Abstract
The genetic basis for sporadic immunodeficiency in patients with 22q11.2 distal deletion syndrome is unknown. We report an adult with a type 1 (D-F) 22q11.2 distal deletion syndrome and recurrent severe infections due to herpes zoster virus, presenting mild T cell lymphopenia and diminished frequency of naive CD4<sup>+</sup> T cells, but increased frequencies of central, effector, and terminally differentiated memory T cells. Antigen-specific CD4<sup>+</sup> and CD8<sup>+</sup> T cells to influenza, rotavirus, and SEB were conserved in the patient, but responses to tetanus toxoid were temporarily undetectable. Exomic sequencing identified the c.20_22dupCGG (NM_002745.4) variant in the remaining MAPK1 gene of the patient, which adds 1 alanine to the polyalanine amino-terminal tract of the protein (p.Ala7dup). The mother, unlike the father, was heterozygote for the variant. Western blot analysis with the patient's activated PBMCs showed a 91% reduction in the MAPK1 protein. Further studies will be necessary to determine whether or not the variant present in the remaining MAPK1 gene of the patient is pathogenic.
Keywords: Immunodeficiency, MAPK1, T lymphocytes, 22q11.2 distal (D–F) deletion syndrome
Established Facts
Two patients with 22q11.2 distal deletion syndrome type 1 (with a D–F deletion which involves the MAPK1 gene, but not the CRKL gene) have had recurrent infections.
The immune response of these patients has not been characterized.
The genetic basis for this immunodeficiency is unknown.
Novel Insights
We report a case study of an adult with a type 1 (D–F) 22q11.2 distal deletion syndrome and recurrent severe infections due to herpes zoster virus, presenting mild T cell lymphopenia, diminished frequency of naive CD4+ T cells, but increased frequencies of central, effector, and terminally differentiated memory T cells.
Exomic sequencing identified the c.20_22dupCGG (NM_002745.4) variant in the remaining MAPK1 gene of the patient, which adds 1 alanine to the polyalanine amino-terminal tract of the protein (p.Ala7dup). Western blot analysis with the patient's activated PBMCs showed a 91% reduction in the MAPK1 protein.
The chromosome 22q11.2 region comprises 8 (A-H) well-defined segmental duplication blocks, known as low copy repeats (LCRs) that promote genetic deletions/duplications due to nonallelic homologous recombination [Shaikh et al., 2007; Burnside, 2015; Bengoa-Alonsoet al., 2016]. These deletions/duplications have been classified as proximal, central, and distal [Burnside, 2015]. The common proximal 22q11.2 deletion syndrome (22q11.2PDS) is characterized by a 3-Mb deletion, spanning from LCR22-A to LCR22-D, and presents clinically as DiGeorge syndrome with typical T cell immunodeficiency [Burnside, 2015; Bengoa-Alonsoet al., 2016]. In contrast, patients with central deletions (LCRs B-D) and distal deletions (LCRs D-H) have more subtle phenotypes, characterized by mild developmental delay, some facial dysmorphism, cardiac and neurological alterations, and delayed growth, which may overlap with those of patients with proximal deletions [Beddow et al., 2011; Tan et al., 2011]. Deletions of distal intervals of the 22q11.2 genomic region are categorized into 3 different subtypes [Burnside, 2015]: type I distal deletions are mainly de novo and involve C-E, D-E, and D-F regions; type II deletions include patients with deletions in E-F regions, and type III deletions comprise any patient with involvement of the SMARCB1/INI gene.
To date, of the over 45 individuals reported with 22q11.2 distal deletion syndrome (22q11.2DDS) [Rauch et al., 2005; Shaikh et al., 2007; Ben-Shachar et al., 2008; Verhagen et al., 2012; Molck et al., 2013; Mikhail et al., 2014; Rump et al., 2014; Burnside, 2015; Bengoa-Alonso et al., 2016], only 9 of them (20%) had some degree of immunodeficiency or recurrent infections, and the immune response of these patients is poorly characterized.
This report describes a 53-year-old adult patient with a diagnosis of type I (D-F deletion) 22q11.2DDS, and a medical history of recurrent and severe herpes zoster virus (HZV) infections. We compared the immunological response of this patient with that of an adult patient with a 22q11.2PDS.
Clinical Report
Clinical data for patients with 22q11.2PDS and 22q11.2DDS are summarized in Table 1, and relevant immunological laboratory findings are presented in Table 2. The patient with 22q11.2DDS was referred to our outpatient Genetics Clinic in 2011 at the age of 46 years because of a history of 8 episodes of HZV infection, starting in 2002 when he was 37 years old. After 2011 to date, he has had at least 3 more episodes of HZV. He had a history of global developmental delay, showed mild cognitive impairment with learning difficulties, and hypothyroidism treated with Levothyroxine. Vaccination records against influenza, Streptococcuspneumoniae, TT, or hepatitis B were unavailable.
