Abstract
Background
Long non-coding RNAs compose an important level of epigenetic regulation in normal physiology and disease. Despite the plethora of publications of lncRNAs in human cancer, the landscape is still unclear.
Methods
Microarray analysis in 44 NSCLC paired specimens was followed by qPCR-based validation in 29 (technical) and 38 (independent) tissue pairs. Cross-validation of the selected targets was achieved in 850 NSCLC tumours from TCGA datasets.
Results
Twelve targets were successfully validated by qPCR (upregulated: FEZF1-AS1, LINC01214, LINC00673, PCAT6, NUTM2A-AS1, LINC01929; downregulated: PCAT19, FENDRR, SVIL-AS1, LANCL1-AS1, ADAMTS9-AS2 and LINC00968). All of them were successfully cross validated in the TCGA datasets. Abnormal DNA methylation was observed in the promoters of FENDRR, FEZF1-AS1 and SVIL-AS1. FEZF1-AS1 and LINC01929 were associated with survival in the TCGA set.
Conclusions
Our study provides through multiple levels of internal and external validation, a comprehensive list of dysregulated lncRNAs in NSCLC. We therefore envisage this dataset to serve as an important source for the lung cancer research community assisting future investigations on the involvement of lncRNAs in the pathogenesis of the disease and providing novel biomarkers for diagnosis, prognosis and therapeutic stratification.
Subject terms: Non-small-cell lung cancer, Long non-coding RNAs
Background
Long non-coding RNAs (lncRNAs) comprise a heterogeneous group of transcripts >200 nucleotides with no protein coding capacity. In relation to their topography of the closest protein-coding gene, lncRNAs have been broadly classified into: sense, antisense, bidirectional, intronic and intergenic lncRNAs.1 They are largely transcribed by RNA polymerase II, spliced, 5′ capped, polyadenylated and regulated at multiple levels similarly to the mRNAs.2 GENCODE release v29 reports 16,066 lncRNA transcripts.3 Although the function of the majority of lncRNAs is still largely unknown, evidence reveals their participation in many regulatory pathways,4 including chromatin organisation, transcriptional regulation, post-transcriptional and post-translational processing.5 These functions are exerted by acting as guides, tethers, decoys, scaffolds, competitive endogenous RNAs and miRNA sponges.4 In general, lncRNAs show lower expression levels, significantly shorter transcript half-lives and higher cell and tissue specificity than mRNAs.1,6,7
Dysregulation of lncRNA expression appears to be extensive in cancer, where they act either as oncogenes or tumour repressors.8–10 In the past few years, several pan-cancer transcriptomic analyses have been carried out after thorough realignment and reannotation of thousands of RNA-seq libraries retrieved from TCGA, providing an overview of lncRNA transcriptional alterations in different human cancers. These studies suggest that (a) lncRNAs are frequently dysregulated in tumours, (b) many of these alterations are highly tumour-specific and (c) they are often associated with other cancer-related (epi)genetic alterations.9,10
Lung cancer is the most frequent cause of cancer-related deaths, and non-small cell lung cancer accounts for the majority of the reported cases.11 Early screening and improved therapies are critical unmet clinical needs12 and the field of lncRNAs has attracted a lot of interest in an effort to identify novel diagnostic biomarkers and therapeutic targets. Analysis of publicly available RNA-seq data indicated 111 intergenic lung cancer-associated lncRNAs that were differentially expressed between tumour and adjacent normal lung tissue.13 Furthermore, following a pan-cancer analysis of RNA-seq data from seven additional TCGA solid tumour types, 58 of these were shown to be specific to lung cancer. To date, there are a number of additional studies analysing publicly available lncRNA expression data in lung cancer14–16 and fewer attempting validation in independent sample sets.17–19 However, despite the plethora of published papers, the lncRNA landscape in NSCLC is far from comprehensive, mainly because of the limited independent validation, inconsistency of targets and sample sets. In this study, we undertook a lncRNA transcriptome-wide microarray approach and performed both technical and biological validation of a number of highly differential expressed lncRNA targets. Qualified targets from this process were cross validated in TCGA datasets. Expression was also tested in a series of NSCLC cell lines. Finally, we investigated the DNA methylation status of those dysregulated lncRNAs bearing CpG islands in their promoters.
Methods
Patients and samples
A set of 82 NSCLC patients from the Liverpool Lung Project (LLP) were used for expression analysis. All patients were recruited, following voluntary informed consent, from Liverpool Heart and Chest Hospital, in accordance with the Declaration of Helsinki. Ethical approval was obtained from the Liverpool Central Research Ethics Committee (ref 97/141). Frozen tissues were macro-dissected to ensure >70% tumour content. For DNA methylation analysis, we utilised DNAs from 129 tumour and 36 normal samples available from previous studies.20,21 Demographic and clinical characteristics of the cancer patients are summarised in Supplementary Table S1.
Analysis of the Cancer Genome Atlas expression data
Clinical and gene expression data was downloaded from the Genomics Data Commons (GDC) data portal using GDC Data Transfer Tool from the following The Cancer Genome Atlas (TCGA) datasets: TCGA-LUAD (Lung adenocarcinoma; N = 585) and TCGA-LUSC (Lung squamous cell carcinoma; N = 504). Patients with prior malignancies, subject to prior treatment and/or synchronous tumours were removed from our analysis. The main demographic and clinical characteristics of the TCGA patients analysed are summarised in Supplementary Table S2. Normalised gene expression values (FPKM-UQ; Fragments Per Kilobase of transcript per Million mapped reads upper quartile) were employed.
Cell lines and growth conditions
Thirteen NSCLC cell lines (A549, Calu-1, Calu-3, Calu-6, COR-L23, DMS53, H358, H2073, HTB-59, HTB-182, LUDLU-1, SK-LU-1 and SK-MES-1) and a non-tumorigenic human foetal lung fibroblast cell line (IMR-90) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM)/Ham’s Nutrient Mixture F-12 (1: 1) containing 10% foetal bovine serum (Sigma-Aldrich, Gillingham, UK) at 37 °C and 5% CO2. All cell lines were mycoplasma tested (e-Myco™ plus Kit, iNtRON Biotechnology, Gyeonggi-do, Korea) and authenticated (GenePrint 10 System, Promega, Southampton, UK).
