Abstract
Background
Uveal melanoma (UM) is the most common intraocular tumour in adults. Currently there are different tests available to determine the risk of UM for metastasis, among which include BRCA1-associated protein-1 (BAP1) immunohistochemistry (IHC) and gene expression profiling (GEP). BAP1 is a deubiquitylating enzyme (DUB) that has tumour suppressor activity, the loss of which therefore is associated with higher risk for tumour growth and metastasis.
Objectives
To compare and correlate the prognostic significance of BAP1-IHC staining patterns and GEP in the prediction of UM's risk for metastasis.
Methods
This is a retrospective chart review with prospective follow-up of patients with primary UM who underwent enucleation from the year 2008 to 2018. Clinical history, histopathologic findings, GEP classification, BAP1-IHC of the formalin-fixed paraffin-embedded tissues, and follow-up data for metastasis were collected and statistically analysed.
Results
A total of 30 enucleated eyes with UM were included in the study. All class 1a tumours had high nuclear BAP1 expression and all class 2 had low nuclear BAP1 expression. Fifty percent of the class 1b tumours had low nuclear BAP1 expression. Among the tumours with low nuclear BAP1 expression, 68% developed metastasis, while 9% developed metastasis among high nuclear expression. Fifty-five percent developed metastasis in tumours with high cytoplasmic expression and 42% for low cytoplasmic expression. Predictive values for metastasis (positive predictive value, negative predictive value) are as follows: (1) nuclear BAP1-IHC (68%, 91%), (2) cytoplasmic BAP1-IHC (55%, 58%), and (3) GEP (73%, 80%). Nuclear BAP1-IHC and GEP had the same accuracy rate of 77% and cytoplasmic BAP1-IHC had an accuracy of 57%.
Conclusion
Low nuclear BAP1-IHC strongly correlates with GEP class 2 and was equally accurate in the prediction of metastasis.
Keywords: BAP1, Gene expression profile, Uveal melanoma, Enucleated eyes
Introduction
Uveal melanoma (UM) is the most common intraocular tumour in adults with a reported annual incidence in the USA of 5.2 cases per million [1, 2]. The 5- and 15-year disease-related mortality of UM is 30 and 45%, respectively [3, 4]. Treatments for metastatic UM are currently being developed and prognostic tests for early detection of a patient's metastatic risk is an important step in management.
There are several prognostic tests available including fluorescence in situ hybridization for the detection of loss of chromosome 3 and gene expression profile (GEP), which have both been shown to be highly correlated with the risk for metastasis [4, 5, 6, 7, 8, 9, 10]. Chromosomal analysis and GEP require laboratory resources that are relatively expensive and may not be universally available. Immunohistochemical (IHC) staining, on the other hand, is a common procedure that is relatively inexpensive and available in most pathology laboratories worldwide.
The BRCA-1 associated protein 1 (BAP1) gene is located at chromosome 3p21.3 and is essential for the preservation of melanocyte identity. A depletion of this gene product (BAP1 protein) will therefore result in UM assuming a stem cell-like phenotype [11, 12]. Mutations in the BAP1gene coding for BAP1 protein have been detected in 84% of metastasizing UM [13]. It has previously been shown that the absence of nuclear BAP1 protein expression detected by IHC is at least as good as the BAP1mutation analysis with Sanger sequencing in the prediction of metastasis [14, 15, 16]. In addition, previous studies have shown good correlation of nuclear BAP1-IHC in fine needle aspiration specimens with GEP classification [17]. The purpose of this study was to correlate GEP with nuclear and cytoplasmic BAP1-IHC and to determine their accuracy in the prediction of metastasis in a series of patients who had primary enucleation for UM.
Materials and Methods
Enucleated eyes submitted with a diagnosis of malignant melanoma obtained at the L.F. Montgomery Laboratory, Emory University, between January 1, 2008, and August 2, 2018, were retrieved. Patients' medical records were then reviewed for demographics, clinical findings, histopathologic findings, and GEP (Decision Dx-UM; Castle Biosciences Inc., Phoenix, AZ, USA). Inclusion criteria were as follows: histologically proven UM, available GEP classification, and availability of formalin-fixed paraffin-embedded tissue samples for BAP1-IHC. Exclusion criteria were as follows: insufficient specimen for BAP1-IHC, no available GEP classification, and prior history of brachytherapy. After reviewing the medical records, 56 enucleated eyes with UM and GEP classification were available, 26 of which were excluded due to the following: prior brachytherapy before enucleation (n = 2), referred cases from outside hospital paraffin blocks not available (n = 22), specimen with 100% necrotic tumour (n = 1), and insufficient specimen for BAP1-IHC (n = 1). Thirty cases met the inclusion criteria and were collected for this study. Microscopic examination of specimens was conducted and graded by 2 independent pathologists (G.S. and H.G.) in a masked fashion. Slides were deidentified, and both pathologists were masked for the GEP classification and patient outcome. Follow-up data were obtained in telephone interviews with patients or relatives after verbal consent (S.P.).
