Abstract
We previously reported dysregulated expression of liver-derived messenger RNA (mRNA) and long noncoding RNA (lncRNA) in patients with advanced fibrosis resulting from nonalcoholic fatty liver disease (NAFLD). Here we sought to identify changes in mRNA and lncRNA levels associated with activation of hepatic stellate cells (HSCs), the predominant source of extracellular matrix production in the liver and key to NAFLD-related fibrogenesis. We performed expression profiling of mRNA and lncRNA from LX-2 cells, an immortalized human HSC cell line, treated to induce phenotypes resembling quiescent and myofibroblastic states. We identified 1964 mRNAs (1377 upregulated and 587 downregulated) and 1460 lncRNAs (665 upregulated and 795 downregulated) showing statistically significant evidence (FDR ≤0.05) for differential expression (fold change ≥|2|) between quiescent and activated states. Pathway analysis of differentially expressed genes showed enrichment for hepatic fibrosis (FDR = 1.35E-16), osteoarthritis (FDR = 1.47E-14), and axonal guidance signaling (FDR = 1.09E-09). We observed 127 lncRNAs/nearby mRNA pairs showing differential expression, the majority of which were dysregulated in the same direction. A comparison of differentially expressed transcripts in LX-2 cells with RNA-sequencing results from NAFLD patients with or without liver fibrosis revealed 1047 mRNAs and 91 lncRNAs shared between the two datasets, suggesting that some of the expression changes occurring during HSC activation can be observed in biopsied human tissue. These results identify lncRNA and mRNA expression patterns associated with activated human HSCs that appear to recapitulate human NAFLD fibrosis.
Keywords: Hepatic stellate cells, Hepatic fibrosis, Cirrhosis, Nonalcoholic fatty liver disease, Long noncoding RNA
Highlights
-
•
Molecular mechanisms underlying HSC activation in the development of NASH fibrosis remain incompletely understood.
-
•
1964 mRNAs and 1460 lncRNAs were differentially expressed between quiescent and activated states.
-
•
Pathway analysis of differentially expressed RNAs associated with HSC activation showed enrichment for hepatic fibrosis.
-
•
Changes in expression of many RNAs that accompany HSC activation are mirrored in advanced fibrosis in NASH patients.
1. Introduction
Nonalcoholic fatty liver disease (NAFLD), an oftentimes progressive condition distinguished by hepatic steatosis, is the most common cause of chronic liver disease worldwide [1]. Approximately 20% of NAFLD patients develop a severe form of the disease known as nonalcoholic steatohepatitis (NASH), which is characterized by inflammation and hepatocyte ballooning degeneration and that can be accompanied by liver fibrosis [2]. The progression from NAFLD to NASH is associated with factors such as oxidative stress [3], pro-inflammatory cytokines [4,5], and immune response [6,7]. Clinical outcomes for NASH patients with hepatocyte injury, liver inflammation, and fibrosis are markedly worse compared to those with simple steatosis and are associated with greater liver-related morbidity and mortality [8,9], yet no pharmaceutical therapies are approved for the treatment of NASH. A subset of NASH patients with mild fibrosis will develop advanced fibrosis or cirrhosis [10] and eventually hepatocellular carcinoma [11]. NASH is the second leading indication for liver transplantation in the United States [12], and is predicted to be the most common indication within the next decade if current trends of increasing NASH prevalence and decreasing hepatitis C virus (HCV) infection continue [13].
Hepatic stellate cells (HSCs) play a critical role in the development of liver fibrosis [14], a condition characterized by excessive extracellular matrix (ECM) production [15]. Under physiologic conditions, HSCs store vitamin A within the perisinusoidal space [16]. In response to hepatic injury, quiescent HSCs are transformed to myofibroblasts, which secrete cytokines and other molecules to attenuate damage to the liver. HSC activation is characterized by loss of intracellular lipid droplets and increased production of alpha-smooth muscle actin (ACTA2), collagen type 1, alpha 1 (COL1A1), and other components of the ECM. Following resolution of injury, activated HSCs are removed through apoptosis and inactivation [17]. In the presence of chronic hepatic inflammation, however, activated HSCs continue to produce ECM, a process which can eventually lead to hepatic scarring. Many signal transduction pathways, including transforming growth factor beta [18] and Wnt signaling pathways [19], have been linked to HSC activation.
While the progression of NAFLD from steatosis to fibrosis has been modeled as a “multi-hit” process [20] that includes both environmental and genetic determinants [21], the molecular factors underlying its development and progression remain incompletely understood. Several studies have profiled differences in gene expression between different stages of disease, such as steatohepatitis versus steatosis and normal liver [22], mild fibrosis and septal fibrosis [23], low versus high levels of steatosis [24], and mild versus advanced fibrosis [25]. In our own work, we compared hepatic gene expression between samples with lobular inflammation and those with advanced fibrosis, and identified 34 differentially expressed transcripts [26]. Differences in expression levels of many of these genes were consistent with findings reported in microarray and quantitative RT-PCR studies [22,27] and replicated in comparisons of activated versus quiescent LX-2 cells [26].
A number of studies have implicated long noncoding RNAs (lncRNAs) in the development and progression of NAFLD [28]. LncRNAs are multifunctional molecules that contribute to diverse biological activities such as protein localization, structural support, and regulation of miRNA levels [[29], [30], [31], [32]]. In human studies, differential hepatic lncRNA expression was observed in a comparison of five NAFLD patients and five healthy individuals [33], 48 NASH patients, 11 patients with simple steatosis, and 23 healthy controls [34], and 24 individuals with normal liver histology, 53 NAFLD patients with lobular inflammation, and 63 NAFLD patients with severe fibrosis [35]. A role for lncRNAs in NAFLD fibrosis is further supported by work showing differential expression between quiescent and activated states in human and rat HSCs, including many lncRNAs associated with anti-fibrotic mechanisms [[36], [37], [38]]. Several studies have demonstrated that lncRNAs contribute to HSC activation through mechanisms involving extracellular matrix remodeling, TGF-β signaling, and regulation of the Notch pathway [36,[39], [40], [41]]. Together, these results suggest that lncRNAs not only participate in the transition of quiescent HSCs to an activated myofibroblast phenotype, but may also contribute to the development and progression of hepatic fibrosis in NAFLD patients. However, results from human, animal, and in vitro studies remain preliminary, and more work is needed to better understand the molecular mechanisms underlying hepatic fibrogenesis, the cell type and stage in disease pathogenesis associated with dysregulated lncRNAs, and the importance of the expression and molecular function behind candidate lncRNAs.
To extend our findings in whole tissue [26,35] and complement previous work in human and rat HSCs [[36], [37], [38],42], we performed microarray-based lncRNA and mRNA profiling using LX-2 cells, a well-characterized, immortalized human HSC cell line that retains many important features of primary HSCs [43]. We observed differential expression of both lncRNAs and mRNAs resulting from LX-2 cell activation and identified a network of co-expressed lncRNAs and nearby mRNAs, suggesting shared mechanisms of transcriptional regulation. A subset of lncRNAs and mRNAs showing differential expression in LX-2 cells were also dysregulated in liver samples from NAFLD patients with biopsy-proven fibrosis [26,35], providing a cellular context for fibrogenic mechanisms that may occur in early stages of disease.
2. Methods
2.1. Cell culture
LX-2 cells (Merck Millipore; Billerica, MA) were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 2% fetal bovine serum (FBS). Cell line authentication was performed using short tandem repeat (STR) profiling (Cell Line Genetics; Madison, Wi), which confirmed the presence of a single cell line and alleles matching the known DNA fingerprint [44]. Cells were grown in T-75 flasks (Corning Life Sciences; Corning, NY) containing 12 mL cell culture medium and placed at 37 °C in a Hera Cell 5% CO2 incubator (Thermo Fisher Scientific; Waltham, MA). Culture medium was replaced the first day after seeding, and then every 72 h. In preparation for treatment with MDI solution, LX-2 cells were seeded at 0.5 × 106 cell/well on 6-well culture dishes (VWR International; Radnor, PA) and serum-starved overnight. Cell culture medium was aspirated and replaced with DMEM, 10% FBS, and MDI solution [0.5 mM isobutylmethylxanthine, 1 μM dexamethasone, and 167 nM insulin (Sigma-Aldrich; St. Louis, MO)] for 72 h to induce a state resembling biological quiescence [45]. General morphology was observed with a light microscope, lipid accumulation was assessed with Oil Red O dye (Sigma-Aldrich), and levels of alpha smooth muscle actin (ACTA2) were measured by western blot analysis.
