Abstract
Thymic natural killer T (NKT)2 cells are a subset of invariant NKT cells with PLZFhiGATA3hiIL-4+. The differentiation of NKT2 cells is not fully understood. In the present study, we report an important role of TRAF3-interacting protein 3 (TRAF3IP3) in the functional maturation and expansion of committed NKT2s in thymic medulla. Mice with T-cell-specific deletion of TRAF3IP3 had decreased thymic NKT2 cells, decreased IL-4-producing peripheral iNKTs, and defects in response to α-galactosylceramide. Positive selection and high PLZF expression in CD24+CD44− and CCR7+CD44− immature iNKTs were not affected. Only CD44hiNK1.1− iNKTs in Traf3ip3−/− mice showed reduced expression of Egr2, PLZF, and IL-17RB, decreased proliferation, and reduced IL-4 production upon stimulation. This Egr2 and IL-4 expression was augmented by MEK1/ERK activation in iNKTs, and TRAF3IP3 at the trans-Golgi network recruited MEK1 and facilitated ERK phosphorylation and nuclear translocation. LTβR-regulated bone marrow-derived nonlymphoid cells in the medullary thymic microenvironment were required for MEK/ERK activation and NKT2 maturation. These data demonstrate an important functional maturation process in NKT2 differentiation that is regulated by MEK/ERK signaling at the trans-Golgi network.
Keywords: TRAF3IP3, NKT2 cells, MEK/ERK signaling, Functional maturation
Subject terms: NKT cells, Lymphopoiesis
Introduction
Type I invariant natural killer T (iNKT) cells are a group of CD1d-restricted T lymphocytes that play critical roles in regulating both innate and adaptive immunity.1,2 Among various iNKT subsets, NKT2 cells are defined as PLZFhiGATA3hiIL-17RBhiCD122− cells that express IL-4 at steady state.3–5 Upon stimulation, NKT2 cells quickly produce high levels of IL-4 and IL-13, and moderate levels of IFN-γ.6,7 In the thymus, NKT2 cells are required for the expansion of Eomes-expressing memory-like CD8 T cells, egress of conventional thymocytes from the thymic perivascular space into the circulation, and regulation of medullary thymic epithelial cell (mTEC) differentiation.8–13 In the periphery, NKT2 cells can suppress Th1-mediated tissue damage and promote B-cell differentiation and antiviral antibody production.14,15 The high levels of IL-4 and IL-13 produced by NKT2 cells in the mucosal area also play an important role in the pathogenesis of ulcerative colitis and allergen-induced airway hyperactivity.16,17
The differentiation of NKT2 cells in the thymus is not fully understood. iNKT cells are positively selected by thymic cortex-resident CD4 and CD8 double-positive (DP) thymocytes presenting glycolipids in the context of CD1d molecules. According to the conventional linear maturation model, the post-selected CD24hiCD69+CD44loNK1.1− (stage 0 or ST0) cells downregulate their CD24 and CD69 levels and differentiate into CD24loCD44loNK1.1− (ST1) cells. ST1 cells then upregulate CD44 (CD44+NK1.1−, ST2) and eventually acquire NK1.1 (CD44+NK1.1+, ST3). IL-4-expressing NKT2 cells can be found in ST1 and ST2 cells.2 As key transcription factors were identified in iNKT subsets, such as PLZF and GATA3, in NKT2 cells, a transcription factor-based lineage differentiation model was also developed.2 Wang et al.13 further used the expression of CCR7 to define the precursor cells of these iNKT subsets, NKTp (CD24lo/−CCR7+CD44lo), which can either reside in or emigrate from the thymus and differentiate into various effector iNKT subsets. Thus, iNKT cells at ST1 and ST2 may contain both NKTp and terminally differentiated CCR7−IL-4+ NKT2 cells. The results from single-cell RNA sequencing showed that CD27hiCD4+CD24− iNKT cells with preferential expression of multiple NKT2-specific genes had high heterogeneity and may contain several differentiation states.18 These results indicate that more work is needed to characterize the key events and regulation involved in NKTp-to-NKT2 differentiation.
The fate determination of NKT2 cells has been reported to require strong TCR signaling during positive selection to induce the highest level of the signature transcription factor PLZF.19–21 iNKT cells with Plzf deficiency had defects in NKT2 development as well as failure to upregulate CD44 and undergo proliferation/effector differentiation.22 However, this high expression of PLZF and likely positive selection in the thymic cortex are not sufficient to drive the full differentiation program of NKT2 cells, as the premature egress of post-selected CD24+PLZFhi (ST0) iNKT cells in CD69−/− mice severely affects the differentiation of NKT2 cells but not NKT1 or NKT17 cells.23 Signals from the thymic medullary microenvironment have been reported to regulate iNKT differentiation/maturation. For instance, transplantation with thymus grafts devoid of Relb-dependent mTECs selectively reduces CD44+ iNKT cell numbers without affecting post-selected ST0 and ST1 cells.24–27 Supplementation with IL-15-IL-15R complexes only increased the number of NKT1-enriched CD44+NK1.1+ ST3 cells.13,24 Thus, the roles of thymic medulla on the differentiation/maturation of NKT2/17-enriched CD44+NK1.1− ST2 cells remain elusive.
Recently, the transmembrane protein TRAF3-interacting protein 3 (TRAF3IP3) was found to regulate the positive selection of conventional T cells by promoting MEK/ERK signaling at the Golgi.28 Whether this pathway is required for iNKT-positive selection is under debate.29–32 We thus made our initial goal to investigate whether TRAF3IP3 is also involved in the positive selection of iNKT cells. Surprisingly, we did not observe altered iNKT positive selection in Traf3ip3fl/flCd4Cre mice. Instead, defects in the differentiation of IL-4-producing CD44+ NKT2 cells were found. The molecular mechanisms and contribution of the medullary thymic microenvironment to NKT2 differentiation were then investigated.
Materials and methods
Mice
Traf3ip3fl/fl mice and Cd4-Cre transgenic mice were kindly provided by Professors Hongshan Zhao (Peking University, China) and Lilin Ye (Army Medical University, China), respectively. LTβR-deficient (Ltbr−/−), Ltbrfl/fl, Cd11c-Cre, Ccr2−/−, and Batf3−/− mice were kindly provided by Professors Mingzhao Zhu (Chinese Academy of Sciences, China), Yu Zhang (Peking University Health Science Center, China), and Li Wu (Tsinghua University). C57BL/6 congenic mice (CD45.1+ and CD45.2+) were purchased from Peking University (Beijing, China). All mice were on a C57BL/6 background and were used at 8–12 weeks of age. The animals were kept in a specific pathogen-free facility at Peking University (Beijing, China). The experimental procedures on the use and care of animals were approved by the Ethics Committee of Peking University.