Histopathologic Assessment
After enucleation, all specimens were formalin fixed and embedded in paraffin. A 10-mm-thick pupil optic nerve section including the central portion of the melanoma was processed using vacuum infiltration with Tissue-Tek® VIP® 6 (Sakura Seiki Co., Ltd., Nagano, Japan). A 4-μm-thick tissue section from the paraffin-embedded specimen was then mounted onto glass slides and stained with haematoxylin-eosin and periodic acid-Schiff according to a routine protocol with an automated stainer (Tissue-Tek® DRSTM, Sakura). Histopathologic features including cell type (mixed, epithelioid, or spindle), largest basal diameter, tumour thickness, scleral invasion, extrascleral extension, rupture of Bruch's membrane, optic nerve invasion, and vortex vein invasion were evaluated using a light microscope (Olympus BHTU; Olympus Corporation, Tokyo, Japan).
Immunohistochemical Staining
A 4-μm-thick tissue section of the existing formalin-fixed paraffin-embedded pupil optic blocks of the UM cases underwent BAP1-IHC with an automated IHC staining system (Leica Bond-III automated system; Leica Microsystems, Chicago, IL, USA) using the immunoperoxidase staining method with red chromogen. The manufacturer's protocol was followed. Briefly, the steps were as follows: (1) deparaffinizing of the sections in xylene for 3 changes, (2) hydration of the section gradually through graded alcohols, (3) heat-induced antigen retrieval, (4) incubation of sections for 20 min with BAP1 antibody (clone sc-28383,1:40 dilution; Santa Cruz Biotechnology, Dallas, TX, USA), and (5) incubation with haematoxylin counterstain.
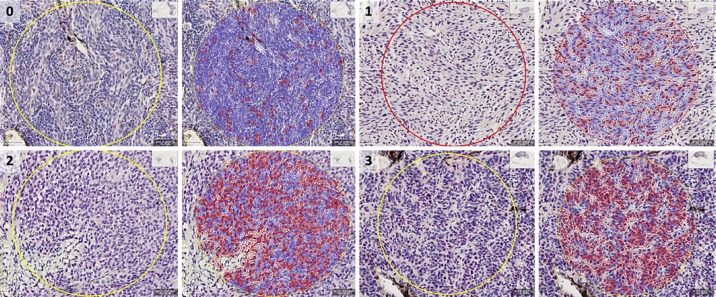
The BAP1-stained tissue sections were screened and graded based on a previous study [18]. Two pathologists without prior knowledge of GEP or patient outcome scored each slide independently (G.S. and H.G.). Under low-power magnification (40×), the area in the tumour with the most intense BAP1 staining was selected. The percentage of tumour cells with immunoreactivity was then assessed in 3 high-power fields (200×) using a 4-point scoring system (0 − positive staining in less than 10% of tumour cells per high-power field, 1 − positive staining between 10 and 33% of tumour cells per high-power field, 2 − positive staining between 33 and 66% of tumour cells per high-power field, and 3 − positive staining in 67% of tumour cells per high-power field) as shown in Figure 1 [18]. The immunoreactivity in the nucleus and cytoplasm was evaluated separately. The mean score from the 3 high-power fields was then used for classification. The BAP1 expression was classified in a binary system into high and low reactivity. Tumours with a mean score of >1 are classified as high, while tumours with a mean score of ≤1 are classified as low. Any discrepancies between the two pathologists were resolved by a consensus discussion.
Fig. 1.
Grading scale of BRCA1-associated protein-1 (BAP1) immunoreactivity using red chromogen in tumour cell nuclei (slides digitally scanned at 200×). Using QuPath, tumour cells were outlined with either red (nuclear positive) or blue (nuclear negative). 0 − positive staining in less than 8% of tumour cells per high-power field; 1 − positive staining in 30% of tumour cells per high-power field; 2 − positive staining in 59% of tumour cells per high power field; and 3 − positive staining in 80% of tumour cells per high-power field. Scale bar, 100 μm.