RNA isolation
Total RNA was extracted from both tissue (10 × 10 μm thick) and cell lines using the Direct-zol mini kit (ZymoResearch, Irvine, CA, USA). RNA concentration and purity were determined on a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Hemel Hempstead, UK). RNA integrity was determined using the RNA 6000 Nano Lab-on-a-Chip kit on a Bioanalyzer 2100 (Agilent Technologies, Warrington, UK).
lncRNA array profiling
lncRNA profiling was performed on 44 normal-tumour pairs, using an Agilent Custom 8×60K expression array (AMADID: 047718) designed by the GENCODE consortium based on the Gencode v15 catalogue of human lncRNA.
Sample labelling and array hybridisation were performed according to the Two-Color Microarray-Based Gene Expression Analysis—Low Input Quick Amp Labeling—protocol version 6.7 (Agilent Technologies, Warrington, UK). Tumour samples were labelled with Cy3 and Cy5 was used with normal tissue. The labelled cRNA was hybridised to the Agilent SurePrint G3 gene expression 8×60K microarray and scanned on an Agilent G2565CA microarray scanner at 100% R + G PMT, 3 μm resolution and 20 bit TIFF with no XDR. Agilent’s Feature Extraction 11.0.1.1 software was used to analyse acquired array images and extract intensity data. The microarray data have been deposited in the Gene Expression Omnibus (GEO) database (GSE130740).
lncRNA RT–qPCR
Technical validation of the selected targets was performed in 29/44 array samples. Biological validation was undertaken on 38 independent NSCLC normal-tumour pairs. Additionally, the expression levels of the validated lncRNA targets were assessed in 14 cell lines. One microgram of total RNA was reverse transcribed (QuantiTect Reverse Transcription Kit, Qiagen, Crawley, UK) according to the manufacturer’s instructions. Quantitative real-time PCR was performed in triplicate using QuantiTect probe PCR mastermix (Qiagen, Crawley, UK). Primers and probes as well as the thermal profiles for each lncRNA qPCR assay are provided in Supplementary Table S3. lncRNA expression was quantified using the 2−ΔΔCt method and TBP as endogenous control.
DNA extraction and methylation analysis
DNA has been previously isolated.20,21 Pyrosequencing methylation analysis assays were designed for those lncRNAs harbouring a CpG island in their promoter (i.e. FENDRR, FEZF1-AS1, LIN00673, NUTM2A-AS1, PCAT6, RNF139-AS1, SVIL-AS1). One microgram of DNA was bisulphite-converted (EZ-96 DNA Methylation-Gold Kit, ZymoResearch, Irvine, CA, USA). DNA methylation analysis was performed by pyrosequencing (Qiagen, Crawley, UK) as previously described.20 Primers and PCR thermal profiles for each pyrosequencing assay are provided in Supplementary Table S4.
Statistical analysis
Statistical analysis of the microarray
Raw data were corrected for background noise using the normexp method and normalised using the loess method in Limma.22 Differential expression analysis was carried out on non-control probes using linear models and robust empirical Bayes moderated t-statistics.23 We compared lung tumours to their matched normal tissue to identify NSCLC-specific lncRNA transcriptional signatures. i-values were adjusted to control the false discovery rate (FDR) below 5% using the Benjamini and Hochberg method. Differentially expressed genes were identified using the double-filtering criterion: adjusted p-value (FDR) < 0.01 and an absolute log2 fold-change (absolute log2 fold-change) > 0.5. All statistical analyses were performed with Bioconductor in the R statistical environment.24 For the visualisation of the 44 pairs of tissue (tumour and normal) data were subtracted for Cy3 and Cy5 separately after background correction, normalised using cyclicloess method, log2 transformed and plotted using the coolmap function in Limma.22
Validation of differentially expressed lncRNA by RT-qPCR
lncRNA targets showing the highest differential expression between tumour and normal tissues were selected for further biological validation. Wilcoxon signed rank tests were used to establish statistically significant changes between the paired tumours and adjacent tissues while Mann–Whitney U and Kruskal–Wallis H tests were used to assess possible associations between expression and clinicopathological features in tumours. ROC curves and AUC were calculated for all the validated targets following binary logistic regression analysis. For each lncRNA, the patient cohort was dichotomised into low and high expression subgroups using the relative quantity (RQ) median; patients’ survival was calculated using the log rank test and plotted using Kaplan-Meier curves.
DNA methylation analysis
Hypermethylation thresholds were calculated using the 95% reference interval (mean + 2 × SD) of the normal sample values. Differences between the methylation status (hypermethylated vs normal) of tumours and clinicopathological features were assessed using Chi-square and Fisher’s exact tests.
Analysis of differentially expressed lncRNAs in the TCGA dataset
Mann–Whitney U tests were used to establish statistically significant changes between the tumours and normal tissues. Associations between expression and clinicopathological features in tumours were assessed using Mann–Whitney U and Kruskal–Wallis H tests. Pairwise multiple comparisons were carried out using Dunn’s post hoc test and p-values were adjusted using the Bonferroni error correction. Patients’ survival was calculated as described above.
Results
Discovery of aberrant lncRNA expression profiles in NSCLC
On the Gencode v15 microarray, each of the 17,535 randomly selected protein coding and 22,001 lncRNA transcripts is targeted by two probes. Three samples that were analysed in duplicate and hybridised on separate arrays were used as technical replicates. Technical replicates showed a strong pairwise Pearson correlation coefficient (r ≥ 0.86, p-value < 10–16). An initial comparison between the lncRNA expression profiles of NSCLC tissue and adjacent normal tissue using unsupervised hierarchical clustering analysis showed that samples could be neatly classified based on the expression levels of antisense and lincRNAs (Supplementary Figs. S1C and S1B) as well as protein coding transcripts (Supplementary Fig. S1A). Gene expression analysis identified the following numbers of differentially expressed transcripts in the NSCLC vs normal comparison: 581 (down) and 368 (up) lincRNAs, 541 (down) and 403 (up) antisense RNAs as well as 1992 (up) and 1,448 (down) protein-coding RNAs, (Supplementary Table S5). In this study, we focused only in the validation of the lncRNAs (lincRNA and antisense RNA transcripts).