2.2. RNA extraction and array hybridization
Total RNA was extracted from six biological replicates for each experimental cell group using the RNeasy Kit (Qiagen; Valencia, CA). RNA quantity and quality were measured using the NanoDrop ND-1000 spectrophotometer (ThermoFisher Scientific) and RNA integrity was assessed with the 2100 Bioanalyzer System (Agilent Technologies; Santa Clara, CA). Samples were amplified and transcribed into fluorescent cRNA using a random priming method (Arraystar Flash RNA Labeling Kit [Arraystar; Rockville, MD]). Labeled cRNAs were purified using the RNeasy mini Kit (Qiagen) and the concentration and specific activity of the labeled cRNAs were measured using the NanoDrop ND-1000. One microgram of each labeled cRNA was fragmented and heated according to the manufacturer's protocol and then added to the microarray slides. Slides were incubated for 17 h at 65 °C in an Agilent Hybridization Oven. The microarray used in this study was the Human LncRNA Microarray V4.0 (Arraystar), which detects 40,173 lncRNAs and 20,730 protein-coding mRNAs. After washing and fixing, arrays were scanned using the Agilent DNA Microarray Scanner.
2.3. Data analysis
Array images were analyzed using Agilent Feature Extraction software (version 11.0.1.1). Raw signal intensities were normalized in quantile method using GeneSpring GX v12.1 software package (Agilent Technologies). Low intensity lncRNAs and mRNAs were removed from further data analysis. False discovery rates (FDR) were adjusted from all p-values using the Benjamini-Hochberg method [46] for multiple testing correction. LncRNAs and mRNAs showing statistically significant evidence for differential expression between the two groups were identified using Volcano Plot filtering (fold-change ≥2.0, FDR ≤0.05). Transcripts were considered to be differentially expressed if the FDR was <0.05 and the absolute fold change was ≥2.
2.4. Pathway analysis
To interpret the biological significance of the microarray data, we uploaded the list of differentially expressed genes (untreated vs. MDI-treated; FDR ≤0.05 and absolute fold change ≥2) containing gene identifiers, expression values, and FDR values into the Ingenuity Pathway Analysis (IPA) software (Qiagen). The “core analysis” function was applied to identify canonical pathways, upstream regulators, and gene networks relevant to the differentially expressed transcripts. Gene identifiers were mapped to corresponding gene objects using the Ingenuity Pathway Knowledge Base. In the comparisons of microarray data with RNA-sequencing data -details of this data set, including patient samples, RNA sequencing experiments, and raw data processing are provided elsewhere [26]- we used the “compare” function as implemented in the IPA software.
2.5. mRNA and lncRNAs co-expression analysis
Differentially expressed lncRNAs were classified in different categories: bidirectional, exon sense-overlapping, intron sense-overlapping, intronic antisense, and natural antisense. We mapped the mRNA genes located within 10 kb of differentially expressed lncRNAs, retaining the mRNAs/lncRNAs pairs both showing differential expression. Then, we counted the pairs showing same or opposing log2 fold change direction, computing the absolute frequency for each lncRNA category.
2.6. Measurement of individual transcripts using quantitative RT-PCR (qPCR)
Total RNA was extracted from six biological replicates from treated and untreated LX-2 cells using the RNeasy Mini Kit (Qiagen). All RNA preparations were diluted to 5 μg/μl. The Taqman RNA-to-Ct 1-Step kit (ThermoFisher Scientific) was used to convert RNA to cDNA according to the protocol, followed by RT-qPCR analysis using Taqman Gene Expression Assays and the QuantStudio 6 Flex (Life Technologies; Carlsbad, CA). We used glyceraldehyde 3-phosphate dehydrogenase (GAPDH) for normalization of expression and the -ΔΔCt method to determine the fold-change of RNA expression. A two-tailed t-test was used to assess statistical significance. Primer sequences for all qPCR assays are available upon request.
2.7. RNA extraction from liver wedge biopsies
Liver wedge biopsies were obtained from individuals enrolled in the Bariatric Surgery Program at the Geisinger Clinic Center for Nutrition and Weight Management [47]. Details of the study population can be found elsewhere [[48], [49], [50]]. All study participants provided written informed consent for research, which was conducted according to The Code of Ethics of the World Medical Association (Declaration of Helsinki). The Institutional Review Boards of Geisinger Health System, Translational Genomics Research Institute, and Temple University School of Medicine approved the research protocol. Liver tissue was cut into ~5–10 mg pieces and added to sterile microcentrifuge tubes (Next Advance; Troy, NY) with RLT Plus lysis buffer containing β-mercaptoethanol (Qiagen), and homogenized in a Bullet Blender Gold homogenizer (Next Advance) for 12–15 min. Total RNA was extracted from homogenized lysate using the RNeasy Mini Kit (Qiagen) and quantified by the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). Quantitative PCR was performed as described above.
3. Results
3.1. Characteristics of cells
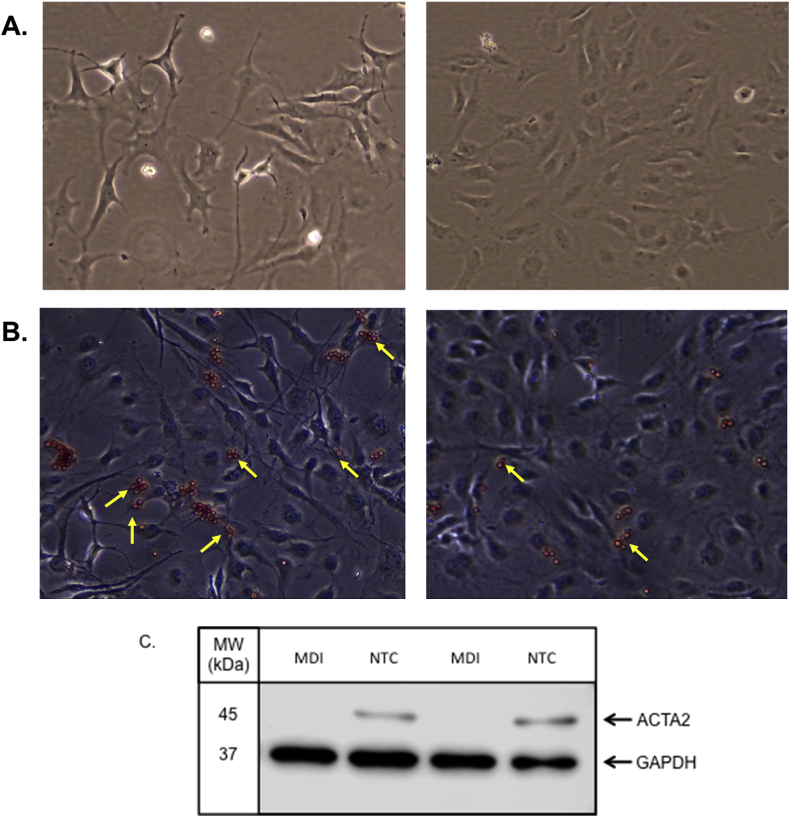
LX-2 cells were grown in plastic culture plates and treated with MDI to induce a state resembling biological quiescence [45,51] or left untreated to achieve a myofibroblast phenotype. As shown in Fig. 1A, untreated cells showed a large, flat appearance due to loss of lipid droplets, while MDI-treated cells acquired a smaller cell body with a spindle-shaped phenotype, consistent with published observations [52]. Oil Red O staining was higher in LX-2 cells treated with MDI (Fig. 1B), indicating a greater accumulation of lipid droplets, and levels of ACTA2 protein were lower in these cells compared to those left untreated (Fig. 1C).
Fig. 1.
Differential characteristics of untreated LX-2 cells versus those treated with MDI. Light microscopy images from A) LX-2 cells treated with MDI for 72 h and LX-2 cells grown under normal culture conditions (NTC) for 72 h. All imaging was performed at 10X using a light microscope. B) Oil Red O staining of LX-2 cells treated with MDI and cells grown under normal conditions. Yellow arrows depict cells showing representative staining. C) protein levels of alpha smooth muscle actin (ACTA2) in MDI-treated cells and untreated cells (NTC) were detected using western blot analysis. Membranes were incubated with primary antibodies directed against ACTA2 (anti-mouse 1:500 dilution; Invitrogen) or GAPDH (anti-rabbit 1:1000; Cell Signaling Technology), washed in 1X TBS-Tween buffer and then incubated with secondary antibodies conjugated to horseradish peroxidase (1:3000; Cell Signaling Technology) for 1 h at room temperature. Membranes were incubated with Clarity Western ECL Blotting Substrate (Bio-Rad; Hercules, CA) according to the manufacturer's instructions. Protein bands were imaged using the Odyssey FC imaging system (LI-COR Biotechnology; Lincoln NE).