Gene Expression Profiling
GEP classification was determined by submitting a fresh fine needle aspiration biopsy specimen collected during enucleation to Castle Biosciences The manufacturer's protocol for specimen submission was followed. In brief: (1) the contents of the needle hub were transferred into one of the two RNAse-free cryovials provided; (2) using the same needle hub, extraction buffer from the second cryovial was aspirated and expelled into the first vial; (3) the first vial containing the specimen was then placed in a specimen bag and frozen immediately at −80°C; and (4) the specimen was placed in the insulated container/box provided with dry ice and shipped to Castle Biosciences (Submission Instruction Ver.10.1)
Statistical Methods
Descriptive characteristics of patients and tumours included in the study were examined and reported (Table 1). The Mann-Whitney U test was used to test the null hypothesis in non-parametrically distributed group-level data, and Fisher's exact test in two-by-two tables. Cohen's kappa statistic (κ) was calculated for inter-method agreement. Binary and multiple logistic regressions were used for estimation of hazard for metastasis. Metastasis-free survival was calculated with the Kaplan-Meier method, and the log-rank (Mantel-Cox) test of equality of survival distributions was applied. Differences with p < 0.05 were considered significant, all p values being two-sided. All statistical analyses were performed using IBM SPSS statistics version 25 (Armonk, NY, USA).
Table 1.
Clinicopathological features in 30 enucleated primary uveal melanoma cases
| Gender, n (%) | |
| Male | 16 (53) |
| Female | 14 (47) |
| Age at diagnosis, years | |
| Mean (SD) | 62.0 (15.7) |
| Median (range) | 64 (24–92) |
| Tumour category, n (%) | |
| 1 | 2 (7) |
| 2 | 5 (17) |
| 3 | 15 (50) |
| 4 | 8 (27) |
| Tumour thickness, mm | |
| Mean (SD) | 8.8 (4.1) |
| Median (range) | 9.0 (0.8–17.4) |
| Largest basal diameter, mm | |
| Mean (SD) | 16.5 (4.8) |
| Median (range) | 18.0 (1.4–22.5) |
| Histology, n (%) | |
| Mixed | 26 (87) |
| Epithelioid | 4 (13) |
| GEP classification, n (%) | |
| 1a | 7 (23) |
| 1b | 8 (27) |
| 2 | 15 (50) |
| Nuclear BAP1 stain, n (%) | |
| High | 11 (37) |
| Low | 19 (63) |
| Cytoplasmic BAP1 stain, n (%) | |
| High | 11 (37) |
| Low | 19 (63) |
| Metastasis, n (%) | |
| Yes | 14 (47) |
| No | 16 (53) |
| Melanoma-specific mortality, n (%) | |
| Yes | 9 (30) |
| No | 21 (70) |
| Follow-up, months | |
| Mean (SD) | 30.3 (23) |
| Median (range) | 26.0 (1–95) |
Data in percentage may total to more than 100% due to rounding up of the integers. GEP, gene expression profiling; BAP1, BRCA1-associated protein-1.
Results
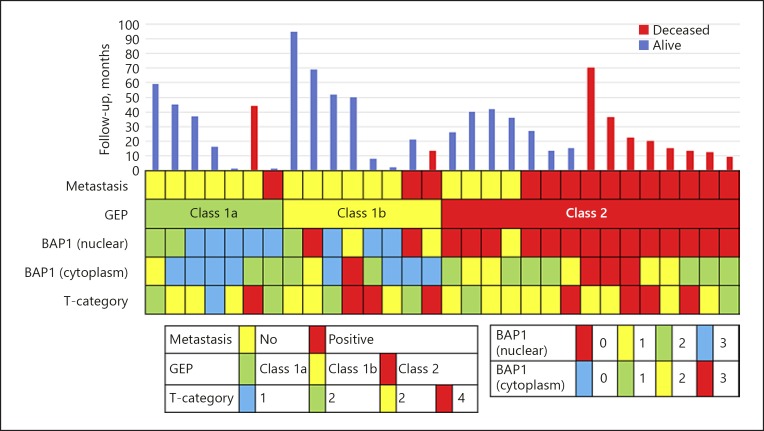
A total of 30 UM patients met the inclusion criteria for this study. Sixteen (53%) were male and 14 (47%) female. The mean age was 62 years (SD 16) with a range of 24–92 years. Based on the GEP classification, 7/30 (23%) cases were class 1a, 8/30 (27%) were class 1b, and 15/30 (50%) were class 2. Mean age at diagnosis of class 1a (n = 7) was 51 years (SD 14), 68 years (SD 14) for class 1b (n = 8), and 65 years (SD 15) for class 2 (n = 15). The mean and median follow-up was 30 (SD 23) and 26 months, respectively, with a range of 0.5–95 months. Histopathologic examination showed that 26 cases (87%) were of the mixed cell type and 4 (13%) were epithelioid cell type. We classified tumour size based on the anatomic extent of the primary tumour according to the 8th edition of the American Joint Committee on Cancer (AJCC) TNM classification for UM [19]. Detailed clinicopathological data are provided in Figure 2 and Table 1.
