAIR - to better define extent of tumor beyond the MRI enhancing component; 3) Increasing the specificity for differentiating regions of HGG recurrence vs. post-treatment radiation effects (e.g., PsP, RN) within MRI enhancing lesions that are used for response assessment following standard chemo-radiation therapy. In subsequent sections of this paper, we introduce the major advanced imaging techniques, and their clinical applications in relation to intratumoral heterogeneity. We focus on the most published and clinically accessible techniques that are commonly available for most imaging practices. These include Diffusion Weighted Imaging (DWI), Diffusion Tensor Imaging (DTI), Dynamic Susceptibility Contrast (DSC)-MRI, Dynamic Contrast Enhanced (DCE)-MRI, MR Spectroscopy (MRS), as well as Positron Emission tomography (PET). We also discuss some of the advanced methods of imaging analysis, including texture analysis, machine learning, and mechanistic modeling, which have been used to develop predictive multi-parametric image-based models, as part of the emerging field of radiomics/radiogenomics. In particular, we discuss the role of radiogenomics in resolving the intratumoral genetic heterogeneity of HGG, and how this can potentially augment the paradigm of individualized oncology.
Dynamic Susceptibility Contrast (DSC) Perfusion MRI
The DSC-MRI technique measures the transient decrease in brain signal intensity - most commonly on T2*W images - after the intravenous bolus administration of gadolinium-based contrast agent (GBCA) to compute relative cerebral blood volume (rCBV) for all image voxels. Measures of rCBV correlate directly with microvessel volume,22,23 which remains distinct from the information on BBB integrity (and vessel leakage) from T1+C images. In other words, rCBV can be measured for all parts of the tumor and surrounding brain, irrespective of enhancement or non-enhancement on T1+C images. In general, high-grade angiogenic tumors (e.g., GBM, anaplastic astrocytomas) exhibit higher microvessel volume and corresponding higher rCBV on DSC-MRI compared to low-grade tumors, normal brain, and non-tumoral processes like inflammation and post-treatment effect. This fundamental principle underlies a number of clinically impactful applications in neuro-oncology, which include distinction between high-grade and low-grade gliomas,24–26,14,27 prediction of malignant degeneration of low-grade tumors28, distinction of tumor recurrence from non-tumoral post-treatment effects (e.g., pseudoprogression, radiation necrosis), quantification of tumor cell density and extent12,29–31,12,29–31, prediction of prognosis and overall survival 27,32–35, and non-invasive characterization of molecular/genomic profiles.36–40
Dynamic Contrast Enhanced (DCE) Perfusion MRI
The DCE-MRI technique measures the dynamic signal increases on T1W images after intravenous GBCA administration to measure vascular characteristics, most notably vessel permeability, through the metric k-trans. While conventional T1+C images detect whether vascular leakage (from BBB disruption) is present, DCE measures how quickly the leakage occurs. Pharmacokinetic modeling (commonly employing a 2-compartment approach) is used to calculate k-trans as a quantitative measure of vascular permeability (from BBB disruption).41 This provides a complement to the qualitative assessment of BBB disruption on T1+C images, and has been correlated with tumor grade and response assessment.42–44 However, compared to DSC-MRI (which can be applied to both non-enhancing and enhancing tumor), the use of DCE-MRI is limited primarily to the enhancing tumor segment, due to the dependence on BBB disruption and vascular leakage. As such, application of DCE-MRI for assessing the non-enhancing tumor components remains limited. In clinical practice, DSC-MRI represents the most common method for assessing tumor perfusion characteristics.45
Diffusion-weighted imaging (DWI)
DWI measures the random microscopic (Brownian) motion of water molecules through the application of dephasing and rephasing gradients (in 3 orthogonal planes) during MRI acquisition. Retention or loss of tissue signal - which relates to the bulk movement of water molecules - is used to calculate quantitative diffusion metrics, typically either apparent diffusion coefficient (ADC) or mean diffusivity (MD).41 In the context of tumor imaging, restricted diffusion (i.e., low ADC and/or low MD) has been shown by some groups to correlate with high tumor cellularity, presumably due to high cellular packing relative to low volume of extracellular water.46,47 However, other studies have reported contradictory correlations, with high diffusion metrics corresponding to high tumor cellularity.30,48 As such, these studies in total introduce uncertainty as to how DWI metrics should be used to quantify tumor cellularity. Also, a number of other pathophysiologic processes (e.g., infarct, hemorrhage, necrosis, infection, post-treatment change) can also manifest as restricted diffusion, which can further confound interpretation and applicability.41
Diffusion Tensor Imaging (DTI)
DTI (like DWI) measures microscopic water diffusion in tissue, but does so in a greater number of orthogonal planes compared to DWI. Specifically, at least 6 planes (or directions) are required by DTI, to allow the use of a mathematical second order diffusion tensor to calculate the directionality (or anisotropy) of water diffusion, most commonly summarized through the metric fractional anisotropy (FA).49 While isotropic diffusion refers to the equal, unconstrained motion of water in all directions, anisotropic diffusion refers to directionally dependent movement of water, which will be relatively more restricted perpendicular to (rather than parallel to) specific microstructural boundaries. 50 In the brain, application of DTI and FA values relate to the inherent, directionally-dependent flow of water molecules along normal white matter fiber tracts. The white matter tracts, as represented through FA values, can be deviated or disrupted by tumor, depending on growth patterns and local infiltration.51 A number of studies have compared FA values with image-localized biopsies to identify imaging correlates of regional tumor quantity and extent of invasion.8,29,48,52
Proton (H1) MR Spectroscopy (MRS)
(H1) MRS measures specific tissue metabolites that can inform of underlying pathophysiologic and molecular processes. For instance, choline [Cho]-containing compounds are primarily located within cell membranes, with elevated [Cho] levels (relative to internal controls) suggesting high cell membrane turnover, indicative of cellular proliferation and tumor growth. Lactate [Lac] indicates the presence of tissue necrosis, which can relate to either tumoral necrosis or post-treatment effect (i.e., radiation necrosis). Despite the clinical utility of MRS, its use in the setting of evaluating intratumoral heterogeneity can be limited by the requirement of large volumes of interest (ideally ~ 1cc) to provide adequate signal to noise ratio (SNR) for the metabolic spectra.53 Even with multi-voxel techniques that can improve spatial resolution, evaluation of intratumoral heterogeneity can be challenging, such as distinguishing admixed regions of tumor recurrence and post-treatment effect.54 Despite this limitation, MRS remains highly clinically applicable for evaluating more homogeneous conditions and disease states. For instance, recent advances in MRS have enabled non-invasive detection of molecular signatures, specifically mutations in Isocitrate dehydrogenase (IDH), which are presumably homogeneously expressed across the entire tumor. This has been accomplished through detection of increased levels of the metabolite 2-hydroxyglutarate (2HG), which is elevated in the presence of IDH-mutant tumors, but normal in IDH-wildtype tumors.55,56
Positron Emission Tomography (PET)
PET imaging is a nuclear medicine method that detects the anatomic distribution of radiolabeled molecules based on tissue-specific metabolic activity and/or accumulation. PET imaging requires the injection of a particular metabolic substrate (e.g., amino acid, glucose analog) which has been linked with a positron-emitting isotope (e.g.,11C, 18F). The utility of PET imaging for neuro-oncology is impacted by the choice of metabolic substrate as well as the PET radioisotope. For instance, 11C-MET (11C-methyl-L-methionine) has been widely studied in neuro-oncology for its increased uptake in brain tumors; however, the short half-life of 11C (~20 min) has restricted its use to only medical centers with an onsite cyclotron.57 For those reasons, 18F-based radiotracers have gained greater clinical adoption due to the longer half-life (~110 min), which increases clinical feasibility. 18F-FDG (2–18F-fluorodeoxyglucose) is the most widely used clinical PET method for evaluation of neoplasms outside of the CNS; however, 18F-FDG has limited applicability to detect uptake in brain tumors due to the high baseline levels of glucose metabolism in normal brain.57 Radiotracers that combine 18F with amino acid analogs have to date shown the greatest utility in neuro-oncology, including 18F-FDOPA (3,4-dihydroxy-6–18F fluoro-L-phenylalanine) and 18F-FET (O-(2–18F-fluoroethyl)- L-tyrosine).7,58 The advantages of these radiotracers include: 1) high accumulation within tumor cells, which has been linked in part to plasma membrane transporters such as LAT1;59 low background activity in normal brain, which facilitates detection of tumoral tissue on imaging;60 and 3) the ability to cross an intact BBB, which aids evaluation of non-enhancing tumor.61 While radiotracers such as 18F-FLT (18F-3’-deoxy-3’-fluorothymidine) have shown high correlation with tumor indices such as proliferation and grade,62 18F-FLT has limited applicability for defining the extent of non-enhancing tumor due to the inability to cross an intact BBB.63 While PET requires separate image acquisition beyond that for MRI, the additional complementary information on tumor metabolism likely provides an important adjunct to routine clinical imaging.