Table 1.
Genetic tests and clinical findings for patients with 22q11.2PDS and 22q11.2DDS
| 22q11.2 proximal deletion syndrome | 22q11.2 distal deletion syndrome | |
|---|---|---|
| Age | 30 years | 49 years |
| Sex | F | M |
| Genetic tests | ||
| Karyotyping | 46,XX | 46,XY |
| MLPA | Deletion in LCR A–D | Deletion in LCR D–F |
| aCGH | arr 22q11.21 (18,648,866 – 21,465,659)×1 Deletion of 2.8 Mb in 22q11.21 region |
Deletion of 1.8 Mb in 22q11.21q11.23 region |
| Clinical data | ||
| Neurodevelopment | Speech delay | Global developmental delay, IQ 72 |
| Heart defects | Atrial septal defect | None |
| Skeletal defects | Klippel Feil disorder, short stature | None |
| Orofacial defects | Submucous cleft palate | None |
| Infections | Chronic supurative otitis media, chronic sinusitis, keratitis, several episodes of pneumonia, pulmonary hypertension secondary to lung fibrosis, recurrent urinary tract infections | Ocular HZV infection, recurrent HZV infection in left hemibody (at least 11 episodes) |
| Ear | Bilateral conductive hearing loss | Left sensorineural hearing loss |
| CNS | Symptomatic focal epilepsy | Arachnoid cyst |
| Endocrine | Hypothyroidism, Hypogonadism | Hypothyroidism |
| Others | Hypersplenism that required surgical spleen remotion, Von Willebrand type II disease, granulomatous liver disease, epilepsy | Postherpetic neuropathy, IV–VI cranial nerve paralysis |
Results of genetic tests and clinical findings were obtained from medical records. aCGH, array comparative genomic hybridization; CNS, central nervous system; HZV, herpes zoster virus; LCR, low copy repeats; MLPA, multiplex ligation-dependent probe amplification.
Table 2.
Immunological laboratory findings of patients with 22q11.2PDS and 22q11.2DDS
| 22q11.2 proximal deletion syndrome | 22q11.2 distal deletion syndrome | |
|---|---|---|
| Leukocytes, cells/µL | 10,340 (4,500 – 10,000) | 4,200 (4,500 – 10,000) |
| CD3+, cells/µL | 1,018 (959 – 2,577) | 473 (959 – 2,577) |
| CD4+, cells/µL | 533 (544 – 1,663) | 271 (544 – 1,663) |
| CD8+, cells/µL | 270 (272 – 932) | 164 (272 – 932) |
| IgA, mg/dL | <1 (70 –320) | 830 (70 – 400) |
| IgM, mg/dL | 82 (50 – 240) | 154.3 (40 – 230) |
| IgG, mg/dL | 840 (640 – 1,350) | 1,212.8 (700 – 1,600) |
| IgE, mg/dL | ND | 2.3 (0 – 100) |
| IgD, mg/dL | ND | 44.8 (0 – 152.7) |
Absolute numbers of leukocytes and CD3+, CD4+ and CD8+ T cell are shown with reference values in parentheses. Serum immunoglobulin levels are shown with reference values in parentheses. For the patient with 22q11.2PDS, serum Ig levels are secondary to treatment with exogenous immunoglobulin. ND, not done.
The patient's HZV infection episodes were characterized by painful blisters, located on the left hemibody (thoracic and limb dermatomes) most of the time, lasting for at least 2 months (range from 2 to 6 months). Episodes were not self-limited and only resolved with medical interventions, consisting of long oral treatment with Acyclovir and Lidocaine patches, and required hospitalization 4 times. Involvement of the right cranial nerves III, IV, ophthalmic branch of V, and VII was also reported once. HIV and syphilis infections were excluded. Rheumatology studies to exclude primary vasculitis and systemic sclerosing disease were negative, including anti-nuclear antibodies, anti-dsDNA, anti-centromere, anti-neutrophilic cytoplasmic, anti-Scl-70, and anti-cardiolipin. Complement serum levels (C3 and C4) were normal. Low transient CD3+, CD4+, and CD8+ T cell counts were observed (Table 2 shows the values obtained when the other immunological experiments were performed). In repeated measures, the CD4+ T cell average count was 346 cells/μL (range from 271 to 454 cell/μL, in n = 4 assays). Serum levels of immunoglobulins (IgM, IgG, and IgE) were in normal ranges for age, but IgA was mildly elevated (Table 2). Frequencies of CD4+CD127-CD25+ and CD4+FoxP3+CD25+ Treg determined in our lab seemed comparable to those of a healthy control evaluated simultaneously, while, as previously described, frequencies of these cells in the patient with 22q11.2PDS seem diminished (data not shown).