Validation of differentially expressed lncRNAs
The selection criteria for the validation step included the p-value, fold-change and the clarity of annotation of the sequences. Regarding the latter, validation was performed only for targets where the probe sequence matched the location or Ensembl_ID indicated in the array manifest. For the validation step, we selected fourteen targets across a range of adjusted p-values (7.6 × 10–3 to 2.7 × 10–23) in order to assess the range of confidence in this microarray dataset.
The selected lncRNAs for technical and biological qPCR-based validation (Supplementary Table S5) were FEZF1-AS1, LINC01214, LINC00673, PCAT6, NUTM2A-AS1, RNF139-AS1, LINC01929, HMGA1P4 (upregulated) and PCAT19, FENDRR, SVIL-AS1, LANCL1-AS1, ADAMTS9-AS2 and LINC00968 (downregulated).
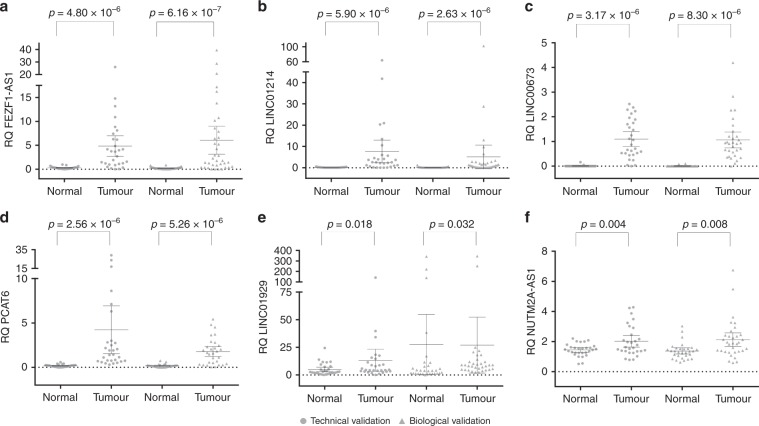
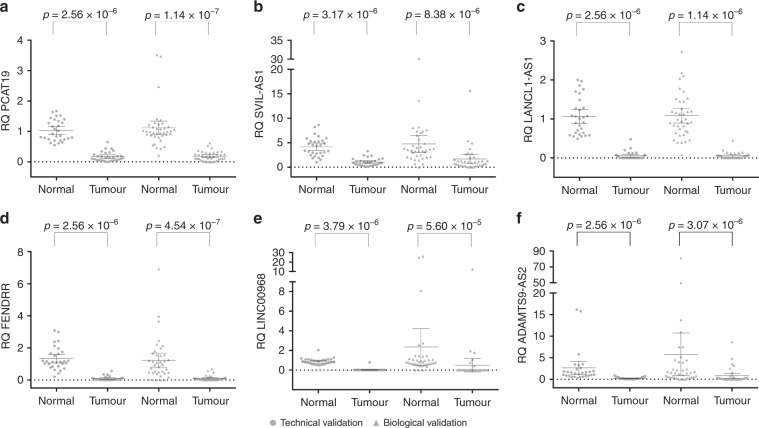
Out of the eight over-expressed lncRNAs identified on the microarray, six were confirmed as upregulated in lung tumours compared with paired normal tissue both in the technical and biological sets (Fig. 1). HMGA1P4 showed no significant difference in expression between tumour and normal paired samples in any of the validation sets (p > 0.05 in both), whereas RNF139-AS1 showed insignificant expression difference in the technical validation set (p = 0.325) and borderline difference in the biological validation set (p = 0.03). All six selected under-expressed lncRNA targets were significantly differentially expressed in lung tumours compared with paired normal lung in both sets (Fig. 2).
Fig. 1. Overexpressed lncRNAs in NSCLC.
Scatterplot diagrams demonstrating the expression levels of lncRNAs FEZF1-AS1 (a), LINC01214 (b), LINC00673 (c), PCAT6 (d), LINC01929 (e) and NUTM2A-AS1 (f), which are overexpressed in lung tumours compared to adjacent normal lung tissue. Means are also indicated along with the 95% confidence intervals. Technical validation values are given by circles, while biological independent validation values are given as triangles. p-values are derived from Wilcoxon ranked tests. RQ relative quantity.
Fig. 2. Downregulated lncRNAs in NSCLC.
Scatterplot diagrams demonstrating the expression levels of lncRNAs PCAT19 (a), SVIL-AS1 (b), LANCL1-AS1 (c), FENDRR (d), LINC00968 (e) and ADAMTS9-AS2 (f), which are downregulated in lung tumours compared to adjacent normal lung tissue. Means are also indicated along with the 95% confidence intervals. Technical validation values are given by circles, while biological independent validation values are given as triangles. p-values are derived from Wilcoxon ranked tests. RQ relative quantity.
In order to estimate the efficiency of each validated target to discriminate between normal and tumour tissue, binary logistic regression analysis was performed, and ROC curves were constructed (Supplementary Fig. S2). Expression of LANCL1-AS1 appeared to discriminate best (AUC = 0.995, 95% CI: 0.987–1), whilst assessment of NUTM2A-AS1 was the least informative (AUC = 0.689, 95% CI: 0.594–0.784).
Correlation between clinical data and the expression of the validated lncRNAs
qPCR-based expression was analysed in relation to the epidemiological and clinicopathopathological parameters. No association with age or histology was observed, while a number of borderline associations between individual lncRNA expression and certain clinicopathological parameters were noticed (Table 1). Higher expression of FEZF1-AS1 (p = 0.03) and ADAMTS9-AS2 (p = 0.03) was observed in males compared to females. In addition, the expression of LINC00968 (p = 0.03) was significantly higher in patients with lymph node involvement (N1/N2 stages when compared with N0), while the opposite relationship was found for LINC01929 (p = 0.05). Finally, PCAT6 (p = 0.02) and LANCL1-AS1 (p = 0.01) expression was significantly lower in poorly differentiated tumours when compared to well and moderately differentiated ones.