3.2. Differentially expressed lncRNAs and mRNAs in LX-2 cells
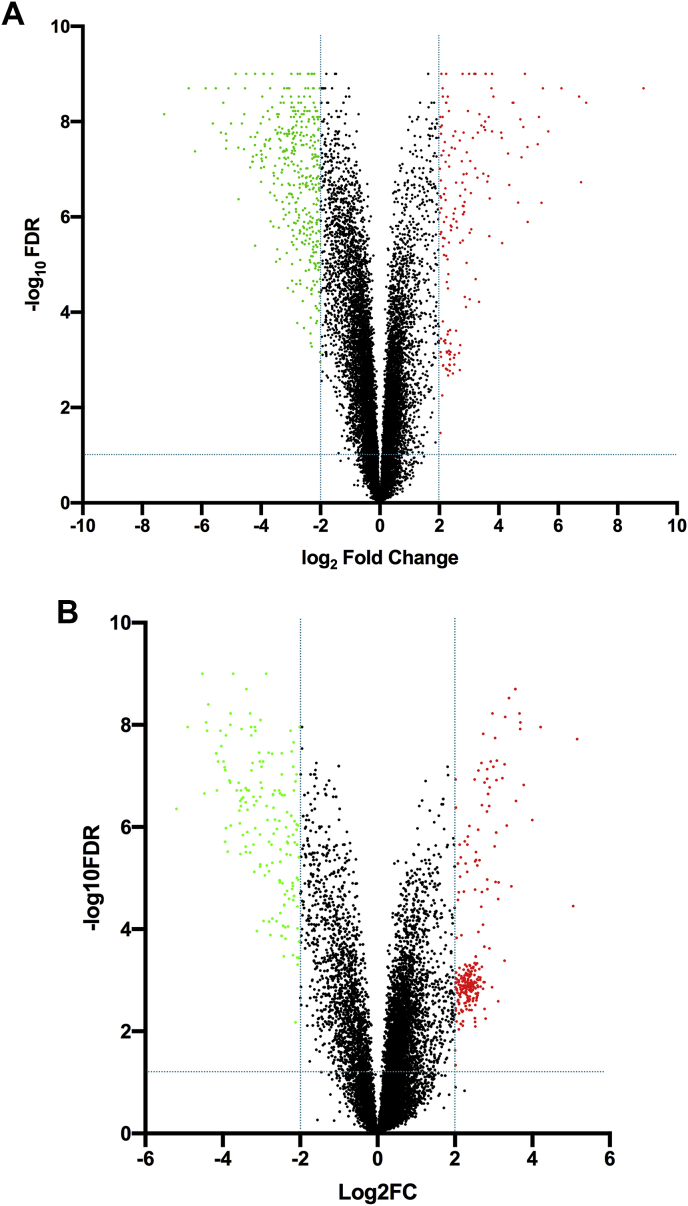
Initial analyses showed that 26,515 (66%) lncRNAs and 15,355 (74%) mRNAs were detected in LX-2 cells. The mean normalized intensity was greater in mRNAs (normalized intensity = 685) compared to lncRNAs (normalized intensity = 113), consistent with other studies [53,54]. In comparisons of MDI-treated and untreated LX-2 cells, 8553 mRNAs were identified, 8311 of which could be mapped. We filtered this dataset using an absolute fold change ≥2 and FDR ≤0.05 and observed 1964 transcripts showing evidence for differential expression between the two experimental conditions (Fig. 2A), corresponding to 1377 upregulated and 586 downregulated transcripts. The top twenty dysregulated mRNAs are shown in Table 1, while the complete list of dysregulated genes can be found in Supplemental Table S1.
Fig. 2.
Differentially expressed transcripts in treated LX-2 cells. Volcano plot filtering between MDI-treated and untreated LX-2 cells was performed to identify differentially expressed A) mRNAs and B) lncRNAs between experimental groups. False discovery rates (FDR) were adjusted from all p-values for multiple testing corrections. The thresholds were fold-change ≥ |2.0| and FDR ≤0.05. Plots were constructed by plotting the FDR p-value (-log10) on the y-axis and the expression fold-change (log2) between the two groups on the x-axis. Data points in red represent upregulated transcripts, while those in green represent downregulated transcripts. Black-colored data points represent RNAs not showing statistically significant evidence for differential expression between the two treatment groups. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Top dysregulated mRNAs in activated LX-2 cells.
| ID | Gene Symbol | Fold Change | FDR | MDI groupa | Untreated groupa |
|---|---|---|---|---|---|
| NM_002421 | MMP1 | 153.70 | 7.00E-09 | 3.71 | 10.97 |
| NM_033066 | MPP4 | 86.59 | 2.00E-09 | 3.24 | 9.46 |
| NM_001077693 | ECSCR | 74.82 | 4.20E-08 | 2.92 | 9.15 |
| NM_133262 | ATP6V1G3 | 58.35 | 2.00E-09 | 2.54 | 8.40 |
| NM_002275 | KRT15 | 49.21 | 1.10E-08 | 3.02 | 8.64 |
| NM_002838 | PTPRC | 46.01 | 2.00E-09 | 2.52 | 8.04 |
| NM_001017373 | SAMD3 | 44.28 | 0.00E+00 | 2.67 | 8.14 |
| NM_016356 | DCDC2 | 40.77 | 1.70E-08 | 5.14 | 10.49 |
| NM_020152 | MAP3K7CL | 38.65 | 4.00E-09 | 5.86 | 11.13 |
| NM_014210 | EVI2A | 38.28 | 0.00E+00 | 2.52 | 7.78 |
| NM_005525 | HSD11B1 | −2684.89 | 0.00E+00 | 14.54 | 3.15 |
| NM_005420 | SULT1E1 | −1177.90 | 0.00E+00 | 12.89 | 2.69 |
| NM_001037132 | NRCAM | −468.64 | 2.00E-09 | 13.88 | 5.01 |
| NM_002612 | PDK4 | −137.45 | 0.00E+00 | 14.97 | 7.87 |
| NM_006006 | ZBTB16 | −123.07 | 4.00E-09 | 12.74 | 5.79 |
| NM_000240 | MAOA | −108.92 | 1.87E-07 | 10.65 | 3.88 |
| NM_002029 | FPR1 | −103.98 | 3.00E-09 | 10.32 | 3.62 |
| NM_024420 | PLA2G4A | −69.06 | 2.00E-09 | 12.09 | 5.98 |
| ENST00000326958 | AC026703.1 | −66.04 | 0.00E+00 | 14.29 | 8.25 |
| NM_001145161 | UBE2QL1 | −50.59 | 1.60E-08 | 10.56 | 4.90 |
Normalized intensity.
In our lncRNA analyses, 1460 lncRNAs showed statistically significant evidence (FDR ≤0.05) for an absolute fold change ≥2 in expression (Fig. 2B). Of these, 665 showed increased and 795 showed decreased levels in activated cells (Supplemental Table S2). The top twenty dysregulated lncRNAs that could be mapped are shown in Table 2.
Table 2.
Top dysregulated, mapped lncRNAs in activated LX-2 cells.
| ID | Gene Symbol | Fold Change | FDR | MDI groupa | Untreated groupa |
|---|---|---|---|---|---|
| ENST00000522718 | LINC01605 | 84.16 | 0.00E+00 | 2.65 | 9.05 |
| NR_105006 | LINC01111 | 79.68 | 2.00E-09 | 4.42 | 10.74 |
| ENST00000444114 | LINC01638 | 47.29 | 0.00E+00 | 6.95 | 12.52 |
| ENST00000452578 | RP11_20J153 | 36.63 | 4.42E-07 | 3.41 | 8.60 |
| ENST00000445974 | RP11_437J191 | 21.51 | 9.00E-09 | 2.52 | 6.94 |
| ENST00000566394 | RP11_553K85 | 17.95 | 3.60E-08 | 2.63 | 6.80 |
| NR_033987 | LINC02393 | 17.74 | 1.93E-07 | 3.12 | 7.27 |
| ENST00000507388 | RP11_1E61 | 17.38 | 5.20E-08 | 2.52 | 6.64 |
| ENST00000430529 | LINC01865 | 16.70 | 1.30E-08 | 2.65 | 6.71 |
| NR_110271 | LINC01812 | 16.40 | 2.60E-08 | 2.52 | 6.55 |
| NR_026860 | LINC00473 | −35.65 | 1.90E-08 | 11.40 | 6.24 |
| NR_120623 | TCERG1L-AS1 | −12.88 | 1.20E-08 | 10.02 | 6.33 |
| uc001hpe.1 | LOC102723834 | −10.11 | 9.40E-07 | 8.69 | 5.35 |
| NR_125801 | PACERR | −8.29 | 1.19E-05 | 7.89 | 4.84 |
| NR_003679 | HAND2-AS1 | −7.57 | 5.20E-08 | 10.37 | 7.45 |
| NR_038328 | TEKT4P2 | −7.11 | 7.40E-08 | 12.94 | 10.11 |
| NR_131986 | KCCAT198 | −6.93 | 5.65E-03 | 6.70 | 3.90 |
| NR_026880 | MGC12916 | −6.44 | 5.60E-08 | 10.09 | 7.40 |
| NR_126347 | LINC01036 | −6.08 | 1.14E-04 | 5.41 | 2.81 |
| NR_046655 | BRWD1-AS1 | −5.99 | 1.97E-03 | 8.35 | 5.76 |
Normalized intensity.