MRI image Texture Analysis
Classically, correlative studies comparing imaging features with tissue benchmarks (e.g., histologic, molecular/genetic) have employed either qualitative descriptions (e.g., presence or absence of MRI enhancement), or have compared individual quantitative metrics (e.g., rCBV, ADC) in univariate fashion. This has yielded robust correlations for certain clinical scenarios, such as the use of DSC-MRI to distinguish tumor from post-treatment effects (e.g., pseudoprogresion, radiation necrosis).64,65 Yet, for other scenarios, such as the prediction of genetic status, the imaging-tissue correlations may not be readily evident by qualitative visual inspection, or may not be adequately represented by simple statistical features (e.g., mean, standard deviation). This has motivated imaging researchers to extract more sophisticated quantitative imaging metrics through techniques such as texture analysis. Specifically, MRI spatially encodes signal intensity values for all voxels comprising each image. The textural patterns between voxel intensities and their surrounding neighbors can provide further insight to tissue microstructure and phenotypic heterogeneity within the local environment.12,37
Some common texture algorithms include Gray Level Co-Occurrence Matrix (GLCM), Local Binary Patterns (LBP), and Gabor features (GF). Gray Level Co-Occurrence Matrix (GLCM) provides detailed gray scale data by describing the angular relationships and distances between neighboring image voxels with similar gray scale intensities.66 Commonly used in texture analysis, GLCM uses second order statistics of the distribution of gray-scale intensity level within a region of interest (ROI). Each element in the co-occurrence matrix shows how often a pair of intensity levels is seen in a configuration defined by a certain radius and angle. Local Binary Patterns (LBP) provides highly discriminatory rotational and illumination invariant structural information by labeling each image voxel (in binary fashion) as higher or lower intensity compared with neighboring voxels.67 Highly cited as a method for texture description, LBP evaluates the intensity distribution of the set of points within a certain radius of each voxel in an ROI. Gabor features (GF), originally introduced by Dennis Gabor 68, are generated through a linear filter used for texture analysis. The technique employs multiplication of a cosine/sine wave with a Gaussian window to identify specific frequencies in image voxel patterns across a specific direction within an image. The methodology has been extended to allow 2-dimensional (2-D) and 3-dimensional (3-D) measurements.69,70
Image-based modeling approaches
In this section, we introduce common data-driven Machine-Learning (ML) approaches that support radiomics/radiogenomics predictive modeling of intratumoral heterogeneity. We also describe alternative mechanistic modeling methods that can complement data-driven ML.
Machine-Learning (ML) models
With advances in imaging technology and molecular/genetic profiling techniques, the complexity of clinical data will continue to rise. Integrating these complementary sources of data often requires advanced modeling approaches (compared to more simplistic univariate statistical correlations). Machine Learning (ML) represents a data-driven approach to identifying meaningful patterns and correlations from often complex data sources. ML models use training data as inputs for the model to “learn” the patterns or associations between those data, and subsequently predict on new instances using the learned patterns from model training. The use of ML algorithms has enabled the emerging field of radiogenomics, which utilizes inputs from image features (often from texture analysis) and genetic profiles (e.g., EGFR amplification status, PTEN deletion status), to train predictive models that inform of genetic status using the image features alone. This same concept can be applied to predict other clinical scenarios by modifying the inputs and re-defining the predicted outputs (e.g., using MRI features to predict tumor cell density). To build the input-output relationships, many ML algorithms can be potentially used. Here, we have highlighted several common choices.
Support Vector Machines (SVM)
SVM was originally developed as a binary classification model, which constructs a hyperplane in a high- or infinite-dimensional feature space to separate samples belonging to two classes71. SVM exploits a kernel approach such that samples do not actually need to be projected to the high- or infinite-dimensional feature space but the computation can be done in the original feature space. Different kernels can be chosen for optimizing performance. SVM has been extended for multi-class classification and regression problems. SVM can identify both linear and non-linear relationships between image features and genetic/cellular markers, depending on the choice of the kernel.
Tree ensembles
This usually refers to a class of methods based on trees, such as decision tree72, bagging73, boosting74, and random forest.75 A decision tree is constructed by recursively partitioning the sample space using image features. The feature used for each partition and the splitting point of the feature for partitioning the present tree into a left and a right branch are determined by optimizing a criterion such as the Gini index.76 Because using a single tree is ambitious and has the risk of missing the optimal predictive model, more advanced methods introduce randomness through bootstrapping the samples or using subspaces of features for improved performance such as bagging, boosting, and random forest. Because more than one predictive model is built in these methods, there is an ensemble of models for which the predictive result for a sample needs to be combined by approaches such as majority vote. Tree ensembles are non-linear models and provide greater interpretability for the reasoning process of how the prediction result is reached through involving a series of features.
Sparse regressions
This refers to a class of regression-based models that are particularly crafted to handle high-dimensional image features. Different from ordinary regression, sparse regression models include penalties of various forms on the regression coefficients to reduce the model complexity. The most well-known model is lasso77, which uses a L1-penalty whose effect is to make small coefficients to be exactly zero and therefore to only keep coefficients representing strong relationship between image features and the genetic/cellular marker. Other penalties such as L2 and L21 are also commonly used, leading to popular models such as elastic net78, fused lasso79, and graph-regularized lasso.80
Mechanistic modelling
Mechanistic models offer a valuable complement to the data-driven approach of machine learning (ML), by providing biological principles and constraints that can improve clinical interpretability. In contrast to ML approaches, mechanistic models begin by making assumptions about the physical system of interest, writing equations capturing these assumptions, and then fitting parameters of these equations to the observed system. Mechanistic models have been used to study gliomas since the mid 1990s. The most dominant model, still in use today, captures the diffuse infiltration and proliferation of the glioma cells and is referred to as the Proliferation-Invasion (PI) model.81,82 This is a relatively simple partial differential equation model with two key parameters D, the diffusion rate, and rho, the proliferation rate. The solution of this model is a traveling wave that travels at some velocity and with some slope, both related to the two key parameters. By working in spherical symmetry and assuming the T1+C abnormality represents regions exhibiting 80% of the tumor cell carrying capacity and above and the T2/FLAIR regions correspond to 16% and above, one can parameterize this model in a patient-specific way given just the standard imaging. These image-based growth kinetics, and the intuition they provide, have proven statistically significantly prognostic in many ways including survival, response to radiation, benefit from resection, and defining response metrics.