Bone marrow reconstitution
CD45.1+ WT or CD45.2+ TRAF3IP3KO recipients were subjected to lethal irradiation (900 rads). Six hours later, WT (CD45.1+) and TRAF3IP3KO (CD45.2+) bone marrow cells were mixed at a 1:1 ratio, and a total of 5 × 106 cells were injected intravenously into recipient mice. The mixed bone marrow chimeric mice were analyzed 8 weeks after transplantation. Lethally irradiated CD45.1+ WT or CD45.2+Ltbr−/− recipients were reconstituted with Ltbr−/− (CD45.2+) or WT (CD45.1+) bone marrow cells (5 × 106), respectively. The recipients were analyzed 8 weeks later.
iNKT cell isolation
Single-cell suspensions of thymocytes were incubated with anti-CD8 (3.155) monoclonal antibody and complement (guinea pig sera) to remove CD8+ cells. After two cycles of killing and removal of dead cells by density centrifugation, the cells were stained for antibodies against TCRβ, CD24, CD44, NK1.1, and CD1d-tet. Total iNKT cells or iNKT cells at ST0–3 were sorted using FACS Aria II (BD Biosciences, San Diego, CA, USA). The purification of TCRβ+CD1d-tet+ iNKT subsets by flow cytometry6 was performed as follows: NKT1, CD122+, NKT2, CD122−CD4+, NKT17, CD122−CD4−.
In vitro cytokine production
Cells isolated from the thymus of WT and TRAF3IP3KO mice were stimulated in a 24-well plate with 100 ng/ml PMA, 1 μg/ml ionomycin, and 3 μg/ml Brefeldin A in complete medium (Rosewell Park Memorial Institute 1640, 10% fetal bovine serum). Cells were incubated for 4 h at 37 °C before cytokine detection by intracellular staining.
Murine iNKT cell-driven hepatitis
To induce acute experimental hepatitis, a single intravenous injection of α-Galcer (2 μg in 200 μL phosphate-buffered saline) was administered to each mouse. Two hours later, the levels of serum IFN-γ and IL-4 were measured by enzyme-linked immunosorbent assay. Liver and spleen cells were cultured in the presence of Brefeldin A for 3 h, and the percentages of IFN-γ+, IL-4+, and IL-17A+ iNKT cells were measured by flow cytometry. Twenty hours after α-Galcer injection, the levels of serum alanine transaminase (ALT) and aspartate transaminase (AST) were measured using an automatic analyzer (Mindray, Shenzhen, China). For histopathology, livers were excised and fixed overnight in 10% neutral formalin solution and embedded in paraffin. Sections of 4 μm were prepared and stained with Hematoxylin and Eosin.
Retroviral production and transduction
Constitutively activated mouse MEK1 (MEK1DD, S218D/S222D), c-Maf, or Traf3ip3 genes were cloned and inserted into the retrovirus backbone pMSCV-ubc-EGFP. These vectors together with the helper vector pCL-Eco were co-transfected into HEK 293T cells using Lipofectamine 2000 (Invitrogen). The supernatant was harvested 48 h later. For retroviral transduction, DN32.D3 cells were suspended in retroviral supernatant in the presence of 4 μg/ml polybrene (Sigma-Aldrich, St Louis, MO, USA) and then spin-infected at 1500 × g for 2 h at 32 °C. Retroviral supernatants were then replaced with fresh culture medium after transduction. After overnight culture, the cells were spin-infected again and cultured for an additional 48 h before being used for the experiment.
Statistical analysis
The statistical analysis of the results was performed using GraphPad Prism 6 software (San Diego, CA). An unpaired or two-tailed paired Student’s t test was used to evaluate the significance of the differences between two groups. Data are presented as the mean ± SEM. A p value < 0.05 was considered significant (*p < 0.05, **p < 0.01, ***p < 0.005).
Additional experimental procedures are described in the supplemental Materials and Methods.
Results
TRAF3IP3 deficiency does not affect the positive selection of iNKT cells
To investigate whether TRAF3IP3 is involved in the positive selection of iNKT cells, we used Traf3ip3f/fCd4Cre (TRAF3IP3KO) mice (Fig. S1A). Consistent with the findings by Zou et al.,28 reduced CD4 and CD8 single-positive conventional T cells were observed in the thymus of TRAF3IP3KO mice (Fig. S1B, C). In contrast, the percentages and numbers of total iNKT cells or iNKT cells at various developmental stages were comparable between TRAF3IP3KO and wild-type (WT) mice (Fig. 1a, Fig. S1D). Similar levels of TCRβ, CD5, and CD1d tetramer loaded with the glycolipid PBS57 were also found in KO and WT iNKT cells (Fig. S1E). We then examined DP thymocytes for the expression of CD1d, SLAMF1, and SLAMF6, which are known to be essential in the positive selection of iNKT cells. As shown in Fig. 1b, no significant differences were found between TRAF3IP3KO and WT DP thymocytes. The presentation of CD1d/glycolipid complexes by WT and KO DP thymocytes was further compared by incubating DP thymocytes with DN32.D3 iNKT hybridoma cells in the presence or absence of α-galactosylceramide (α-GalCer). Twenty-four hours later, comparable levels of IL-2 production were detected in the samples with WT and KO thymocytes (Fig. 1c). We also examined the positive selection of iNKT cells in mixed bone marrow chimeras in vivo. CD45.2+ TRAF3IP3KO and CD45.1+ WT bone marrow cells at a 1:1 ratio were co-injected into lethally irradiated CD45.2+ TRAF3IP3KO or CD45.1+ WT mice. Eight weeks after reconstitution, the KO (CD45.2+) to WT (CD45.2−) ratio of total thymocytes, total iNKT cells, and iNKT cells at various developmental stages were similar, irrespective of WT or KO recipients (Fig. 1d and Fig. S1F, G). This finding indicates that positive selection of iNKT cells is grossly unaffected by TRAF3IP3 deficiency.