Fig. 2.
Summary of clinicopathological data for patients and tumours included in this study.
Correlation of BAP1 Expression with GEP and Risk of Metastasis
Among class 1a, 7/7 (100%) had high nuclear BAP1 expression, 1/7 (14%) had high cytoplasmic BAP1 expression, and 6/7 (86%) had low cytoplasmic BAP1 expression. In class 1b, 4/8 (50%) had high nuclear BAP1 expression, 4/8 (50%) had low nuclear BAP1 expression, 6/8 (75%) had low cytoplasmic BAP1 expression, and 2/8 (25%) had high cytoplasmic BAP1 expression. Among class 2, 15/15 (100%) had low nuclear BAP1 expression, 8/15 (53%) had high cytoplasmic BAP1 expression, and 7/15 (47%) had low cytoplasmic BAP1 expression. Among cases with low nuclear BAP1 expression, 13/19 (68%) developed metastasis. Among cases with high nuclear BAP1 expression, 1/11 (9%) developed metastasis and 1 had metastasis secondary to skin melanoma. Metastasis developed in 1/7 (14%) among class 1a, 2/8 (25%) among class 1b, and 11/15 (73%) among class 2. With regard to patient death, among class 1a, 1/7 (14%) died due to metastasis to the stomach from skin melanoma. The remaining deaths − 1/8 (13%) in class 1b, 8/15 (53%) in class 2, 9/19 (47%) among cases with low nuclear BAP1, and 5/11 (45%) among cases with high cytoplasmic BAP1 − occurred due to liver metastasis from UM.
On the one hand, comparing GEP class 1 (a and b) or 2 and nuclear BAP1 (high or low) in a two-by-two table shows a substantial agreement between GEP class 2 and low nuclear BAP1 (κ = 0.73). Nuclear BAP1 expression was significantly lower in GEP class 2 versus class 1 tumour (Fisher's exact test p < 0.001). On the other hand, comparing GEP class 1 (a and b) or 2 and cytoplasmic BAP1 (high or low) in a two-by-two table shows a fair agreement between GEP class 2 and high cytoplasmic BAP1 expression (κ = 0.33). Cytoplasmic BAP1 was higher, but not significantly higher, in GEP class 2 versus class 1 tumour (Fisher's exact test p = 0.1281). The sensitivity (Sn) and specificity (Sp) of the diagnostic tests for detecting the risk of metastasis are as follows: GEP classification (Sn = 79%, Sp = 75%), BAP1-IHC (Sn = 93%, Sp = 63%), and cytoplasmic BAP1-IHC (Sn = 43%, Sp = 69%). The positive predictive value of low nuclear BAP1 expression was 69% (95% CI: 53–81%), and the negative predictive value was 91% (95% CI: 60–99%). For high cytoplasmic BAP1 expression, the positive and negative predictive values were 55% (95% CI: 32–76%) and 58% (95% CI: 44–71%), respectively. By dichotomizing GEP classes into 1 (no distinction between 1a or 1b) or 2, the positive predictive value of GEP 2 was shown to be 73% (95% CI: 53–87%), and the negative predictive value was 80% (95% CI: 59–92%). Nuclear BAP1-IHC and GEP had the same accuracy rate of 77% while cytoplasmic BAP1-IHC had an accuracy of 56.67%.