Specific clinical challenges that arise from intratumoral heterogeneity (and how imaging can address these challenges)
We review the pertinent literature and focus on those published studies employing image-localized biopsies to resolve the spatial intratumoral heterogeneity of glioma.
Differentiating high-grade vs. low-grade glioma regions in non-enhancing gliomas
Non-enhancing gliomas (i.e., those lacking BBB disruption) present unique challenges for surgical targeting during diagnostic biopsy. While non-enhancement generally suggests low-grade tumor, approximately one-third of nonenhancing gliomas are in fact malignant.13–15 Due to intratumoral heterogeneity, these high-grade components can actually co-exist with low-grade tumor within the same non-enhancing lesion. And lack of a T1+C enhancing target makes it difficult to reliably localize those high-grade components using conventional MRI alone. All of these factors can contribute to a reported 30% incidence of sampling error and misdiagnosis (i.e., undergrading) of non-enhancing high-grade gliomas.83 To address this, Maia et al. compared DSC-MRI measures of rCBV with histologic grade from image-localized biopsies in a cohort of non-enhancing gliomas, including multiple biopsies from the same tumor.14 They found that low-grade diffuse astrocytomas exhibited significantly lower rCBV compared to high-grade anaplastic astrocytomas, which they later showed to relate to increased VEGF expression 84 They also found that other types of low-grade gliomas (e.g., oligodendrogliomas) can demonstrate mildly higher rCBV compared with low-grade astrocytomas, but not as high as anaplastic astrocytomas. They concluded that DSC-MRI provides a clinically valuable method for guiding surgical targeting to identify potential high-grade components in non-enhancing gliomas. These findings were supported in a prospective study by Chaskis et al, in which they used DSC-MRI rCBV maps to assist MRI-guided biopsies in 55 glioma patients.83 They found that rCBV improved target selection, independently from T1+C enhancement, for discriminating high-grade from low-grade gliomas. Figure 2 shows an example how DSC-MRI rCBV maps, superimposed intraoperatively with conventional MRI, can improve the diagnostic yield and recovery of high-grade tumor, from otherwise non-specific, non-enhancing gliomas. DSC-MRI measures of rCBV have been shown to demonstrate malignant transformation from low-grade to high-grade tumor up to 1 year prior to the development of contrast enhancement.28 Pöpperl et al. evaluated 18F-FET PET imaging in 54 glioma cases using image-guided biopsies to differentiate tumors based on histologic grade. The cohort included both enhancing and non-enhancing tumors. While maximum standardized uptake value (SUV) showed adequate accuracy for differentiating low-grade from high-grade tumors (sensitivty 71%, specificity 85%), they found that evaluation of dynamic uptake values could improve diagnostic performance (sensitivity 94%, specificity 100%).58
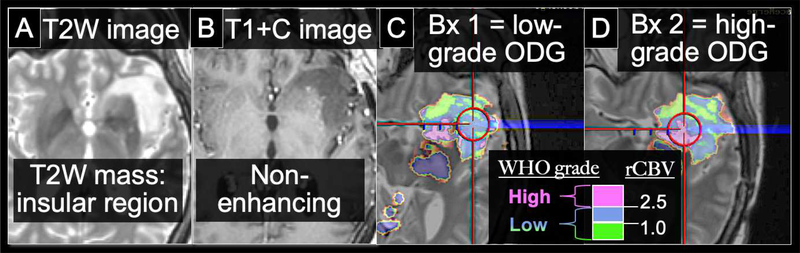
Figure 2: 40 y/o male with 2 separate biopsies for a mass suspicious for low-grade glioma.
The suspected low-grade glioma appears as a (A) T2W hyperintense, expansile mass in the left insular region with (B) no appreciable enhancing focus,. (C, D) On DSC-MRI rCBV maps that have been thresholded and color-coded, pink regions indicate high rCBV above 2.5. Green indicates low rCBV below 1.0, and Blue indicates moderate rCBV between 1 to 2.5. Two separate biopsies were taken from the patient’s tumor. Biopsy #1 within the lesion (C) was taken from a moderate rCBV region (blue) and yielded low grade (Grade 2) Oligodendroglioma on histopathology. Biopsy #2, from a high rCBV region (pink), revealed a high-grade (Grade 3) component with MIB-1 of 19%, consistent with elevated proliferative indices. The rCBV threshold of 2.5 remains consistent with the study by Maia et al. (reference #15) to separate high- vs. low-grade oligodendrogliomas.
Differentiating regions of HGG from non-tumoral edema and defining tumor extent beyond regions of MRI enhancement
Non-enhancing tumor can account for a substantial proportion of overall tumor burden for many GBM tumors. Because surgical resection favors T1+C enhancing volume, and because T2W imaging lacks the capability to distinguish infiltrating tumor from vasogenic edema, the non-enhancing tumor component is often left unresected after surgery and submaximally dosed by radiation therapy. Localizing the regional extent and relative abundance of non-enhancing tumor could help refine image-guided targeting and treatment. These issues underscore the motivation for many groups to develop better image-based methods that can quantify and optimally treat the non-enhancing tumor segment of GBM.
Multiple independent studies have correlated various advanced MRI metrics with regional tumor cell density using image-localized biopsies from the enhancing and non-enhancing components of glial tumors. Across multiple independent studies, the most consistent correlations have been observed with DSC-MRI. Measures of rCBV have shown consistently low-to-moderate positive correlations between rCBV and tumor cell density across 4 different image-localized biopsy studies.29–31,85 For DTI, the reported correlations have been slightly more variable. Two separate studies reported strong negative correlations between anisotropic diffusion (i.e., FA) and regional tumor cell density from image-localized biopsies, suggesting that tumor growth disrupts the integrity of white matter fiber tracts.48,86 However, a third study reported a strongly positive correlation between FA and regional tumor cell density, which contradicts the previous studies.52 Discrepancies have also been observed with reported DWI correlations, which have been split between negative and positive correlations between ADC and/or MD and regional tumor cell density from corresponding image-localized biopsies.30,46–48 Based on these studies, if choosing one imaging technique, DSC-MRI measures of rCBV appear to provide the most consistent correlation with tumor cell density. However, this correlation is low-to-moderate at best, which suggests the limitations in relying on a single imaging metric to reliably predict regional tumor cell content.
Recognizing this gap, several groups have developed predictive models that incorporate multiple complementary imaging features, in combination, to improve the predictive performance for quantifying regional tumor cell density. Hu et al. employed multi-parametric MRI (including rCBV, MD, FA) and texture analysis to train an ML model that predicted the binary classification of high- vs. low-tumor content (≥80% vs. <80% tumor) within corresponding image-localized biopsies. The model achieved 85% cross-validation accuracy, and 82% accuracy in a separate validation set.12 Other groups have developed ML models to predict tumor cell density on a continuous scale, rather than using a binary approach. Durst et al. employed an ML model that incorporated multiple MRI contrasts (including rCBV, MD, FA) and achieved strong correlations (r=0.75) in the training set between predicted and actual tumor cell density from image-localized biopsies. Similarly, Chang et. al reported a multi-parametric model (including ADC) that also achieved strong correlation (r=0.74) in the training set.