Table 1.
Correlation between clinical data and the expression of the validated lncRNAs.
| N | Relative quantity (RQ) | Mann–Whitney test | ||||
|---|---|---|---|---|---|---|
| Mean | SD | U-score | p-value | |||
| Gender | ||||||
| FEZF1-AS1 | Male | 49 | 6.49 | 8.31 | 268 | 0.03 |
| Female | 17 | 2.72 | 3.68 | |||
| ADAMTS9-AS2 | Male | 50 | 0.66 | 1.51 | 255 | 0.03 |
| Female | 16 | 0.11 | 0.16 | |||
| Lymph node involvement | ||||||
| LINC01929 | N(−) | 33 | 31.63 | 74.00 | 339 | 0.05 |
| N(+) | 29 | 8.00 | 7.39 | |||
| LINC00968 | N(−) | 34 | 0.20 | 0.50 | 435 | 0.03 |
| N(+) | 32 | 0.38 | 2.18 | |||
| Differentiation | ||||||
| PCAT6 | Poor | 18 | 2.03 | 3.85 | 212 | 0.02 |
| Well-Moderate | 38 | 3.60 | 5.83 | |||
| LANCL1-AS1 | Poor | 20 | 0.02 | 0.05 | 266 | 0.01 |
| Well-Moderate | 41 | 0.07 | 0.11 | |||
SD standard deviation.
Only the expression of LINC01214 and FENDRR appeared to have an effect in survival (Supplementary Fig. S3). Longer survival was observed among the patients with high LINC01214 expression level (mean survival time: 73.24 months, 95% CI: 52.12–94.35) compared to low LINC01214 level (mean survival time: 41.14 months, 95% CI: 23.22–59.05, log rank p = 0.031). Likewise, patients with high FENDRR expression levels showed a better outcome (mean survival time: 75.29 months, 95% CI: 49.90–100.69) than those with low FENDRR expression levels (mean survival time: 43.71 months, 95% CI: 28.65–58.78, log rank p = 0.021).
Cross-validation of selected targets with TCGA datasets
Out of the 14 lncRNAs selected for validation in our study, 13 were confirmed as being differentially expressed between tumour and normal tissue in the TCGA dataset (Supplementary Figs. S4 and S5). The only significant difference between our dataset and TCGA was for RNF139-AS1; only a borderline expression difference was seen in our biological validation set (p = 0.03), but it was significantly higher in tumours compared to normal tissues in the TCGA set (p = 1.27 × 10–22).
A number of significant associations were observed between tumour histology and the expression of the validated lncRNAs (Supplementary Fig. S6). Adenocarcinomas showed a significantly higher expression for PCAT19 (p = 4.14 × 10–18), FENDRR (p = 0.011), LANCL1-AS1 (p = 1.26 × 10–4), LINC00968 (p = 3.97 × 10–12) and ADAMTS9-AS2 (p = 1.92 × 10–44) when compared to squamous cell carcinomas. On the other hand, the expression of LINC00673 (p = 6.42 × 10–30), PCAT6 (p = 1.01 × 10–7), NUTM2A-AS1 (p = 3.69 × 10–5) and LINC01929 (p = 3.26 × 10–6) was significantly higher in squamous cell carcinomas than in adenocarcinomas.
Tumour stage significantly associated with the expression of LINC00673 (p = 0.005), LINC01929 (p = 0.002), PCAT19 (p = 0.001), FENDRR (p = 8.33 × 10–6), SVIL-AS1 (p = 0.037), LANCL1-AS1 (p = 8.69 × 10–7), LINC00968 (p = 2.32 × 10–7) and ADAMTS9-AS2 (8.29 × 10–6) as determined by Kruskal–Wallis test (Supplementary Fig. S7). Dunn’s post hoc pairwise test revealed that median expression level was significantly lower in T1 compared to T2 stage tumours for LINC00673 (p = 0.004) and LINC01929 (p = 0.001). SVIL-AS1 median expression was significantly lower in T1 compared to T3 NSCLC tumours (p = 0.027). On the other hand, for PCAT19, FENDRR, LANCL1-AS1, LINC00968 and ADAMTS9-AS2 the median expression level was significantly higher in T1 compared to T2 (p-values ≤ 0.001) and in T1 compared to T3 stage tumours (p-values ≤ 0.01) (Supplementary Fig. S7).
Regarding nodal stage (Supplementary Fig. S8), the expression of LINC00673 (p = 0.021) was significantly higher in patients with lymph node involvement (N1/N2 stages when compared with N0), while the opposite relationship was found for LANCL1-AS1 (p = 0.001), LINC00968 (p = 0.009) and ADAMTS9-AS2 (p = 0.003).
The expression of FEZF1-AS1 and LINC01929 appeared to be related to survival in the TCGA dataset (Supplementary Fig. S9). A trend for longer survival was observed among the patients with high FEZF1-AS1 level (mean survival time: 92.65 months, 95% CI: 74.46–110.84) compared to the low FEZF1-AS1 expression group (mean survival time: 76.25 months, 95% CI: 62.81–89.69, log rank p = 0.040). In addition, patients with low LINC01929 expression levels had better outcome (mean survival time: 95.38 months, 95% CI: 79.03–111.73) than those with high expression (mean survival time: 67.68 months, 95% CI: 59.63–75.73, log rank p = 0.022).