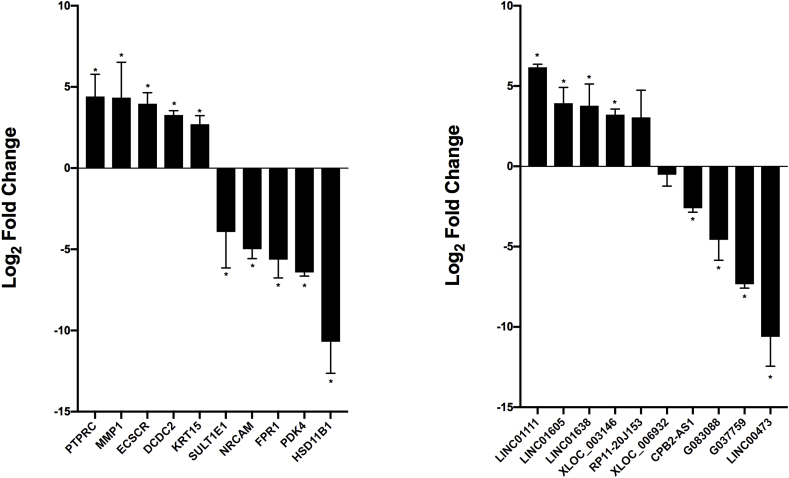
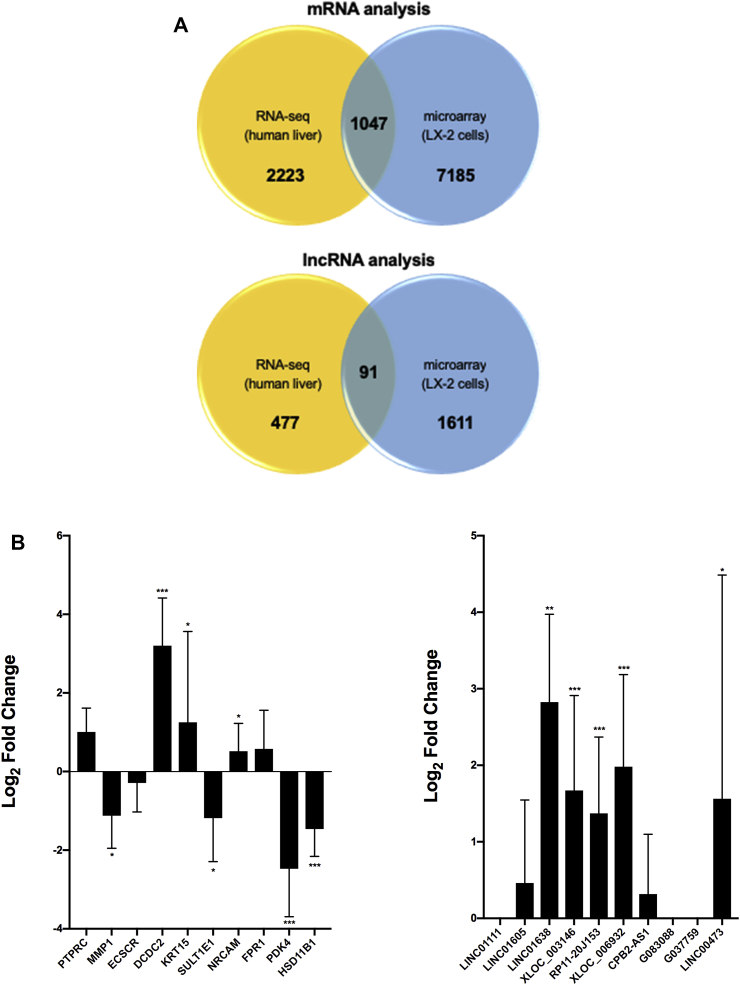
We selected twenty differentially expressed RNAs for analysis using an orthogonal platform to verify the accuracy of the microarray findings. All assayed lncRNAs and mRNAs showed differential expression between treated and untreated LX-2 cells in directions consistent with the array results (Fig. 3). Of these, LINC01111 was the most upregulated (6.2-fold) and G037759 the most downregulated (10.6-fold) in untreated cells; while PTPRC was the most upregulated (4.4-fold) and HSD11B1 the most downregulated (10.7-fold) mRNA.
Fig. 3.
Validation of differentially expressed RNAs using an orthogonal platform. Results are shown for mRNAs and lncRNAs in untreated versus MDI-treated LX-2 cells. Expression levels were normalized against GAPDH. All experiments were performed in triplicate. Data are expressed as mean ± standard deviation. *P ≤ 0.05, **P ≤ 0.001, and ***P ≤ 0.0001.
3.3. Enriched biological pathway analysis of differentially expressed genes
Of the 1964 dysregulated mRNAs, 1896 (1340 upregulated and 556 downregulated) could be mapped using the IPA software. To determine the biological significance of these 1896 genes, we identified the most significant canonical pathways involved in LX-2 activation using IPA. We found overlap of 74 canonical pathways (P < 0.01) associated with hepatic fibrosis, EMT transition, and osteoarthritis. The most significant canonical pathways are shown in Table 3. The top biological functions connected with the dysregulated mRNAs were cellular movement (553 molecules; p-value range: 7.52E-11 - 6.58E-45), cellular development (646 molecules; p-value range: 7.96E-11 – 7.13E-25), cell death and survival (621 molecules; p-value range: 4.48E-11 – 2.22E-24), cellular growth and proliferation (649 molecules; p-value range: 7.96E-11 – 1.86E-21), and cell-to-cell signaling and interaction (431 molecules; p-value range: 8.71E-11 – 1.58E-19).
Table 3.
Top canonical pathways associated with LX-2 activation.
| Pathway | # mol/totala | p-valueb | Molecules |
|---|---|---|---|
| Hepatic fibrosis | 54/186 | 1.35E-16 | TGFBR1, IL6, IL1R2, VEGFA, TGFB1, LAMA1, MYL4, SERPINE1, IL1RAP, TNFRSF11B, CXCL8, COL4A1, COL2A1, VEGFC, IGFBP5, MMP2, COL21A1, COL6A3, COL13A1, ACTA2, IGFBP3, CD14, AGTR1, COL3A1, IL1A, ICAM1, COL4A5, FN1, COL4A6, FGF2, COL4A3, EGF, CCL5, COL4A2, COL15A1, PGF, COL1A2, COL16A1, CCL2, EDN1, TIMP1, TGFB2, LBP, MMP1, EGFR, IL4, COL6A2, EDNRB, COL12A1, IL1R1, FGF1, IL1B, COL9A2, MMP9 |
| Osteoarthritis | 55/212 | 1.47E-14 | TGFBR1, RARRES2, MMP3, PTHLH, IL1R2, VEGFA, ITGA3, TGFB1, CNMD, ADAMTS5, IL1RAP, ADAMTS4, CASP10, ITGA4, CXCL8, DDIT4, DCN, COL2A1, ITGA5, VEGFC, TCF3, NKX3-2, S1PR3, TLR2, FZD5, HTRA1, JAG1, CASP5, TCF4, FN1, FRZB, FGF2, CASP4, HIF1A, PGF, GLIS1, PRKAA2, CASP1, CASP8, MMP1, TIMP3, EPAS1, ITGA2, GREM1, TCF7L1, IL1R1, CEBPB, WNT3A, PTH1R, IL1B, WISP1, PTGS2, GLI1, P2RX7, MMP9 |
| Axonal guidance signaling | 83/501 | 1.09E-09 | GAB2, ADAMTS8, MYL10, WNT3, MMP3, PIK3R1, MMP16, CXCL12, ADAM11, ADAMTS2, VEGFA, ITGA3, EPHB1, PLCE1, SEMA3D, MAPK3, PIK3CG, ABLIM3, MYL4, PLCB1, IRS2, FRS2, ADAMTS5, ADAMTS4, ITGA4, PAPPA, NGEF, FES, ITGA5, VEGFC, L1CAM, MMP2, PLCL2, ADAMTS9, PIK3R3, ADAMTS6, ADAM12, ARHGEF6, PRKCH, PIK3CD, FZD5, EPHA2, GNAL, NRP1, MME, ERAP2, MMP7, NTF3, RGS3, BMP3, EGF, ADAM33, EPHA4, PLXNA2, NGF, PGF, EFNB2, GLIS1, RHOD, SDC2, ADAM19, TUBA3C/TUBA3D, SEMA3B, RASSF5, PLCD4, MMP1, UNC5C, ADAMTS15, EPHB4, PLXNC1, ITGA2, SLIT2, ROBO3, PLXND1, SEMA3A, WNT3A, NTRK3, EPHA5, SEMA3C, SEMA4B, GLI1, MMP9, WNT5A |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis | 60/327 | 4.86E-09 | SOCS1, GAB2, MMP3, WNT3, PIK3R1, CXCL12, IL6, CCND1, IL18R1, IL1R2, VEGFA, CAMK2D, PLCE1, TGFB1, MAPK3, PIK3CG, PLCB1, IRS2, IL1RAP, FRS2, PRSS3, ADAMTS4, TNFRSF11B, CXCL8, VEGFC, PLCL2, IL37, TCF3, IL7, TLR2, PIK3R3, FZD5, PIK3CD, PRKCH, TCF4, IL1A, ICAM1, FN1, FRZB, FGF2, CEBPD, CCL5, PGF, IRAK1, CCL2, TLR3, PLCD4, MMP1, MAP2K7, MYD88, IL15, DAAM1, IKBKE, IL1R1, TCF7L1, CEBPB, PRSS1, WNT3A, IL1B, WNT5A |
| Cardiac hypertrophy signaling | 80/498 | 9.0E-09 | GAB2, MAP3K15, LIF, TGFBR1, WNT3, TGFBR3, PIK3R1, PDE3A, IL6, IL18R1, IL1R2, ITGA3, PLCE1, PDE6A, CAMK2D, PDE7B, TGFB1, MAPK3, PIK3CG, PLCB1, IRS2, FRS2, ITGA4, IL2RB, TNFRSF11B, FGF19, CXCL8, PDE2A, ITGA5, PLCL2, IL37, PDE4B, ITPR1, PDE1C, PIK3R3, ADRA2A, CD70, ITPR3, IFNLR1, PRKCH, PIK3CD, FZD5, AGTR1, TG, FGF5, IL1A, IL17RD, FGF2, IL22RA1, MAP3K5, PDE1A, FGF13, IL17B, HAND2, ADRB1, EDN1, TGFB2, PDE4D, IL27RA, PLCD4, ACVR1C, IL4, MAP2K7, EDNRB, PDE9A, IL15, ITGA2, RPS6KA5, IKBKE, IL1R1, FGF1, NPPB, PRKG1, WNT3A, MYOCD, PDE5A, IL1B, FGF20, PTGS2, WNT5A |
| Sperm motility | 44/225 | 8.08E-08 | TYRO3, PLA2R1, LMTK3, EPHA4, PDE1A, STYK1, EPHB1, PLCE1, TXK, PLA2G5, PLCB1, KIT, PDE4D, PLCD4, EGFR, PDE2A, EPHB4, MAP2K7, FES, PLA2G4C, PLA2G1B, PLCL2, ITPR1, PDE4B, ROR1, AXL, FRK, CNGA1, DDR1, PDE1C, NPPB, TEC, PLA2G4A, PRKG1, ABL2, FLT3LG, NTRK3, ITPR3, EPHA5, RARRES3, PRKCH, EPHA2, CATSPER1, PAFAH1B3 |
| Bladder cancer signaling | 25/97 | 2.49E-07 | MMP7, MMP3, FGF2, SUV39H1, MMP16, EGF, CCND1, FGF13, PGF, VEGFA, MAPK3, MMP1, FGF19, EGFR, MMP19, CXCL8, DAPK1, VEGFC, MMP2, RPS6KA5, FGF1, CDKN1A, FGF20, MMP9, FGF5 |
| Regulation of EMT | 39/201 | 5.32E-07 | GAB2, TCF4, SNAI2, TGFBR1, WNT3, FGF2, PIK3R1, PARD6B, EGF, HIF1A, PARD6A, SMURF1, FGF13, MAML1, TGFB1, PIK3CG, MAPK3, TGFB2, IRS2, FRS2, EGFR, FGF19, MAP2K7, ESRP2, MMP2, TCF7L1, TCF3, FGF1, PIK3R3, GSC, WNT3A, CDH12, FGF20, FZD5, PIK3CD, JAG1, MMP9, FGF5, WNT5A |