Survival Analysis
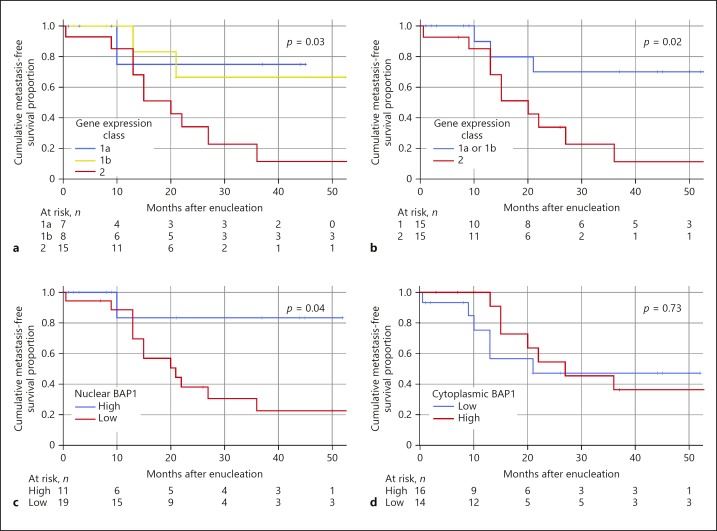
Binary logistic regression analysis confirmed that both nuclear BAP1 expression and gene expression class, but not cytoplasmic BAP1 expression or other clinicopathological parameters, individually increased the odds of developing metastatic disease (Table 2a). A multivariate logistic regression model showed that neither nuclear BAP1 expression nor gene expression were significant as independent predictors of metastasis (Table 2b). In Kaplan-Meier analysis, patients had significantly shorter metastasis-free survival if their tumours had gene expression class 2 versus class 1a and/or 1b (log-rank p = 0.02, p = 0.03; Fig. 3a, b) and low nuclear BAP1 expression (log-rank p = 0.04; Fig. 3c) but not with high cytoplasmic BAP1 expression (log-rank p = 0.73; Fig. 3d).
Table 2.
Binary and multivariate logistic regression analyses
| a Binary logistic regression analysis of predictors of metastatic risk in uveal melanoma | ||
| HR (95% CI) | p value | |
| Age | 1.2 (0.8–1.8) | 0.46 |
| Sex | 0.4 (0.1–1.5) | 0.18 |
| LBD | 1.0 (0.6–1.5) | 0.98 |
| Tumour thickness | 1.0 (0.7–1.6) | 0.86 |
| AJCC T category | ||
| 1 | Reference | |
| 2 | 0.7 (0.03–18.1) | 0.81 |
| 3 | 0.7 (0.04–12.8) | 0.79 |
| 4 | 1.7 (0.07–38.0) | 0.75 |
| Nuclear BAP1 stain | ||
| High | Reference | |
| Low | 21.7 (2.2–210.1) | <0.01 |
| Cytoplasmic BAP1 stain | ||
| Low | Reference | |
| High | 2.2 (0.6–9.6) | 0.29 |
| Gene expression class | ||
| 1a | Reference | |
| 1b | 2.0 (0.1–28.4) | 0.61 |
| 2 | 16.5 (1.5–183.1) | 0.02 |
| b Multivariate logistic regression model evaluating metastatic risk | ||
| OR (95% CI) | p value | |
| Nuclear BAP1 stain | 12.4 (0.4–413.2) | 0.16 |
| Gene expression class | 1.5 (0.2–11.2) | 0.7 |
LBD, largest basal diameter; AJCC, American Joint Committee on Cancer [19]; T, primary tumour; BAP1, BRCA1-associated protein-1; HR, hazard ratio; OR, odds ratio.
Fig. 3.
Kaplan-Meier cumulative metastasis-free survival proportion after enucleation. a Patients with tumours with gene expression class 2 (red) versus 1a (blue) and 1b (yellow) (log-rank p for trend = 0.03). b Patients with tumours with gene expression class 2 (red) versus class 1a or 1b (blue, log-rank p = 0.02). c Patients with tumour with low nuclear BAP1 expression (red) versus high (blue, log-rank p = 0.04). d Patients with tumours with low cytoplasmic BAP1 expression (blue) versus high (red, log-rank p = 0.73).
