Abstract
Wnt signaling has been implicated as a driver of prostate cancer-related osteoblast differentiation, and previous studies have linked modifications in Wnt function with the induction of tumor metastasis. A unique aspect of prostate cancer bone metastases in mouse models is their relative predilection to the hindlimb (femur) compared to the forelimb (humerus). Comparative gene expression profiling was performed within the humerus and femur from non–tumor-bearing mice to evaluate differences in the microenvironments of these locations. This revealed the relative overexpression of the Wnt signaling inhibitors WIF1 and SOST in the humerus compared to the femur, with increased WNT5A expression in femur bone marrow, suggesting a coordinated upregulation of Wnt signals within the femur compared to the humerus. Conditioned medium (CM) from bone marrow stromal cells (HS-5 cells) was used to mimic the bone marrow microenvironment, which strongly promoted prostate cancer cell invasion (3.3-fold increase in PC3 cells, P < .05; 7-fold increase in LNCaP cells, P < .05). WNT5A shRNA knockdown within the CM-producing HS-5 cells significantly decreased PC3 (56%, P < .05) and LNCaP (60%, P < .05) cell invasion. Similarly, preincubation of CM with WIF1 significantly blocked LNCaP cell invasion (40%, P < .05). shRNA-mediated knockdown of the Wnt receptors FZD4 and FZD8 also strongly inhibited tumor cell invasion (60% inhibition shFZD4, P < .05; 63% shFZD8, P < .05). Furthermore, small molecule inhibition of JNK, which is an important component of the noncanonical Wnt signaling pathway, significantly inhibited CM-mediated tumor invasion. Overall, this study reveals a role for Wnt signaling as a driver of prostate cancer bone metastatic tropism and invasion.
Introduction
Skeletal metastases are a major cause of morbidity and mortality in advanced prostate cancer, as 90% of patients who die of metastatic prostate cancer have bone metastasis [1]. Furthermore, these bone metastases occur more commonly in the axial (central) skeleton than the appendicular skeleton [2]. This tendency of prostate cancer cells to spread preferentially to bone is driven by both tumor-specific characteristics and alterations in host microenvironments, and understanding the signaling mechanisms that drive this tropism may have important clinical implications for the treatment of men with metastatic disease [3].
Aberrant Wnt signaling has been implicated as a critical driver in numerous cancers, including those of the breast, brain, colon, lung, oropharynx, ovary, skin, and prostate, suggesting its role as a regulator of carcinogenesis [4], [5], [6], [7], [8], [9], [10], [11]. However, the Wnt pathway has more recently been established as a key signaling pathway in advanced prostate cancer [12], [13], [14], [15], [16], [17]. Wnt signaling has been suggested to impact prostate cancer biology through the regulation of the androgen receptor (AR), propagation of prostate cancer stem cells, promotion of osteoblastic metastasis, and resistance to antiandrogen therapy [15], [18], [19], [20], [21]. Furthermore, next-generation sequencing studies involving patients with castrate-resistant prostate cancer (CRPC) have identified frequent genomic alterations in the Wnt pathway, whereas Wnt pathway modifications are virtually absent in hormone-naive primary prostate cancer [22]. Importantly, we recently reported that metastatic CRPC patient circulating tumor cell expression of WNT5A, BMP7, and AURKA independently predicts overall survival [17].
Wnt signaling occurs via the following two distinct pathways: the canonical, β-catenin–dependent pathway and the noncanonical, β-catenin–independent pathway. It consists of many proteins involved in cell proliferation, differentiation, polarity, and cell fate determination during embryogenesis [23], [24]. Canonical Wnt signaling is regulated by intracellular accumulation of β-catenin, which results in downstream transcriptional co-activation of a host of key genes involved in cellular development. The noncanonical pathway can be further divided into the planar cell polarity pathway and the Ca2+/PKC pathway, which regulates cellular organization, cytoskeleton restructuring, and cell polarity and often signals via the JNK pathway [25], [26]. For some tumor cells, when acquiring an invasive phenotype, noncanonical Wnt signaling is activated via WNT5A. Wnt ligands, such as WNT5A, bind to the Frizzled (FZD) family of G-protein coupled receptors and co-receptors, including ROR, Ryk, and PTK. Downstream, these interactions can lead to the release of intracellular calcium and upregulation of vimentin and snail (SNAI1), and facilitate a transition to an invasive, mesenchymal phenotype [11], [23], [25].