Expression of the validated lncRNAs in NSCLC cell lines
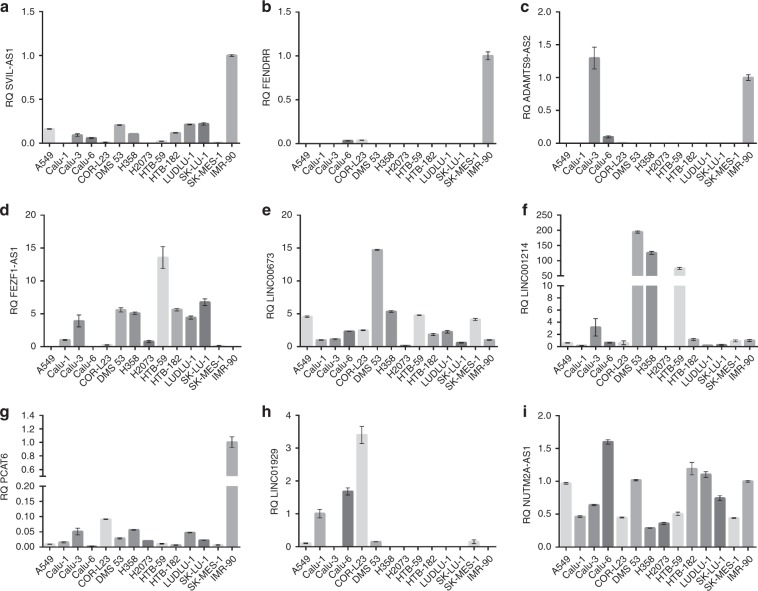
The expression profiles of the 12 validated lncRNAs were also assessed in a panel of 13 lung cancer cell lines in comparison to a normal lung fibroblast line (IMR-90). Expression of the 6 downregulated targets in lung cancer tissue was significantly reduced (SVIL-AS1, FENDRR, ADAMTS9-AS2) or completely absent (PCAT19, LANCL1-AS1 and LINC00968) (Fig. 3) in lung cancer cell lines while positive controls (pooled normal lung tissue RNA) performed as expected in these experiments.
Fig. 3. Bar chart diagrams showing the expression levels of the validated lncRNAs in NSCLC cell lines and a normal lung fibroblast cell line (IMR90), which was used as a calibrator (RQ = 1) for the analysis, with the exception of FEZF1-AS1 and LINC01929, which are not expressed in IMR90.
In this case Calu-1 was used as calibrator. All downregulated lncRNAs in lung cancer tissue were reduced (SVIL-AS1 (a), FENDRR (b), ADAMTS9-AS2 (c)) or completely undetected (PCAT19, LANCL1-AS1 and LINC00968, data not shown). Upregulated in lung cancer tissue lncRNAs did not necessarily follow the same pattern in cell lines. FEZF1-AS1 (d), LIN00673 (e) and LINC01214 (f) were higher in NSCLC cell lines, PCAT6 (g) and LINC01929 (h) were lower in NSCLC cell lines compared to IMR90, while NUTM2A-AS1 (i) was highly variable. Error bars represent standard error of the mean. RQ relative quantity.
All the lncRNAs found upregulated in lung cancer tissue were widely expressed in the lung cancer cell lines analysed, however, not all of them followed the tissue pattern. When compared with the non-tumorigenic IMR-90, the expression of FEZF1-AS1, LIN00673 and LINC01214 was markedly higher in most lung cancer cell lines; the expression of PCAT6 and LINC01929 was strikingly lower and no clear trend was observed for the expression of NUTM2A-AS1 (Fig. 3).
Methylation of lncRNA loci in NSCLC patients
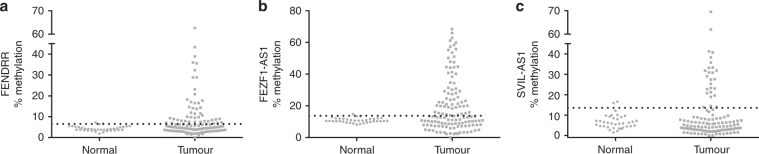
Only 7 out of the 14 lncRNAs selected from the microarray phase harbour a CpG island in their promoter. Pyrosequencing analysis demonstrated that PCAT6, RNF139-AS1, LINC00673 and NUTM2A-AS1 had <5% DNA methylation levels in both normal or tumour tissue, which is usually classified as noise.25 In contrast, variable methylation levels were observed for the remaining three targets. Hypermethylation in tumours (defined as > 95% reference range of normal values) was observed in 45/126 (35.7%) of tumours for FENDRR, 61/128 (47.7%) for FEZF1-AS1 and 26/128 (20.3%) for SVIL-AS1 (Fig. 4). DNA and corresponding RNA was available for 42 common samples between the two (expression and methylation analysis) sets. No direct correlation was established between RNA expression and hypermethylation status in any of these three genes.
Fig. 4. DNA methylation of lncRNA promoters in NSCLC.
Scatterplot diagrams demonstrating the DNA methylation levels of lncRNAs FENDRR (a), FEZF1-AS1 (b) and SVIL-AS1 (c) in lung tumours compared to adjacent normal lung tissue. The horizontal dotted line in each diagram defines the 95% reference range (= mean + 2 × Standard deviation) of the normal tissues. Tumour samples with values over this line are classified as hypermethylated.
Association between clinical data and the methylation of the validated lncRNAs
FENDRR hypermethylation was more frequent in adenocarcinomas (56.9%) than in squamous cell carcinomas (21.6%, p = 5.5 × 10–5) while an opposite borderline trend was found for FEZF1-AS1 (37.3% vs 55.3%, p = 0.05, Table 2). Non-significant trends were found between, FEZF1-AS1 (p = 0.08) and SVIL-AS1 (p = 0.06) hypermethylation and the differentiation status of the tumours, while no associations were obtained between hypermethylation of any promoter and age, gender and nodal stage.
Table 2.
Association between clinical data and hypermethylation status of FENDRR and FEZF1-AS1.
| N | Hypermethylated (%) | Pearson χ2 test p-value | ||
|---|---|---|---|---|
| Histology | ||||
| FENDRR | AdenoCa | 51 | 29 (56.9%) | 5.5 × 10–5 |
| SqCCa | 74 | 16 (21.6%) | ||
| FEZF1-AS1 | AdenoCa | 51 | 19 (37.3%) | 0.05 |
| SqCCa | 76 | 42 (55.3%) | ||
| Differentiation | ||||
| FEZF1-AS1 | Poor | 36 | 12 (34.3%) | 0.08 |
| Well-Moderate | 87 | 45 (51.7%) | ||
| SVIL-AS1 | Poor | 35 | 11 (31.4%) | 0.06 |
| Well-Moderate | 87 | 14 (16.1%) | ||
AdenoCa adenocarcinoma, SqCCa squamous cell carcinoma.