Discussion/Conclusion
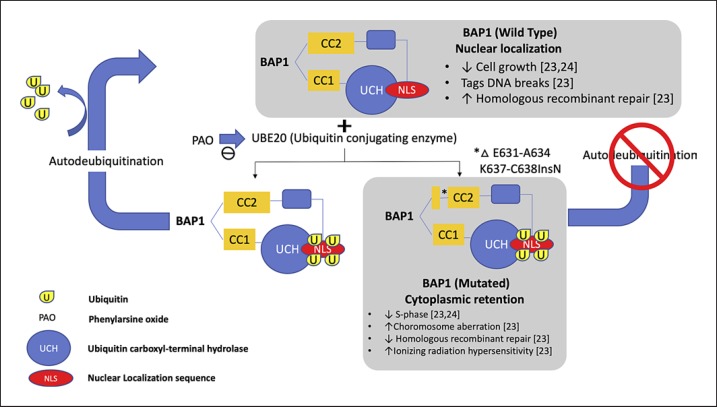
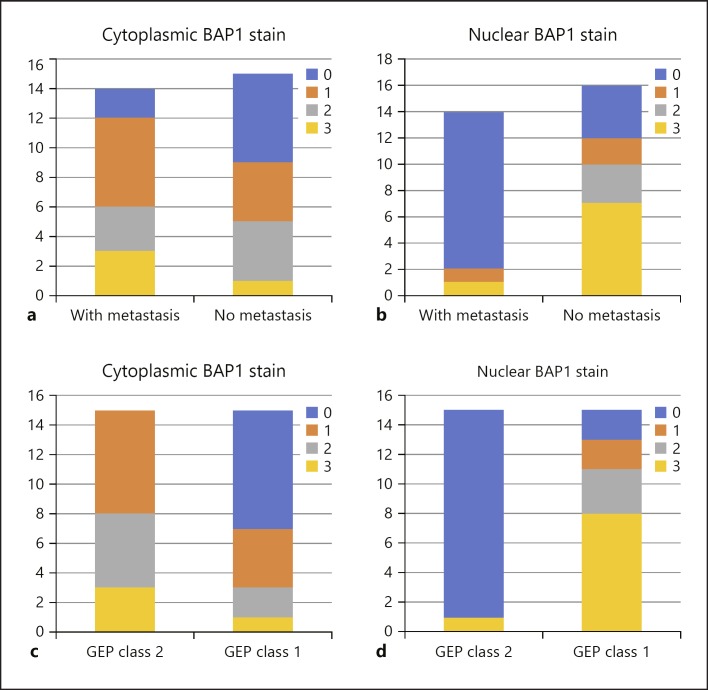
Previous studies have shown that GEP class 2 tumours are strongly associated with monosomy 3, which suggests that loss of one copy of the said chromosome may unmask the mutant gene on the remaining copy [13]. Since the BAP1 gene is located in chromosome 3, a mutation in this gene would therefore suggest an increase in metastatic risk as previously described [20]. BAP1 is a ubiquitin carboxyl-terminal hydrolase (UCH), a subfamily of deubiquinating enzyme (DUB). This was first isolated through yeast two hybrid screening for BRCA1 RING finger domain by Jensen et al. [21] in 1998. DUB functions to cleave ubiquitin from proteins and molecules. BAP1 protein has 3 domains: (1) N-terminal: UCH − this functions to remove the ubiquitin; (2) C-terminal: UCH 37-like domain (ULD) which are binding domains for both BRCA1 and BARD1 forming a tumour suppressor heterodimeric complex and 2 nuclear localization sequences (NLS) (Lys-128) − this sequence tags a protein for import into the cell nucleus; and (3) linker region: host cell factor C1 binding domain, which interacts with histone-modifying complexes during cell divisions [22, 23, 24]. Mashtalir et al. [25] has previously described the BAP1 localization via the NLS. The ubiquitin conjugation enzyme (UBE20) mediates monoubiquination of the NLS of the BAP1 protein. With an intact BAP1gene, BAP1 is able to autodeubiqutinize the ubiquitins that are attached to its NLS domain and hence can function and localize itself properly inside the nucleus. In UM, as described by Harbour et al. [13], class 2 UM were found to have frame deletion of the coiled-coil C-terminal strand (CC2) of the BAP1 gene specifically in E631-A634 and K637-C638InsN. With this mutation, BAP1 protein cannot autodeubiqutinize the ubiquitin found in the NLS and will therefore localize itself in the cytoplasm and lose its tumour suppression activity (Fig. 4) [25]. Previous studies have shown that only tumours with low nuclear BAP1 have good correlation with increased risk of metastasis [18]. However, in our study, interestingly we noted a slight increase in cytoplasmic BAP1 stain in class 2 tumours as well as in cases with metastasis as shown in Figure 5, which suggests that there is indeed a loss of nuclear localization of BAP1 protein in mutated BAP1 gene. However, due to the small sample size and short follow-up time, we have not been able to show a significant difference between them.
Fig. 4.