number of genes identified in dataset/total number participating in pathway.
calculated using Fisher's exact test.
Of the 1460 differentially expressed lncRNAs, 530 (383 upregulated and 147 downregulated) could be mapped using IPA. In contrast to the enriched pathway analyses for differentially expressed mRNAs, only seven canonical pathways were identified for lncRNAs, and only one molecule was found in each pathway (data not shown). These results may reflect the relative dearth of lncRNA annotation compared to mRNAs.
3.4. Associated network functions of differentially expressed genes
We identified 25 significant networks associated with the dysregulated genes in LX-2 cell activation. These networks were scored based on the number of genes participating in a given network. The top networks and associated functions were 1) carbohydrate metabolism, nucleic acid metabolism, small molecule biochemistry (33 molecules; Fig. 4); 2) embryonic development, organismal development, tissue development (33 molecules); 3) endocrine system disorders, organ morphology, organismal development (32 molecules); and 4) developmental disorders, hereditary disorders, metabolic disease (31 molecules). Analysis of regulator effector networks revealed fibronectin 1 (FN1) to be the top regulator associated with the migration of connective tissue cells. For the lncRNA analysis, three significant networks and associated functions were identified: 1) cell morphology, hair and skin development and function, cancer (17 focus molecules); 2) organismal development, cellular compromise, cell cycle (11 focus molecules); 3) DNA replication, recombination and repair, cellular assembly and organization, cellular function and maintenance (8 focus molecules).
Fig. 4.
Top network associated with carbohydrate metabolism, nucleic acid metabolism, and small molecule biochemistry in differentially expressed mRNAs. Pathway analysis was performed using the “core analysis” function in IPA. This plot depicts the top network identified.
3.5. Expression of genes located near differentially expressed lncRNAs are also dysregulated
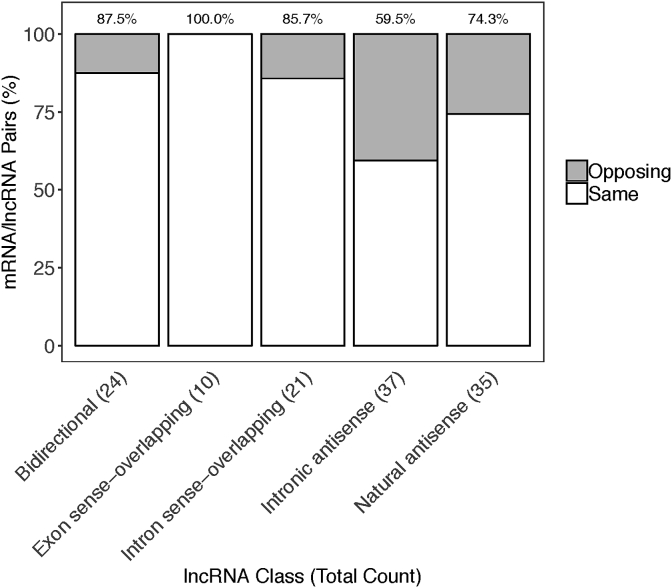
Because lncRNAs are characterized by their location relative to nearby protein-coding genes [55], we examined the relationship between differentially expressed lncRNAs and nearby mRNA transcripts. We identified 127 differentially expressed lncRNA/nearby mRNA pairs. The distribution of the transcriptional relationships included 24 bidirectional, 10 exon sense overlapping, 21 intron sense overlapping, 36 intronic antisense, and 35 natural antisense lncRNAs (Supplemental Table S3). Overall, 76% of differentially expressed lncRNA/nearby mRNA pairs showed expression changes in the same direction, with the greatest matching (100%) occurring for exon sense-overlapping lncRNAs and the lowest matching (59.5%) occurring for intronic antisense lncRNAs (Fig. 5).
Fig. 5.
mRNA/lncRNA interaction. Barplot representing the proportion of lncRNAs having same (white) or opposite (grey) log2 fold change direction with close mapping mRNAs. The total number of lncRNA/mRNA pairs for each lncRNA category is depicted on the x-axis, and the percentage of lncRNA/mRNA pairs with same fold change direction is shown at the top of the plot.
3.6. Differentially expressed lncRNAs and mRNAs are dysregulated in NAFLD-related fibrosis
LX-2 cells are a transformed cell line and the reversion to quiescence applied here is not expected to fully recapitulate HSC activation in vivo [44]. Therefore, we sought to assess whether expression of differentially expressed mRNAs and lncRNAs identified using our LX-2 model was altered in human hepatic fibrosis. We first compared the datasets of differentially expressed mRNAs and lncRNAs obtained in LX-2 cells with those obtained by RNA-sequencing using biopsied liver tissue from NAFLD patients with normal (N = 24) or fibrotic (N = 53) histology [26,35]. We identified 1047 mRNAs and 91 lncRNAs shared between the two datasets (Fig. 6A). The majority of overlapping dysregulated RNAs showed downregulated expression in LX-2 activation and NAFLD fibrosis; the top downregulated mRNAs and lncRNAs are shown in Table 4. We next performed qPCR analysis to measure expression of a subsample of transcripts in NAFLD patients with advanced fibrosis and normal liver histology (Fig. 6B). We observed increased expression of DCDC2, KRT15, PTPRC, LINC01638, LINC01605, XLOC_003146, and RP11_20J153 and decreased expression of HSDB11, PDK4, SULT1E1, consistent with results obtained in LX-2 cells. Interestingly, all of the lncRNAs downregulated in LX-2 cell activation showed increased hepatic expression in fibrotic liver.
Fig. 6.
Identification of differentially expressed RNAs shared between LX-2 model and human liver samples from NAFLD patients with fibrosis. A) Intersection of filtered (FDR ≤0.05) and mapped datasets of differentially expressed mRNAs and lncRNAs obtained in LX-2 cells and biopsied liver tissue from NAFLD patients with normal (N = 24) or fibrotic (N = 53) histology [26,35]. B) Quantitative PCR analysis using hepatic RNA from NAFLD patients with normal liver histology (n = 10) and severe fibrosis (n = 10). Transcript levels were normalized to GAPDH, which showed invariant expression. Data are presented as relative log2 fold-change. A Mann-Whitney U test was used to assess statistical significance. *P ≤ 0.05, **P ≤ 0.001, and ***P ≤ 0.0001.
