Abstract
Biomarkers to predict risk of liver fibrosis in subjects with nonalcoholic fatty liver disease, a common risk factor for hepatocellular carcinoma, would allow for early preventive interventions. We sought to characterize bile acid profiles associated with liver fibrosis in subjects from the community‐based Cameron County Hispanic Cohort, a population in South Texas with high rates of nonalcoholic fatty liver disease, liver fibrosis and hepatocellular carcinoma. Plasma bile acid levels were measured in 390 subjects. These subjects were screened with liver elastography, detecting significant liver fibrosis in 58 subjects and steatosis in 186 subjects. Unsupervised clustering of the bile acid profiles revealed five clusters that differed by liver fibrosis, liver steatosis, liver injury, age and gender, identifying these parameters as major determinants of circulating bile acid changes. Total bile acid levels were significantly higher in subjects with fibrosis, with chenodeoxycholic acid displaying the greatest increase among individual bile acids. The primary conjugated bile acids, glycocholic and glycochenodeoxycholic acids, displayed the strongest association with fibrosis by logistic regression. High lithocholic acid levels were strongly associated with advanced fibrosis. In contrast, deoxycholic acid and total unconjugated secondary bile acids were positively associated with steatosis, whereas relative glycoursodeoxycholic acid abundance was negatively associated. Milk and yogurt intake notably contributed to fibrosis‐associated bile acid changes. In addition, multiple families within the Firmicutes phylum, Prevotellaceae, and Bacteroides species in stool significantly correlated with fibrosis‐associated and steatosis‐associated bile acid parameters, suggesting that the gut microbiome contributes to bile acid changes in the context of liver disease. Conclusion: Circulating bile acid levels were markedly but differently changed in liver fibrosis and steatosis in a high‐risk Mexican‐American population.
Within the community‐based Cameron County Hispanic Cohort, a population in South Texas with high rates of nonalcoholic fatty liver disease, liver fibrosis and hepatocellular carcinoma, circulating bile acid levels were markedly but differently changed in liver fibrosis and liver steatosis. In addition, milk and yogurt intake, as well as several gut microbiota, contributed to fibrosis‐associated bile acid changes.
Abbreviations
- ALT
alanine aminotransferase
- AOR
adjusted odds ratio
- AST
aspartate aminotransferase
- CA
cholic acid
- CAP
controlled attenuation parameter
- CCHC
Cameron County Hispanic Cohort
- CDCA
chenodeoxycholic acid
- CI
confidence interval
- DCA
deoxycholic acid
- ELISA
enzyme‐linked immunosorbent assay
- GCA
glycocholic acid
- GCDCA
glycochenodeoxycholic acid
- GDCA
glycodeoxycholic acid
- GLCA
glycolithocholic acid
- GUDCA
glycoursodeoxycholic acid
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HSDH
hydroxysteroid dehydrogenase
- LCA
lithocholic acid
- LSM
liver stiffness measurement
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- OR
odds ratio
- PAM
Partitioning Around Medoids
- TCA
taurocholic acid
- TCDCA
taurochenodeoxycholic acid
- TDCA
taurodeoxycholic acid
- TLCA
taurolithocholic acid
- TUDCA
tauroursodeoxycholic acid
- UDCA
ursodeoxycholic acid
Nonalcoholic fatty liver disease (NAFLD) ranges from simple steatosis to the more severe nonalcoholic steatohepatitis (NASH). Liver fibrosis commonly arises from NAFLD and is the primary determinant of mortality in NAFLD in the United States.( 1 ) NAFLD is also a common risk factor for hepatocellular carcinoma (HCC).( 2 ) NAFLD is closely associated with metabolic comorbidities, including obesity and diabetes; subsequently, the incidence of NAFLD, NASH, liver fibrosis, and HCC are expected to continue increasing in the United States, due to the epidemics of obesity and diabetes.( 3 , 4 )
Mexican Americans are an expanding population in the United States with a high prevalence of obesity, diabetes, liver steatosis, and liver fibrosis.( 5 , 6 , 7 ) Although HCC incidence is higher in males than females, the incidence in both male and female Mexican Americans is double that of their non‐Mexican‐American white counterparts,( 4 ) at 19.7 and 7.8 new cases per 100,000, respectively, in Mexican Americans versus 10.3 and 3.6 new cases in non‐Mexican‐American whites. A disproportionately large fraction of HCC cases are of NASH etiology in Mexican Americans.( 8 ) Furthermore, Mexican Americans in South Texas have higher HCC incidence than Mexican Americans living elsewhere in the United States.( 9 , 10 ) Biomarkers that predict the risk of liver fibrosis in the context of NASH would allow for early preventive interventions in this high‐risk population.
Although dysregulation of hepatic bile acid synthesis and increased circulating bile acid levels have been documented in the context of NAFLD and NASH,( 11 , 12 , 13 , 14 , 15 ) bile acid changes during liver fibrosis development remain to be characterized. It is also unknown whether bile acid profiles are different between races and ethnicities. We therefore aimed to characterize the bile acid profiles associated with liver fibrosis in subjects from the Cameron County Hispanic Cohort (CCHC). CCHC is a population‐based Mexican‐American cohort in South Texas, with high rates of obesity, NAFLD, and HCC.( 16 , 17 , 18 , 19 )
Methods
Study Participants
The study includes 390 randomly selected CCHC participants enrolled between March 1, 2016, and June 19, 2018. Written informed consent was obtained from each participant for their clinical records to be used in this study. The study protocol was approved by the Committee for the Protection of Human Subjects of the University of Texas Health Science Center at Houston. All participants underwent a comprehensive clinical exam, detailed health history, and demographic interview. Hepatitis C virus (HCV) antibodies and hepatitis B virus (HBV) surface antigen (HBsAg) were assayed using the Ortho HCV Version 3.0 enzyme‐linked immunosorbent assay (ELISA) Test System (Ortho‐Clinical Diagnostics, Raritan, NJ) and HBsAg ELISA Kit (Abnova, Taipei, Taiwan), respectively. Participants who were positive for HCV or HBV were excluded from the study. Fasting blood samples were analyzed for metabolic and lipid panels at a Clinical Laboratory Improvement Amendments–certified laboratory, except insulin, which was measured in the laboratory by ELISA assay (Mercodia, Uppsala, Sweden). Obesity was defined as body mass index (BMI) ≥ 30. Prediabetes was defined as either fasting blood glucose of 100‐125 mg/dL or hemoglobin A1c (HbA1c) of 5.7%‐6.4% with no history of diabetic medication. Diabetes was defined as fasting blood glucose ≥ 126 mg/dL, HbA1c ≥ 6.5%, or history of diabetic medication. The homeostasis model assessment (HOMA) was calculated as glucose (mg/dL)/18 × insulin (mU/L)/22.5. Abnormal aspartate aminotransferase (AST) was defined as > 33 U/L, and abnormal alanine aminotransferase (ALT) was defined as > 40 U/L for males and > 31 U/L for females. Heavy drinking was defined as weekly alcohol consumption of > 20 g/day for men and > 10 g/day for women; moderate drinking was defined as nonzero weekly consumption that did not reach the criteria for heavy drinking. Former smoking was defined as lifetime consumption of ≥ 100 cigarettes, plus no smoking at the time of survey. Current smoking was defined as lifetime consumption of ≥ 100 cigarettes, plus smoking at the time of survey. Information on autoimmune liver disease status is not available for this cohort.