Schematic representation of the localization of wild-type and mutated BAP1 protein. BAP1 protein normally localizes itself in the cell nucleus due to the presence of the nuclear localization sequence (NLS). In the presence of an ubiquitin conjugating enzyme (UBE20), NLS will be covered with ubiquitin localizing the BAP1 protein in the cell's cytoplasm. Wild-type BAP1 protein has a capability to remove the ubuiquitin and hence is able to enter back into the nucleus. Mutated BAP1 protein loses its autodeubiquitination property hence it remains in the cytoplasm.
Fig. 5.
Nuclear and cytoplasmic BAP1 stain grading in relation to metastasis and gene expression profile (GEP). a Cytoplasmic BAP1 staining grade in relation to patients' metastatic status. b Nuclear BAP1 staining grade in relation to patients' metastatic status. c Cytoplasmic BAP1 staining grade in relation to GEP classification. d Nuclear BAP1 staining grade in relation to GEP classification.
In our study, we have determined that nuclear BAP1-IHC highly correlates with GEP classification, similar to the results reported from previous studies [17, 26]. In addition, we have shown a fair agreement between high cytoplasmic BAP1 stain among class 2 tumours. This coincides with previous studies which have correlated cytoplasmic localization of BAP1 protein in the presence of mutation in the BAP1 gene [25]. The importance of cytoplasmic staining patterns in most tumour cells, such as UM, still remains unclear. Though cytoplasmic BAP1 staining in itself cannot accurately predict a patient's risk of metastasis, it should still be interpreted carefully. Nuclear BAP1-IHC and GEP have shown that both methods can independently predict increased odds of developing metastatic disease, with the same accuracy for the prediction of metastasis from enucleated eye specimens.
Limitations to the study include the small sample size and the fact that data were collected retrospectively from one institution only. The low number of events produced wide confidence intervals and prevented the use of median survival in Kaplan-Meier analysis. The estimated means are consequently limited to the longest survival times within each category. Additionally, our findings should not be generalized to material obtained by other means, i.e., fine needle aspiration biopsies. Indeed, a large proportion of patients with UM will never undergo enucleation, and manual assessments of IHC stains are also subjective by nature and may suffer from poor inter-observer reproducibility.
In conclusion, our results indicate that BAP1-IHC may be used as an alternative to gene expression classification for evaluating the prognosis for metastasis in enucleated eyes with UM. This is especially attractive from a global perspective in places where IHC and examination under light microscopy may be the only technical resources available. Also, it is important to note the probable significance of BAP1 protein localization and its correlation to its function as tumour suppressor and risk for metastasis.
Statement of Ethics
The study was performed according to the tenets of the Declaration of Helsinki. It was approved by the Emory University Institutional Review Board and the Clinical and Translational Review Committee of the Emory Winship Cancer Institute (IRB00105948, EU4462).
Disclosure Statement
The authors have no conflicts of interest to declare.
Author Contributions
The conception and design of the study were developed by H.G., T.R.S., and G.S. Data analysis was performed by G.S. and T.R.S. Data acquisition was done by T.R.S. and S.P. The paper was written by T.R.S. and G.S, and reviewed by H.G.
Acknowledgements
This work was supported in part by a Departmental Grant from Research to Prevent Blindness, Inc. (P30EY0630, CA176001). The authors would also like to thank Dr. Yang Hua for her help.