Table 4.
Top overlapping dysregulated RNAs in LX-2 cells and human liver.
| ID | Gene symbol | RNA class | Log2 fold change |
|
|---|---|---|---|---|
| LX-2 cells | NASH fibrosis | |||
| NM_001025200 | CTRB2 | mRNA | −1.39 | −6.66 |
| NM_001001676 | LCN9 | mRNA | −1.56 | −6.14 |
| NM_001042506 | PABPC1L2B | mRNA | −1.61 | −6.07 |
| NM_001300814 | KRT78 | mRNA | −1.26 | −5.78 |
| NM_198691 | KRTAP10-1 | mRNA | −1.80 | −5.71 |
| LINC00364 | LINC00364 | lncRNA | −1.53 | −7.32 |
| LINC00922 | LINC00922 | lncRNA | −1.40 | −6.85 |
| LINC01048 | LINC01048 | lncRNA | −1.70 | −6.77 |
| LINC00658 | LINC00658 | lncRNA | −1.58 | −6.62 |
| MIR137HG | MIR137HG | lncRNA | −1.38 | −6.29 |
4. Discussion
The presence of liver fibrosis in NAFLD is associated with increased liver-related morbidity and mortality [56] and a major risk factor for the development of hepatocellular carcinoma [57]. Advanced liver fibrosis represents the end-stage pathology evolving from a number of pathogenic mechanisms [58]. Among these, HSCs, which play a major role in ECM maintenance in the liver, are important contributors to hepatic fibrogenesis and morphological changes induced in these cells as a result of hepatic stress have been associated with dysregulated expression of protein-coding genes [[36], [37], [38],42], lncRNAs [36,37], and miRNAs [59]. However, a limited understanding of the relationship between altered HSC expression patterns and the development of liver fibrosis in NAFLD patients persists. Here, we identified a set of mRNAs and lncRNAs that discriminated between induced phenotypic states of LX-2 cells and validated a subset of these in liver tissue from NAFLD patients with fibrosis. These results are the first to provide biological overlap between transcriptional changes occurring in an in vitro model of HSC activation and liver tissue from NAFLD patients.
LncRNAs are known to influence expression of genes located in proximity (cis-acting) or elsewhere (trans-acting) through interactions with DNA, RNA, and protein [60]. Some of the mechanisms by which lncRNAs regulate gene expression include direct interaction with other nucleic acids by complementary base-pairing, recruitment of transcription factors or chromatin-modifying complexes to DNA targets, formation of heterogeneous nuclear ribonucleoprotein complexes, and sequestering of RNA-binding proteins and microRNAs through decoy action [61]. Compared with protein-coding genes, lncRNAs show stronger tissue-specific patterns of expression [62], and these transcripts may associate with other co-expressed RNAs to produce similar phenotypic effects [53]. While expression levels of lncRNAs are generally lower than mRNAs [63], emerging single cell analyses reveal abundant expression in individual cells compared to bulk tissue [64,65], suggesting that analysis of whole tissues may average gene expression signatures of many different cell types and mute expression patterns in individual cells. This observation may explain why expression patterns of lncRNAs in whole tissue were less likely to replicate cellular profiles compared to mRNAs.
Patterns of mRNA expression identified in LX-2 cells followed a similar distribution in fibrotic liver compared to expression of lncRNAs, and all lncRNAs showing validated downregulation in LX-2 cells were upregulated in fibrotic liver compared to normal tissue. These findings may be due to cell-type-specific expression patterns in whole tissue analyses, a characteristic of lncRNAs [66], or may represent bona fide differences in expression between acute HSC activation versus established fibrosis. These data highlight the need for analysis of single cells obtained in vivo.
To our knowledge, five studies, using different experimental designs and cell models, have profiled mRNA or lncRNA changes accompanying HSC activation. The first investigation applied oligonucleotide microarrays to identify expression patterns of protein-coding genes in immortalized human hepatic stellate cells (LI90 cells) cultured on Matrigel to induce a quiescent phenotype [42]. While differential expression of 3350 transcripts was found between activated and quiescent LI90 cells, myocardin (MYOCD) emerged as a key candidate involved in the HSC activation process and its expression was increased in activated LI90 cells, rat primary HSCs, and liver from a dimethylnitrosamine-induced rat model of hepatic fibrosis. RNA-mediated knockdown of MYOCD expression in LI90 cells and activated rat primary HSCs corresponded with decreased levels of ACTA2 and COL1A1 [42]. In the current study, we also observed upregulation of MYOCD in activated LX-2 cells in parallel with increased ACTA2 expression, providing further support that this protein contributes to or accompanies HSC transition to a myofibroblastic phenotype.
Two independent investigations profiled gene expression using primary HSCs isolated from Sprague-Dawley rats [36,38]. In the first study, an array-based approach was used to identify >2000 differentially expressed (≥2-fold change) protein-coding genes in HSCs obtained from normal untreated animals and cultured for either one day (quiescent) or seven days (activated) [38]. The major finding from this work showed that the Wnt5a signaling pathway participated in the activation of rat HSCs, with increased Wnt5a expression in activated versus quiescent states and in livers from rats treated with carbon-tetrachloride (CCl4) to induce hepatic fibrosis. Suppression of Wnt5a expression also resulted in impaired proliferation and downregulation of COL1A1 and TGFB1 [38]. The second study performed RNA sequencing using primary rat HSCs cultured for either two (quiescent) or seven (activated) days and identified 529 and 24 differentially expressed mRNAs and lncRNAs, respectively [36]. The authors identified a network of 632 matched lncRNA-mRNA pairs consisting of 24 and 122 differentially expressed lncRNAs and mRNAs, respectively, with NONRATT0139819.2 being the lncRNA showing the highest correlation with co-expressed mRNAs. NONRATT0139819.2 was found to lie adjacent to the gene encoding lysyl oxidase (Lox) and both transcripts were co-expressed during activation of HSC-T6 rat and fibrotic liver tissue from CCl4-treated animals. Together, the studies conducted in Sprague-Dawley rats identified two distinct pathways contributing to HSC activation. Interestingly, although the two studies utilized an almost identical cell model of HSC activation, the microarray-based investigation identified 2566 differentially expressed (1396 upregulated and 1170 downregulated) genes in culture-activated HSCs, while the RNA-sequencing approach reported only 529 differentially expressed (155 upregulated and 374 downregulated) genes. The reason for this disparity in the number of detected genes is not clear, and the published results available from both studies were not provided in a format allowing for a direct comparison of the two datasets.
In a fourth study, Zhou et al. [41] sought to define the repertoire of lncRNAs expressed in human HSCs. The researchers used RNA-sequencing to profile lncRNAs in human fetal HSCs in response to treatment with transforming growth factor beta (TGF-β), a known activator of HSCs, and identified 381 loci whose expression was altered by TGF-β treatment. Of these, 195 lncRNAs were found to be directly affected by TGF-β signaling based on SMAD3 occupancy, indicating that TGF-β may contribute to HSC function and fibrogenesis. The fifth study reported more recently by Li et al. [37] performed RNA-sequencing on human-derived HSCs treated with 5 mM valproic acid for four days to achieve a quiescent phenotype. The authors identified 1686 and 3763 differentially expressed mRNAs and lncRNAs, respectively, the majority of which were upregulated in activated cells. One of the main findings from this work was the identification of three lncRNAs associated with the genes encoding connective tissue growth factor (CTGF), fibroblast growth factor 2 (FGF2), and netrin 4 (NTN4), all of which have been implicated in processes related to HSC activation or liver fibrosis.
Although these studies shared a fundamental goal of identifying transcriptomic changes accompanying HSC activation, albeit using variations on a common experimental approach, we found surprisingly little overlap among the reported findings. This observation could be due to unexpected effects of different treatment conditions to induce biological quiescence in cultured cells or to disparities in transcriptomic responses between human- and rodent-derived cells. Immortalized human HSCs cultured under different conditions to achieve phenotypic states resembling biological quiescence or myofibroblastic activation may not replicate molecular processes that occur during HSC activation in human hepatic fibrogenesis, which we recognize as a potential limitation of the current study. The cells used here, LX-2 cells, were generated by immortalization of HSCs isolated from a normal human donor using the SV40 large T antigen followed by selective culture [43]. These cells have been extensively characterized, are homogeneous, and exhibit critical features of primary HSCs, including cytokine signaling, retinoid metabolism, and fibrogenesis. The induction of quiescence in these cells by physical or chemical means is well established [43,67] and readily assessed by markers of ECM and intracellular lipid accumulation. For example, treatment of LX-2 cells with transforming growth factor beta (TGF-beta) induces proliferation and expression of ECM components. Stimulation with TGF-beta (final concentration 2 ng/mL) for 48 h increased mRNA and protein expression of alpha-SMA, collagen type 1, PDGF, TIMP1, and TIMP2 compared to untreated LX-2 cells [68]. Other studies have reported similar trends [[69], [70], [71], [72]], suggesting that TGF-beta activates LX-2 cells to a greater extent than simply culturing cells on a stiff surface. In agreement with this, we validated lncRNA and mRNA expression patterns identified in MDI-treated LX-2 cells in LX-2 cells grown on Matrigel, which provides a soft support suitable for induction of quiescence (data not shown). More importantly, a significant number of differentially expressed coding and noncoding transcripts were observed in human liver, indicating that transcriptomic changes occurring during myofibroblastic transition in LX-2 cells are shared in advanced NAFLD fibrosis. None of the lncRNAs previously identified using ex vivo or in vitro approaches have been assessed in human NAFLD fibrosis. Therefore, one of the key strengths of the current study is the validation of numerous differentially expressed lncRNAs and mRNAs in patients with advanced NAFLD fibrosis.