Fig. 1.
TRAF3IP3 deficiency did not affect the positive selection of iNKT cells. a Flow cytometry analysis of iNKT cells (stained with PBS57-loaded CD1d tetramer (CD1d-tet) and TCRβ) in the thymus. The percentage and number of iNKT cells were calculated. Three independent experiments were performed with 5–8 mice in each group. b The expression of CD1d, SLAMF1, and SLAMF6 in WT and TRAF3IP3KO DP thymocytes. Data are representative of three independent experiments. c DN32.D3 NKT hybridoma cells were co-cultured with either WT or TRAF3IP3KO DP thymocytes in the absence or presence of 100 ng/ml α-GalCer for 24 h. The supernatant was then collected and measured for IL-2 production by ELISAs. The experiment was repeated twice. d WT (CD45.1+) and TRAF3IP3KO (CD45.2+) bone marrow cells were mixed at a 1:1 ratio and injected into lethally irradiated CD45.1+ WT recipients. Eight weeks later, the ratios of CD45.2+-to-CD45.2− (KO-to-WT) in total thymocytes, total iNKT cells, and iNKT cells at various stages (ST0–1, CD44loNK1.1−; ST2, CD44+NK1.1−; and ST3, CD44+NK1.1+) are shown. Three chimeric mice were analyzed. e–i Flow cytometry analysis of Egr2, c-Myc, CD127, BrdU, Ki67, and Annexin V in ST0 (CD24+CD44lo) iNKT cells from WT and TRAF3IP3KO mice. BrdU incorporation analysis was performed after 24 h of intraperitoneal BrdU injection (1 mg). At least three independent experiments were performed. Student’s t test was used for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant
During positive selection, the Egr family members are one of the earliest transcription factors induced by TCR signaling.29,33,34 Compared with WT controls, TRAF3IP3KO iNKT cells at ST0, a stage at which iNKT cells have just undergone positive selection, showed similar levels of Egr2 staining (Fig. 1e). Positive selection is also followed by a phase of massive proliferation that depends on c-Myc and IL-7.35–37 c-Myc and its target gene expression were elevated in TRAF3IP3−/− Treg cells.38 However, the levels of c-Myc and CD127 in KO ST0 iNKT cells were comparable to their WT controls (Fig. 1f, g). Similar levels of ST0 cell proliferation and survival were also observed in KO and WT iNKT cells (Fig. 1h, i, Fig. S1H–I). Together, our data demonstrated that positive selection and early development of iNKT cells were not affected in TRAF3IP3-deficient mice.
TRAF3IP3 is critical for NKT2 cell effector function
We next determined whether iNKT subsets and effector functions were affected in TRAF3IP3KO mice. Compared with WT thymi, the percentage and number of PLZFhiRORγt− NKT2 cells were significantly lower, whereas those of PLZFloRORγt− NKT1 and PLZFintRORγt+ NKT17 cells were similar in TRAF3IP3KO cells (Fig. 2a). NKT2 cells can also be identified by surface expression of IL-17RB and CD27.2,3 We found reduced frequencies of IL-17RB+CD27+ or IL-17RB+CD4+ iNKT cells in KO thymi (Fig. 2b). Upon PMA and ionomycin stimulation, TRAF3IP3KO iNKT cells showed significantly fewer IL-4+IFN-γ− and IL-4+IFN-γ+ cells, more IL-4−IFN-γ+ cells, and comparable IL-17+ cells than WT cells (Fig. 2c, d). Because activated NKT1 cells could secrete IFN-γ and IL-4 simultaneously, we further purified NKT1 (CD122+), NKT2 (CD122−CD4+), and NKT17 (CD122−CD4−) subsets6 from WT and TRAF3IP3KO mice. As shown in Fig. 2e and S2A, only KO NKT2 cells produced less IL-4 upon stimulation. The low percentages of IL-4+ cells in WT and KO NKT1 populations were similar. These results suggest that TRAF3IP3KO mice have reduced NKT2 cells in the thymus.
Fig. 2.
NKT2 differentiation and function were impaired in TRAF3IP3KO mice. a Flow cytometry analysis of NKT1 (PLZFloRORγt−), NKT2 (PLZFhiRORγt−), and NKT17 (PLZFintRORγt+) cells in thymic CD1d-tet+TCRβ+CD24− iNKT cells obtained from WT and TRAF3IP3KO mice. The percentage and number of NKT1, NKT2, and NKT17 cells were calculated. Four independent experiments were performed with 5–7 mice in each group. b Comparison of IL-17RB+CD27+ or IL-17RB+CD4+ NKT2 (CD1d-tet+TCRβ+) cells in WT and TRAF3IP3KO thymi. Data are representative of three independent experiments. c–d Intracellular staining of IFN-γ, IL-4, and IL-17A in thymic CD24− iNKT cells after 4 h of PMA and ionomycin stimulation. Data are representative of 3–5 independent experiments. e CD122+ NKT1, CD122−CD4+ NKT2, and CD122−CD4− NKT17 cells were purified by flow cytometry and stimulated with PMA and ionomycin. The intracellular staining of IL-4 was performed 4 h later. f Analysis of serum IFN-γ and IL-4 levels by ELISAs. WT and TRAF3IP3KO mice were stimulated with α-GalCer via intravenous injection. The sera were collected 2 h later. Two independent experiments were performed with three mice in each group. g–h Flow cytometry analysis of IFN-γ+, IL-4+, and IL-17A+ peripheral iNKT cells 2 h after α-GalCer injection. Data are representative of three independent experiments. i Quantitative PCR analysis of Il13 transcription in purified CD1d-tet+TCRβ+ liver iNKT cells 3 h after α-GalCer injection. Four mice were analyzed. j Representative H&E staining of liver sections obtained from α-GalCer-stimulated WT and TRAF3IP3KO mice. The scale bar is 50 μm. k Quantification of serum ALT and AST obtained from α-GalCer-stimulated mice. Data are representative of two independent experiments with 4–5 mice in each group
We further examined the effector functions of peripheral iNKT cells by stimulating the mice with α-GalCer via intravenous injection. Compared with the serum samples obtained from WT mice, those from TRAF3IP3KO mice had much lower levels of IL-4 (Fig. 2f). Intracellular staining further showed that KO mice had fewer IL-4+IFN-γ+ and IL-4+IFN-γ− iNKT cells in the liver and spleen (Fig. 2g). The serum level of IFN-γ and the percentages of IFN-γ+IL-4− and IL-17+ iNKT cells were comparable between WT and KO mice (Fig. 2f–h). In addition to IL-4, IL-13 expression was reduced in KO iNKT cells (Fig. 2i). IL-4 produced by iNKT cells was shown to induce liver damage in α-GalCer-stimulated mice.39 Compared with WT controls, TRAF3IP3KO mice showed less severe liver damage as measured by histology and ALT/AST levels (Fig. 2j, k). These thymic and peripheral iNKT cell results demonstrated that TRAF3IP3KO mice have defects in NKT2 cell effector functions.