Liver Elastography
For all subjects, trained operators obtained controlled attenuation parameter (CAP) measurements (dB/m) for liver steatosis, and liver stiffness measurements (LSMs) (kiloPascals) for liver fibrosis, using vibration‐controlled transient elastography (either FibroScan 502 Touch or FibroScan 530 Compact; Echosens, Waltham, MA) with automatic probe selection and a total of 10 valid measures. Presence of liver steatosis was defined as CAP > 281. Significant fibrosis was defined as LSM > 7.1 kPa, whereas advanced fibrosis and cirrhosis were defined as LSM > 8.8 kPa and > 11.8 kPa, respectively.
Dietary Intake Information
During participant visits, dietary intake for the day before the interview was recorded using a diet questionnaire adapted from the School Physical Activity and Nutrition survey. Dietary groups were calculated from the sum of individual foods, whereas composite scores for unhealthy and healthy eating indices were calculated based on past studies using the School Physical Activity and Nutrition survey and CCHC data.( 20 , 21 ) The specific foods included in each group and index are provided in Supporting Table S1.
Bile Acid Measurements
Plasma samples were collected during participant visits. Samples were sent to Metabolon (Durham, NC) for analysis of absolute bile acid concentrations by liquid chromatography tandem mass spectrometry. The bile acid panel included measurements for 15 major primary and secondary acids, as well as their glycine and taurine conjugates: cholic acid (CA), glycocholic acid (GCA), taurocholic acid (TCA), chenodeoxycholic acid (CDCA), glycochenodeoxycholic acid (GCDCA), taurochenodeoxycholic acid (TCDCA), deoxycholic acid (DCA), glycodeoxycholic acid (GDCA), taurodeoxycholic acid (TDCA), lithocholic acid (LCA), glycolithocholic acid (GLCA), taurolithocholic acid (TLCA), ursodeoxycholic acid (UDCA), glycoursodeoxycholic acid (GUDCA), and tauroursodeoxycholic acid (TUDCA). For measurements below the limit of quantitation, the measurement was replaced by half of the lower limit of quantitation.
Stool DNA Extraction and Bacterial 16S ribosomal RNA Sequencing
Stool samples were collected from 123 CCHC subjects using the OMNIgene GUT stool collection kit (DNA Genotek, Ontario, Canada). Samples were shipped to the MD Anderson Cancer Center at room temperature and stored at −80°C until analysis. Sample preparation and 16S ribosomal RNA (rRNA) sequencing were performed at the MD Anderson Cancer Center Microbiome Core Facility. Briefly, bacterial genomic DNA was extracted using the QIAamp Fast DNA stool mini Kit (Qiagen, Hilden, Germany). The V4 region of the bacterial 16S rDNA gene was amplified by polymerase chain reaction (PCR) (approximately 400 bp), from 100 ng of purified genomic stool DNA, using primers 515F (5′‐ AATGATACGGCGACCACCGAGATCTACACGCTXXXXXXXXXXXXTATGGTAATTGTGTGYCAGCMGCCGCGGTAA‐3′, where XXXXXXXXXXXX is an index sequence for multiplexing libraries) and 806R (5′‐ CAAGCAGAAGACGGCATACGAGATAGTCAGCCAGCCGGACTACNVGGGTWTCTAAT‐3′). The PCR conditions used were 98°C for 3 minutes, followed by 27 cycles of 98°C for 50 seconds, 55°C for 30 seconds, 72°C for 30 seconds, then a final 72°C for 5 minutes. Libraries were purified using Zymo I‐96 column purification and analyzed on the Agilent 4200 Tapestation system (Agilent, Santa Clara, CA). The barcoded amplicons were pooled in equal concentrations and quantified by Qubit fluorometer (Invitrogen, Carlsbad, CA); molarity was calculated based on amplicon size. The sequencing run was performed using 250 bp paired‐end on the Illumina MiSeq platform using custom primers (Read1 seq primer: 5′‐TATGGTAATTGTGTGYCAGCMGCCGCGGTAA‐3′; Read2 seq primer: 5′‐AGTCAGCCAGCCGGACTACNVGGGTWTCTAAT‐3′; and index primer: AATGATACGGCGACCACCGAGATCTACACGCT). Sequencing data from paired‐end reads were de‐multiplexed and split in QIIME.( 22 ) Merging of paired‐end reads to create consensus sequences was done by VSEARCH v7, allowing up to 10 mismatched. The cluster_otus command, an implementation of the UPARSE algorithm, was used to perform 97% related operational taxonomic unit (OTU) clustering. Denoising was done by the unoise3 command. OTUs were then subjected to taxonomy assignment using the Mothur with Silva database. A phylogenetic tree was built with the FastTree method in the QIIME package.
Statistical Analyses
Partitioning Around Medoids (PAM) clustering was performed in R (version 3.5.1; R Foundation for Statistical Computing, Vienna, Austria) to cluster samples based on their individual bile acid profiles. Consensus clustering was run to determine cluster number and class membership by stability evidence, using 80% item resampling, 100% feature resampling, a maximum evaluated k of 10, 1,000 resamplings, and PAM clustering algorithm following 1 − Spearman correlation distances. The consensus cumulative distribution function plot and change in area under the curve indicated that a cluster number of five or more was appropriate; subsequently, subjects were clustered into five groups based on PAM clustering. Logistic regression was performed to determine clinical differences between each bile acid cluster versus the rest of the study group, and differences between each grouping of high and low bile acid abundance. To that end, odds ratios and 95% confidence intervals for each clinical parameter were generated, adjusted by age and gender. Differences in bile acid parameters between fibrotic and nonfibrotic subjects was assessed by the Mann‐Whitney U test, with P < 0.05 considered statistically significant. Principal component analysis was performed with the Euclidian‐based distances matrix, generated using log10‐transformed abundance of the 15 individual bile acids. Redundancy analysis was performed using the RDA function in the Vegan package for R. Log10‐transformed abundances of the 15 individual bile acids were used as the response variables; log10‐transformed diet parameters were used as the explanatory variables. Analysis of variance–like, permutation‐based tests were used to assess the significance of the model and of each constrained axis, as well as the marginal effects of each explanatory variable. Mantel's test using 9,999 permutations was used to assess the association between bile acid profiles and microbiome profiles of the same subjects. The test was performed on the Euclidean‐based distance matrix of bile acid profiles, and on the weighted UniFrac‐based distance matrix of microbiome profiles. Spearman's test was used to assess correlation between bacterial taxa and bile acid parameters.