Multiplex ligation-dependent probe amplification (MLPA) on DNA extracted from the patient's blood was performed, and showed a reduced peak signal of 22q11.2 probes LCRD (LCR D-F), demonstrating a type 1 distal deletion of the 22q11.2 locus. This result was confirmed by aCGH, which showed a heterozygous deletion of 1.83 Mb in the 22q11.21q11.23 region (breaking points chr22:21454661-23301036, GRCh38) that compromises 21 genes, including MAPK1, BCR, and 3 microRNA genes.
Whole-exome DNA sequencingof the trio (father, mother, and proband) was performed by the company Biotecnología y genética (BTG) in Bogota, Colombia with an Agilent V kit. Exomes were analyzed bioinformatically for variants of clinical significance with emphasis on immune-related genes. The exomes of the father and the mother presented 39/78 and 26/67, respectively, - good quality variants in the region of the deletion that are heterozygous - while, as expected from the MLPA and aCGH studies, the proband had 0/47 heterozygous variants in the region, indicating that the deletion was de novo in the proband. Two immune-related heterozygous variants were found in the proband: 1 in exon 35 of the C5 gene: c.4358A>G (p.Tyr1453Cys) and 1 in exon 1 of the IKBKB gene: c.82C>T (p.Pro28Ser). The first variant inherited from the mother is classified as pathogenic, and the second, inherited from the father, is classified as of uncertain significance. Since both variants are present in a heterozygous state in the healthy father and mother, they are improbably related to the clinical features of the patient. The same can be said of the only other nonimmune heterozygous pathogenic variant inherited from the father observed in the OPLAH gene: NM_ 017570.4 c.3265G>A (p.Val1089Ile). In addition, we evidenced 22 variants in 10 genes in the region of the deletion of the patient. We focused our attention on one of these variants in the MAPK1 gene reported as c.20_22dupCGG (NM_002745.4) that adds 1 alanine to the amino-terminal polyalanine region of the protein (p.Ala7dup). This variant was assigned to rs770506771; however, it was recently merged into rs751880548 in the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/rs751880548, consulted in May 2019). In the gnomAD browser (http://gnomad-old.broadinstitute.org/variant/22-22221708-C-CCCG, consulted in May 2019) the variant has an allele frequency of 0.004251 (considering both exomes and genomes) in the total population. The population with the highest allele frequency is the Latino population (0.05439), and only 1 homozygote individual has been reported in the “Other” population category. The in silico algorithm MutationTaster (http://www.mutationtaster.org/cgi-bin/MutationTaster/MutationTaster69.cgi?sequence_snippet=CGGCGGCGGCGG[/CGG]CGGCGG&transcript_stable_id_text=ENST00000215832&gene=MAPK1&sequence_type=CDS&nt_alignment=1, consulted in May 2019) classifies the variants as a polymorphism. The Varsome engine [Kopanos et al., 2018] (https://varsome.com/variant/hg38/rs770506771, consulted in May 2019) classifies the variant as of uncertain significance by using the criteria of the American College of Medical Genetics and Genomics (ACMGG) [Richards et al., 2015]. Of note, this engine ascribes the PP3 criteria of the ACMGG to the variant when using the GRCh37 due to one pathogenic prediction from GERP (vs. no benign predictions) suggesting that the variant is localized in an evolutionarily conserved part of the gene. The Variant Effect Predictor program classifies the variant as having a moderate impact (http://www.ensembl.org/Homo_sapiens/Tools/VEP/Results?db=core;tl=cVKAGm0GPSepuhLn-5281545, consulted in May 2019). The mother, unlike the father, was heterozygote for the variant. All other variants in the region of the deletion are reported with minor allele frequencies greater than 0.05 and are probably polymorphisms or present in noncoding regions. The above are available upon request.
Materials and Methods
Flow Cytometry Analyses
Frequencies of T cells that proliferate and produce cytokines in response to SEB, TT, or PBS were determined as previously reported [Martinez et al., 2016]. The assay lasts for 5 days, and T cell proliferation is determined by CFSE dilution with a simultaneous intracellular cytokine assay [Martinez et al., 2016]. In a subset of experiments, PBMCs were similarly stimulated with SEB, TT, or PBS for 10 h at 37°C, with10 μg/mL Brefeldin A (Sigma), added for the last 5 h of the incubation [Parra et al., 2014]. Cells were then fixed, permeabilized, and stained as for the 5-day assay but with the addition of antibodies against the differentiation markers CCR7, PECy7 and CD45RA PE. Samples were acquired on an LSR Fortessa (BD Biosciense) and analyzed using FlowJo v.10.5.0.