TRAF3IP3 regulates NKT2 cells in a cell-intrinsic manner
To examine whether the reduced NKT2 cells in TRAF3IP3KO mice are a cell-autonomous defect, the KO or WT recipients reconstituted with mixed bone marrow chimera were again analyzed. In either type of recipient, the percentages of PLZFhiRORγt−, IL-4+IFN-γ−, and IL-4+IFN-γ+ cells were significantly lower in KO than in WT donor-derived cells (Fig. 3, Fig. S2B, C). The ratios of IL-4−IFN-γ+ NKT1 and IL-17+ NKT17 cells were comparable. This finding indicates that the defects in NKT2 cells are owing to cell-intrinsic mechanisms.
Fig. 3.
TRAF3IP3 regulated NKT2 differentiation in a cell-intrinsic manner. WT (CD45.1+) and TRAF3IP3KO (CD45.2+) bone marrow cells were mixed at a 1:1 ratio and injected into lethally irradiated CD45.1+ WT a–b or CD45.2+ TRAF3IP3KO c–d recipients. a–b Flow cytometry analysis of iNKT subsets including PLZFloRORγt−, PLZFhiRORγt−, and PLZFintRORγt+ a and cytokine production upon stimulation b in thymic iNKT cells obtained from WT recipients. c–d iNKT subsets c and cytokine production d analysis of thymic iNKT cells obtained from TRAF3IP3KO recipients. Data are representative of three chimeric mice in each group
TRAF3IP3 deficiency results in impaired expansion and maturation of NKT2 cells
To investigate how NKT2 differentiation is affected, cell survival was first measured, and similar ratios of Annexin V+ iNKT cells were found between WT and KO thymi (Fig. 4a). In contrast, TRAF3IP3KO iNKT cells showed a selective reduction of proliferation in CD44+NK1.1− (ST2) or CCR7−CD44+NK1.1− cells (Fig. 4b). The expansion of immature CD44loCD24− (ST1), CCR7+CD24− (NKTp), or CCR7−CD44lo cells and mature NK1.1+ ST3 cells was similar between WT and KO mice (Fig. 4b). These results suggest that the expansion of CD44+NK1.1− iNKT cells is specifically impaired in TRAF3IP3KO mice.
Fig. 4.
TRAF3IP3KO mice had defects in NKT2 cell expansion and maturation. a Flow cytometry analysis of Annexin V+ cells in WT and TRAF3IP3KO thymic iNKT cells. The experiment was repeated three times. b Comparison of cell proliferation (Ki67 and BrdU staining) in various WT and KO NKT subpopulations (ST1–3 and CCR7+, CCR7−CD44lo, and CCR7−CD44+). Three independent experiments were performed. c Intracellular staining of IL-4 in CD44lo and CD44+ thymic iNKT cells after 4 h of PMA and ionomycin simulation. The experiment was repeated three times. d–e Flow cytometry analysis of GATA3 d and PLZF e expression in WT and KO thymic iNKT cells at various stages. f Comparison of PLZF and RORγt expression in CD44+NK1.1− (ST2) iNKT cells in WT and KO mice. g–h Flow cytometry analysis of Egr2 and c-Maf expression in WT and KO thymic iNKT cells at various stages. i Comparison of c-Maf expression in WT and KO PLZFhi NKT2 cells and RORγt+ NKT17 cells. j Flow cytometry analysis of IL-17RB and SLAMF6 in WT and KO iNKT cells at various stages. Data represent three independent experiments with at least three mice in each group
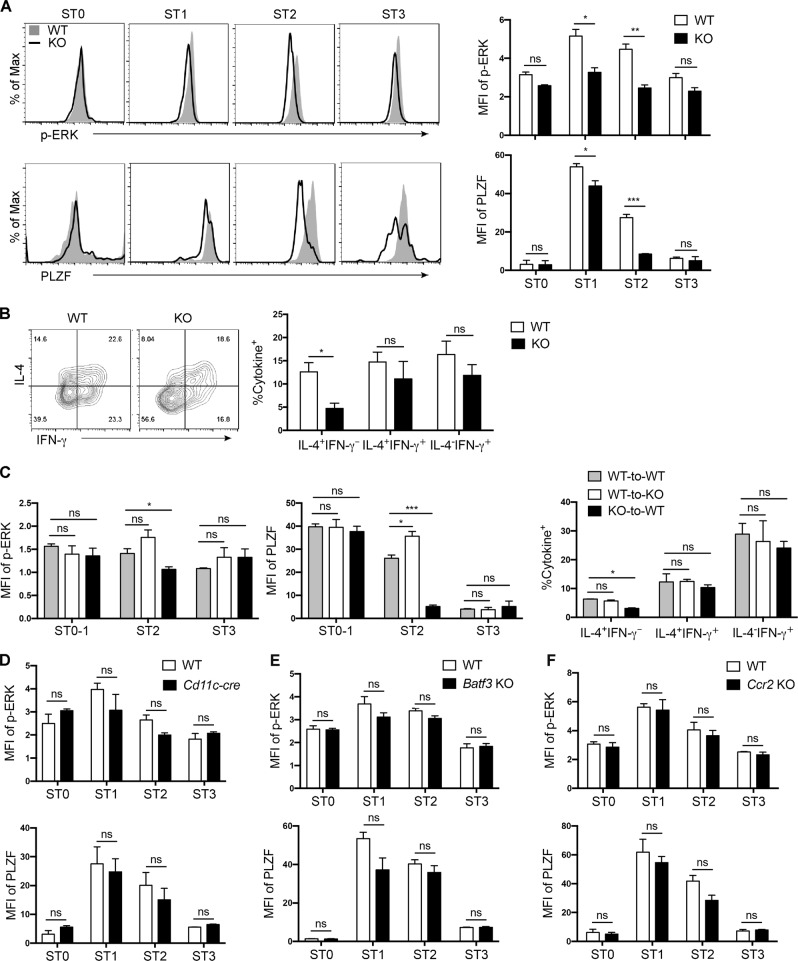
We also examined IL-4-producing capability and its regulation of transcription factors in iNKT cells at various developmental stages. Upon stimulation, a clear IL-4+ population measured by flow cytometry could only be found in CD44+ but not CD44lo iNKT cells (Fig. 4c). Significantly reduced IL-4+ cells were observed in KO CD44+ iNKTs. High expression of PLZF and GATA3 determines the IL-4-producing capability of thymic NKT2 cells. Compared with WT cells, TRAF3IP3KO iNKT cells had lower levels of GATA3 at both ST1 and ST2 (Fig. 4d, Fig. S2D). PLZF was substantially upregulated at ST1 and somewhat downregulated at ST2 in WT iNKT cells. ST0 and ST3 cells had the lowest levels of PLZF (Fig. 4e). In TRAF3IP3KO iNKT cells, the expression of PLZF was reduced specifically at ST2 (Fig. 4e). Thus, within the CD44+NK1.1− (ST2) TRAF3IP3KO iNKT population, fewer PLZFhiRORγt− (NKT2) cells and more PLZFloRORγt− cells were observed (Fig. 4f). The frequencies of RORγt+ (NKT17) cells were similar between WT and KO ST2 iNKTs. As the levels of T-bet and CD122 expression were similar between WT and KO iNKT cells, irrespective of developmental stage (Fig. S2E), the increased PLZFloRORγt− cells in CD44+ KO iNKTs may contain “poor NKT2” cells that fail to maintain a high level of PLZF expression.