Results
Bile Acid Profiles Identify Five Subject Clusters
Plasma samples were obtained from 390 participants in the CCHC who were screened with FibroScan for liver fibrosis and steatosis. Participants positive for HCV or HBV were excluded. The demographic and clinical parameters of all study participants are given in Supporting Table S2. Briefly, the age average was 51.9, and 134 subjects (34.4%) were male. Among all participants, 188 (48.3%) were obese, 100 (26%) had diabetes, and 186 (47.7%) had liver steatosis (CAP > 281). In addition, 102 (26.2%) were moderate drinkers and 24 (6.2%) were heavy drinkers. The study cohort included 52 subjects (13.3%) with significant liver fibrosis (kPa > 7.1), of which 27 subjects (6.9%) had advanced liver fibrosis (kPa > 8.8) and 19 subjects (4.9%) had cirrhosis (kPa > 11.8). Subjects with significant fibrosis were more likely to have liver steatosis (37 [71.2%]) and to be obese (34 [65.4%]), diabetic (24 [46.2%]), and heavy drinkers (5 [9.6%]). A total of 15 bile acids were measured in the plasma of these 390 participants (Fig. 1). The most abundant bile acid was GCDCA (mean = 321.4 ng/mL), followed by DCA (mean = 190.1 ng/mL). With the exception of TLCA and TUDCA, all were detected above the detection limit in more than 50% of samples.
Fig. 1.
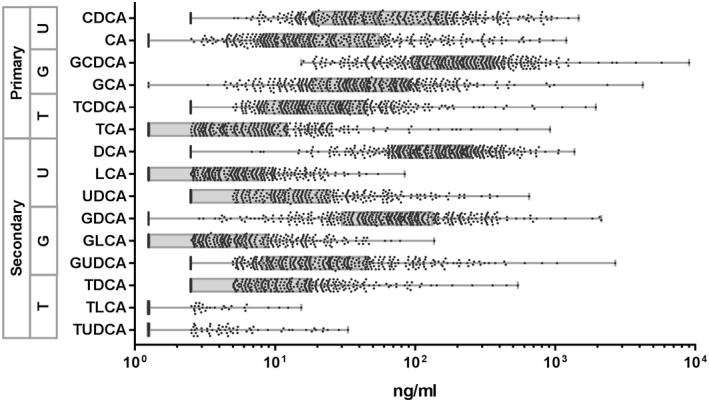
Plasma levels of individual bile acids. Dots represent individual subjects. Box represents the range between first and third quartiles. Line within the boxes represents the median values. Whiskers denote the minimum and maximum values. Abbreviations: G, glyco‐conjugated; T, tauro‐conjugated; U, unconjugated.
Consensus clustering with PAM of the 390 samples based on bile acid profiles revealed five clusters (clusters A‐E) (Fig. 2A). Cluster A was the most distinct, followed by cluster B, whereas clusters C, D, and E were more similar to each other. Separation of clusters was also observed by principal component analysis of all subjects, based on individual bile acid profiles (Fig. 2B). Subjects in clusters D and C had healthier liver function compared with all study participants. Subjects in cluster D were older, with a median age of 55.8 compared with 51.9 in the rest of the study population (P = 0.01). Only 4.9% of subjects in cluster D had abnormal AST levels compared with 17% in the rest of the study population (odds ratio [OR] = 0.25, 95% confidence interval (CI) = 0.09‐0.71, P = 0.01), and 22% had abnormal ALT levels compared with 36.3% in the rest of the study population (OR = 0.49, 95% CI = 0.28‐0.88, P = 0.016). Similarly, cluster C was associated with lower ALT (P = 0.023) and lower AST, although not significantly. Subjects in cluster C were more likely to be female (OR = 3.38, 95% CI = 1.38‐8.25, P = 0.007), to have a lower BMI (OR = 0.92, 95% CI = 0.86‐0.98, P = 0.013), and to be never drinkers and never smokers (80.5% and 85.4% compared with 66.2% and 66.2% in the rest of the study population, respectively). However, subjects in cluster C had significantly higher cholesterol, high‐density lipoprotein and low‐density lipoprotein, and most (55%) were prediabetic. Subjects in cluster B were significantly younger (P = 0.004) and had lower rates of metabolic disease, although not significantly. Interestingly, while AST and ALT levels in cluster B subjects were comparable to the overall study cohort, the NAFLD score was significantly lower at −1.9 compared with −1.5 in the rest of the study population (OR = 0.80, 95% CI = 0.64‐1.00, P = 0.045). Subjects in clusters A and E had significantly higher frequencies of abnormal FibroScan results, but remarkably, liver fibrosis and liver steatosis did not cluster together. Subjects in cluster A had the highest frequency of significant liver fibrosis (16.4% compared with 12.1%), and the highest LSM average at 7.1 kPa compared with the overall 5.1 kPa in the study population (P = 0.017). Cluster A also had higher daily alcohol intake (4.7 g/day compared with 2.4 g/day in the rest of the study population, P = 0.035) and a higher frequency of heavy drinkers (9.1% compared with 14%), although this did not reach significance. On the other hand, subjects in cluster E had the highest frequency of subjects with liver steatosis (58.8%) (OR = 1.88, 95% CI = 1.21‐2.93, P = 0.005) and the highest average FibroScan CAP score of 285.9 compared with 270.2 in the rest of the study population (P = 0.024). Twenty‐two percent of subjects in cluster E had abnormal AST compared with 12.0% in the rest of the study population (OR = 1.85, 95% CI = 1.03‐3.31, P = 0.04). It is also important to note that subjects in cluster E were significantly younger (OR = 0.98, 95% CI = 0.97‐1.00, P = 0.018), more likely to be male (OR = 1.88, 95% CI = 1.20‐2.95, P = 0.006), to be moderate drinkers (OR = 1.78, 95% CI = 1.09‐2.90, P = 0.021), and to have high HOMA scores (OR = 1.08, 95% CI = 1.00‐1.16, P = 0.038). Triglyceride levels were also high in subjects of cluster E. A summary of the demographic and clinical parameters specifically associated with the five clusters is provided in Supporting Table S3. In conclusion, liver fibrosis was the major contributor of bile acid changes in blood. Other contributors included age, liver injury, liver steatosis, and gender (Fig. 2B). Remarkably, obesity, diabetes, and metabolic diseases are only marginally associated with bile acid profiles.
Fig. 2.
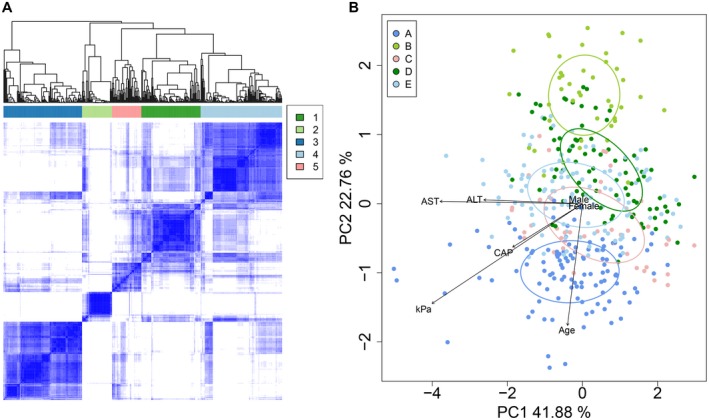
Consensus clustering with PAM identified five groups of subjects based on bile acid profiles. (A) Similarity matrix for the five clusters of subjects. (B) Principal component analysis based on log10 bile acid abundances shows separation of the five clusters. Ellipses were drawn using the SD of point scores. Selected clinical variables were fitted onto the plot using the envfit function in the Vegan package.