Western Blot for MAPK1 and MAPK3 (ERK2 and ERK1, Respectively)
Ficoll gradient purified PBMCs from the patient with 22q11.2DDS and a healthy control volunteer were stimulated with PMA (Sigma) 40 nM, as previously described [Chow et al., 2001]. Briefly, PBMCs were resuspended in RPMI plus FBS 10% at 1×106/mL and treated with PMA 40 nM during 5 and 10 min at 37°C 5% CO2. Samples containing 20 µg of total protein were separated by gel electrophoresis using pre-cast gels and transferred to PVDF membranes using the Mini Trans-Blot Electrophoresis Transfer Cell (Bio-Rad). Total MAPK1/MAPK3 protein was detected using a mouse monoclonal antibody (specific for ERK1/ERK2 clone 216703) and phospho-specific rabbit polyclonal antibody - Phospho-ERK1 (T202/Y204)/ERK2 (T185/Y187) - obtained from Bio-Techne (catalog Nos. MAB1576 and AF1018, respectively). A secondary antibody labeled with horseradish peroxidase was used for detection by chemoluminescence, as recommended by the manufacturer (ECL Thermo scientific). Densitometry of each protein normalized to β-tubulin was calculated using the ImageJ software (developed by NIH).
Results
Frequencies of Proliferating and Cytokine-Producing T Cells in Response to SEB, Influenza, TT, and Rotavirus
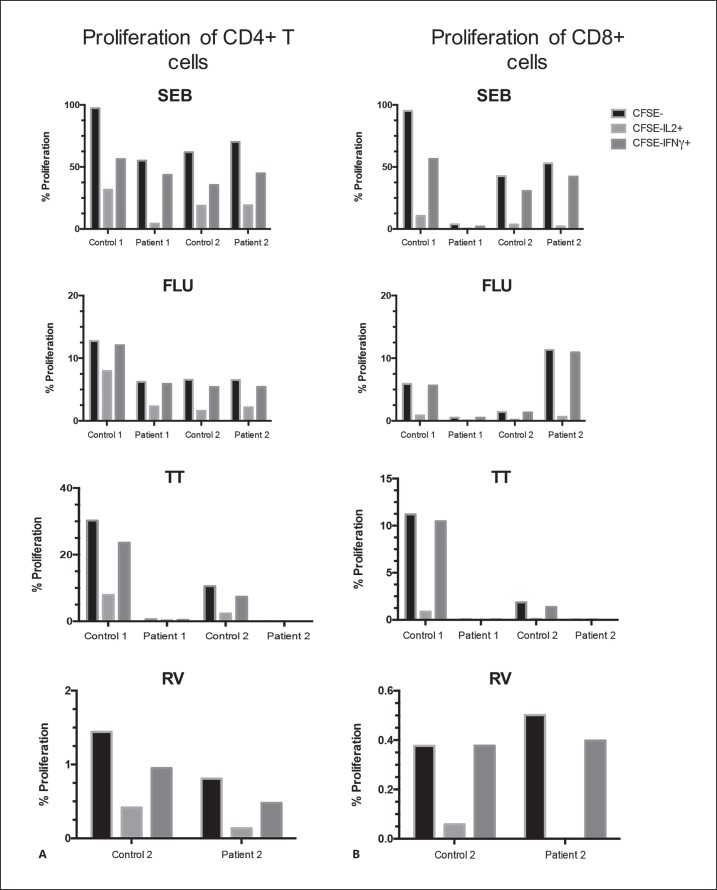
The CD4+ and CD8+ T cell responses against SEB, influenza virus, TT, and rotavirus were evaluated with an assay that simultaneously measures T cell proliferation and production of cytokines (IFNγ, IL-2, IL-10, or IL-13), as previously described [Martinez et al., 2016]. Two healthy control volunteers with the same sex and similar age were simultaneously assessed. Only differences in the frequencies of proliferating cells that produce IFNγ or IL-2 were noted between the patients and controls (Fig. 1). While the patient with 22q11.2PDS had diminished T cell response to all evaluated antigens, in both CD4+ and CD8+ T cells (especially in those producing IL-2), the patient with 22q11.2DDS had comparable levels of CD4+ and CD8+ T cells responding to SEB, influenza vaccine, and rotavirus antigens, but response to TT was undetectable (Fig. 1A, B). This response against TT has been notable since, although evidence of previous TT vaccination in this patient could not be documented; he was most probably vaccinated with TT as a child.
Fig. 1.
Proliferation and intracellular cytokine T cells assays in response to polyclonal SEB stimulation, influenza, TT, and rotavirus. Frequency of CD4+ (A) and CD8+ T cells (B) that proliferate (CFSE-), producing IL-2 (CFSE-IL-2+) or IFNγ (CFSE-IFNγ+) of the patients and their respective matched controls after stimulation with SEB, influenza (FLU), TT, and rotavirus (RV). Patient 1 and patient 2 correspond to the patients with 22q11.2PDS and 22q11.2DDS, respectively.