The results obtained in the current study demonstrate that changes in expression of lncRNAs and mRNAs that accompany myofibroblastic activation of HSCs are mirrored in advanced NASH fibrosis. Future investigations, including functional characterization of dysregulated transcripts and lncRNA-mRNA co-expressed networks, will be important to extend these findings.
Author contributions
Study concept and design: JKD; acquisition of samples and data: XW, BD, DW, IP, AH, XC, ATP, CDS; analysis and interpretation of data: JKD and DW; drafting of the manuscript: JKD; critical revision of the manuscript for important intellectual content: JKD, GSG, DW, AH, XC, ATP, CDS; obtained funding: JKD, GSG.
Declaration of competing interest
CDS receives grant and consulting support from Ethicon Endo-Surgery. The remaining authors declare no competing financial and/or non-financial interests in relation to the work.
Acknowledgements
This study was funded in part by NIDDK DK088231.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100753.
Contributor Information
Glenn S. Gerhard, Email: gsgerhard@temple.edu.
Bethany Davis, Email: bdavis@tgen.org.
Xiumei Wu, Email: xwu@tgen.org.
Amanda Hanson, Email: ahanson@tgen.org.
Danielle Wilhelmsen, Email: dwilhelmsen@tgen.org.
Ignazio S. Piras, Email: ipiras@tgen.org.
Christopher D. Still, Email: cstill@geisinger.edu.
Xin Chu, Email: xchu@geisinger.edu.
Anthony T. Petrick, Email: atpetrick@geisinger.edu.
Johanna K. DiStefano, Email: jdistefano@tgen.org.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Younossi Z., Tacke F., Arrese M., Chander Sharma B., Mostafa I., Bugianesi E. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69(6):2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 2.Spengler E.K., Loomba R. Recommendations for diagnosis, referral for liver biopsy, and treatment of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Mayo Clin. Proc. 2015;90(9):1233–1246. doi: 10.1016/j.mayocp.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koek G.H., Liedorp P.R., Bast A. The role of oxidative stress in non-alcoholic steatohepatitis. Clin. Chim. Acta. 2011;412(15–16):1297–1305. doi: 10.1016/j.cca.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Jorge A.S., Andrade J.M., Paraiso A.F., Jorge G.C., Silveira C.M., de Souza L.R. Body mass index and the visceral adipose tissue expression of IL-6 and TNF-alpha are associated with the morphological severity of non-alcoholic fatty liver disease in individuals with class III obesity. Obes. Res. Clin. Pract. 2018;12(Suppl 2):1–8. doi: 10.1016/j.orcp.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Liu W., Baker R.D., Bhatia T., Zhu L., Baker S.S. Pathogenesis of nonalcoholic steatohepatitis. Cell. Mol. Life Sci. 2016;73(10):1969–1987. doi: 10.1007/s00018-016-2161-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrese M., Cabrera D., Kalergis A.M., Feldstein A.E. Innate immunity and inflammation in NAFLD/NASH. Dig. Dis. Sci. 2016;61(5):1294–1303. doi: 10.1007/s10620-016-4049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heymann F., Tacke F. Immunology in the liver - from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 2016;13(2):88–110. doi: 10.1038/nrgastro.2015.200. [DOI] [PubMed] [Google Scholar]
- 8.Matteoni C.A., Younossi Z.M., Gramlich T., Boparai N., Liu Y.C., McCullough A.J. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116(6):1413–1419. doi: 10.1016/s0016-5085(99)70506-8. [DOI] [PubMed] [Google Scholar]
- 9.Rafiq N., Bai C., Fang Y., Srishord M., McCullough A., Gramlich T. Long-term follow-up of patients with nonalcoholic fatty liver. Clin. Gastroenterol. Hepatol. : Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2009;7(2):234–238. doi: 10.1016/j.cgh.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Singh S., Allen A.M., Wang Z., Prokop L.J., Murad M.H., Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin. Gastroenterol. Hepatol. 2015;13(4):643–654. doi: 10.1016/j.cgh.2014.04.014. e1-9; quiz e39-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White D.L., Kanwal F., El-Serag H.B. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin. Gastroenterol. Hepatol. 2012;10(12):1342–13459 e2. doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cholankeril G., Wong R.J., Hu M., Perumpail R.B., Yoo E.R., Puri P. Liver transplantation for nonalcoholic steatohepatitis in the US: temporal trends and outcomes. Dig. Dis. Sci. 2017;62(10):2915–2922. doi: 10.1007/s10620-017-4684-x. [DOI] [PubMed] [Google Scholar]
- 13.Charlton M.R., Burns J.M., Pedersen R.A., Watt K.D., Heimbach J.K., Dierkhising R.A. Frequency and outcomes of liver transplantation for nonalcoholic steatohepatitis in the United States. Gastroenterology. 2011;141(4):1249–1253. doi: 10.1053/j.gastro.2011.06.061. [DOI] [PubMed] [Google Scholar]
- 14.Friedman S.L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008;88(1):125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman S.L. Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N. Engl. J. Med. 1993;328(25):1828–1835. doi: 10.1056/NEJM199306243282508. [DOI] [PubMed] [Google Scholar]
- 16.Hendriks H.F., Verhoofstad W.A., Brouwer A., de Leeuw A.M., Knook D.L. Perisinusoidal fat-storing cells are the main vitamin A storage sites in rat liver. Exp. Cell Res. 1985;160(1):138–149. doi: 10.1016/0014-4827(85)90243-5. [DOI] [PubMed] [Google Scholar]
- 17.Friedman S.L. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 2000;275(4):2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 18.Gressner A.M., Weiskirchen R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-beta as major players and therapeutic targets. J. Cell Mol. Med. 2006;10(1):76–99. doi: 10.1111/j.1582-4934.2006.tb00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang F., Parsons C.J., Stefanovic B. Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation. J. Hepatol. 2006;45(3):401–409. doi: 10.1016/j.jhep.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Younossi Z.M. Review article: current management of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 2008;28(1):2–12. doi: 10.1111/j.1365-2036.2008.03710.x. [DOI] [PubMed] [Google Scholar]
- 21.Day C.P. Genes or environment to determine alcoholic liver disease and non-alcoholic fatty liver disease. Liver Int. 2006;26(9):1021–1028. doi: 10.1111/j.1478-3231.2006.01323.x. [DOI] [PubMed] [Google Scholar]
- 22.Starmann J., Falth M., Spindelbock W., Lanz K.L., Lackner C., Zatloukal K. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PloS One. 2012;7(10) doi: 10.1371/journal.pone.0046584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asselah T., Bieche I., Laurendeau I., Paradis V., Vidaud D., Degott C. Liver gene expression signature of mild fibrosis in patients with chronic hepatitis C. Gastroenterology. 2005;129(6):2064–2075. doi: 10.1053/j.gastro.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Wruck W., Kashofer K., Rehman S., Daskalaki A., Berg D., Gralka E. Multi-omic profiles of human non-alcoholic fatty liver disease tissue highlight heterogenic phenotypes. Sci. Data. 2015;2:150068. doi: 10.1038/sdata.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moylan C.A., Pang H., Dellinger A., Suzuki A., Garrett M.E., Guy C.D. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 2014;59(2):471–482. doi: 10.1002/hep.26661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerhard G.S., Legendre C., Still C.D., Chu X., Petrick A., DiStefano J.K. Transcriptomic profiling of obesity-related nonalcoholic steatohepatitis reveals a core set of fibrosis-specific genes. J. Endocr. Soc. 2018;2(7):710–726. doi: 10.1210/js.2018-00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bieche I., Asselah T., Laurendeau I., Vidaud D., Degot C., Paradis V. Molecular profiling of early stage liver fibrosis in patients with chronic hepatitis C virus infection. Virology. 2005;332(1):130–144. doi: 10.1016/j.virol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Hanson A., Wilhelmsen D., DiStefano J.K. The role of long non-coding RNAs (lncRNAs) in the development and progression of fibrosis associated with nonalcoholic fatty liver disease (NAFLD) Noncoding RNA. 2018;4(3) doi: 10.3390/ncrna4030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran V.A., Perera R.J., Khalil A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan B., Wang Z. Long noncoding RNA: its physiological and pathological roles. DNA Cell Biol. 2012;31(Suppl 1):S34–S41. doi: 10.1089/dna.2011.1544. [DOI] [PubMed] [Google Scholar]