Bile Acids Associated With Liver Fibrosis and Liver Steatosis
A significant increase in total bile acid levels was observed in subjects with fibrosis (median = 1340 ng/mL) compared to subjects with no fibrosis (median = 721.3 ng/mL) (fold change = 1.86, P < 0.001). This effect was largely due to primary bile acids (median for fibrosis = 787.4 ng/mL and for no fibrosis = 396.3 ng/mL, fold change = 1.99, P < 0.001). Accordingly, the primary/secondary bile acid ratio was also increased (median for fibrosis = 1.69 and for no fibrosis = 1.27, fold change = 1.33, P = 0.017). To further determine the specific bile acids associated with liver fibrosis, subjects were then grouped into quartile 4 (Q4) versus quartiles 1‐3 (Q1‐Q3) for each bile acid parameter for binary logistic regression. After adjusting for age and gender, a strong positive association between liver fibrosis (kPa > 7.1) and high total bile acid levels (Q4) was observed (adjusted odds ratio [AOR] = 4.40, 95% CI = 2.37‐8.18, P < 0.001) (Fig. 3). The association was even stronger with liver cirrhosis (kPa > 11.8) (AOR = 7.41, 95% CI = 2.71‐20.27, P < 0.001). Although this association was observed for both total primary and total secondary bile acids, the effect was stronger for primary bile acids, especially in the context of cirrhosis (AOR = 7.53, 95% CI = 2.76‐20.55, P < 0.001 compared with AOR = 2.62, 95% CI = 1.02‐6.71, P = 0.045). Among the five bile acid groups (CDCA, CA, DCA, LCA, and UDCA), UDCA was the only one not contributing to the association between higher bile acid levels and liver fibrosis. Among individual bile acids, the strongest association with liver fibrosis was observed for the glyco‐conjugated forms of CA (GCA) and CDCA (GCDCA) (AOR = 3.14, 95% CI = 1.70‐5.79, P < 0.001 and AOR = 3.26, 95% CI = 1.76‐6.01, P < 0.001; AOR = 7.91, 95% CI = 2.88‐21.71, P < 0.001 and AOR = 4.74, 95% CI = 1.83‐12.27, P = 0.001, respectively, for advanced fibrosis) (Fig. 3). The association between GCA and GCDCA and fibrosis remained when the data were further adjusted for total bile acid levels. High levels of LCA were particularly associated with advanced liver fibrosis (kPa > 8.8; AOR = 5.16, 95% CI = 2.07‐12.88, P < 0.001). The association again remained when adjusted by total bile acid levels.
Fig. 3.
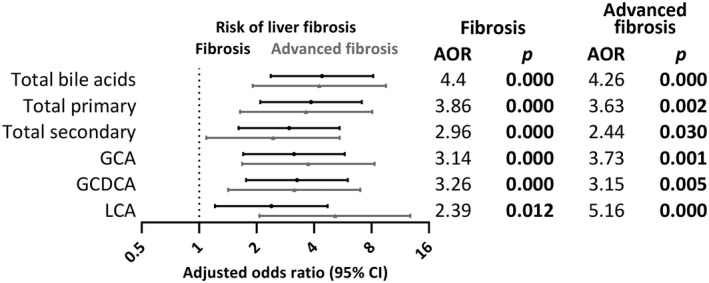
Forest plot of significant associations of high bile acid abundance (Q4) expression with liver fibrosis. Black lines represent AORs for fibrosis (kPa > 7.1), and gray lines represent AORs for advanced fibrosis (kPa > 8.8).
In contrast to liver fibrosis, total bile acids levels were not significantly increased in subjects with liver steatosis with medians of 842.2 ng/mL and 677.8 ng/mL in subjects with and without steatosis, respectively (fold change = 1.24, P < 0.063). With logistic regression, after adjusting for age and gender, a significant positive association between liver steatosis and high expression (Q4) was observed for total unconjugated secondary bile acids (AOR = 1.80, 95% CI = 1.12‐2.88, P = 0.015), largely due to DCA (AOR = 1.88, 95% CI = 1.17‐3.01, P = 0.009) (Fig. 4). High relative amounts of GUDCA were negatively associated with steatosis (AOR = 0.55, 95% CI = 0.35‐0.89, P = 0.014) (Fig. 4).
Fig. 4.
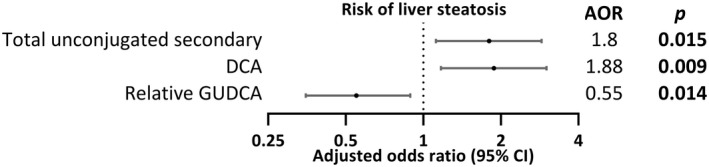
Forest plot of significant associations of high bile acid abundance (Q4) expression with liver steatosis.
Taken together, these results suggest that important overall changes in circulating bile acids were observed in the context of fibrosis, but only limited specific changes were observed in the context of steatosis with no overlap between fibrosis‐associated and steatosis‐associated profiles. High abundance of the glyco‐conjugated primary bile acids GCA and GCDCA displayed the strongest association with fibrosis. Accordingly, total bile acids, total primary bile acids, and the individual bile acids GCA and GCDCA displayed the strongest positive correlations with FibroScan LSM (r = 0.225, r = 0.234, r = 0.252, and r = 0.211, respectively; P < 0.001) (Fig. 5).
Fig. 5.
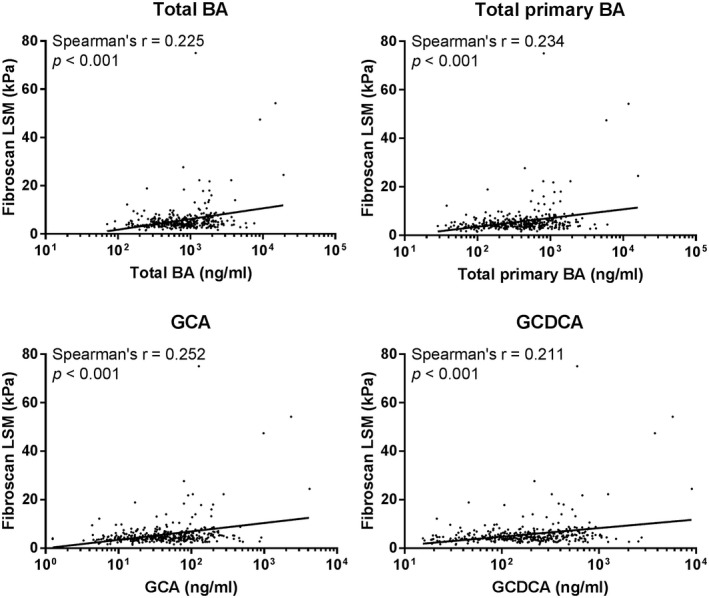
Spearman's correlations between selected bile acid parameters and FibroScan LSM. Abbreviation: BA, bile acid.