References
- 1.Bakalian S, Marshall JC, Logan P, Faingold D, Maloney S, Di Cesare S, et al. Molecular pathways mediating liver metastasis in patients with uveal melanoma. Clin Cancer Res. 2008 Feb;14((4)):951–6. doi: 10.1158/1078-0432.CCR-06-2630. [DOI] [PubMed] [Google Scholar]
- 2.Aronow ME, Topham AK, Singh AD. Uveal Melanoma: 5-Year Update on Incidence, Treatment, and Survival (SEER 1973-2013) Ocul Oncol Pathol. 2018 Apr;4((3)):145–51. doi: 10.1159/000480640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kujala E, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003 Nov;44((11)):4651–9. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- 4.Plasseraud KM, Wilkinson JK, Oelschlager KM, Poteet TM, Cook RW, Stone JF, et al. Gene expression profiling in uveal melanoma: technical reliability and correlation of molecular class with pathologic characteristics. Diagn Pathol. 2017 Aug;12((1)):59. doi: 10.1186/s13000-017-0650-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010 Jul;12((4)):461–8. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damato B, Duke C, Coupland SE, Hiscott P, Smith PA, Campbell I, et al. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology. 2007 Oct;114((10)):1925–31. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Decatur CL, Ong E, Garg N, Anbunathan H, Bowcock AM, Field MG, et al. Driver Mutations in Uveal Melanoma: Associations With Gene Expression Profile and Patient Outcomes. JAMA Ophthalmol. 2016 Jul;134((7)):728–33. doi: 10.1001/jamaophthalmol.2016.0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correa ZM, Augsburger JJ. Independent Prognostic Significance of Gene Expression Profile Class and Largest Basal Diameter of Posterior Uveal Melanomas. Am J Ophthalmol. 2016;162:g20-7–e1. doi: 10.1016/j.ajo.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Worley LA, Onken MD, Person E, Robirds D, Branson J, Char DH, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res. 2007 Mar;13((5)):1466–71. doi: 10.1158/1078-0432.CCR-06-2401. [DOI] [PubMed] [Google Scholar]
- 10.Griewank KG, van de Nes J, Schilling B, Moll I, Sucker A, Kakavand H, et al. Genetic and clinico-pathologic analysis of metastatic uveal melanoma. Mod Pathol. 2014;27((2)):175–83. doi: 10.1038/modpathol.2013.138. [DOI] [PubMed] [Google Scholar]
- 11.Koopmans AE, Verdijk RM, Brouwer RW, van den Bosch TP, van den Berg MM, Vaarwater J, et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod pathology. 2014;27((10)):1321–30. doi: 10.1038/modpathol.2014.43. [DOI] [PubMed] [Google Scholar]
- 12.Matatall KA, Agapova OA, Onken MD, Worley LA, Bowcock AM, Harbour JW. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer. 2013 Aug;13((1)):371. doi: 10.1186/1471-2407-13-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010 Dec;330((6009)):1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Nes JA, Nelles J, Kreis S, Metz CH, Hager T, Lohmann DR, et al. Comparing the Prognostic Value of BAP1 Mutation Pattern, Chromosome 3 Status, and BAP1 Immunohistochemistry in Uveal Melanoma. Am J Surg Pathol. 2016 Jun;40((6)):796–805. doi: 10.1097/PAS.0000000000000645. [DOI] [PubMed] [Google Scholar]
- 15.Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014 Sep;111((7)):1373–80. doi: 10.1038/bjc.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah AA, Bourne TD, Murali R. BAP1 protein loss by immunohistochemistry: a potentially useful tool for prognostic prediction in patients with uveal melanoma. Pathology. 2013 Dec;45((7)):651–6. doi: 10.1097/PAT.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 17.Glasgow BJ, McCannel TA. Correlation of Immunocytochemistry of BRCA1-associated Protein-1 (BAP1) With Other Prognostic Markers in Uveal Melanoma. Am J Ophthalmol. 2018 May;189:122–6. doi: 10.1016/j.ajo.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szalai E, Wells JR, Ward L, Grossniklaus HE. Uveal Melanoma Nuclear BRCA1-Associated Protein-1 Immunoreactivity Is an Indicator of Metastasis. Ophthalmology. 2018 Feb;125((2)):203–9. doi: 10.1016/j.ophtha.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kivela TS, Grossniklaus HE, et al. AJCC Cancer Staging Manual. 8th ed. New York: Springer; 2017. [Google Scholar]
- 20.Patrone S, Maric I, Rutigliani M, Lanza F, Puntoni M, Banelli B, et al. Prognostic value of chromosomal imbalances, gene mutations, and BAP1 expression in uveal melanoma. Genes Chromosomes Cancer. 2018 Aug;57((8)):387–400. doi: 10.1002/gcc.22541. [DOI] [PubMed] [Google Scholar]
- 21.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998 Mar;16((9)):1097–112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 22.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39((3 Pt 2)):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 23.Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci USA. 2014 Jan;111((1)):285–90. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa H, Wu W, Koike A, Kojima R, Gomi H, Fukuda M, et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 2009 Jan;69((1)):111–9. doi: 10.1158/0008-5472.CAN-08-3355. [DOI] [PubMed] [Google Scholar]
- 25.Mashtalir N, Daou S, Barbour H, Sen NN, Gagnon J, Hammond-Martel I, et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol Cell. 2014 May;54((3)):392–406. doi: 10.1016/j.molcel.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 26.van Essen TH, van Pelt SI, Versluis M, Bronkhorst IH, van Duinen SG, Marinkovic M, et al. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br J Ophthalmol. 2014 Dec;98((12)):1738–43. doi: 10.1136/bjophthalmol-2014-305047. [DOI] [PMC free article] [PubMed] [Google Scholar]