Bone is the preferred site of prostate cancer metastatic spread, and previous studies have focused on the molecular mechanisms of this tropism, suggesting a complex interplay occurs between tumor cells, osteoblasts, and osteoclasts in modulating the bone microenvironment [3], [27]. We previously modeled the anatomic distribution of bone metastasis using in vivo murine models of human prostate cancer cell metastasis and demonstrated that the majority of animals that develop skeletal metastases have either spinal lesions or lesions in the bones of the hindlimb (femur) in contrast to the primarily axial distribution of metastases in humans. Lesions develop in the bones of the forelimb (humerus) much less frequently, implying a difference in the bone microenvironment of these regions. We reported that GAS6 is partially responsible for this phenomenon [28]. To further understand the mechanism of this phenomenon, we used microarrays to assess the difference in gene expression between the forelimb and hindlimb bone. Given that the relative importance of the bone marrow and endosteum in creating the metastatic niche is still not well understood, we assessed gene expression separately in these two compartments. We observed lower expression of the Wnt inhibitors SOST and WIF1 in the hindlimb compared with the forelimb and hypothesize that Wnt signaling may hold significant insight into advanced prostate cancer metastasis.
Materials and Methods
Microarray Analysis
Nine CB17 severe combined immunodeficiency (SCID) mice (4-6 weeks old) were randomly split into three groups and sacrificed. The femur and humerus bones were separated, and the top and bottom caps of the bone were removed with a blade and centrifuged at 10,000 rpm for 2 minutes at 4°C. Bone marrow cells were collected for mRNA extraction, and the supernatant was collected for cytokine analysis. The bone caps were flushed with cold phosphate-buffered saline to remove residual bone marrow cells, and TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) was used to dissolve the bone endosteum layer in the medullary cavity for mRNA extraction. Total RNA was isolated and sent to the University of Michigan Sequencing Core for gene expression microarray analysis. This analysis was based on Mouse Gene ST 1.1 strip arrays that were processed at the University of Michigan microarray facility using an Ambion WT Expression Kit (Thermo Fisher Scientific). This array included 28,000 coding transcripts and 7000 noncoding transcripts. Genes with the highest fold changes were examined, and genes associated with mechanisms that are relevant to prostate cancer were the primary focus of this study. Gene expression was calculated using a robust multiarray average [29].
Human Tumor Xenograft Models
Four- to 6-week-old male CB17 SCID mice were procured from the University of Michigan breeding colony. Subcutaneous tumors were established at both sides of the dorsal flank of the mice. At the end of the studies, the mice were sacrificed, and the hindlimb and forelimb were harvested. The University of Michigan University Committee on the Use and Care of Animals approved all in vivo studies. For the VCaP tumor model, 3 × 106 VCaP cells in serum-free medium with 50% Matrigel (BD Biosciences) were injected subcutaneously into the dorsal flank on both sides of the mice.
Enzyme-Linked Immunosorbent Assay (ELISA)
Bone marrow aspirates were obtained from 10 men with clinically localized prostate cancer before surgery. The aspirates were drawn from the pubic bone, and the samples were transferred to 7.5 ml EDTA (purple cap) tubes and shipped overnight to The University of Michigan on wet ice. Normal marrow samples (n = 3; All Cells, Alameda, CA) were drawn from the posterior superior iliac crest of paid donors, transferred to heparinized tubes, and shipped overnight on wet ice. ELISAs were performed on bone marrow plasma using a commercial Wnt5a ELISA kit from LSBio (cat# LS-F6742, Seattle, WA). Bone marrow plasma was diluted twice in dilution buffer that was provided with the kit prior to assaying. The assays were performed in triplicate, and the results were compared with standard curves obtained using human recombinant Wnt5a that was provided with the kit.
Cell Lines and Culture Conditions
Cell lines were maintained using standard conditions. Specifically, PC3 and LNCaP cell lines [American Type Culture Collection (ATCC), Manassas, VA] were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific). All of the cell lines were grown at 37°C in a 5% CO2 cell culture incubator. To ensure identity, the cell lines were genotyped at the University of Michigan Sequencing Core using Profiler Plus (Applied Biosystems, Foster City, CA) and compared with the short tandem repeat profiles of respective cell lines available in the STR Profile Database (ATCC). All of the cell lines were routinely tested and found to be free of Mycoplasma contamination.
Reverse-Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from the cell lines using an RNeasy Plus Mini Kit (Qiagen, Netherlands). RNA was reverse transcribed to cDNA using 250 ng of total RNA in a final volume of 30 μl using random primers with a Verso cDNA Synthesis Kit (Thermo Fisher Scientific). According to the manufacturer's protocol, the reverse transcription was performed at 42°C for 30 minutes and then at 95°C for 2 minutes. Real-Time PCR analysis was performed using a standard protocol from Power SYBR-Green (Applied Biosystems, Foster City, CA). All protocols were performed according to the manufacturer's instructions. The 2−ΔΔCt values were separately normalized to those of actin and tubulin. Primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). The primer sequences used for the studies are provided (Supplementary Figure 1). The qPCR assays and data collection were performed using an ABI 7900 instrument (Applied Biosystems, Thermo Fisher Scientific). Each sample was analyzed in triplicate.