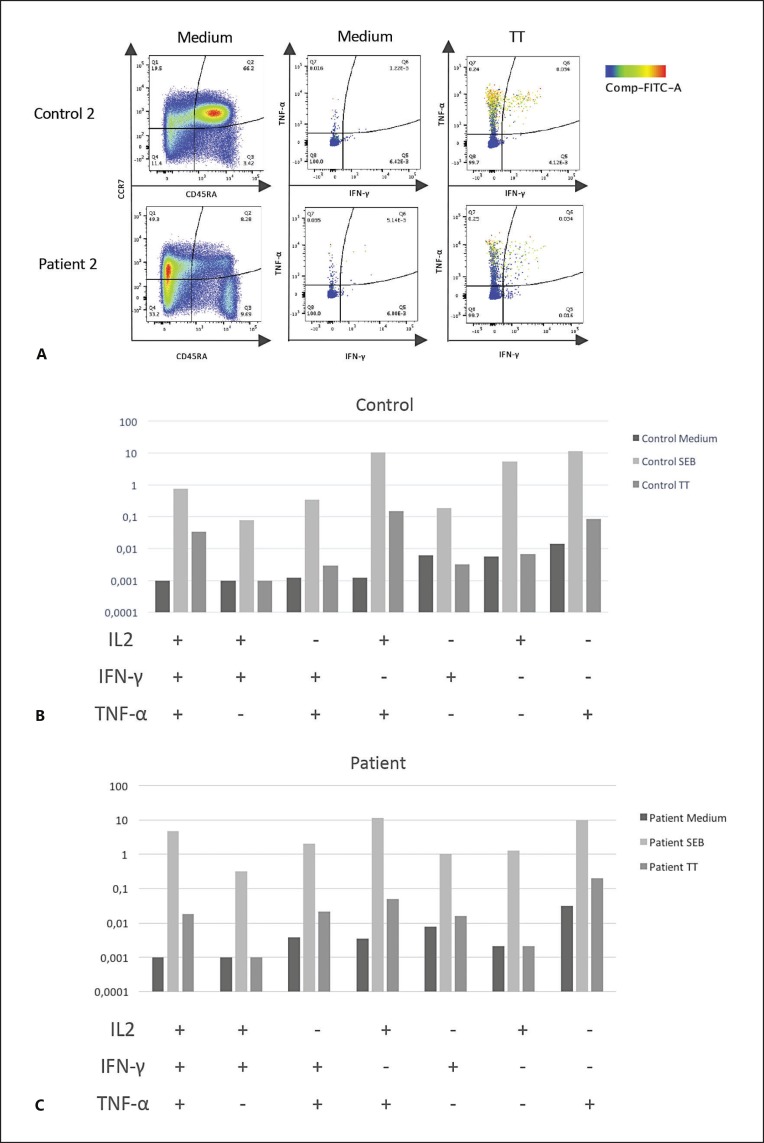
To determine if the T cells of the patient with 22q11.2DDS could be evidenced after TT vaccination, the CD4+ and CD8+ T cell responses against SEB and TT were evaluated 2 weeks after TT vaccination with a short-term intracellular cytokine (IFNγ, IL-2, or TNFα) producing assay, which simultaneously measures the differentiation state of the T cells (CCR7 and CD45RA) [Parra et al., 2014]. For comparison, T cells from a healthy volunteer boosted with TT 3 years previously were simultaneously studied. As can be seen in Figure 2, compared with the control volunteer, this patient had a marked decrease in the frequencies of naive (CCR7+, CD45RA+) T cells, with relative increases in central (CCR7+, CD45RA-), effector (CCR7-, CD45RA-), and terminally differentiated (CCR7-, CD45RA+) CD4+ T cells. In both, the patient and control, comparable frequencies of cytokine-producing CD4+ (Fig. 2) and CD8+ T cells (data not shown) were observed in response to TT and SEB (Fig. 2). As in the control volunteer, the TT-specific CD4+ T cell response of the patient included cells simultaneously producing the 3 cytokines (Fig. 2C).
Fig. 2.
Phenotype and frequencies of cytokine-producing CD4+ T cells in response to SEB and TT of the patient with 22q11.2DDS two weeks after TT vaccination, in comparison with T cells from a healthy control. A Left dot plots show frequencies of live CD3+CD4+ T cells expressing CD45RA and CCR7. Middle and right dot plots represent frequencies of live CD3+CD4+ T cells producing IFNγ and TNFα (with the expression of IL-2 being represented for each event by the color bar on the right) after stimulation with control medium or TT. B Boolean analysis of CD4+ T cells that produce IL-2, IFN-γ, or TNF-α, individually or in combination, after stimulation with medium, SEB, and TT in the control volunteer (top) and patient (bottom).