Egr2 directly activates PLZF transcription in iNKT cells.40,41 c-Maf expression regulated by PLZF could restore the production of IL-4 and IL-10 in PLZF−/− iNKT cells.41 Consistent with PLZF expression, both Egr2 and c-Maf showed a specific reduction in KO ST2 iNKT cells. (Fig. 4g, h, Fig. S2F, G). We also examined c-Maf expression in PLZFhi NKT2 and PLZFintRORγt+ NKT17 subsets as ST2 contains both of these subsets, and c-Maf could also promote IL-17 expression in peripheral iNKT cells.42 Compared with WT cells, a lower level of c-Maf was found in TRAF3IP3KO NKT2 but not NKT17 cells (Fig. 4i). Specific downregulation of Egr2 and PLZF was also observed in TRAF3IP3KO iNKT cells when CCR7-CD44+NK1.1− terminally differentiated cells were gated (Fig. S3A, B). IL-17RB expression can also define NKT2 and IL-4 production. SLAMF6 was recently reported as one of the NKT2 signature molecules.18 As shown in Fig. 4j and Fig. S3C, D, lower levels of IL-17RB and SLAMF6 were found in ST2 but not ST1 iNKT cells in KO mice. Together, these data suggest that TRAF3IP3 is involved in regulating the expansion and functional maturation of CD44+ NKT2 cells.
MEK1/ERK activation facilitates NKT2 functional maturation
The ERK cascade regulates GATA3 stability, Il4 transcription, and Th2 differentiation in mature T cells,43,44 whereas TRAF3IP3 has been shown to promote MEK/ERK signaling in DP thymocytes.28 We thus measured MEK/ERK activation in iNKT cells. In WT cells, the phosphorylation of MEK and ERK was higher at ST1–2 than at ST0 (Fig. 5a). TRAF3IP3KO iNKT cells, however, showed reduced MEK and ERK activation at both ST1 and ST2 (Fig. 5a). Similarly, lower levels of phosphorylated ERK were observed in CCR7−CD44lo and CCR7−CD44+NK1.1− iNKT cells in KO mice (Fig. 5b). The levels of MEK/ERK activation in post-selected ST0 and CCR7+ NKTp cells were comparable between WT and KO mice (Fig. 5a, b). In contrast, WT and KO iNKT cells at all stages showed similar levels of SLP-76 phosphorylation (Fig. S3E).
Fig. 5.
Activation of the MEK/ERK signaling pathway promoted NKT2 maturation. a Flow cytometry analysis of phosphorylated MEK (p-MEK) and p-ERK in WT and TRAF3IP3KO thymic iNKT cells at various stages. b Comparison of p-ERK expression in CCR7+ and CD24−CCR7− iNKT subpopulations between WT and KO mice. c Enhanced ERK phosphorylation in DN32.D3 cells infected with MEK1DD (Ser218 to Asp, Ser221 to Asp)-expressing retrovirus. d Overexpression of MEK1DD increased Egr2 transcription (left) and protein expression (right) in DN32. D3 cells. e Overexpression of MEK1DD promoted Il4 transcription (left) and protein expression (right) upon PMA and ionomycin stimulation. Data are representative of three independent experiments
To examine whether MEK/ERK activation contributes to NKT2 maturation, DN32. D3 cells were transduced with retrovirus expressing a constitutively active form of MEK1 (MEK1DD).28 Compared with the cells with WT MEK, those with MEK1DD showed augmented ERK activation and elevated Egr2 and Il4 transcription and protein expression (Fig. 5c–e). The expression of c-Maf was also higher in the cells transduced with MEK1DD (data not shown). These data suggest that MEK/ERK activation at ST1 (CD44lo) likely facilitates the functional maturation of NKT2 cells.
TRAF3IP3 at the trans-Golgi network promotes MEK recruitment and subsequent ERK activation
We next investigated how TRAF3IP3 regulates MEK/ERK activity in iNKT cells. In DN32.D3 cells that were transduced with TRAF3IP3-expressing retrovirus, ERK phosphorylation and nuclear translocation were substantially elevated in the presence of anti-CD3 antibody (Fig. 6a, b). An interaction of TRAF3IP3 with MEK1 was detected in DN32.D3 cells (Fig. 6c). As the transmembrane protein TRAF3IP3 was found at Golgi in conventional T cells and lysosomes in regulatory T cells,28,38 we then used confocal microscopy to examine the cellular localization of TRAF3IP3 in iNKT cells. As shown in Fig. 6d, 70% of TRAF3IP3 colocalized with the trans-Golgi network (TGN) marker TGN46, whereas only 30% overlapped with the lysosomal marker LAMP1 (Fig. 6d). We also found overlapping distributions of MEK and TRAF3IP3, MEK, and TGN46 in WT iNKT cells (Fig. 6e, f). In TRAF3IP3KO iNKT cells, reduced colocalization of MEK and TGN46 was observed (Fig. 6f). These data indicated that TRAF3IP3 at the trans-Golgi network promotes MEK1 recruitment and subsequent ERK phosphorylation and nuclear translocation in iNKT cells.