Western Blot Analysis
Cells were lysed using radioimmunoprecipitation assay buffer (Sigma-Aldrich, St. Louis, MO) and supplemented with a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Proteins were separated using a NuPAGE 4-12% Bis-Tris Gel kit (Lift Technologies, Carlsbad, CA). Specific monoclonal antibodies against β-Catenin (1:1000 dilution, cat#: 8480), non-phospho-β-catenin (1:1000 dilution, cat#: 19807), JNK (1:1000 dilution, cat#: 9252), p-JNK (1:1000 dilution, cat#: 4668), WIF1 (1:1000 dilution, cat#: 9652), and Wnt5a/b (1:1000 dilution, cat#: 2530) were obtained (Cell Signaling Technology, Inc., Danvers, MA). The horseradish peroxidase (HRP)–conjugated β-actin antibody (1:50,000 dilution, cat#: sc-47778 HRP, Santa Cruz, Dallas, TX) and all primary antibodies were incubated overnight at 4°C. The secondary antibody was HRP-conjugated goat anti-rabbit IgG (1:5000 dilution; 7074P2) (Cell Signaling Technology, Inc., Danvers, MA). An enhanced chemiluminescence chromogenic substrate was used to visualize the bands and recorded with Ultra-High Contrast Western blotting film (Thomas Scientific, Swedesboro, NJ).
Cell Transfection
The WIF1 expression vector (pLenti-C-Myc-DDK-WIF1) was purchased from Origene (Cat# RC205256, Rockville, MD). 293T cells were used to overexpress the WIF1 protein. Briefly, 293T cells were seeded in antibiotic-free DMEM supplemented with 10% FBS the day before transfection. A total of 10 μg of plasmid DNA was diluted with 700 μl of Opti-MEM Medium (Thermo Fisher Scientific), and 20 μl of Lipofectamine 2000 (Thermo Fisher Scientific) was diluted with 700 μl of Opti-MEM Medium. The diluted DNA was added to diluted Lipofectamine 2000, and the mixture was incubated at room temperature for 5 minutes. The DNA-lipid complex was added to cells and incubated for 2 days. The cell lysate and supernatant were collected for gene expression analysis and invasion assay.
Lentivirus Infection
To suppress gene expression, lentiviruses with shRNA against WNT5A, FZD4 and FZD8 were prepared by the University of Michigan Vector Core. Lentivirus against WNT5A was used to infect human bone marrow stromal cells (HS-5 cells, ATCC). Lentiviruses against FZD4 and FZD8 were used to infect human prostate cancer cells (PC3 cells, ATCC). The knockdown efficiency of target genes was evaluated either by Western blot or qPCR.
Transwell Invasion Assay
Cell invasion assays were carried out using two methods. First, FluoroBiok Transwell chambers with inserts of 8-mm pore size (cat# 351152, Corning Inc., Corning, NY) were used. Briefly, Growth Factor Reduced Matrigel (cat# 354230, BD Biosciences, Bedford, MA) was diluted to 200 μg/ml, added to a Transwell (100 μl) in a 24-well plate (cat#353504, Corning Inc.), and incubated in an incubator for 4 hours to allow the Matrigel to set. Prostate cancer cell lines (PC3 and LNCaP cells) were serum-starved for 18 hours before the invasion experiments were performed. The cells were trypsinized, counted, and resuspended in serum-depleted medium (RPMI 1640, 1% FBS) containing 10% concentrated (10×) conditioned medium (CM). The bottoms of the 24 wells were filled with complete medium (RPMI 1640, 10% FBS). The cells were allowed to invade for 48 hours. Then, Calcein AM dye (C3100MP, Thermo Fisher Scientific) was used to stain the live cells. Fluorescence images were obtained with an Olympus IX70 microscope (Olympus, Center Valley, PA), and the fluorescence intensity was scanned.
For the second method, clear-bottom Transwell plates (cat# 3422, Corning Inc.) were used, and the aforementioned protocol was performed. Cells that invaded through the membrane to the underside of the Transwell membrane were fixed with 4% formalin, stained with Crystal violet for 10 minutes, imaged, and counted.