MAPK1 Protein Levels in the Patient with 22q11.2DDS
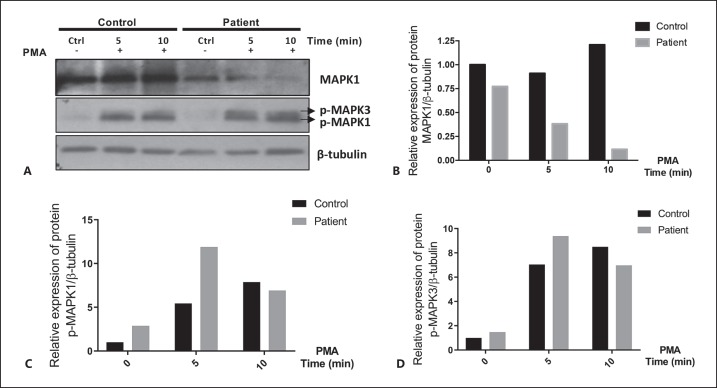
To evaluate if the expression of the MAPK1 protein was reduced in cells from the patient with 22q11.2DDS, his PBMCs and those from a healthy control were stimulated or unstimulated with PMA [Chow et al., 2001], and cell extracts were studied by Western blot for MAPK1/MAPK3 protein. In the Western blot, only 1 band was seen in our experiments, corresponding to MAPK1 (Fig. 3A) [Napolitano et al., 2018]. Unstimulated PBMCs from the patient had a 33% reduction in MAPK1 protein compared to PBMCs from a healthy volunteer (Fig. 3A, B). After stimulation, the relative reduction in MAPK1 protein of the patient, compared with the control volunteer, was 58% at 5 min and 91% at 10 min (Fig. 3A, B). In contrast, levels of phosphorylated MAPK1 and MAPK3 were comparable, except for a 2-fold increase in phosphorylated MAPK3 protein in the patient at 5 min after stimulation (Fig. 3A, C, D).
Fig. 3.
A-D MAPK1 protein from activated PBMCs is decreased in the patient with 22q11.2DDS. A PBMCs from the patient with 22q11.2DDS and the control volunteer were unstimulated or stimulated with PMA for 5 and 10 min. MAPK1/MAPK3 protein was detected using the total ERK2/ERK1 mouse monoclonal antibody and phospho-specific rabbit polyclonal antibodies. A secondary antibody labeled with horseradish peroxidase was used for detection by chemoluminescence. Densitometry of total MAPK1 (B), phosorylated MAPK1 (C), and MAPK3 (D) were normalized to β-tubulin.
Discussion and Conclusion
To our knowledge, the patient with 22q11.2DDS (type 1 D-F deletion) presented here is the first case with reported recurrent severe HZV infections [Burnside, 2015]. Most patients with 22q11.2DDS have been reported to have mild immunodeficiency, ear infections, or recurrent upper respiratory tract infections [Rump et al., 2014; Burnside, 2015; Bengoa-Alonso et al., 2016]. The majority of patients with recurrent infections and type 1 distal deletions have had C to D/E deletions that involve the CRKL gene (associated with immunodeficiency) [Burnside, 2015; Giacomelli et al., 2016]. To our knowledge, only 2 patients with type 1 D-F deletions (that do not involve the CRKL gene), such as the patient with 22q11.2DDS presented here, had signs of immunodeficiency: Pseudomonas aeruginosa sepsis accompanied by T cell deficiency in one case [Rauch et al., 1999] and undetermined recurrent infections in the other [Rauch et al., 2005]. Notably, HZV recurrence in immunocompetent patients occurs between 1 and 6% of the cases [Shiraki et al., 2017], and these are more severe and frequent in immunocompromised patients, such as those with HIV infection [Blank et al., 2012].
The patient with 22q11.2DDS had normal frequencies of Treg cells (data not shown), and only T cell response after stimulation with TT was greatly diminished or absent (Fig. 1). Nevertheless, 2 weeks after receiving the TT vaccine, the frequencies of cytokine-producing T cells in response to SEB and TT were comparable to a control volunteer (Fig. 2A, middle and right, and Fig. 2B). Thus, the T cells of this patient seem to have a normal response to SEB and can respond to TT. Remarkably, the distribution of CD4+ T cells expressing CCR7 and CD45RA were markedly different in this patient, having an importantly diminished frequency of naive T cells and increased frequencies of central, effector, and terminally differentiated memory T cell subsets (Fig. 2A, left). These results suggest that the patient with 22q11.2DDS has an alteration in the process of T cell differentiation.