Gaw et al. demonstrated the feasibility of incorporating mechanistic models with machine-learning (ML) to improve the biological interpretability and predictive performance of ML-based models.87 They developed a hybrid model integrating ML and the mechanistic (PI) model, using image-localized biopsies in the 18 patient GBM cohort from Hu et al.12 This hybrid ML-PI model improved the predictive performance (r=0.84) for quantifying tumor cell density (as a continuous variable) compared to ML alone (r=0.52). The strength of ML derives from its ability to integrate complex arrays of MRI and tissue data (including patient-specific histologic inputs) to inform model development.12,29 This approach is strengthened further by using spatially-matched MRI and image-localized biopsies as inputs for model training. Yet, ML is inherently limited in the ability to generalize beyond these localized data inputs. As such, integration of mechanistic modeling approaches would help to integrate the spatial relationships of neighboring biopsies or to generalize predicted outputs between biopsy locations, by providing constraints from grounded biological principles. This can also beneficially affect the feasibility of predicted outputs (e.g., tumor growth/invasion along brain edges). The mechanistic PI approach offers biological inferences and spatial contiguity that ideally complement data-driven ML.
The aforementioned studies have all taken a “one-model-fits-all” approach that uses a training cohort of patient data to develop a single model, which would then be applied uniformly to all future prospective patients. Such models would lack the capability to account or adjust for potential interpatient variabilities in image-tissue correlations, which could degrade the model fit for outlier cases. To address this potential confound, Hu et al. developed an ML model using Transfer Learning to further improve predictive performance.29 This approach uses training cohort data to develop a template model, which can then be transferred and optimized for each individual patient using their own MRI and biopsy data. Following cross validation, the model significantly improved the correlation (r=0.88) between predicted and actual tumor cell density across the entire cohort of image-localized biopsies (n=82), compared to the one-model-fits-all approach. Interestingly, this correlation increased further (r=0.94) when focussing on only those biopsies from the non-enhancing invasive edge (n=33). This work offers proof of concept for developing individualized patient-specific models to optimize the predictive performance for image-based applications such as dosimetric radiation treatment planning.
PET imaging has also shown utility in identifying the presence of HGG tumor and helping to define tumor extent beyond MRI enhancement. For this application, amino acid radiotracers have shown the greatest utility, due to the ability to cross an intact BBB. For instance, Pafundi et al. evaluated 18F-FDOPA PET imaging and MRI in 10 glioma patients (23 image-localized biopsies). They compared SUV of 18F-FDOPA within T2W/FLAIR abnormal regions, beyond MRI enhancement, and found strong correlation with both the presence of tumor and histologic features of aggressiveness (e.g., cell density, proliferative indices).7 Stockhammer et al. evaluated 22 non-enhancing glioma patients using 18F-FET PET imaging 60. They found that cell density and vascular density correlated strongly with 18F-FET uptake, despite lack of MRI enhancement.
Differentiating regions of HGG recurrence from post-treatment radiation effects (e.g., PsP, RN) for image-based response assessment
Conventional contrast-enhanced MRI guides response assessment for essentially all HGG patients and clinical trials worldwide. Specifically, the T1+C enhancing volume is used as a surrogate of recurrent tumor burden. However, there is an increasing awareness of non-tumoral post-treatment radiation effects (PTRE) – namely pseudoprogression and radiation necrosis – that can exactly mimic tumor recurrence on conventional MRI.19,41,88,89 While tumor recurrence signals treatment failure, PTRE represents a positive response to treatment with a good prognosis. This distinction can be further complicated by the histologic admixture between HGG tumor and PTRE, which can impact both diagnosis and prognostication, depending on the relative histologic burden of each entity.54,65,90,91 And in the case of surgical biopsy, regional heterogeneity and resulting sampling errors can negatively affect the diagnostic confirmation of tumor vs. PTRE, as well as the adequacy of tumor content for molecular and genomic profiling.3 These issues underscore the importance of improving image-based response assessment, particularly in the context of intratumoral heterogeneity.
Over the past decade, DSC-MRI has emerged as a clinically valuable and accessible tool to distinguish tumor recurrence from PTRE within otherwise non-specific T1+C enhancing lesions.92 A meta-analysis by Patel et al. has highlighted a continually expanding body of literature showing consistently higher rCBV values in HGG tumor compared to PTRE.21,41,92–94,95 Yet, variability in reported rCBV thresholds across different studies can generate confusion about prospective clinical guidelines, which underscores the need to not only standardize DSC-MRI methodology, but to also employ spatially accurate validation methods that can address the confounds of intratumoral histologic heterogeneity. First, not all studies employ histologic validation, as some rely instead on serial imaging (in lieu of histology) to classify MRI lesions as tumor recurrence (serially enlarging) vs. post-treatment effect (serially regressing).96,97 This approach has pitfalls given that post-treatment effects can serially enlarge over time (mimicking growth of tumor), and even regressing lesions can contain components of indolent tumor. Second, across those studies employing histologic validation, rCBV thresholds could be affected by the variability in histologic criteria used to define HGG tumor vs post-treatment effect. For instance, some studies have required that surgical samples contain at least 20% tumor to be classified as tumor 98, while other studies have used lower histologic thresholds.64,65,93,99 Finally, most studies have employed non-localizing methods of image correlation, which could promote tissue sampling error and potential discrepancies between the biopsy location and the analyzed portions of the MRI enhancing lesion.92
To overcome the challenges of intratumoral heterogeneity and tissue sampling error, Hu et al. undertook a series of studies employing image-localized biopsies and spatially matched DSC-MRI to identify an rCBV threshold that could accurately separate HGG tumor samples from those with PTRE.22,64,65,99 They found that the rCBV threshold of 1.0 (when normalized against contralateral white matter) could separate HGG recurrence from post-treatment changes with 96% accuracy (100% specificity, 92% sensitivity).64 This accuracy improved further when evaluating only patients with GBM.65 Moreover, this rCBV threshold could be used to classify each image voxel as containing either tumor or PTRE, which provides voxel-based maps of spatial distribution for regional recurrence vs. post-treatment change (Figure 3). Quantifying the abundance of tumor voxels, relative to PTRE voxels, defines the metric Fractional Tumor Burden (FTB), which correlates with histologic tumor burden and overall survival.65 This provides a clinically validated and accessible tool for response assessment, which has since been independently validated by Prah et al. using image-localized biopsies and spatially matched rCBV. 21
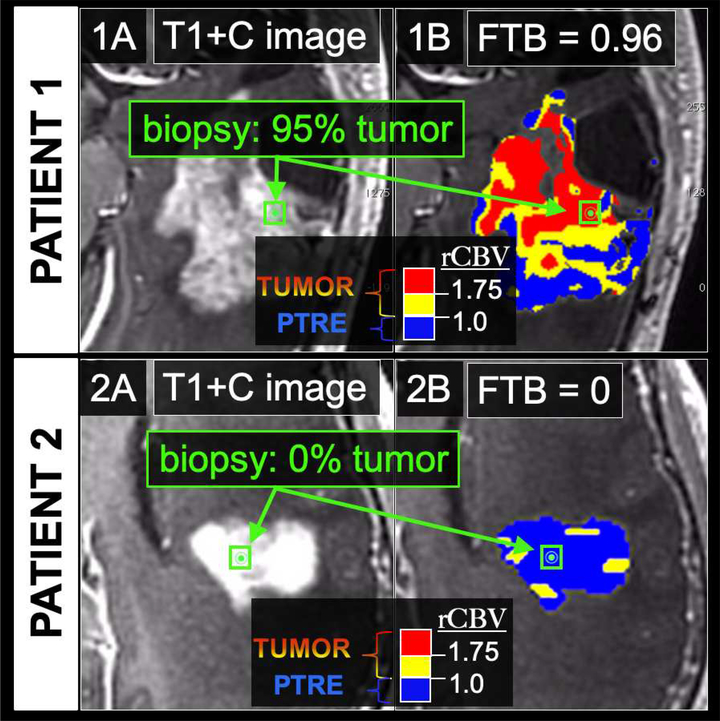
Figure 3: Two separate GBM patients status post standard adjuvant chemo-radiation therapy undergoing surgical biopsy for suspected recurrence.