Statistical Analysis
The data are presented as the mean ± standard error of the mean. In total, three replicate wells were examined per assay, and each experiment was performed in triplicate. Statistical comparisons between groups were determined by a one-tailed Student's t test. We also utilized the Michigan Portal for the Analysis of next-generation sequencing data (http://mipanda.org) to access and analyze high-throughput RNA sequencing data from PC3 cells. Expression heatmaps were generated using Morpheus software (MIT/Broad Institute). All data were analyzed using Prism. P < .05 was considered to indicate a statistically significant difference.
Results
Gene Expression Analysis of Hindlimb and Forelimb Bone Using Microarray
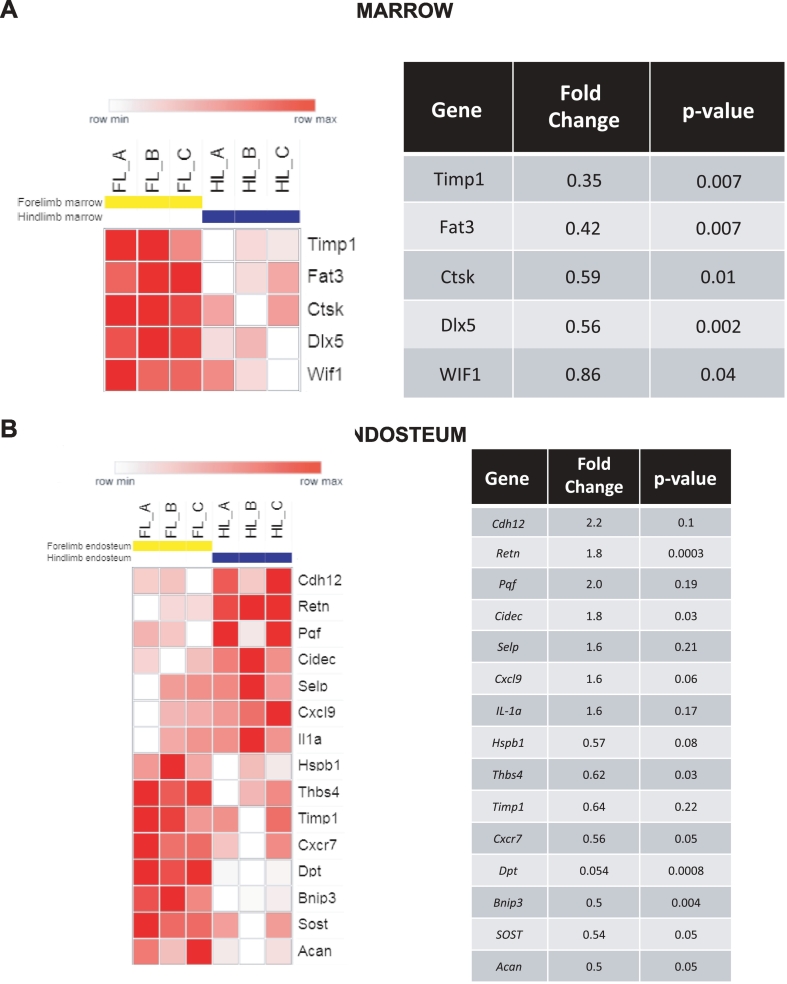
To evaluate if potential gene expression differences between the bone microenvironment in the hindlimb and the forelimb impact prostate cancer tropism, bone marrow samples and matched endosteum samples from non–tumor-bearing SCID mice were evaluated with gene expression microarrays as described in the methods. We found that proteins previously shown to be involved in tumor cell proliferation and metastasis, such as placental growth factor (PGF) and P-selectin (SELP), were expressed at higher levels in the hindlimb bone endosteum. In addition, IL-1a was expressed at higher levels in the hindlimb compared to the forelimb (Figure 1). This cytokine has been shown to stimulate the expression of epidermal growth factor (EGF) and fibroblast growth factor (FGF), which are potential drivers of prostate cancer invasion, growth, and progression to androgen independence.
Figure 1.
Differential expression of metastasis- and invasion-related genes between the mouse hindlimb and forelimb. Microarray data are presented as expression in the hindlimb relative to the forelimb. Each column of the heat map represents one mouse. (A) Differential expression in the marrow fraction (cells removed from bone by flushing). (B) Differential expression in the endosteal fraction (retained cells).