- 33.Sun C., Liu X., Yi Z., Xiao X., Yang M., Hu G. Genome-wide analysis of long noncoding RNA expression profiles in patients with non-alcoholic fatty liver disease. IUBMB Life. 2015;67(11):847–852. doi: 10.1002/iub.1442. [DOI] [PubMed] [Google Scholar]
- 34.Atanasovska B., Rensen S.S., van der Sijde M.R., Marsman G., Kumar V., Jonkers I. A liver-specific long noncoding RNA with a role in cell viability is elevated in human nonalcoholic steatohepatitis. Hepatology. 2017;66(3):794–808. doi: 10.1002/hep.29034. [DOI] [PubMed] [Google Scholar]
- 35.Leti F., Legendre C., Still C.D., Chu X., Petrick A., Gerhard G.S. Altered expression of MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates CXCL5 in hepatic stellate cells. Transl. Res. : J. Lab. Clin. Med. 2017;190:25–39 e21. doi: 10.1016/j.trsl.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo C.J., Xiao X., Sheng L., Chen L., Zhong W., Li H. RNA sequencing and bioinformatics analysis implicate the regulatory role of a long noncoding RNA-mRNA network in hepatic stellate cell activation. Cell. Physiol. Biochem. 2017;42(5):2030–2042. doi: 10.1159/000479898. [DOI] [PubMed] [Google Scholar]
- 37.Li X.Q., Ren Z.X., Li K., Huang J.J., Huang Z.T., Zhou T.R. Key anti-fibrosis associated long noncoding RNAs identified in human hepatic stellate cell via transcriptome sequencing analysis. Int. J. Mol. Sci. 2018;19(3) doi: 10.3390/ijms19030675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong W.J., Hu L.J., Jian Y.C., Wang L.J., Jiang M., Li W. Wnt5a participates in hepatic stellate cell activation observed by gene expression profile and functional assays. World J. Gastroenterol. 2012;18(15):1745–1752. doi: 10.3748/wjg.v18.i15.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu F., Jiang Z., Chen B., Dong P., Zheng J. NEAT1 accelerates the progression of liver fibrosis via regulation of microRNA-122 and Kruppel-like factor 6. J. Mol. Med. (Berl.) 2017;95(11):1191–1202. doi: 10.1007/s00109-017-1586-5. [DOI] [PubMed] [Google Scholar]
- 40.Zhang K., Han X., Zhang Z., Zheng L., Hu Z., Yao Q. The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFbeta and Notch pathways. Nat. Commun. 2017;8(1):144. doi: 10.1038/s41467-017-00204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou C., York S.R., Chen J.Y., Pondick J.V., Motola D.L., Chung R.T. Long noncoding RNAs expressed in human hepatic stellate cells form networks with extracellular matrix proteins. Genome Med. 2016;8(1):31. doi: 10.1186/s13073-016-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimada H., Ochi T., Imasato A., Morizane Y., Hori M., Ozaki H. Gene expression profiling and functional assays of activated hepatic stellate cells suggest that myocardin has a role in activation. Liver Int. 2010;30(1):42–54. doi: 10.1111/j.1478-3231.2009.02120.x. [DOI] [PubMed] [Google Scholar]
- 43.Xu L., Hui A.Y., Albanis E., Arthur M.J., O'Byrne S.M., Blaner W.S. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54(1):142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiskirchen R., Weimer J., Meurer S.K., Kron A., Seipel B., Vater I. Genetic characteristics of the human hepatic stellate cell line LX-2. PloS One. 2013;8(10) doi: 10.1371/journal.pone.0075692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y., Liu X., Zhou Q., Huang C., Meng X., Xu F. Silent information regulator 1 (SIRT1) ameliorates liver fibrosis via promoting activated stellate cell apoptosis and reversion. Toxicol. Appl. Pharmacol. 2015;289(2):163–176. doi: 10.1016/j.taap.2015.09.028. [DOI] [PubMed] [Google Scholar]
- 46.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 47.Wood G.C., Chu X., Manney C., Strodel W., Petrick A., Gabrielsen J. An electronic health record-enabled obesity database. BMC Med. Inf. Decis. Making. 2012;12(1):45. doi: 10.1186/1472-6947-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leti F., Malenica I., Doshi M., Courtright A., Van Keuren-Jensen K., Legendre C. High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease-related fibrosis. Transl. Res.: J. Lab. Clin. Med. 2015;166(3):304–314. doi: 10.1016/j.trsl.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DiStefano J.K., Kingsley C., Craig Wood G., Chu X., Argyropoulos G., Still C.D. Genome-wide analysis of hepatic lipid content in extreme obesity. Acta Diabetol. 2015;52(2):373–382. doi: 10.1007/s00592-014-0654-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerhard G.S., Benotti P., Wood G.C., Chu X., Argyropoulos G., Petrick A. Identification of novel clinical factors associated with hepatic fat accumulation in extreme obesity. J. Obes. 2014;2014:368210. doi: 10.1155/2014/368210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.She H., Xiong S., Hazra S., Tsukamoto H. Adipogenic transcriptional regulation of hepatic stellate cells. J. Biol. Chem. 2005;280(6):4959–4967. doi: 10.1074/jbc.M410078200. [DOI] [PubMed] [Google Scholar]
- 52.Anthony B., Allen J.T., Li Y.S., McManus D.P. Hepatic stellate cells and parasite-induced liver fibrosis. Parasites Vectors. 2010;3(1):60. doi: 10.1186/1756-3305-3-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guttman M., Garber M., Levin J.Z., Donaghey J., Robinson J., Adiconis X. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCullough A.J. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin. Liver Dis. 2004;8(3):521–533. doi: 10.1016/j.cld.2004.04.004. (viii) [DOI] [PubMed] [Google Scholar]
- 57.Ghouri Y.A., Mian I., Rowe J.H. Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017;16:1. doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altamirano-Barrera A., Barranco-Fragoso B., Mendez-Sanchez N. Management strategies for liver fibrosis. Ann. Hepatol. 2017;16(1):48–56. doi: 10.5604/16652681.1226814. [DOI] [PubMed] [Google Scholar]
- 59.Guo C.J., Pan Q., Cheng T., Jiang B., Chen G.Y., Li D.G. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009;276(18):5163–5176. doi: 10.1111/j.1742-4658.2009.07213.x. [DOI] [PubMed] [Google Scholar]
- 60.Chen L.L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016;41(9):761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 61.DiStefano J.K. The emerging role of long noncoding RNAs in human disease. Methods Mol. Biol. 2018;1706:91–110. doi: 10.1007/978-1-4939-7471-9_6. [DOI] [PubMed] [Google Scholar]
- 62.Kapusta A., Kronenberg Z., Lynch V.J., Zhuo X., Ramsay L., Bourque G. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9(4) doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 64.Liu S.J., Nowakowski T.J., Pollen A.A., Lui J.H., Horlbeck M.A., Attenello F.J. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shalek A.K., Satija R., Adiconis X., Gertner R.S., Gaublomme J.T., Raychowdhury R. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ransohoff J.D., Wei Y., Khavari P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018;19(3):143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olsen A.L., Bloomer S.A., Chan E.P., Gaca M.D., Georges P.C., Sackey B. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301(1):G110–G118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang K.H., Qian Z.J., Ryu B., Karadeniz F., Kim D., Kim S.K. Hepatic fibrosis inhibitory effect of peptides isolated from navicula incerta on TGF-beta1 induced activation of LX-2 human hepatic stellate cells. Prev. Nutr. Food Sci. 2013;18(2):124–132. doi: 10.3746/pnf.2013.18.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta P., Sata T.N., Yadav A.K., Mishra A., Vats N., Hossain M.M. TGF-beta induces liver fibrosis via miRNA-181a-mediated down regulation of augmenter of liver regeneration in hepatic stellate cells. PloS One. 2019;14(6) doi: 10.1371/journal.pone.0214534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Y.F., Zhang Q., Cheung P.Y., Shi L., Fong C.C., Zhang Y. Effects of rhDecorin on TGF-beta1 induced human hepatic stellate cells LX-2 activation. Biochim. Biophys. Acta. 2006;1760(11):1587–1595. doi: 10.1016/j.bbagen.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 71.Tee J.K., Peng F., Tan Y.L., Yu B., Ho H.K. Magnesium isoglycyrrhizinate ameliorates fibrosis and disrupts TGF-beta-mediated SMAD pathway in activated hepatic stellate cell line LX2. Front. Pharmacol. 2018;9:1018. doi: 10.3389/fphar.2018.01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan B., Chen Y., Wu Z., Zhang L., Zhuang Y., Zhao X. Proteomic profiling of human hepatic stellate cell line LX2 responses to irradiation and TGF-beta1. J. Proteome Res. 2019;18(1):508–521. doi: 10.1021/acs.jproteome.8b00814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.