The deletion observed in the patient with type 1 (D-F) 22q11.2DDS excludes the CRKL gene, and, to our knowledge, the gene (or genes) responsible for the immunodeficiency which has been reported in these patients is unknown. Of the 21 genes present in this region, MAPK1 and BCR are potential candidates for this role [Busca et al., 2016]. MAPK1 seems most attractive because it is implicated in the developmental and neurological alterations of patients with 22q11.2DDS [Fagerberg et al., 2013]. Also, Erk1 (MAPK3) and Erk2 (MAPK1) play redundant roles in the development of murine hematopoietic progenitor cells, and, when both genes are absent, the development of myeloid cells and T and B lymphocytes is impaired [Chan et al., 2013]. Besides, the Erk2-/- mice have alterations in T cell development [Chan et al., 2013], and these mice also have decreased Th1 differentiation and enhancement of Foxp3+ Treg [Chang et al., 2012]. The latter findings support the hypothesis that decreased levels of the MAPK1 protein observed in the patient's stimulated cells are related to his T cell lymphopenia and clinical immunodeficiency.
The variant we found in MAPK1 increases the length of the poly(A) repeat in the amino-terminal region of the protein by 1 alanine. Alteration in the poly(A) tract of proteins may be important for their nuclear export [Li et al., 2017] and cause aggregation of the proteins, altering their stability [Pelassa et al., 2014], which is in agreement with our results (Fig. 3A, B) showing that upon activation of the cells, the MAPK1 protein seems to be unstable. It is worth noting that the expansion of a single alanine in a poly(A) repeat tract in the ARX gene has been associated with cognitive disability [Grønskov et al., 2004]. However, the relatively high allele frequency of the c.20_22dupCGG (NM_002745.4) variant (0.05439 in the Latino population) and the fact that a homozygous individual for this variant has been reported in gnomAD seem to be at odds with the variant being pathogenic. Besides, this variant is at present classified as of uncertain significance according to the ACMGG criteria [Richards et al., 2015].
In summary, for the first time, we report a patient with a type 1 22q11.2 distal deletion with T cell lymphopenia, altered T cell differentiation, low protein levels of MAPK1 after activation, and severe recurrent HZV infections. Further studies will be necessary to determine if the c.20_22dupCGG (NM_002745.4) variant present in the remaining MAPK1 gene of the patient is pathogenic.
Statement of Ethics
This study was approved by the Ethics Committee of the School of Medicine of Pontificia Universidad Javeriana, Bogota, Colombia and was done in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). Written informed consent, approved by the Ethics Committee, was obtained from all participants.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
This study was funded by the Pontificia Universidad Javeriana (ID PPTA 6435 and ID PROY 6542).
Author Contributions
D.H. and M.P. performed flow cytometry experiments and contributed to writing the paper. K.P. and L.R. performed Western blot experiments and contributed to writing the paper. A.S., M.G., A.P., R.Q., R.O., J.A.R., and J.C.P. followed the patient, obtained clinical information for the paper, and contributed to writing the paper. J.C.P., J.A., L.R., and M.F. wrote and obtained funding for the project, directed the work, and wrote the paper.
Acknowledgment
We would like to thank the patients and healthy volunteers that donated blood for the experiments reported.
References
- 1.Beddow RA, Smith M, Kidd A, Corbett R, Hunter AG. Diagnosis of distal 22q11.2 deletion syndrome in a patient with a teratoid/rhabdoid tumour. Eur J Med Genet. 2011;54:295–298. doi: 10.1016/j.ejmg.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, et al. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008 doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bengoa-Alonso A, Artigas-Lopez M, Moreno-Igoa M, Cattalli C, Hernandez-Charro B, Ramos-Arroyo MA. Delineation of a recognizable phenotype for the recurrent LCR22-C to D/E atypical 22q11.2 deletion. Am J Med Genet A. 2016;170:1485–1494. doi: 10.1002/ajmg.a.37614. [DOI] [PubMed] [Google Scholar]
- 4.Blank LJ, Polydefkis MJ, Moore RD, Gebo KA. Herpes zoster among persons living with HIV in the current antiretroviral therapy era. J Acquir Immune Defic Syndr. 2012;61:203–207. doi: 10.1097/QAI.0b013e318266cd3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnside RD. 22q11.21 Deletion syndromes: a review of proximal, central, and distal deletions and their associated features. Cytogenet Genome Res. 2015;146:89–99. doi: 10.1159/000438708. [DOI] [PubMed] [Google Scholar]