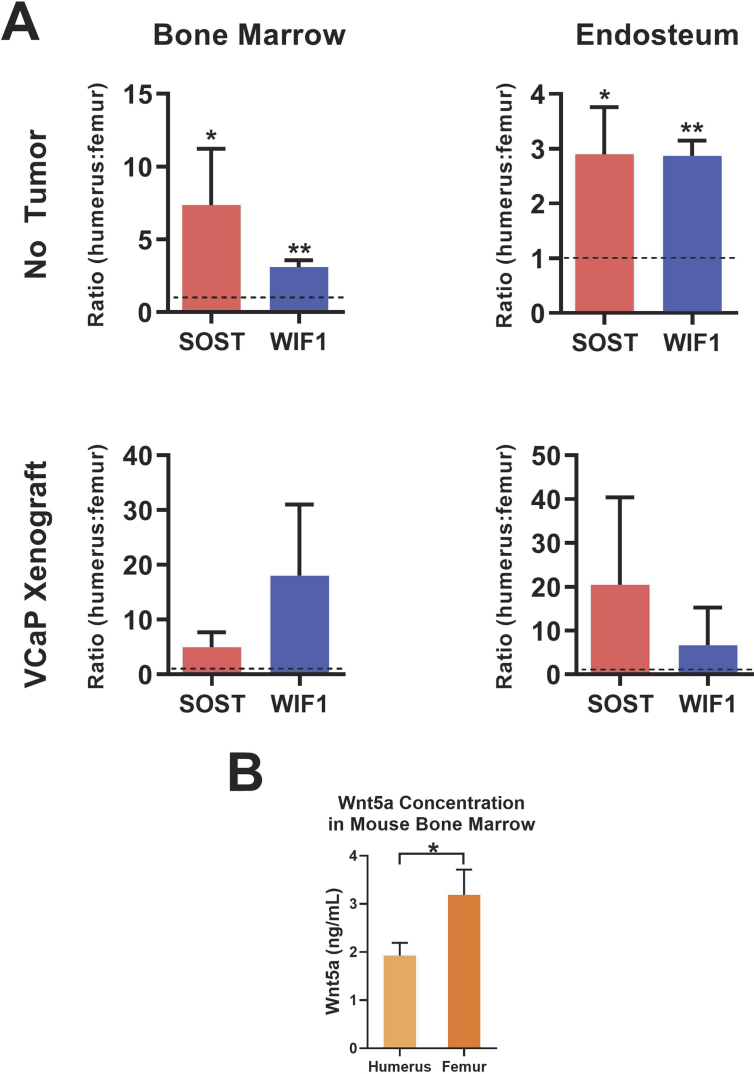
In contrast, the Wnt signaling inhibitors SOST and WIF1 were expressed at lower levels in the hindlimb compared to the forelimb, suggesting the possibility of increased Wnt signals within sites of preferential metastasis. To further evaluate the differential expression of these genes, real-time PCR was used to assess WIF1 and SOST expression in the bone marrow in SCID mice with or without subcutaneous xenograft tumors (VCaP cells). WIF1 and SOST expression in the bone marrow and endosteum was elevated in the forelimb compared to the hindlimb in both tumor-naive mice (P < .05) and in those with implanted xenografts (Figure 2A). These results suggest a higher level of baseline Wnt signaling inhibition in the forelimb, implicating upregulation of Wnt signals in the hindlimb that may regulate preferential prostate cancer metastatic spread. To further evaluate this hypothesis, ELISA was performed to quantify WNT5A expression within the bone marrow of SCID mice. WNT5A expression appeared to be nearly two-fold higher in the hindlimb than in the forelimb (Figure 2B, P < .05).
Figure 2.
Increased expression of Wnt signaling inhibitors in the forelimb compared to the hindlimb. (A) RT-qPCR of WIF1 and SOST expression in the bone marrow and endosteum of SCID mice with and without subcutaneous xenograft tumors (VCaP cells). The expression ratio of SOST of WIF1 expression in the forelimb relative to the expression in the hindlimb is shown in Figure 2A. Significant differences (*P < .05; **P < .01) are relative to an expression ratio of 1 (dotted line). (B) ELISA of WNT5A expression in the humerus and femur bone marrow of SCID mice.