Bold values indicate statistical significance p-values.
Discussion
The discovery of long non-coding RNAs has revolutionised biological research, from basic gene function to clinical exploitation as disease specific biomarkers or therapeutic targets. The lncRNA transcriptome constitutes a whole new level of regulation in cell physiology. The promise of utilisation of lncRNAs in translational research and clinical management will rely on validation of differential gene expression levels associated with clinically relevant disease states and outcomes. Although there is still a lot to learn about lncRNA biology, there are already an increasing number of attempts to address lncRNA detection in body fluids to assist lung cancer diagnosis.26
The existing lncRNA profiling data in lung cancer to date is substantial. This includes high-throughput approaches13,14,16–18 and targeted studies.27–31 However, despite the plethora of reports, the landscape is less clear than one would hope. The main reason is the marked diversity of approaches and validation methods. In addition, with the exception of studies mining existing data in the public domain, most studies utilise limited discovery sample sets, which leads to high FDR.
In this study, we aimed to strengthen the existing data and contribute to a more comprehensive landscape of lncRNA dysregulation in NSCLC. We used a comparatively large discovery set in microarray analysis (44 pairs) and performed both technical (29 pairs) and independent biological (38 pairs) validation of targets by RT-qPCR, in order to identify lncRNA profiles significantly distinguishing NSCLCs tissue from paired normal lung tissue.
Following microarray data analysis and having set the desired thresholds (adjusted p-value (FDR) < 0.01 and absolute log2 fold-change > 0.5) we selected targets for validation with different p-values and fold-change values, in order to obtain a representative estimation of validity for our microarray analysis. An important selection factor was gene annotation and verification of probe sequences against locus sequence. For validation, we only selected among targets for which we had cross-referenced the hybridisation probe in the microarray with the annotated gene name in the databases (NCBI Genbank and ENSEMBL). Twelve out of the 14 targets selected for qPCR validation (six up- and six down-regulated in tumours) were successfully validated to demonstrate a strong and consistent difference of expression between NSCLC tissue and normal lung, suggesting a role in lung cancer pathogenesis. Interestingly, the two targets that failed in validation did not represent the highest p-values in the microarray analysis, indicating that even 10–2 adjusted p-values can be trusted. Surprisingly, a target with a very low p-value in microarray analysis (10–19) failed to validate, probably indicating potential problems with probe specificity and/or target annotation.
To the best of our knowledge, from the 12 validated lncRNAs, this is the first study where LANCL1-AS1, LINC01214, LINC01929, NUTM2A-AS1, PCAT19 and SVIL-AS1 are shown to be aberrantly expressed in cancer, while ADAMTS9-AS2, FENDRR, FEZF1-AS1, LINC00673, LINC00968 and PCAT6 have been previously reported to be dysregulated in lung and other types of cancer. Interestingly, our analysis of the available TCGA datasets demonstrated that all the 12 targets show significant differential expression in NSCLC, increasing further the confidence in this result. Apart from the validation of the RNF139-AS1, the main difference between our dataset and TCGA is centred around clinicopathological associations. A substantial number of associations of these lncRNA targets were found in TCGA, while we only found a few associations of borderline significance. This is not surprising, as TCGA datasets contained 850 tumours as opposed to 67 for our set used for clinicopathological comparisons. The clinicopathological associations revealed within the TCGA dataset point to future research in establishing the potential prognostic value of these targets. In addition, no major associations were found between lncRNA expression and overall survival, with the exception of a few statistical trends (0.02 < p < 0.05) for FEZF1-AS1 and LINC01929 in TCGA dataset and LINC01214 and FENDRR in our dataset. The differences in this finding probably reflects the size difference of the two datasets. Nevertheless, one has to consider that overall survival in lung cancer is a challenging parameter as the cause of death is frequently related to comorbidities and/or therapy related toxicity.