- 6.Busca R, Pouyssegur J, Lenormand P. ERK1 and ERK2 Map kinases: specific roles or functional redundancy? Front Cell Dev Biol. 2016;4:53. doi: 10.3389/fcell.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan G, Gu S, Neel BG. Erk1 and Erk2 are required for maintenance of hematopoietic stem cells and adult hematopoiesis. Blood. 2013;121:3594–3598. doi: 10.1182/blood-2012-12-476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang CF, D'Souza WN, Ch'en IL, Pages G, Pouyssegur J, Hedrick SM. Polar opposites: Erk direction of CD4 T cell subsets. J Immunol. 2012;189:721–731. doi: 10.4049/jimmunol.1103015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow S, Patel H, Hedley DW. Measurement of MAP kinase activation by flow cytometry using phospho-specific antibodies to MEK and ERK: potential for pharmacodynamic monitoring of signal transduction inhibitors. Cytometry. 2001;46:72–78. doi: 10.1002/cyto.1067. [DOI] [PubMed] [Google Scholar]
- 10.Fagerberg CR, Graakjaer J, Heinl UD, Ousager LB, Dreyer I, et al. Heart defects and other features of the 22q11 distal deletion syndrome. Eur J Med Genet. 2013;56:98–107. doi: 10.1016/j.ejmg.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Giacomelli M, Kumar R, Soresina A, Tamassia N, Lorenzini T, et al. Reduction of CRKL expression in patients with partial DiGeorge syndrome is associated with impairment of T-cell functions. J Allergy Clin Immunol. 2016;138:229–240. doi: 10.1016/j.jaci.2015.10.051. [DOI] [PubMed] [Google Scholar]
- 12.Grønskov K, Hjalgrim H, Nielsen IM, Brøndum-Nielsen K. Screening of the ARX gene in 682 retarded males. Eur J Hum Genet. 2004;12:701–705. doi: 10.1038/sj.ejhg.5201222. [DOI] [PubMed] [Google Scholar]
- 13.Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Ng NK, Koon AC, Chan HY. Expanded polyalanine tracts function as nuclear export signals and promote protein mislocalization via eEF1A1 factor. J Biol Chem. 2017;292:5784–5800. doi: 10.1074/jbc.M116.763599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez E, Castañeda D, Jaramillo S, Iregui A, Quiñonez T, et al. Altered immune parameters correlate with infection-related hospitalizations in children with Down syndrome. Hum Immunol. 2016;77:594–599. doi: 10.1016/j.humimm.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Mikhail FM, Burnside RD, Rush B, Ibrahim J, Godshalk R, et al. The recurrent distal 22q11.2 microdeletions are often de novo and do not represent a single clinical entity: a proposed categorization system. Genet Med. 2014;16:92–100. doi: 10.1038/gim.2013.79. [DOI] [PubMed] [Google Scholar]
- 17.Molck MC, Vieira TP, Sgardioli IC, Simioni M, Dos Santos AP, et al. Atypical copy number abnormalities in 22q11.2 region: report of three cases. Eur J Med Genet. 2013;56:515–520. doi: 10.1016/j.ejmg.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Napolitano G, Esposito A, Choi H, Matarese M, Benedetti V, et al. mTOR-dependent phosphorylation controls TFEB nuclear export. Nat Commun. 2018;9:3312. doi: 10.1038/s41467-018-05862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parra M, Herrera D, Jácome MF, Mesa MC, Rodríguez LS, et al. Circulating rotavirus-specific T cells have a poor functional profile. Virology. 2014:468–470. doi: 10.1016/j.virol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Pelassa I, Corà D, Cesano F, Monje FJ, Montarolo PG, Fiumara F. Association of polyalanine and polyglutamine coiled coils mediates expansion disease-related protein aggregation and dysfunction. Hum Mol Genet. 2014;23:3402–3420. doi: 10.1093/hmg/ddu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rauch A, Pfeiffer RA, Leipold G, Singer H, Tigges M, Hofbeck M. A novel 22q11.2 microdeletion in DiGeorge syndrome. Am J Hum Genet. 1999;64:659–o666. doi: 10.1086/302235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch A, Zink S, Zweier C, Thiel CT, Koch A, et al. Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet. 2005;42:871–876. doi: 10.1136/jmg.2004.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards S, Aziz N, Bale S, Bick D, Das S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rump P, de Leeuw N, van Essen AJ, Verschuuren-Bemelmans CC, Veenstra-Knol HE, et al. Central 22q11.2 deletions. Am J Med Genet. 2014;A164A:2707–2723. doi: 10.1002/ajmg.a.36711. [DOI] [PubMed] [Google Scholar]
- 25.Shaikh TH, O'Connor RJ, Pierpont ME, McGrath J, Hacker AM, et al. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–491. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiraki K, Toyama N, Daikoku T, Yajima M, Miyazaki Dermatologist. Society Herpes zoster and recurrent herpes zoster. Open Forum Infect Dis4:ofx007. 2017 doi: 10.1093/ofid/ofx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan TY, Collins A, James PA, McGillivray G, Stark Z, et al. Phenotypic variability of distal 22q11.2 copy number abnormalities. Am J Med Genet. 2011;A155A:1623–1633. doi: 10.1002/ajmg.a.34051. [DOI] [PubMed] [Google Scholar]
- 28.Verhagen JM, Diderich KE, Oudesluijs G, Mancini GM, Eggink AJ, et al. Phenotypic variability of atypical 22q11.2 deletions not including TBX1. Am J Med Genet. 2012;A158A:2412–2420. doi: 10.1002/ajmg.a.35517. [DOI] [PubMed] [Google Scholar]