Effects of Diet on Fibrosis‐Associated or Steatosis‐Associated Bile Acid Profiles
Redundancy analysis was performed to assess the relationship between the different diet groups and variation in bile acid profiles associated with fibrosis and steatosis. The selected diet variables explained 4.8% of the variation observed (P = 0.001), with the first two axes displaying 3.0% (P = 0.002) and 1.2% (P = 0.284) of the total variation (Fig. 6). Notably, a significant association with bile acid profile variation (P < 0.01) was observed for total milk and yogurt (P = 0.001), total protein (P = 0.004), as well as for the unhealthy eating index (P = 0.006). Total milk and yogurt showed the highest contribution to variation at 1.82%, and the direction of arrows in the plot indicated a negative correlation between total milk and yogurt and levels of the fibrosis‐associated bile acids GCDCA and GCA. Total protein showed a contribution to variation of 1.17%, and the direction of the arrows in the plot indicated a negative correlation between total protein and levels of the steatosis‐negatively associated bile acid GUDCA.
Fig. 6.
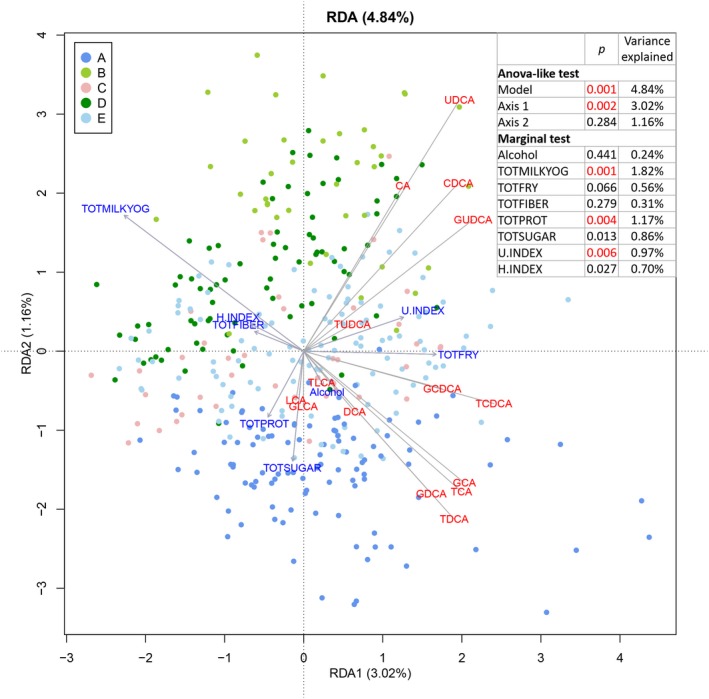
Triplot of the redundancy analysis in 376 subjects, after exclusion of those with missing variables. The first two canonical axes are shown. Explanatory variables (dietary components) are shown in blue, and response variables (log10‐transformed bile acids) are shown in red. Abbreviations: ANOVA, analysis of variance; H.INDEX, healthy eating index; RDA, redundancy analysis; TOTFIBER, total fiber; TOTFRY, total fried foods; TOTMILKYOG, total milk and yogurt; TOTPROT, total protein; TOTSUGAR, total sugar; U.INDEX, unhealthy eating index. P values <0.01 are shown in red.
Gut Microbiome Composition Affects Bile Acid Profiles Associated With Liver Fibrosis or Steatosis
We next sought to determine whether the bile acid changes associated with liver fibrosis or steatosis correlated with specific stool microbiome composition. Stool was collected from 123 of the 390 study subjects, excluding those with a history of antibiotic or proton pump inhibitor use within the month before collection. Following bacterial 16S rRNA sequencing, Mantel's test was conducted to assess the correlation between the distance matrix based on bile acid profiles and the distance matrix based on microbiome composition within the same subjects. Both absolute concentrations of the 15 individual bile acids and relative abundances were assessed. Although not significant, the microbiome‐based distance matrix correlated more strongly with variations in relative bile acid composition than variations in absolute bile acid concentrations (P = 0.308 vs. P = 0.525, respectively). Correlations were improved when evaluating the relative abundance of unconjugated bile acids alone (P = 0.166).
Although overall microbiome composition was not closely associated with circulating bile acid profiles, we next sought to identify specific microbiome changes associated with the identified bile acid parameters of interest: total bile acids, total primary bile acids, GCDCA, GCA and LCA for liver fibrosis, and DCA, total unconjugated secondary bile acids, and relative GUDCA for liver steatosis. Spearman's correlation between each bacterial taxa (from phylum to species level) and the eight bile acid parameters of interest were determined; only taxa with ≥ 0.1% abundance in at least half of the samples were included.
A total of 57 significant bile acid‐microbiome correlations from 29 bacterial taxa were identified (|r| ≥ 0.2 and P < 0.05) (Fig. 7). Notably, the fibrosis‐associated parameters total bile acids, total primary bile acids, GCDCA, and GCA shared a similar correlation pattern with members of the firmicutes phylum. In particular, a negative correlation was observed with members of the Rikenellaceae and Christensenellaceae family, as well as unclassified Roseburia species and Ruminococcaceae UCG‐005. Conversely, a positive correlation was observed for the Peptostreptococcaceae family and the Romboutsia genus within it. All four parameters were also positively correlated with the bile‐resistant Sutterella. LCA showed a distinct correlation pattern. A negative correlation was observed with Enterobacteriaceae, whereas a positive correlation was observed with members of the Ruminococcaceae family and Sutterella.
Fig. 7.