Wnt5a/FZD Knockdown in the Bone Microenvironment Decreases Prostate Cancer Cell Invasion
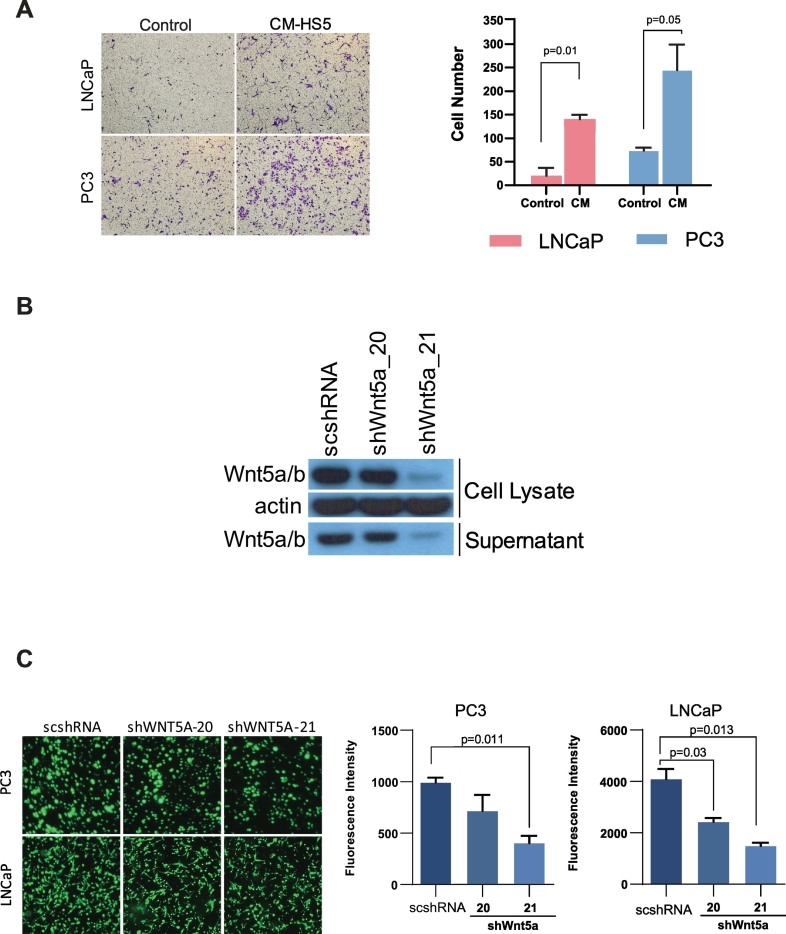
To further assess the impact of the microenvironment on the regulation of prostate cancer metastasis, we used HS-5 cells to simulate components of the bone marrow microenvironment. HS-5 cells are immortalized stromal cells from human marrow and secrete significant levels of growth factors, including granulocyte-colony stimulating factor (G-CSF), granulocyte-macrophage-colony stimulating factor (GM-CSF), and macrophage-colony stimulating factor (M-CSF) [30]. CM was collected from HS-5 cells and added to LNCaP and PC3 cells in culture for invasion assays. Both LNCaP and PC3 cells showed a significant increase in invasion capability after the addition of 10% CM (Figure 3A).
Figure 3.
LNCaP and PC3 cell invasion increased after the addition of HS-5 cell CM. (A) LNCaP and PC3 cell invasion increased after the addition of CM from HS-5 immortalized stromal cells. (B) The efficiency of shRNA-mediated A/B knockdown by Western blot. (C) LNCaP and PC3 cell invasion in the presence of HS-5 CM. Decreased cell invasion was observed in the presence of shRNA-mediated WNT5A knockdown in PC3 (P = .011) and LNCaP (P = .013) cells.
Next, we aimed to determine the extent to which signaling through WNT5A in the CM could be responsible for this increase in cell line invasiveness. To test this, we performed shRNA-mediated WNT5A knockdown within the CM donor HS-5 cells. Efficiency of shRNA knockdown was measured by Western blotting (Figure 3B), and LNCaP and PC3 cell invasion was evaluated with addition of HS-5 cell CM with and without presence of shRNA-mediated WNT5A knockdown. A significant decrease in tumor cell invasion was observed after WNT5A knockdown in CM (Figure 3C). This implicates Wnt signaling as a potential driver of cell invasiveness in the bone microenvironment.