In patient 1, (1A) T1+C images demonstrate a large heterogeneously enhancing mass concerning for tumor recurrence. The green dot depicts the stereotactic location of the biopsy specimen. (1B) On the coregistered FTB map, which is superimposed on the T1+C image, blue voxels correspond to predicted PTRE regions with low rCBV ≤ 1.0. The yellow (1.75 ≥ rCBV > 1.0) and red (rCBV > 1.75) voxels correspond to predicted tumor regions. The FTB metric is defined as the percentage of both yellow and red voxels relative to all voxels within the green ROI (green box) around the biopsy location (green dot). The FTB for the biopsy measured 0.96 (i.e., 96% of the voxels were predicted as tumor), which correlated with histologic quantification of 95% tumor from the spatially matched biopsy specimen. In patient 2, (2A) the T1+C image again demonstrates a large heterogeneous mass concerning for tumor recurrence. However, (2B) the FTB map shows an abundance of blue voxels consistent with predominant PTRE, with an FTB measurement of 0 (i.e., 0% of the voxels were predicted as tumor) within the ROI (green box) around the biopsy location (green dot). This correlated with the histologic findings of post-treatment effect, with no visible tumor within the spatially matched biopsy specimen.
Role of radiogenomics in resolving intratumoral genetic heterogeneity in HGG, and how this potentially augments the paradigm of individualized oncology
While the prognosis for GBM remains poor with standard treatment, genomic profiling offers the potential to improve outcomes through more personalized therapies.100 In particular, individualized oncology seeks to drive optimal treatment decisions based on each patient’s genetic diagnosis and the unique drug sensitivities of each tumor.101 Unfortunately, this potential benefit has yet to be realized in any meaningful way, due in large part to GBM’s profound intratumoral heterogeneity and the confounding issues of tissue sampling.102 Specifically, each GBM tumor actually comprises multiple genetically distinct subpopulations with differing sensitivities, such that genetic targets from one biopsy location may not accurately reflect those from other parts of the same tumor.103 Tissue sampling errors are magnified by the fact that surgical targeting favors MRI enhancing tumor components but leaves behind residual subpopulations within the non-enhancing tumor segment.37,104,105 Ironically, these uncharacterized residual subpopulations represent the primary targets of adjuvant therapy and the main source of recurrence, but may harbor genetic drug targets that remain “unknown”, even after gross total resection.86,106
The emerging field of radiogenomics has shown the feasibility of using MRI-based signatures to predict underlying genetic status, and the potential to apply these predictions to inform clinical decisions in the context of individualized oncology. As an example, radiogenomics models that can predict the amplification status of receptor tyrosine kinases (e.g., EGFR, PDGFRA) within the residual tumor segment could help stratify which patients would benefit from adjuvant targeted drug therapies, given the abundance of clinically tested and available inhibitors on market.101 Historically, most published radiogenomics studies have employed non-localizing correlations, which assume homogeneous expression of these genetic targets, whereby a single representative genetic profile and imaging signature are used to summarize the entire tumor as a whole.36,38,40,106–111 Unfortunately, these non-localizing studies remain incapable of resolving GBM’s intratumoral heterogeneity, particularly within the non-enhancing tumor populations, given that most biopsies, by clinical convention, originate from MRI enhancement.106,112 Further, MRI contrast-enhancement itself (whether on T1W or T2W/FLAIR imaging) due to BBB disruption inherently lacks the specificity to distinguish many of the genetically distinct tumor populations that can reside within a single tumor.37 This is due to the fact that genetic heterogeneity can exist even amongst different regional biopsy samples from the same MRI enhancing segment. This genetic heterogeneity has also been observed within the non-enhancing tumor segment.37
To address this challenge of intratumoral heterogeneity, Hu et al. undertook the first study of its kind using spatially resolved tissue sampling to develop predictive models of regional genetic heterogeneity in GBM. To accomplish this, they employed a combination of image-localized biopsies, spatially matched MRI and texture features, and machine-learning (ML) methodology. They identified MRI signatures at the voxel-level that could classify the status of key GBM driver genes throughout different regions within a single GBM tumor, including the non-enhancing invasive edge.37 Importantly, this offers a clinically viable solution for non-invasively diagnosing the potentially unique drug targets within the residual unresected tumor segment, which remains a persisting challenge due to the inability to routinely surgically sample this region (Figures 4 and 5). Barajas et. al also undertook an image-localized biopsy study, which confirmed that enhancing and non-enhancing tumor segments often express different gene expression and imaging signatures, further validating the importance of spatially resolving the unique genetic targets within the non-enhancing tumor segment.106
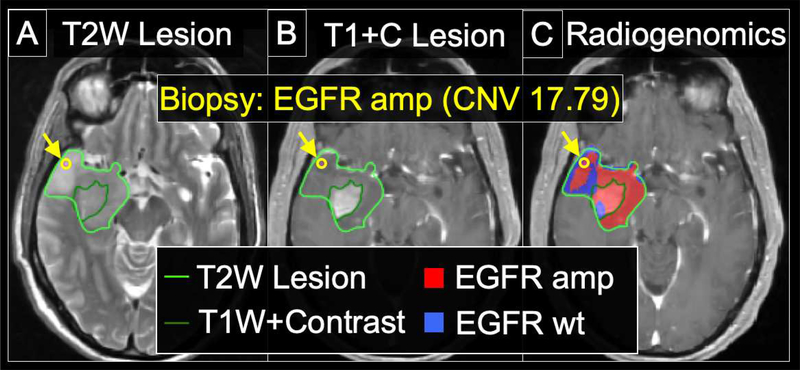
Figure 4: Radiogenomics map demarcating regions of amplification (amp) for Epidermal Growth Factor Receptor (EGFR) in a 63 y/o Male with primary GBM.
(A) The lesion is shown on the T2W image, with the margins demarcated by the bright green outline. The location of the biopsy (which was subsequently genetically profiled) is shown by the yellow arrow and yellow circle. (B) The central enhancing component is outlined by the dark green line on the T1+C image. The biopsy location (yellow arrow, yellow circle) is again shown, originating from the peripheral T2W non-enhancing component of tumor. (C) The radiogenomics map shows predicted regions of EGFR amplification (red) and non-amplified EGFR wildtype (wt) (blue), within the T2W region of interest (bright green outline) around the tumor. The radiogenomics map prediction of EGFR amplification (red) for the biopsy location corresponds with the elevated copy number variant (CNV) of 17.79, confirming EGFR amplification.