Fig. 6.
TRAF3IP3 at the trans-Golgi network promoted MEK recruitment and subsequent ERK activation. a–b Overexpression of TRAF3IP3 promoted ERK phosphorylation a and nuclear translocation b in anti-CD3-activated DN32.D3 cells. Data are representative of two independent experiments. c DN32.D3 cells were infected with Flag-TRAF3IP3-expressing retrovirus. The interaction of MEK1 with TRAF3IP3 was measured by immunoprecipitation and western blotting. The experiment was repeated twice, and representative images are shown. d Analysis of subcellular localization of TRAF3IP3 in WT iNKT cells by fluorescence confocal microscopy. TGN46, trans-Golgi network marker, LAMP1, lysosome marker. e Confocal analysis of the colocalization of MEK and TRAF3IP3 in WT iNKT cells. f Confocal analysis of MEK and TGN46 in WT and KO iNKT cells. The experiments were repeated twice, and representative images are shown
The LTβR-regulated medullary thymic microenvironment promotes MEK/ERK activation and NKT2 maturation
As NKTp cells in the thymus migrate from the cortex to the medulla to continue their iNKT subset differentiation, we next investigated whether the medullary microenvironment provides signals for MEK/ERK activation in CCR7−CD44lo iNKT cells. Altered thymic medulla can be found in mice with lymphotoxin β receptor (LTβR) deficiency that leads to impaired negative selection of conventional T cells and emigration of iNKT cells.10,45 The differentiation of NKT2 was not examined in these mice. We found that iNKT cells in mice with global deletion of Ltbr had selective reduction of ERK phosphorylation and PLZF expression at ST1–2 and decreased IL-4+ cells upon stimulation (Fig. 7a, b), mimicking the phenotype obtained from TRAF3IP3KO mice. LTβR is expressed in a variety of nonlymphoid cells. To determine whether thymic stromal cells or bone marrow-derived nonlymphoid cells regulated by LTβR contribute to NKT2 maturation, lethally irradiated WT or Ltbr−/− recipients were reconstituted with Ltbr−/− or WT bone marrow cells, respectively. As shown in Fig. 7c, similar reductions in ERK phosphorylation, PLZF expression, and IL-4+ cell frequency were found in WT recipients with Ltbr−/− bone marrow cells but not Ltbr−/− recipients with WT bone marrow cells. Bone marrow-derived dendritic cells (DCs) have been shown to be critical for thymic negative selection of iNKT cells and differentiation of NKT17 cells.14,46 We thus used Cd11c-Cre to delete Ltbr in medullary DCs or used Batf3−/− and Ccr2−/− to specifically delete CD8α+ and CCR2+ DCs, respectively. However, none of these mice showed altered PLZF expression or ERK activation in iNKT cells (Fig. 7d–f). These results indicate that LTβR-regulated bone marrow-derived nonlymphoid cells play an important role in MEK/ERK activation and NKT2 maturation. However, the specific cell type that regulates NKT2 maturation awaits further investigation.
Fig. 7.
The LTβR-regulated thymic medullary microenvironment promotes MEK/ERK activation and NKT2 maturation. a Flow cytometry analysis of p-ERK and PLZF in thymic iNKT cells in WT and Ltbr−/− mice. b Intracellular staining of IL-4 in WT and Ltbr−/− iNKT cells upon stimulation. c Expression of p-ERK, PLZF, and IL-4 (after stimulation) in donor-derived thymic iNKT cells. WT-to-KO, lethally irradiated Ltbr−/− mice were reconstituted with WT bone marrow cells. WT-to-WT and KO-to-WT, WT mice were reconstituted with bone marrow cells from WT and Ltbr−/− mice, respectively. The experiments were repeated twice with three mice in each group. d–f Flow cytometry analysis of p-ERK and PLZF in thymic iNKT cells at various stages. iNKT cells obtained from Ltbrfl/flCd11c-Cre d, Batf3−/−e, and Ccr2−/−f mice were compared with those from WT mice. Three mice were in each group
Discussion
The differentiation of iNKT subsets is regulated by a variety of signaling pathways activated at the right stage and at appropriate levels. In the current study, we demonstrated that MEK/ERK activation in CCR7−CD44lo iNKT cells is critical for the functional maturation and expansion of CD44+ NKT2 cells in the thymic medulla. TRAF3IP3 at the trans-Golgi network promotes the recruitment of MEK1 and the subsequent phosphorylation and nuclear translocation of ERK during NKT2 maturation.
Thymic PLZFhi NKT2 cells with positive Il4 transcripts can be found in both CD44lo ST1 and CD44+ ST2 cells.3–5,13 The difference between and the relationship of ST1 and ST2 in terms of NKT2 differentiation remains elusive. We found that CD44+ iNKT cells were the only population in the WT thymus that could produce high levels of IL-4 protein upon stimulation. In TRAF3IP3KO thymus, the IL-4-producing capability was impaired. The transcription factors and surface molecules that are closely associated with IL-4 expression, including PLZF, c-Maf, IL-17RB, and SLAMF6, were also specifically reduced in ST2 and CCR7−CD44+ cells. These data demonstrated the importance of committed NKT2 cells (CD24−CCR7−CD44loPLZFhiIl4-transcript+) to undergo a tightly regulated maturation process to expand, upregulate their CD44 expression, and acquire a full capacity of IL-4 protein expression upon activation. This finding further suggests that care should be taken when interpreting the number and percentage of ST2 iNKT cells or PLZFloRORγt− NKT1 cells. iNKT cells at ST2 contain not only terminally differentiated NKT2, NKT17, and immature PLZFloNK1.1−NKT1 cells (Fig. 4f)13 but also “poor NKT2” cells that fail to maintain high PLZF/GATA3 expression and acquire an IL-4-producing capability upon stimulation. Thus, the study of iNKT subset differentiation requires a full evaluation of cytokine production as well as a set of key transcription factors, such as GATA3 and T-bet.