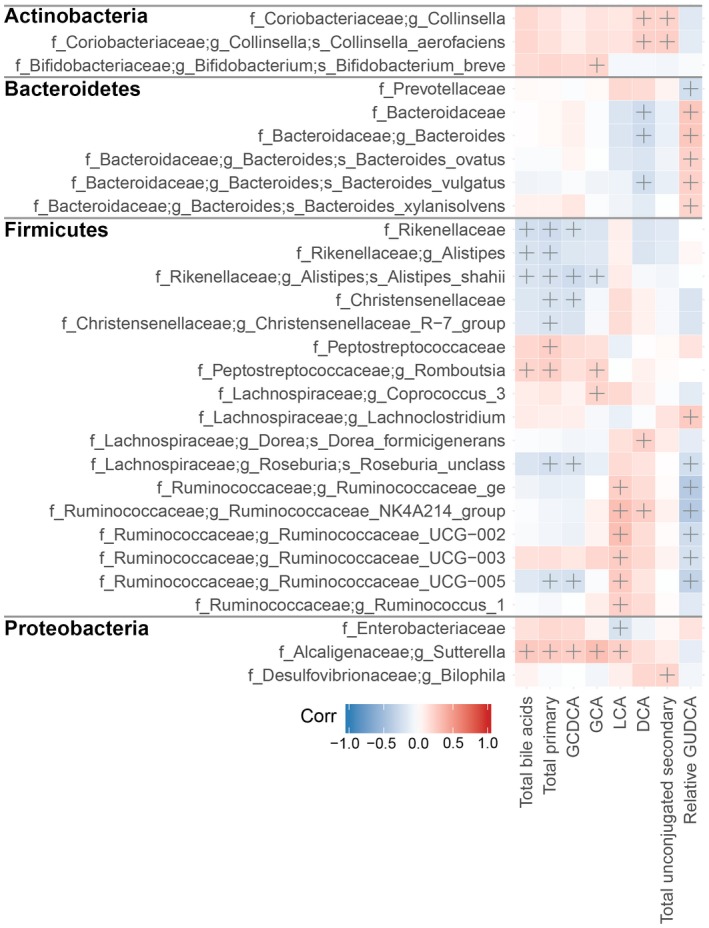
Heatmap showing Spearman's correlations between selected bile acid parameters and stool microbiome features, grouped by phylum (phlyum names in bold). Classifications at the family (f_), genus (g_), and species (s_) levels are shown. Crosses (+) indicate significant correlations (|r| ≥ 0.2 and P < 0.05.
For liver steatosis‐associated bile acid markers, DCA and total unconjugated secondary bile acids showed a positive correlation with Collinsella and Collinsella aerofaciens. In addition, DCA was negatively correlated with members of the Bacteroidaceae family. Finally, relative GUDCA abundance showed a correlation pattern opposite to LCA and DCA, negatively correlating with Prevotellaceae and members of Ruminococcaceae, but positively correlating with members of the Bacteroidaceae family.
Discussion
NASH and liver fibrosis are major risk factors for HCC in the United States. Currently, clinical markers to predict risk of liver fibrosis are lacking. Bile acids are emerging as a potential contributor to NAFLD and NASH, due to their role as signaling molecules, regulating their own synthesis as well as glucose, lipid, and energy metabolism through receptors that have varying affinities to the different bile acids.( 23 ) This study therefore aimed to evaluate whether bile acids may improve the prediction of liver fibrosis in a high‐risk, community‐based Mexican‐American cohort with high rates of NASH. The study cohort, consisting of 390 subjects, is well representative of the overall cohort.( 18 ) PAM clustering of subjects based on their plasma bile acid profiles revealed five distinct clusters that significantly differed by liver fibrosis, liver injury, liver steatosis, age and gender, with liver fibrosis being the major contributor to circulating bile acid changes.
High total bile acid levels showed the strongest association with significant fibrosis among all bile acid parameters. An increase in total bile acids has been reported in multiple studies of NASH,( 11 , 12 ) alcoholic liver disease,( 24 , 25 ) diabetes( 26 , 27 , 28 , 29 , 30 ) and obesity,( 26 , 31 ) suggesting that it is a hallmark of metabolic disease and liver disease. Only a few studies on small numbers of samples have reported bile acid changes in liver fibrosis. Puri et al. reported increased circulating levels GCA and TCA and decreasing secondary/primary ratio in 24 subjects with significant fibrosis (F ≥ 2) compared with 59 subjects with no‐to‐minimal fibrosis (F0‐F1).( 32 ) Similarly, in patients with HCV patients, Cano et al. reported an increase in circulating levels of the primary conjugated bile acids TCDCA, TCA, and GCA in 69 subjects with F ≥ 2 compared to 134 subjects with F0‐F1.( 33 ) Caussy et al. also observed an increase in total bile acids, conjugated CA, and conjugated CDCA with increasing liver fibrosis stage.( 34 ) Finally, Lelouvier et al. reported a decrease in circulating LCA/CDCA, UDCA/CDCA, and TLCA/CDCA ratios in 11 obese subjects with fibrosis compared to 26 obese subjects without fibrosis, concluding that there may be a decreased capability to convert primary bile acids to secondary bile acids.( 35 ) In accordance with these studies, we also observed a significant increase in primary/secondary ratio in subjects with liver fibrosis. We further showed that the glyco‐conjugated primary bile acids GCA and GCDCA were strongly associated with liver fibrosis, whereas LCA had the strongest association with advanced fibrosis.
High levels of the unconjugated secondary bile acids, essentially DCA, and low levels of relative GUDCA were identified to be associated with liver steatosis in this cohort. UDCA and its conjugates are the most hydrophilic bile acids, with well‐known hepatoprotective properties. UDCA is clinically approved for primary biliary cholangitis and dissolution of cholesterol gallstones. Multiple clinical trials have assessed the use of UDCA in NAFLD and NASH, alone or in combination with other therapies.( 36 , 37 , 38 , 39 ) However, studies have so far been in small groups of patients with heterogenous results, requiring additional studies.( 23 , 40 , 41 ) Our observation that low GUDCA is associated with liver steatosis further supports the use of UDCA and its conjugates as a potential therapeutic strategy in NAFLD.
Interestingly, specific dietary parameters were associated with bile acid profiles. Redundancy analysis showed that total intake of milk and yogurt was associated with a healthier bile acid profile with less risk of liver fibrosis, whereas high protein consumption was associated with bile acid profiles observed in liver steatosis. One limitation of our study was that dietary factors were only based on recall from the previous day: Although the assumption is that single‐day intake may somewhat represent long‐term habits, true associations with long‐term habits could not be made. Additional studies to further determine whether a causal relationship exists between these dietary factors and bile acid levels may be useful for future lifestyle interventions.
The contribution of the gut microbiome to bile acid changes was also explored. The gut bacteria mediate deconjugation of bile acids as well as the transformation of CA and CDCA into the secondary bile acids DCA and LCA, respectively, through 7α‐dehydroxylation. Although LCA, DCA, and UDCA are all considered secondary bile acids, UDCA is transformed from CDCA through two‐step epimerization by 7α‐hydroxysteroid and 7β‐hydroxysteroid dehydrogenases (HSDH).( 42 , 43 ) Additionally, UDCA can be converted to LCA by 7β‐dehydroxylation. Although a broad variety of commensal gut bacteria are able to deconjugate bile acids, only a limited group of gut microbes possess 7α‐dehydroxylating and 7β‐dehydroxylating activity required for the conversion of the primary bile acids and UDCA into LCA/DCA, of which the most well characterized are the Clostridium and Eubacterium genera.( 42 , 43 , 44 ) Additionally, members of the Clostridium XIVa and Clostridium XI have been reported to possess 7‐dehydroxylating activity.( 45 ) However, challenges in comparing across previous studies includes the use of different databases for alignment,( 46 , 47 ) updates to the database over time, and reclassification of certain taxa. Although bacteria from these clusters were originally assigned to the Clostridium genus, many have now been reclassified into the families Lachnospiraceae, Peptostreptococcaceae and Ruminococcaceae,( 48 ) all within the Clostridiales order. Our study showed that multiple members of Ruminococcaceae and Lachnospiraceae were positively correlated with LCA and/or DCA, suggesting that changes in their abundance may contribute to the bile acid changes observed in liver fibrosis and steatosis. Additionally, members of the Rikenellaceae and Christensenellaceae families were negatively correlated with the conjugated primary bile acids. Conversely, Peptostreptococcaceae and Romboutsia from within the family were positively correlated with the total and primary bile acids.