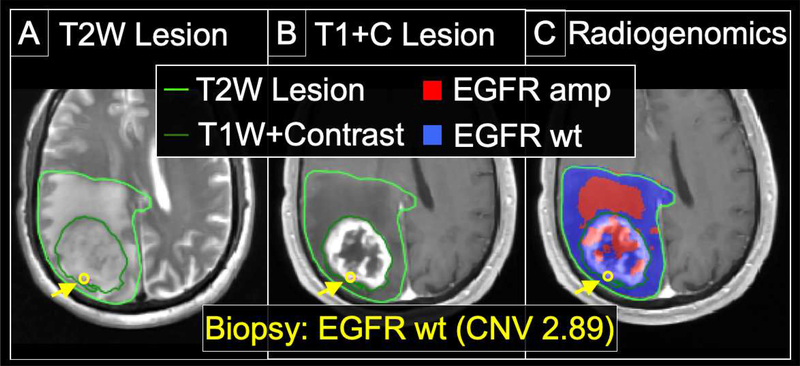
Figure 5: Radiogenomics map demarcating regions of amplification (amp) for Epidermal Growth Factor Receptor (EGFR) in a 67 y/o Male with primary GBM.
(A) The lesion is shown on the T2W image, with the margins demarcated by the bright green outline. The location of the biopsy (which was subsequently genetically profiled) is shown by the yellow arrow and yellow circle. (B) The central enhancing component is outlined by the dark green line on the T1+C image. The biopsy location (yellow arrow, yellow circle) is again shown, originating from the T1+C enhancing component of tumor. (C) The radiogenomics map shows predicted regions of EGFR amplification (red) and non-amplified wildtype (wt) (blue), throughout the entire T2W region of interest (bright green outline). The radiogenomics map prediction of EGFR wildtype (blue) for the biopsy location corresponds with the low copy number variant (CNV) of 2.89 for EGFR, consistent with absence of amplification.
Future directions and the path to improving patient care
Advanced imaging and image-based modeling offer a variety of impactful and clinically feasible solutions to address the challenges brought on by intratumoral heterogeneity in glioma. In the case of DSC-MRI, use of the metric rCBV has already shown far reaching and widely validated applications for surgical targeting and response assessment. Further progress will likely stem from continued work in developing consensus recommendations to standardize both image acquisition and post-processing methods across institutions. And for the still burgeoning field of radiogenomics, clinical assimilation of these predictive models will need to address issues of clinician confidence, to overcome the “black box” stigma of ML and other technologies stemming from Artificial Intelligence. Such efforts to address predictive uncertainty 113 and the biological interpretability 87 of model predictions will provide the needed transparency to facilitate their integration into clinical decision-making. And finally, work will be needed to identify ways to continually improve the standards of clinical care through the use of these advanced imaging
Highlights.
High-grade gliomas like Glioblastoma (GBM) can exhibit profound intratumoral heterogeneity that confounds accurate diagnosis and effective therapy
Conventional MRI routinely guides first-line treatments, like surgery and radiation, but remains incapable of characterizing intratumoral heterogeneity
Advanced imaging techniques, combined with spatially accurate validation methods, can help resolve intratumoral heterogeneity to improve diagnosis and treatment
DSC Perfusion MRI can help distinguish high-grade from low-grade components within otherwise non-specific non-enhancing gliomas
DSC Perfusion MRI can distinguish tumor recurrence from post-treatment effects and help quantify histologic burden of recurrent disease
Machine-learning models employing multi-parametric MRI can resolve the regional heterogeneity of tumor cell density and extent in GBM
Radiogenomics models can resolve the genetically distinct subpopulations and regional drug targets throughout each GBM tumor to guide emerging personalized therapeutic paradigms
Acknowledgments
Grant funding: Funding: NS082609, CA221938, CA220378, Mayo Clinic Foundation, James S. McDonnell Foundation, Ivy Foundation, Arizona Biomedical Research Commission
Footnotes
Potential Conflicts of Interest
US Patents: US8571844B2 (KRS)
US Patent Applications: 15/290,963 (LSH, JL); PCT/US2018/061887 (LSH, AHD, JL, KRS); PCT/US2019/019687 (LSH, JL, KRS)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ostrom QT et al. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-Oncology vol. 19 v1–v88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young JS, Prados MD & Butowski N Using genomics to guide treatment for glioblastoma. Pharmacogenomics 19, 1217–1229 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Prados MD et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro. Oncol. 17, 1051–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkaria JN et al. Is the blood-brain barrier really disrupted in all glioblastomas? - A critical assessment of existing clinical data. Neuro. Oncol. (2017) doi: 10.1093/neuonc/nox175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pafundi DH et al. Biopsy validation of 18F-DOPA PET and biodistribution in gliomas for neurosurgical planning and radiotherapy target delineation: results of a prospective pilot study. Neuro. Oncol. 15, 1058–1067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price SJ et al. Improved delineation of glioma margins and regions of infiltration with the use of diffusion tensor imaging: an image-guided biopsy study. AJNR Am. J. Neuroradiol. 27, 1969–1974 (2006). [PMC free article] [PubMed] [Google Scholar]
- 9.Berens ME & Giese A ‘...those left behind.’ Biology and oncology of invasive glioma cells. Neoplasia 1, 208–219 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldock AL et al. Patient-specific metrics of invasiveness reveal significant prognostic benefit of resection in a predictable subset of gliomas. PLoS One 9, e99057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corwin D et al. Toward patient-specific, biologically optimized radiation therapy plans for the treatment of glioblastoma. PLoS One 8, e79115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu LS et al. Multi-Parametric MRI and Texture Analysis to Visualize Spatial Histologic Heterogeneity and Tumor Extent in Glioblastoma. PLoS One 10, e0141506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott JN, Brasher PMA, Sevick RJ, Rewcastle NB & Forsyth PA How often are nonenhancing supratentorial gliomas malignant? A population study. Neurology 59, 947–949 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Maia ACM Jr, et al. Stereotactic biopsy guidance in adults with supratentorial nonenhancing gliomas: role of perfusion-weighted magnetic resonance imaging. J. Neurosurg. 101, 970–976 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Barker FG et al. Age and the risk of anaplasia in magnetic resonance-nonenhancing supratentorial cerebral tumors. Cancer: Interdisciplinary International Journal of the American Cancer Society 80, 936–941 (1997). [PubMed] [Google Scholar]
- 16.Lee EK, Lee EJ, Kim S & Lee YS Importance of Contrast-Enhanced Fluid-Attenuated Inversion Recovery Magnetic Resonance Imaging in Various Intracranial Pathologic Conditions. Korean J. Radiol. 17, 127–141 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubota T et al. Relationship between contrast enhancement on fluid-attenuated inversion recovery MR sequences and signal intensity on T2-weighted MR images: visual evaluation of brain tumors. J. Magn. Reson. Imaging 21, 694–700 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Brandes AA et al. Disease progression or pseudoprogression after concomitant radiochemotherapy treatment: pitfalls in neurooncology. Neuro. Oncol. 10, 361–367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke JL & Chang S Pseudoprogression and pseudoresponse: challenges in brain tumor imaging. Curr. Neurol. Neurosci. Rep. 9, 241–246 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Hu LS et al. Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro. Oncol. 14, 919–930 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prah MA et al. Spatial discrimination of glioblastoma and treatment effect with histologically-validated perfusion and diffusion magnetic resonance imaging metrics. J. Neurooncol. 136, 13–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu LS et al. Correlations between perfusion MR imaging cerebral blood volume, microvessel quantification, and clinical outcome using stereotactic analysis in recurrent high-grade glioma. AJNR Am. J. Neuroradiol. 33, 69–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathak AP et al. MR-derived cerebral blood volume maps: issues regarding histological validation and assessment of tumor angiogenesis. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 46, 735–747 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Aronen HJ et al. Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology 191, 41–51 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Boxerman JL, Schmainda KM & Weisskoff RM Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am. J. Neuroradiol. 27, 859–867 (2006). [PMC free article] [PubMed] [Google Scholar]