ADAMTS9-AS2 was previously found dysregulated in several cancer types including NSCLC.32,33 Overexpression of ADAMTS9-AS2 reduces proliferation, cell migration, invasion, and induces apoptosis in vitro while restrains tumour growth in vivo.32–34 Recent evidence suggests that ADAMTS9-AS2 might exert its function by ‘sponging’ common microRNAs.33,34 For instance, in NSCLC, ADAMTS9-AS2 represses cancer progression by regulating the miR-223-3p/TGFBR3 axis,33 while in ovarian cancer it regulates the miR-182-5p/FOXF2 pathway.34
A few studies have analysed the role of FENDDR in various cancer types suggesting that FENDRR plays an inhibitory role in tumour progression by regulating cell migration, invasion and metastasis.35–37 FENDRR was found to be downregulated in gastric cancer tissues, which increased cell migration and invasion in vitro through up-regulation of FN1 and MMP2 and MMP9.35 FENDRR expression was also shown to be suppressed in gastric cancer associated fibroblasts (CAFs) by hypermethylation.38 In NSCLC, we have observed that FENDRR expression is reduced in lung tumours compared with paired normal tissue and lung cancer cell lines showed a similar trend. This result is in agreement with a previous study reporting a significant downregulation of FENDRR in NSCLC as well as a potential involvement of FENDRR in regulating epithelial mesenchymal transition (EMT), stemness and metastasis of lung cancer cells via EZH2.37
Our finding on FEZF1-AS1 upregulation in NSCLC is in agreement with previous reports on many tumour types including NSCLC39,40 and breast.41 FEZF1-AS1 exerts regulatory functions at a transcriptional and post-transcriptional level, promoting cell proliferation, migration, and invasion. In fact, it has been reported to modulate the expression of Nanog by sponging miR-30a to promote cancer stemness in breast cancer.41 However, FEZF1-AS1 can also act at a transcriptional level by recruiting chromatin-modifying enzymes to target genes. For instance, FEZF1-AS1 can bind to LSD1 and EZH2 and epigenetically repress E-cadherin in NSCLC39 or p57 in lung adenocarcinomas.40
In our dataset, LINC00673 was overexpressed in lung tumours, which is in agreement with previous reports on NSCLC.42–44 Upregulation of LINC00673 has also been reported in gastric cancer,45,46 having a role in cell proliferation, migration, and invasion. LINC00673 transcripts have been localised both in the nucleus and the cytoplasm,42,44–46 suggesting that it can function at a transcriptional and post-transcriptional level. In NSCLC, Lu et al proposed that LINC00673 promotes TGF-β induced EMT by sponging miR-150-5p and thereby modulating ZEB1 expression indirectly.43 In addition, Roth et al have reported that LINC00673 negatively regulates p53 translation by directly interacting with its 3′UTR leading to senescence bypass.44 LINC00673 can also regulate the expression of target genes epigenetically. Notably, in lung cancer, LINC00673 represses HOXA5 expression through recruiting the epigenetic repressor EZH2 to its promoter region.47 Shi et al observed that LINC00673 epigenetically inhibited NCALD expression through direct binding to LSD1.42 In a similar way, in gastric cancer LINC00673 suppressed KLF4 expression by interacting with EZH2 and DNMT146 and direct binding to LSD1 and EZH2 repressed KLF2 and LATS2 expression.45
Regarding PCAT6, our findings are in line with previous studies reporting its upregulation in various cancer types including NSCLC48 and colon.49 Functional studies have shown that PCAT6 exerts its oncogenic activity, at least partly, by leading chromatin-modifying complexes to specific promoter regions. For instance, in NSCLC, PCAT6 interacts with EZH2 epigenetically, suppressing LATS2,48 while in colon cancer, the association of PCAT6 with EZH2 modulates ARC expression.49
We observed that LINC00968 was significantly downregulated in lung tumours compared with paired normal lung and completely absent in lung cancer cell lines, which is in contrast to a previous study reporting upregulation of LINC00968 in NSCLC.50 However, that study evaluated LINC00968 expression using datasets from the Oncomine database and four NSCLC cell lines, whereas we performed wet validation in 82 NSCLC tissue samples and 13 NSCLC cell lines. We speculate that inconsistency of results may lie with incorrect annotation in the previous bioinformatic analysis.
To explore the molecular mechanisms responsible for the modulated expression observed, we hypothesised that DNA methylation might be one of the mechanisms responsible. Therefore, we undertook DNA methylation analysis in seven of our target genes that were found to bear a CpG island in their promoter. Of those, hypermethylation was detected only in three, namely FENDRR, SVIL-AS1 and FEZF1-AS1. There was a general lack of correlation between expression and DNA methylation in the genes tested. This may be due to the low number of common samples in the two analyses, but it most likely indicates the presence of strong alternative regulation factors besides DNA methylation. FENDRR hypermethylation has previously been shown in gastric CAFs.38 FENDRR promoter methylation demonstrated a significant increase in adenocarcinoma, however this was not the case for FENDRR expression.
This study contributes significantly to the wider knowledge of lncRNA involvement in NSCLC pathogenesis. Its strength lies with the technical and independent biological validation in patient tissues as well as its agreement with the available relevant TCGA datasets. Out of the 12 validated targets, half are shown to be differentially expressed in cancer for the first time, while our results for the other six lncRNAs confirm previous findings. Certainly, there are still a large number of dysregulated candidate lncRNAs in our microarray analysis list that require independent validation. Our data have been made publicly available, therefore open to other research groups to attempt independent validation. To date, the involvement of lncRNAs in NSCLC development is undisputable; however, further studies are needed to establish additional lncRNA targets and to identify their functional role in the pathogenesis of NSCLC.
Supplementary information
Acknowledgements
We would like to thank Dr. Gordon K. Smyth and Dr Ramyar Molania from the Walter & Eliza Hall Institute of Medical Research, Melbourne, Australia for their invaluable assistance in the bioinformatics analysis. We would also want to acknowledge the voluntary contribution of patients with their specimens and information as well as the nursing staff that recruited them in the study.
Author contributions
The study was conceived and designed by T.L. and A.A.S. Methodology was developed by A.A.S., P.P. and T.L. Data acquisition was carried out by A.A.S., B.U., P.P., C.M., L.R., M.P.A.D., J.K.F. Analysis and interpretation of the data was undertaken by A.A.S., N.G.B., M.W.M., T.L. Writing, review, and/or revision of the manuscript was done by A.A.S., N.G.B., B.U., M.P.A.D., J.K.F. and T.L. This research was supervised by T.L.
Ethics approval and consent to participate
Ethical approval was obtained from the Liverpool Central Research Ethics Committee (ref 97/141). All patients were recruited following voluntary informed consent, and the study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Data availability
Microarray data have been deposited in the Gene Expression Omnibus (GEO) database (GSE130740).
Competing interests
The authors declare no competing interests.
Funding information
This research was supported by the Roy Castle Lung Cancer Foundation, UK (Grant no 2014/05/Liloglou) and a Roy Castle Lung Cancer Senior Fellowship (MPAD).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-020-0742-9.