Interestingly, although LCA and DCA exhibited a positive correlation with Prevotellaceae and negative correlation with Bacteroides species, relative GUDCA abundance showed the opposite associations. Relative GUDCA was also negatively correlated with the Ruminococcaceae and Lachnospiraceae families, consistent with their putative 7‐dehydroxylating activity. Genes encoding 7α‐HSDH have been identified in a relatively wide range of bacteria, including Clostridium and Bacteroides species, whereas genes encoding 7β‐HSDH have only been demonstrated in Clostridium absonum, Collinsella aerofaciens, Ruminococcus gnavus, and Ruminococcus torques.( 49 , 50 ) This suggests that the Prevotellaceae, Ruminococcaceae and Lachnospiraceae families may either prevent the production of UDCA from CDCA, or increase conversion of UDCA to LCA. Conversely, Bacteroides species may directly contribute to the transformation of CDCA to UDCA.
There are several limitations to this study. As this is a population‐based cohort with subjects recruited from households, and not from patients enrolled in a clinical setting, none of the subjects received a diagnosis of liver disease. Instead, FibroScan imaging was used as a noninvasive technique to detect the presence of liver steatosis and fibrosis. A strength of such an approach, however, is that this group of volunteers from the community is representative of the Mexican‐American population in South Texas. Another limitation is the cross‐sectional nature of the study; the direct contribution of the observed bile changes to liver fibrosis remains unknown. Follow‐up of subjects without liver fibrosis at the time of blood collection, but with bile acid signatures identified to be associated with liver fibrosis, would determine the biological effects of these bile acid changes on liver fibrosis.
In conclusion, we characterized the circulating bile acid profiles of a Mexican‐American cohort with high prevalence of liver steatosis and fibrosis. Bile acid levels were markedly but differently changed for both conditions, suggesting a contribution to disease progression. These findings may be used as biomarkers for prediction of liver steatosis and fibrosis in this high‐risk population, and inform clinical decisions on surveillance and prevention.
Supporting information
Table S1
Table S2
Table S3
Acknowledgment
The authors would like to thank the CCHC cohort recruitment team, particularly project manager Rocio Uribe and her team, who recruited and documented the participants, and Marcela Morris and Israel Hernandez for the samples and data management. The authors would also like to thank Dr. Robert Jenq and Dr. Tina Chang of the MD Anderson Cancer Center Microbiome Core Facility, for their help with stool specimen processing, sequencing, and analysis.
Financial support: This research is supported in part by the MD Anderson Cancer Center Support Grant (CA016672). J.Q. was supported by a CPRIT Research Training Grant Award (RP170067).
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Dulai PS, Singh S, Patel J, Soni M, Prokop LJ, Younossi Z, et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta‐analysis. Hepatology 2017;65:1557‐1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Llovet JM, Zucman‐Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. [DOI] [PubMed] [Google Scholar]
- 3. Estes C, Anstee QM, Arias‐Loste MT, Bantel H, Bellentani S, Caballeria J, et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016–2030. J Hepatol 2018;69:896‐904. [DOI] [PubMed] [Google Scholar]
- 4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7‐34. [DOI] [PubMed] [Google Scholar]
- 5. Hales CM, Carroll MD, Fryar CD, Ogden CI. Prevalance of Obesity Among Adults and Youth: United States, 2015‐2016 (NCHS Data Brief, No. 288). Hyattsville, MD: National Center for Health Statistics; 2017. [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention . National Diabetes Statistics Report, 2017. Atlanta, GA: Centers for Disease Control and Prevention, U.S. Dept of Health and Human Services; 2017. [Google Scholar]
- 7. Rich NE, Oji S, Mufti AR, Browning JD, Parikh ND, Odewole M, et al. Racial and ethnic disparities in nonalcoholic fatty liver disease prevalence, severity, and outcomes in the United States: a systematic review and meta‐analysis. Clin Gastroenterol Hepatol 2018;16:198‐210.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venepalli NK, Modayil MV, Berg SA, Nair TD, Parepally M, Rajaram P, et al. Features of hepatocellular carcinoma in Hispanics differ from African Americans and non‐Hispanic Whites. World J Hepatol 2017;9:391‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramirez AG, Weiss NS, Holden AE, Suarez L, Cooper SP, Munoz E, et al. Incidence and risk factors for hepatocellular carcinoma in Texas Latinos: implications for prevention research. PLoS One 2012;7:e35573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramirez AG, Munoz E, Holden AE, Adeigbe RT, Suarez L. Incidence of hepatocellular carcinoma in Texas Latinos, 1995–2010: an update. PLoS One 2014;9:e99365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferslew BC, Xie G, Johnston CK, Su M, Stewart PW, Jia W, et al. Altered bile acid metabolome in patients with nonalcoholic steatohepatitis. Dig Dis Sci 2015;60:3318‐3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiao N, Baker SS, Chapa‐Rodriguez A, Liu W, Nugent CA, Tsompana M, et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 2018;67:1881‐1891. [DOI] [PubMed] [Google Scholar]
- 13. Mouzaki M, Wang AY, Bandsma R, Comelli EM, Arendt BM, Zhang L, et al. Bile acids and dysbiosis in non‐alcoholic fatty liver disease. PLoS One 2016;11:e0151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aguilar‐Olivos NE, Carrillo‐Cordova D, Oria‐Hernandez J, Sanchez‐Valle V, Ponciano‐Rodriguez G, Ramirez‐Jaramillo M, et al. The nuclear receptor FXR, but not LXR, up‐regulates bile acid transporter expression in non‐alcoholic fatty liver disease. Ann Hepatol 2015;14:487‐493. [PubMed] [Google Scholar]
- 15. Bechmann LP, Kocabayoglu P, Sowa JP, Sydor S, Best J, Schlattjan M, et al. Free fatty acids repress small heterodimer partner (SHP) activation and adiponectin counteracts bile acid‐induced liver injury in superobese patients with nonalcoholic steatohepatitis. Hepatology 2013;57:1394‐1406. [DOI] [PubMed] [Google Scholar]
- 16. Pan JJ, Fisher‐Hoch SP, Chen C, Feldstein AE, McCormick JB, Rahbar MH, et al. Burden of nonalcoholic fatty liver disease and advanced fibrosis in a Texas Hispanic community cohort. World J Hepatol 2015;7:1586‐1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ha J, Chaudhri A, Avirineni A, Pan JJ. Burden of hepatocellular carcinoma among Hispanics in South Texas: a systematic review. Biomark Res 2017;5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fisher‐Hoch SP, Rentfro AR, Salinas JJ, Perez A, Brown HS, Reininger BM, et al. Socioeconomic status and prevalence of obesity and diabetes in a Mexican American community, Cameron County, Texas, 2004–2007. Prev Chronic Dis 2010;7:A53. [PMC free article] [PubMed] [Google Scholar]