Appropriate ERK activation has been reported to play an important role in the expansion and cytokine production of peripheral iNKT cells. For instance, RASAL3-deficient mice with hyperactivated Ras/ERK and TPL2-deficient mice with hypoactivated ERK all showed severe defects in IL-4 and IFN-γ production in liver iNKT cells upon α-GalCer or ConA stimulation.31,32 Similar results were found in activated splenic iNKT cells in the presence of ERK inhibitor U0126.47 We also observed decreased IL-4+IFN-γ− and IL-4+IFN-γ+ iNKT cells in the liver and spleen of TRAF3IP3KO mice upon α-GalCer stimulation. The percentage of IL-4−IFN-γ+ and IL-17+ iNKT cells or serum IFN-γ was not affected. Our thymic iNKT cell data further indicated that the MEK/ERK signaling pathway has a critical role in promoting the functional maturation of NKT2 cells in the thymic medulla. Thus, ERK phosphorylation was reduced in ST1 and ST2 iNKT cells in TRAF3IP3KO and Ltbr−/− thymi. Overexpression of active MEK1 resulted in augmented ERK activation and increased IL-4 production in iNKT cells. NKT17 cells and immature NK1.1− NKT1 cells can also be found at ST2 (Fig. 4f),13 and the levels of TRAF3IP3 expression were similar among iNKT cells at various stages (data not shown), which further suggests that the expression levels of T-bet/RORγt and IFN-γ/IL-17 are not dependent on MEK/ERK activity.
The kinetics, duration, and strength of ERK signaling are regulated by subcellular localization and interaction with other signaling cascades or anchoring proteins.48 The Golgi apparatus is one such subcellular organelle that serves as a signaling platform to facilitate mitotic Golgi fragmentation and signaling to cytosolic substrates while suppressing MEK/ERK dissociation and ERK translocation to the nucleus.49–51 The data from our experiments indicate that Golgi localization can also have a positive effect on the nuclear functions of ERK. At least in iNKT cells, the TGN-localized transmembrane protein TRAF3IP3 interacts with MEK1 and facilitates ERK phosphorylation and nuclear translocation. The nuclear localized ERK likely promotes at least Egr2 transcription via phosphorylation of the Ets transcription factor ELKs.52
Thus, we demonstrated the importance of a functional maturation step in NKT2 differentiation in thymic medulla. This NKT2 maturation process requires MEK/ERK activation in CD24−CCR7−CD44loNK1.1− iNKT cells and is promoted by TRAF3IP3 at the trans-Golgi network. LTβR-regulated bone marrow-derived nonlymphoid cells in the medullary thymic microenvironment contribute to ERK activation and NKT2 maturation.
Supplementary information
Supplemental materials and supplemental figure legends
Acknowledgements
We thank Lilin Ye (Army Medical University, China) for kindly providing the Cd4-Cre transgenic mice and Li Bai (University of Science and Technology of China, China) for the DN32.D3 cell line. We also thank Professors Yu Zhang, Jun Zhang, Wenling Han (Peking University Health Science Center, China), and Li Wu (Tsinghua University) for critical comments, helpful discussions, and critical reagents. α-GalCer and unloaded and PBS57-loaded CD1d tetramers conjugated with phycoerythrin or allophycocyanin were supplied by the National Institutes of Health Tetramer Core Facility. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0104500), the National Natural Science Foundation of China (81471525, 31671244, 31872734, Q.G., 31872824, H.Z.), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81621001), and the Non-Profit Central Research Institute Fund of Chinese Academy of Medical Sciences, 2018PT31039.
Author contribution
Q.G., X.Z. and H.Z. designed the research, analyzed the data, and wrote the paper. X.Z. performed the research. Y.W., S.Z. and M.L. performed the research. L.C. J.H., R.J., X.S., X.H. and M.Z. contributed reagents and technical support. H.W. helped with flow cytometry. H.Z. edited the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Hounan Wu, Email: wuhounan@bjmu.edu.cn.
Hongshan Zhao, Email: hongshan@bjmu.edu.cn.
Qing Ge, Email: qingge@bjmu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41423-019-0234-0) contains supplementary material.
References
- 1.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat. Immunol. 2015;16:1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 2.Kwon DI, Lee YJ. Lineage differentiation program of invariant natural killer T Cells. Immune Netw. 2017;17:365–377. doi: 10.4110/in.2017.17.6.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watarai H, et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10:e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr. Opin. Immunol. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YJ, et al. Lineage-specific effector signatures of invariant NKT cells are shared amongst gammadelta T, innate lymphoid, and Th cells. J. Immunol. 2016;197:1460–1470. doi: 10.4049/jimmunol.1600643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgiev H, Ravens I, Benarafa C, Forster R, Bernhardt G. Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets. Nat. Commun. 2016;7:13116. doi: 10.1038/ncomms13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YJ, et al. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity. 2015;43:566–578. doi: 10.1016/j.immuni.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franki AS, et al. A unique lymphotoxin {alpha}beta-dependent pathway regulates thymic emigration of V{alpha}14 invariant natural killer T cells. Proc. Natl. Acad. Sci. USA. 2006;103:9160–9165. doi: 10.1073/pnas.0508892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White AJ, et al. A type 2 cytokine axis for thymus emigration. J. Exp. Med. 2017;214:2205–2216. doi: 10.1084/jem.20170271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallabhapurapu S, et al. Rel/NF-kappaB family member RelA regulates NK1.1- to NK1.1+ transition as well as IL-15-induced expansion of NKT cells. Eur. J. Immunol. 2008;38:3508–3519. doi: 10.1002/eji.200737830. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Hogquist KA. CCR7 defines a precursor for murine iNKT cells in thymus and periphery. Elife. 2018;7:e34793. doi: 10.7554/eLife.34793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Pietro C, et al. MicroRNA-133b regulation of Th-POK expression and dendritic cell signals affect NKT17 cell differentiation in the thymus. J. Immunol. 2016;197:3271–3280. doi: 10.4049/jimmunol.1502238. [DOI] [PubMed] [Google Scholar]