References
- 1.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim. Biophys. Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashouri A, Sayin VI, Van den Eynden J, Singh SX, Papagiannakopoulos T, Larsson E. Pan-cancer transcriptomic analysis associates long non-coding RNAs with key mutational driver events. Nat. Commun. 2016;7:13197. doi: 10.1038/ncomms13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 12.Field JK, Duffy SW, Devaraj A, Baldwin DR. Implementation planning for lung cancer screening: five major challenges. Lancet Respir. Med. 2016;4:685–687. doi: 10.1016/S2213-2600(16)30233-8. [DOI] [PubMed] [Google Scholar]
- 13.White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Lin J, Liu T, Chen T, Pan S, Huang W, et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer. 2014;85:110–115. doi: 10.1016/j.lungcan.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci. Rep. 2014;4:6591. doi: 10.1038/srep06591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou M, Guo M, He D, Wang X, Cui Y, Yang H, et al. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J. Transl. Med. 2015;13:231. doi: 10.1186/s12967-015-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Xu Q, Liu F, Ye X, Wang J, Meng X. Identification and validation of long noncoding RNA biomarkers in human non-small-cell lung carcinomas. J. Thorac. Oncol. 2015;10:645–654. doi: 10.1097/JTO.0000000000000470. [DOI] [PubMed] [Google Scholar]
- 18.Feng N, Ching T, Wang Y, Liu B, Lin H, Shi O, et al. Analysis of microarray data on gene expression and methylation to identify long non-coding RNAs in non-small cell lung cancer. Sci. Rep. 2016;6:37233. doi: 10.1038/srep37233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Z, Bai Y, Wang P, Wu Z, Zhou L, Zhong M, et al. Identification of long noncoding RNAs for the detection of early stage lung squamous cell carcinoma by microarray analysis. Oncotarget. 2017;8:13329–13337. doi: 10.18632/oncotarget.14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolaidis G, Raji OY, Markopoulou S, Gosney JR, Bryan J, Warburton C, et al. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res. 2012;72:5692–5701. doi: 10.1158/0008-5472.CAN-12-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bediaga NG, Davies MP, Acha-Sagredo A, Hyde R, Raji OY, Page R, et al. A microRNA-based prediction algorithm for diagnosis of non-small lung cell carcinoma in minimal biopsy material. Br. J. Cancer. 2013;109:2404–2411. doi: 10.1038/bjc.2013.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust Hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016;10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, et al. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br. J. Cancer. 2006;94:561–568. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan L, Zhang L, Fan K, Wang JJ. Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non-small cell lung cancer. Onco Targets Ther. 2017;10:5695–5702. doi: 10.2147/OTT.S149314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 28.Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol. Carcinog. 2015;54(Suppl. 1):E1–E12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 29.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol. Cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol. Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 32.Cao B, Liu C, Yang G. Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed. Pharmacother. 2018;107:185–193. doi: 10.1016/j.biopha.2018.06.146. [DOI] [PubMed] [Google Scholar]
- 33.Liu C, Yang Z, Deng Z, Zhou Y, Gong Q, Zhao R, et al. Upregulated lncRNA ADAMTS9-AS2 suppresses progression of lung cancer through inhibition of miR-223-3p and promotion of TGFBR3. IUBMB Life. 2018;70:536–546. doi: 10.1002/iub.1752. [DOI] [PubMed] [Google Scholar]
- 34.Wang A, Jin C, Li H, Qin Q, Li L. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. Int J. Biol. Macromol. 2018;120:1705–1713. doi: 10.1016/j.ijbiomac.2018.09.179. [DOI] [PubMed] [Google Scholar]
- 35.Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J. Hematol. Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kun-Peng Z, Chun-Lin Z, Xiao-Long M. Antisense lncRNA FOXF1-AS1 Promotes Migration and Invasion of Osteosarcoma Cells Through the FOXF1/MMP-2/-9 Pathway. Int J. Biol. Sci. 2017;13:1180–1191. doi: 10.7150/ijbs.21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao L, Huang Z, Zengli Z, Li H, Chen Q, Yao C, et al. Loss of long noncoding RNA FOXF1-AS1 regulates epithelial-mesenchymal transition, stemness and metastasis of non-small cell lung cancer cells. Oncotarget. 2016;7:68339–68349. doi: 10.18632/oncotarget.11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Najgebauer H, Liloglou T, Jithesh PV, Giger OT, Varro A, Sanderson CM. Integrated omics profiling reveals novel patterns of epigenetic programming in cancer-associated myofibroblasts. Carcinogenesis. 2019;40:500–512. doi: 10.1093/carcin/bgz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC) Biomed. Pharmacother. 2017;95:331–338. doi: 10.1016/j.biopha.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 40.Jin S, Chen S, Ma Y, Yang B, Liu Y. LincRNA FEZF1-AS1 contributes to the proliferation of LAD cells by silencing p57 expression. Oncotarget. 2017;8:103004–103013. doi: 10.18632/oncotarget.21265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Sun L, Zhang Y, Lu G, Li Y, Wei Z. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J. Cell Physiol. 2018;233:8630–8638. doi: 10.1002/jcp.26611. [DOI] [PubMed] [Google Scholar]
- 42.Shi X, Ma C, Zhu Q, Yuan D, Sun M, Gu X, et al. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7:25558–25575. doi: 10.18632/oncotarget.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol. Cancer. 2017;16:118. doi: 10.1186/s12943-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roth A, Boulay K, Gross M, Polycarpou-Schwarz M, Mallette FA, Regnier M, et al. Targeting LINC00673 expression triggers cellular senescence in lung cancer. RNA Biol. 2018;15:1499–1511. doi: 10.1080/15476286.2018.1553481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, et al. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol. Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu YB, et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. 2017;8:95542–95553. doi: 10.18632/oncotarget.20980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma C, Wu G, Zhu Q, Liu H, Yao Y, Yuan D, et al. Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget. 2017;8:32696–32705. doi: 10.18632/oncotarget.16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi X, Liu Z, Liu Z, Feng X, Hua F, Hu X, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187. doi: 10.1016/j.ebiom.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang W, Su G, Huang X, Zou A, Wu J, Yang Y, et al. Long noncoding RNA PCAT6 inhibits colon cancer cell apoptosis by regulating anti-apoptotic protein ARC expression via EZH2. Cell Cycle. 2019;18:69–83. doi: 10.1080/15384101.2018.1558872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Zhou J, Xu YJ, Hu HB. Long non-coding RNA LINC00968 acts as oncogene in NSCLC by activating the Wnt signaling pathway. J. Cell Physiol. 2018;233:3397–3406. doi: 10.1002/jcp.26186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data have been deposited in the Gene Expression Omnibus (GEO) database (GSE130740).