- 19. Jiao J, Watt GP, Lee M, Rahbar MH, Vatcheva KP, Pan JJ, et al. Cirrhosis and advanced fibrosis in Hispanics in Texas: the dominant contribution of central obesity. PLoS One 2016;11:e0150978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reininger B, Lee M, Jennings R, Evans A, Vidoni M. Healthy eating patterns associated with acculturation, sex and BMI among Mexican Americans. Public Health Nutr 2017;20:1267‐1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Velazquez CE, Pasch KE, Ranjit N, Mirchandani G, Hoelscher DM. Are adolescents' perceptions of dietary practices associated with their dietary behaviors? J Am Diet Assoc 2011;111:1735‐1740. [DOI] [PubMed] [Google Scholar]
- 22. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 2010;7:335‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arab JP, Karpen SJ, Dawson PA, Arrese M, Trauner M. Bile acids and nonalcoholic fatty liver disease: molecular insights and therapeutic perspectives. Hepatology 2017;65:350‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bajaj JS, Kakiyama G, Zhao D, Takei H, Fagan A, Hylemon P, et al. Continued alcohol misuse in human cirrhosis is associated with an impaired gut‐liver axis. Alcohol Clin Exp Res 2017;41:1857‐1865. [DOI] [PubMed] [Google Scholar]
- 25. Kakiyama G, Hylemon PB, Zhou H, Pandak WM, Heuman DM, Kang DJ, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol 2014;306:G929‐G937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cariou B, Chetiveaux M, Zair Y, Pouteau E, Disse E, Guyomarc'h‐Delasalle B, et al. Fasting plasma chenodeoxycholic acid and cholic acid concentrations are inversely correlated with insulin sensitivity in adults. Nutr Metab (Lond) 2011;8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12alpha‐hydroxylated bile acids. Diabetes 2013;62:4184‐4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sonne DP, van Nierop FS, Kulik W, Soeters MR, Vilsboll T, Knop FK. Postprandial plasma concentrations of individual bile acids and FGF‐19 in patients with type 2 diabetes. J Clin Endocrinol Metab 2016;101:3002‐3009. [DOI] [PubMed] [Google Scholar]
- 29. Vincent RP, Omar S, Ghozlan S, Taylor DR, Cross G, Sherwood RA, et al. Higher circulating bile acid concentrations in obese patients with type 2 diabetes. Ann Clin Biochem 2013;50(Pt 4):360‐364. [DOI] [PubMed] [Google Scholar]
- 30. Wewalka M, Patti ME, Barbato C, Houten SM, Goldfine AB. Fasting serum taurine‐conjugated bile acids are elevated in type 2 diabetes and do not change with intensification of insulin. J Clin Endocrinol Metab 2014;99:1442‐1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prinz P, Hofmann T, Ahnis A, Elbelt U, Goebel‐Stengel M, Klapp BF, et al. Plasma bile acids show a positive correlation with body mass index and are negatively associated with cognitive restraint of eating in obese patients. Front Neurosci 2015;9:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Puri P, Daita K, Joyce A, Mirshahi F, Santhekadur PK, Cazanave S, et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology 2018;67:534‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cano A, Marino Z, Millet O, Martinez‐Arranz I, Navasa M, Falcon‐Perez JM, et al. A metabolomics signature linked to liver fibrosis in the serum of transplanted hepatitis C patients. Sci Rep 2017;7:10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caussy C, Hsu C, Singh S, Bassirian S, Kolar J, Faulkner C, et al. Serum bile acid patterns are associated with the presence of NAFLD in twins, and dose‐dependent changes with increase in fibrosis stage in patients with biopsy‐proven NAFLD. Aliment Pharmacol Ther 2019;49:183‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lelouvier B, Servant F, Paisse S, Brunet AC, Benyahya S, Serino M, et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: a pilot analysis. Hepatology 2016;64:2015‐2027. [DOI] [PubMed] [Google Scholar]
- 36. Pietu F, Guillaud O, Walter T, Vallin M, Hervieu V, Scoazec JY, et al. Ursodeoxycholic acid with vitamin E in patients with nonalcoholic steatohepatitis: long‐term results. Clin Res Hepatol Gastroenterol 2012;36:146‐155. [DOI] [PubMed] [Google Scholar]
- 37. Mueller M, Castro RE, Thorell A, Marschall HU, Auer N, Herac M, et al. Ursodeoxycholic acid: effects on hepatic unfolded protein response, apoptosis and oxidative stress in morbidly obese patients. Liver Int 2018;38:523‐531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ratziu V, de Ledinghen V, Oberti F, Mathurin P, Wartelle‐Bladou C, Renou C, et al. A randomized controlled trial of high‐dose ursodesoxycholic acid for nonalcoholic steatohepatitis. J Hepatol 2011;54:1011‐1019. [DOI] [PubMed] [Google Scholar]
- 39. Leuschner UF, Lindenthal B, Herrmann G, Arnold JC, Rossle M, Cordes HJ, et al. High‐dose ursodeoxycholic acid therapy for nonalcoholic steatohepatitis: a double‐blind, randomized, placebo‐controlled trial. Hepatology 2010;52:472‐479. [DOI] [PubMed] [Google Scholar]
- 40. Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018;67:328‐357. [DOI] [PubMed] [Google Scholar]
- 41. Xiang Z, Chen YP, Ma KF, Ye YF, Zheng L, Yang YD, et al. The role of ursodeoxycholic acid in non‐alcoholic steatohepatitis: a systematic review. BMC Gastroenterol 2013;13:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jia W, Xie G, Jia W. Bile acid‐microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 2018;15:111‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016;7:22‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006;47:241‐259. [DOI] [PubMed] [Google Scholar]
- 45. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 2016;24:41‐50. [DOI] [PubMed] [Google Scholar]
- 46. Balvociute M, Huson DH. SILVA, RDP, Greengenes, NCBI and OTT ‐ how do these taxonomies compare? BMC Genom 2017;18(Suppl. 2):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Henderson G, Yilmaz P, Kumar S, Forster RJ, Kelly WJ, Leahy SC, et al. Improved taxonomic assignment of rumen bacterial 16S rRNA sequences using a revised SILVA taxonomic framework. PeerJ 2019;7:e6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Galperin MY, Brover V, Tolstoy I, Yutin N. Phylogenomic analysis of the family Peptostreptococcaceae (Clostridium cluster XI) and proposal for reclassification of Clostridium litorale (Fendrich et al. 1991) and Eubacterium acidaminophilum (Zindel et al. 1989) as Peptoclostridium litorale gen. nov. comb. nov. and Peptoclostridium acidaminophilum comb. nov. Int J Syst Evol Microbiol 2016;66:5506‐5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Song C, Wang B, Tan J, Zhu L, Lou D. Discovery of tauroursodeoxycholic acid biotransformation enzymes from the gut microbiome of black bears using metagenomics. Sci Rep 2017;7:45495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RMT, Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome 2019;7:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3