- 26.Knopp EA et al. Glial neoplasms: dynamic contrast-enhanced T2*-weighted MR imaging. Radiology 211, 791–798 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Law M et al. Low-grade gliomas: dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging—prediction of patient clinical response. Radiology 238, 658–667 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Danchaivijitr N et al. Low-grade gliomas: do changes in rCBV measurements at longitudinal perfusion-weighted MR imaging predict malignant transformation? Radiology 247, 170–178 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Hu LS et al. Accurate Patient-Specific Machine Learning Models of Glioblastoma Invasion Using Transfer Learning. AJNR Am. J. Neuroradiol. 40, 418–425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadeghi N et al. Apparent diffusion coefficient and cerebral blood volume in brain gliomas: relation to tumor cell density and tumor microvessel density based on stereotactic biopsies. AJNR Am. J. Neuroradiol. 29, 476–482 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barajas RF, Jr et al. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro. Oncol. 14, 942–954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmainda KM et al. Dynamic susceptibility contrast MRI measures of relative cerebral blood volume as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 multicenter trial. Neuro. Oncol. 17, 1148–1156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boxerman JL, Schmainda KM & Zhang Z Dynamic susceptibility contrast MRI measures of relative cerebral blood volume continue to show promise as an early response marker in the setting of bevacizumab …. Neuro (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Álvarez-Torres MDM et al. Robust association between vascular habitats and patient prognosis in glioblastoma: An international multicenter study. J. Magn. Reson. Imaging (2019) doi: 10.1002/jmri.26958. [DOI] [PubMed] [Google Scholar]
- 35.Boonzaier NR et al. Multiparametric MR Imaging of Diffusion and Perfusion in Contrast-enhancing and Nonenhancing Components in Patients with Glioblastoma. Radiology 284, 180–190 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Ryoo I et al. Cerebral blood volume calculated by dynamic susceptibility contrast-enhanced perfusion MR imaging: preliminary correlation study with glioblastoma genetic profiles. PLoS One 8, e71704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu LS et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro. Oncol. 19, 128–137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta A et al. Pretreatment Dynamic Susceptibility Contrast MRI Perfusion in Glioblastoma: Prediction of EGFR Gene Amplification. Clin. Neuroradiol. 25, 143–150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain R et al. Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology 267, 212–220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tykocinski ES et al. Use of magnetic perfusion-weighted imaging to determine epidermal growth factor receptor variant III expression in glioblastoma. Neuro. Oncol. 14, 613–623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiroishi MS, Boxerman JL & Pope WB Physiologic MRI for assessment of response to therapy and prognosis in glioblastoma. Neuro. Oncol. 18, 467–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Law M et al. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. AJNR Am. J. Neuroradiol. 25, 746–755 (2004). [PMC free article] [PubMed] [Google Scholar]
- 43.Bisdas S et al. Distinguishing recurrent high-grade gliomas from radiation injury: a pilot study using dynamic contrast-enhanced MR imaging. Acad. Radiol. 18, 575–583 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Nakamura H, Murakami R, Hirai T, Kitajima M & Yamashita Y Can MRI-derived factors predict the survival in glioblastoma patients treated with postoperative chemoradiation therapy? Acta radiol. 54, 214–220 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Essig M et al. Perfusion MRI: The Five Most Frequently Asked Technical Questions. American Journal of Roentgenology vol. 200 24–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellingson BM et al. Validation of functional diffusion maps (fDMs) as a biomarker for human glioma cellularity. J. Magn. Reson. Imaging 31, 538–548 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang PD et al. A Multiparametric Model for Mapping Cellularity in Glioblastoma Using Radiographically Localized Biopsies. AJNR Am. J. Neuroradiol. 38, 890–898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stadlbauer A et al. Gliomas: histopathologic evaluation of changes in directionality and magnitude of water diffusion at diffusion-tensor MR imaging. Radiology 240, 803–810 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Le Bihan D et al. Diffusion tensor imaging: concepts and applications. J. Magn. Reson. Imaging 13, 534–546 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Sundgren PC et al. Diffusion tensor imaging of the brain: review of clinical applications. Neuroradiology 46, 339–350 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Mori S et al. Brain white matter anatomy of tumor patients evaluated with diffusion tensor imaging. Ann. Neurol. 51, 377–380 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Beppu T et al. Fractional anisotropy value by diffusion tensor magnetic resonance imaging as a predictor of cell density and proliferation activity of glioblastomas. Surg. Neurol. 63, 56–61; discussion 61 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Peeling J & Sutherland G High-resolution 1H NMR spectroscopy studies of extracts of human cerebral neoplasms. Magn. Reson. Med. 24, 123–136 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Rock JP et al. Correlations between magnetic resonance spectroscopy and image-guided histopathology, with special attention to radiation necrosis. Neurosurgery 51, 912–9; discussion 919–20 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Choi C et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 18, 624–629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi C et al. Prospective Longitudinal Analysis of 2-Hydroxyglutarate Magnetic Resonance Spectroscopy Identifies Broad Clinical Utility for the Management of Patients With IDH-Mutant Glioma. J. Clin. Oncol. 34, 4030–4039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langen K-J, Galldiks N, Hattingen E & Shah NJ Advances in neuro-oncology imaging. Nat. Rev. Neurol. 13, 279–289 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Pöpperl G et al. FET PET for the evaluation of untreated gliomas: correlation of FET uptake and uptake kinetics with tumour grading. Eur. J. Nucl. Med. Mol. Imaging 34, 1933–1942 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Youland RS et al. The role of LAT1 in (18)F-DOPA uptake in malignant gliomas. J. Neurooncol. 111, 11–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stockhammer F, Plotkin M, Amthauer H, van Landeghem FKH & Woiciechowsky C Correlation of F-18-fluoro-ethyl-tyrosin uptake with vascular and cell density in non-contrast-enhancing gliomas. Journal of Neuro-Oncology vol. 88 205–210 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Rapp M et al. Diagnostic performance of 18F-FET PET in newly diagnosed cerebral lesions suggestive of glioma. J. Nucl. Med. 54, 229–235 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Price SJ et al. Imaging regional variation of cellular proliferation in gliomas using 3’-deoxy-3’-[18F]fluorothymidine positron-emission tomography: an image-guided biopsy study. Clin. Radiol. 64, 52–63 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Nowosielski M et al. An intra-individual comparison of MRI, [18F]-FET and [18F]-FLT PET in patients with high-grade gliomas. PLoS One 9, e95830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu LS et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. AJNR Am. J. Neuroradiol. 30, 552–558 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu LS et al. Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro. Oncol. 14, 919–930 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haralick RM, Shanmugam K & Dinstein I Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. SMC-3, 610–621 (1973). [Google Scholar]
- 67.Ojala T, Pietikainen M & Maenpaa T Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 24, 971–987 (2002). [Google Scholar]
- 68.Gabor D Theory of communication. Part 1: The analysis of information. Journal of the Institution of Electrical Engineers - Part III: Radio and Communication Engineering 93, 429–441 (1946). [Google Scholar]