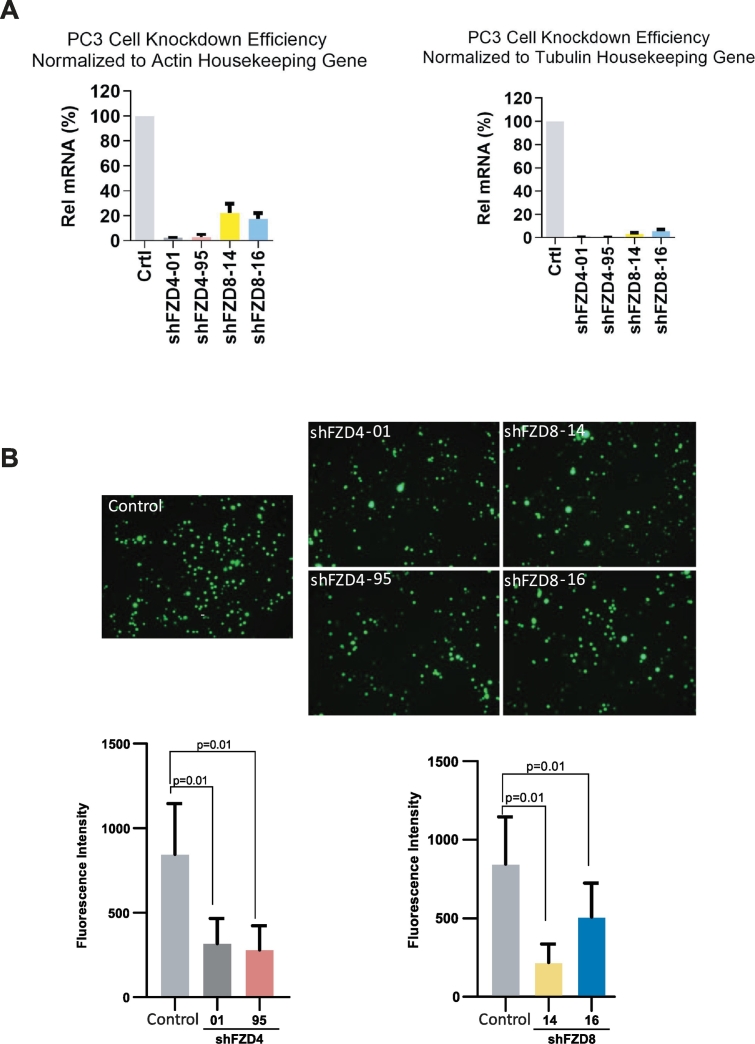
Similarly, we hypothesized that inhibition of downstream receptors of Wnt signaling would also result in decreased cell invasion from the baseline. PC3 transcriptome data were downloaded from the MiPanda website (http://www.mipanda.org), and FZD8 was determined to be the most highly expressed member of the FZD family within PC3 cells [31] (Supplementary Figure 2). Therefore, we performed shRNA-mediated knockdown of the downstream Wnt signaling receptors FZD4 and FZD8 to evaluate the effect on PC3 cell invasion. After confirming the knockdown efficiency (Figure 4A), we found that the addition of both shFZD4 and shFZD8 decreased PC3 cell invasiveness (Figure 4B).
Figure 4.
Knockdown of the downstream Wnt signaling receptors FZD4 and FZD8 results in decreases PC3 cell invasion. (A) Efficiency of shRNA-mediated FZD4 and FZD8 knockdown in PC3 cells. (B) PC3 cell invasion in the presence of HS-5 CM significantly decreased after shRNA-mediated FZD4 (P = .01) and FZD8 (P = .01) knockdown.
WIF1 Overexpression Decreases Prostate Cancer Cell Invasion
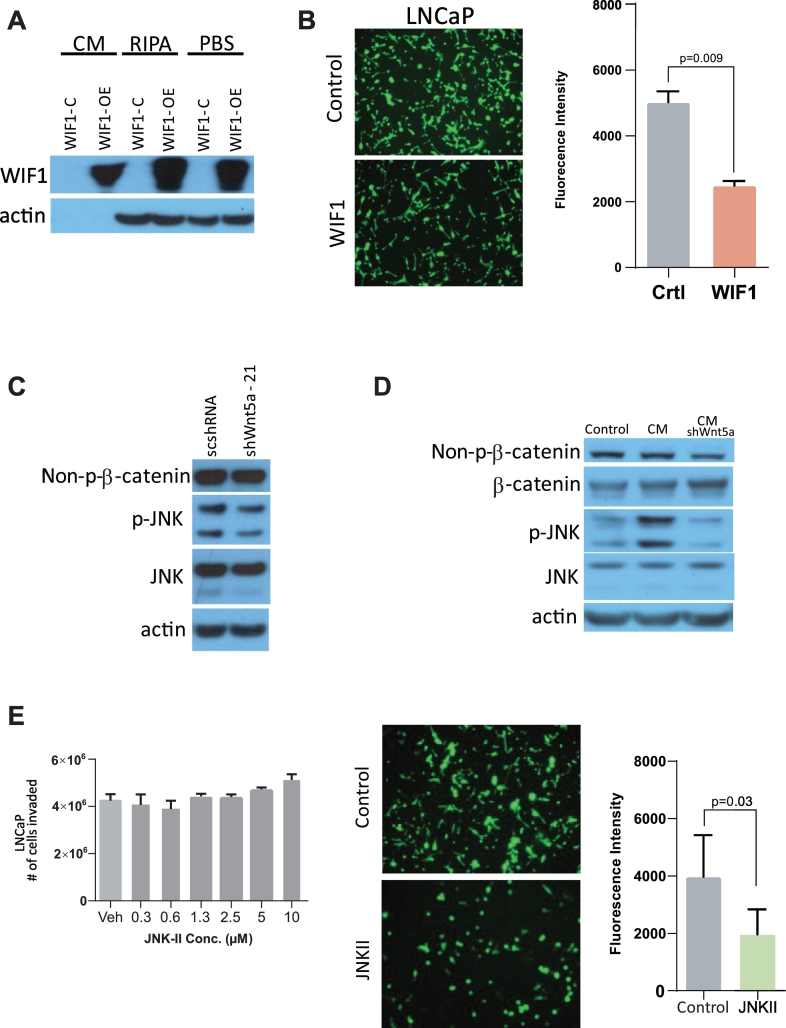
To further confirm the role of Wnt signaling in prostate cancer cell invasion, we overexpressed WIF1, which is an inhibitor of Wnt signaling, in 293T cells and assessed the ability to inhibit prostate cancer cell invasion. CM from WIF1-overexpressing 293T cells was collected, and WIF1 expression was confirmed by Western blot (Figure 5A). CM was then added to LNCaP cells, and an invasion assay was performed. As expected, decreased cell invasion was observed in WIF1-overexpressing LNCaP cells cultured in CM (Figure 5B).