- 15.Gaya M, et al. Initiation of antiviral B cell immunity relies on innate signals from spatially positioned NKT cells. Cell. 2018;172:517–533.e520. doi: 10.1016/j.cell.2017.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Cardell SL. The Yin and Yang of invariant natural killer T cells in tumor immunity-suppression of tumor immunity in the intestine. Front. Immunol. 2017;8:1945. doi: 10.3389/fimmu.2017.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akbari O, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat. Med. 2003;9:582–588. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 18.Engel I, et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 2016;17:728–739. doi: 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz Tleugabulova M, et al. Discrete TCR binding kinetics control invariant NKT cell selection and central priming. J. Immunol. 2016;197:3959–3969. doi: 10.4049/jimmunol.1601382. [DOI] [PubMed] [Google Scholar]
- 21.Zhao M, et al. Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70. Nat. Commun. 2018;9:2627. doi: 10.1038/s41467-018-05095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura MY, et al. CD69 prevents PLZF(hi) innate precursors from prematurely exiting the thymus and aborting NKT2 cell differentiation. Nat. Commun. 2018;9:3749. doi: 10.1038/s41467-018-06283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White AJ, et al. An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J. Immunol. 2014;192:2659–2666. doi: 10.4049/jimmunol.1303057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elewaut D, et al. NIK-dependent RelB activation defines a unique signaling pathway for the development of V alpha 14i NKT cells. J. Exp. Med. 2003;197:1623–1633. doi: 10.1084/jem.20030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J. Exp. Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin C, Zhu M. RelB intrinsically regulates the development and function of medullary thymic epithelial cells. Sci. China Life Sci. 2018;61:1039–1048. doi: 10.1007/s11427-017-9298-3. [DOI] [PubMed] [Google Scholar]
- 28.Zou Q, et al. T cell development involves TRAF3IP3-mediated ERK signaling in the Golgi. J. Exp. Med. 2015;212:1323–1336. doi: 10.1084/jem.20150110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu T, Gimferrer I, Simmons A, Wiest D, Alberola-Ila J. The Ras/MAPK pathway is required for generation of iNKT cells. PLoS ONE. 2011;6:e19890. doi: 10.1371/journal.pone.0019890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borowski C, Bendelac A. Signaling for NKT cell development: the SAP-FynT connection. J. Exp. Med. 2005;201:833–836. doi: 10.1084/jem.20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito S, et al. RASAL3, a novel hematopoietic RasGAP protein, regulates the number and functions of NKT cells. Eur. J. Immunol. 2015;45:1512–1523. doi: 10.1002/eji.201444977. [DOI] [PubMed] [Google Scholar]
- 32.Vyrla D, et al. TPL2 kinase is a crucial signaling factor and mediator of NKT effector cytokine expression in immune-mediated liver injury. J. Immunol. 2016;196:4298–4310. doi: 10.4049/jimmunol.1501609. [DOI] [PubMed] [Google Scholar]
- 33.Lawson VJ, Weston K, Maurice D. Early growth response 2 regulates the survival of thymocytes during positive selection. Eur. J. Immunol. 2010;40:232–241. doi: 10.1002/eji.200939567. [DOI] [PubMed] [Google Scholar]
- 34.Lazarevic V, et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 2009;10:306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dose M, et al. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. USA. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mycko MP, et al. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J. Immunol. 2009;182:4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 37.Tani-ichi S, et al. Interleukin-7 receptor controls development and maturation of late stages of thymocyte subpopulations. Proc. Natl. Acad. Sci. USA. 2013;110:612–617. doi: 10.1073/pnas.1219242110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X, et al. Metabolic control of regulatory T cell stability and function by TRAF3IP3 at the lysosome. J. Exp. Med. 2018;215:2463–2476. doi: 10.1084/jem.20180397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Feng D, Park O, Yin S, Gao B. Invariant NKT cell activation induces neutrophil accumulation and hepatitis: opposite regulation by IL-4 and IFN-gamma. Hepatology. 2013;58:1474–1485. doi: 10.1002/hep.26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seiler MP, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 2012;13:264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gleimer M, von Boehmer H, Kreslavsky T. PLZF controls the expression of a limited number of genes essential for NKT cell function. Front. Immunol. 2012;3:374. doi: 10.3389/fimmu.2012.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu JS, et al. Differentiation of IL-17-producing invariant natural killer T cells requires expression of the transcription factor c-Maf. Front. Immunol. 2017;8:1399. doi: 10.3389/fimmu.2017.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita M, et al. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J. Biol. Chem. 2005;280:29409–29419. doi: 10.1074/jbc.M502333200. [DOI] [PubMed] [Google Scholar]
- 44.Tripathi P, et al. A novel mechanism for ERK-dependent regulation of IL4 transcription during human Th2-cell differentiation. Immunol. Cell Biol. 2012;90:676–687. doi: 10.1038/icb.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu M, Chin RK, Tumanov AV, Liu X, Fu YX. Lymphotoxin beta receptor is required for the migration and selection of autoreactive T cells in thymic medulla. J. Immunol. 2007;179:8069–8075. doi: 10.4049/jimmunol.179.12.8069. [DOI] [PubMed] [Google Scholar]
- 46.Chun T, et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J. Exp. Med. 2003;197:907–918. doi: 10.1084/jem.20021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian J, et al. The differential roles of mTOR, ERK, and JNK pathways in invariant natural killer T-cell function and survival. Inflammation. 2014;37:2013–2019. doi: 10.1007/s10753-014-9933-y. [DOI] [PubMed] [Google Scholar]
- 48.Wainstein E, Seger R. The dynamic subcellular localization of ERK: mechanisms of translocation and role in various organelles. Curr. Opin. Cell Biol. 2016;39:15–20. doi: 10.1016/j.ceb.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Yao Z, Seger R. The ERK signaling cascade--views from different subcellular compartments. Biofactors. 2009;35:407–416. doi: 10.1002/biof.52. [DOI] [PubMed] [Google Scholar]
- 50.Wortzel I, Seger R. The ERK cascade: distinct functions within various subcellular organelles. Genes Cancer. 2011;2:195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev. Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 52.Maurice D, Costello P, Sargent M, Treisman R. ERK signaling controls innate-like CD8(+) T cell differentiation via the ELK4 (SAP-1) and ELK1 transcription factors. J. Immunol. 2018;201:1681–1691. doi: 10.4049/jimmunol.1800704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and supplemental figure legends