- 69.Daugman JG Uncertainty relation for resolution in space, spatial frequency, and orientation optimized by two-dimensional visual cortical filters. J. Opt. Soc. Am. A 2, 1160–1169 (1985). [DOI] [PubMed] [Google Scholar]
- 70.Heeger DJ Model for the extraction of image flow. J. Opt. Soc. Am. A 4, 1455–1471 (1987). [DOI] [PubMed] [Google Scholar]
- 71.Boser BE, Guyon IM & Vapnik VN A Training Algorithm for Optimal Margin Classifiers in Proceedings of the Fifth Annual Workshop on Computational Learning Theory 144–152 (ACM, 1992). [Google Scholar]
- 72.Safavian SR & Landgrebe D A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 21, 660–674 (1991). [Google Scholar]
- 73.Bühlmann P & Yu B Analyzing bagging. Ann. Stat. 30, 927–961 (2002). [Google Scholar]
- 74.Freund Y, Schapire R & Abe N A short introduction to boosting. Journal-Japanese Society For Artificial Intelligence 14, 1612 (1999). [Google Scholar]
- 75.Liaw A, Wiener M & Others. Classification and regression by randomForest. R news 2, 18–22 (2002). [Google Scholar]
- 76.Lerman RI & Yitzhaki S A note on the calculation and interpretation of the Gini index. Econ. Lett. 15, 363–368 (1984). [Google Scholar]
- 77.Tibshirani R Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Series B Stat. Methodol. 58, 267–288 (1996). [Google Scholar]
- 78.Zou H & Hastie T Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 67, 301–320 (2005). [Google Scholar]
- 79.Tibshirani R, Saunders M, Rosset S, Zhu J & Knight K Sparsity and smoothness via the fused lasso. J. R. Stat. Soc. Series B Stat. Methodol. 67, 91–108 (2005). [Google Scholar]
- 80.Cheng W, Zhang X, Guo Z, Shi Y & Wang W Graph-regularized dual Lasso for robust eQTL mapping. Bioinformatics 30, i139–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szeto MD et al. Quantitative metrics of net proliferation and invasion link biological aggressiveness assessed by MRI with hypoxia assessed by FMISO-PET in newly diagnosed glioblastomas. Cancer Res. 69, 4502–4509 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang CH et al. Prognostic significance of growth kinetics in newly diagnosed glioblastomas revealed by combining serial imaging with a novel biomathematical model. Cancer Res. 69, 9133–9140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chaskis C, Stadnik T, Michotte A, Van Rompaey K & D’Haens J Prognostic value of perfusion-weighted imaging in brain glioma: a prospective study. Acta Neurochir. 148, 277–85; discussion 285 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Maia ACM Jr, et al. MR cerebral blood volume maps correlated with vascular endothelial growth factor expression and tumor grade in nonenhancing gliomas. AJNR Am. J. Neuroradiol. 26, 777–783 (2005). [PMC free article] [PubMed] [Google Scholar]
- 85.Price SJ et al. Correlation of MR relative cerebral blood volume measurements with cellular density and proliferation in high-grade gliomas: an image-guided biopsy study. AJNR Am. J. Neuroradiol. 32, 501–506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Price SJ et al. Improved delineation of glioma margins and regions of infiltration with the use of diffusion tensor imaging: an image-guided biopsy study. AJNR Am. J. Neuroradiol. 27, 1969–1974 (2006). [PMC free article] [PubMed] [Google Scholar]
- 87.Gaw N et al. Integration of machine learning and mechanistic models accurately predicts variation in cell density of glioblastoma using multiparametric MRI. Sci. Rep. 9, 10063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brandsma D & van den Bent MJ Pseudoprogression and pseudoresponse in the treatment of gliomas. Curr. Opin. Neurol. 22, 633–638 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Fink J, Born D & Chamberlain MC Pseudoprogression: relevance with respect to treatment of high-grade gliomas. Curr. Treat. Options Oncol. 12, 240–252 (2011). [DOI] [PubMed] [Google Scholar]
- 90.Kim J-H et al. Pathologic diagnosis of recurrent glioblastoma: morphologic, immunohistochemical, and molecular analysis of 20 paired cases. Am. J. Surg. Pathol. 36, 620–628 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Forsyth PA et al. Radiation necrosis or glioma recurrence: is computer-assisted stereotactic biopsy useful? J. Neurosurg. 82, 436–444 (1995). [DOI] [PubMed] [Google Scholar]
- 92.Patel P et al. MR perfusion-weighted imaging in the evaluation of high-grade gliomas after treatment: a systematic review and meta-analysis. Neuro. Oncol. 19, 118–127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barajas RF Jr, et al. Differentiation of recurrent glioblastoma multiforme from radiation necrosis after external beam radiation therapy with dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 253, 486–496 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu LS et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. AJNR Am. J. Neuroradiol. 30, 552–558 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fatterpekar GM, Galheigo D, Narayana A, Johnson G & Knopp E Treatment-related change versus tumor recurrence in high-grade gliomas: a diagnostic conundrum--use of dynamic susceptibility contrast-enhanced (DSC) perfusion MRI. AJR Am. J. Roentgenol. 198, 19–26 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Seeger A et al. Comparison of three different MR perfusion techniques and MR spectroscopy for multiparametric assessment in distinguishing recurrent high-grade gliomas from stable disease. Acad. Radiol. 20, 1557–1565 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Sugahara T et al. Posttherapeutic intraaxial brain tumor: the value of perfusion-sensitive contrast-enhanced MR imaging for differentiating tumor recurrence from nonneoplastic contrast-enhancing tissue. AJNR Am. J. Neuroradiol. 21, 901–909 (2000). [PMC free article] [PubMed] [Google Scholar]
- 98.Gasparetto EL et al. Posttreatment recurrence of malignant brain neoplasm: accuracy of relative cerebral blood volume fraction in discriminating low from high malignant histologic volume fraction. Radiology 250, 887–896 (2009). [DOI] [PubMed] [Google Scholar]
- 99.Hu LS et al. Optimized preload leakage-correction methods to improve the diagnostic accuracy of dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging in posttreatment gliomas. AJNR Am. J. Neuroradiol. 31, 40–48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brennan CW et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bai R-Y, Staedtke V & Riggins GJ Molecular targeting of glioblastoma: Drug discovery and therapies. Trends Mol. Med. 17, 301–312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bonavia R, Inda M-M, Cavenee WK & Furnari FB Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 71, 4055–4060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ene CI & Fine HA Many tumors in one: a daunting therapeutic prospect. Cancer cell vol. 20 695–697 (2011). [DOI] [PubMed] [Google Scholar]
- 104.Marusyk A, Almendro V & Polyak K Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334 (2012). [DOI] [PubMed] [Google Scholar]
- 105.Sottoriva A et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. U. S. A. 110, 4009–4014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barajas RF Jr, et al. Glioblastoma multiforme regional genetic and cellular expression patterns: influence on anatomic and physiologic MR imaging. Radiology 254, 564–576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gutman DA et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 267, 560–569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jain R et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology 272, 484–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Itakura H et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci. Transl. Med. 7, 303ra138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pope WB et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology 249, 268–277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aghi M et al. Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clin. Cancer Res. 11, 8600–8605 (2005). [DOI] [PubMed] [Google Scholar]
- 112.Van Meter T et al. Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn. Mol. Pathol. 15, 195–205 (2006). [DOI] [PubMed] [Google Scholar]
- 113.Hawkins-Daarud A, Johnston SK & Swanson KR Quantifying uncertainty and robustness in a biomathematical model--based patient-specific response metric for glioblastoma. JCO clinical cancer informatics 3, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gatenby RA, Silva AS, Gillies RJ & Frieden BR Adaptive therapy. Cancer Res. 69, 4894–4903 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Louis DN et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]