Figure 5.
Overexpression of WIF1 and JNK inhibition result in decreased prostate cancer cell invasion. (A) Western blot of the efficiency of WIF1 overexpression in 293T cells after the addition of CM, radioimmunoprecipitation buffer, or phosphate-buffered saline). (B) LNCaP cell invasion was significantly decreased after the addition of CM from WIF1-overexpressing 293T cells (P = .009). (C) The addition of shWNT5a-21 resulted in decreased JNK phosphorylation, as indicated by Western blot. (D) HS-5 CM with WNT5A shRNA resulted in decreased JNK phosphorylation, as indicated by Western blot. (E) LNCaP cells with HS-5 CM showed significantly decreased cell invasion in the presence of the JNK II inhibitor (P = .03). The presence of the JNK II inhibitor alone did not affect LNCaP cell invasion.
JNK Is Involved in Wnt Signal Transduction and Can Be Targeted by Inhibition
The JNK family of kinases is composed of key signaling molecules involved in the intracellular propagation of cellular responses. Additionally, JNK has been shown to be activated by WNT5A and is involved in carrying out functions of the noncanonical Wnt pathway [26]. Therefore, we sought to evaluate the role of JNK-mediated Wnt signaling on prostate cancer invasion and evaluate the role of small molecule inhibition of JNK as a viable therapeutic option.
First, to examine JNK involvement in the transduction of autocrine Wnt signaling in prostate cancer cells, we evaluated JNK phosphorylation with and without WNT5A knockdown. WNT5A knockdown via shRNA resulted in modestly decreased levels of p-JNK, consistent with WNT5A activation of JNK (Figure 5C). Importantly, CM from HS-5 cells markedly increased JNK phosphorylation but not the level of active (nonphosphorylated) β-catenin. This effect was completely lost when WNT5A was knocked down by shRNA in the HS-5 cells (Figure 5D). Next, we examined the effect of JNK inhibition on LNCaP cell migration. Addition of a 30-fold dose range of JNK inhibitor to LNCaP cells in the absence of CM did not affect cell invasion. However, cell invasion stimulated by HS-5 CM was significantly inhibited in the presence of the JNK inhibitor (Figure 5E).
Discussion
Historically, investigative efforts have often been “tumor-centric” in an attempt to discover novel molecular drivers of cancer proliferation. However, tumor microenvironmental factors also play a role in dictating cancer cell behavior, and many of these factors have been increasingly well described [32], [33]. Yet, while prostate neoplasms have a striking tendency to metastasize to bone, the mechanisms that account for this bone tropism are still being elucidated. For metastases to occur, prostate cancer cells must disseminate into and adhere to bone marrow matrix components and establish new tumors in the bone microenvironment [34]. This occurs by vascular spread, which is at least partially enhanced by chemoattractants within the bone marrow, suggesting a pivotal role of the bone microenvironment in prostate cancer spread [35], [36], [37]. Furthermore, prior in vivo exploration has shown an unequal prevalence in long bone metastasis, with fewer metastatic foci in the forelimb bones (humerus, radius, and ulna) than in the hindlimb bones (femur, tibia, and fibula) of mice. This was at least partly related to a differential pattern of gene expression in the hindlimb and forelimb bones, with increased expression of GAS6 mRNA, which is a known prostate cancer proliferation inhibitor, isolated from whole bone marrow and osteoblast fractions from the forelimb [28].
In this study, we performed comparative gene expression profiling within the humerus and femur of mice to assess for baseline differences in the microenvironment of these locations. Our study reveals a coordinated upregulation of Wnt signals within the femur versus the humerus, suggesting that prostate cancer bone metastasis may be substantially enhanced under the direction of the Wnt signaling pathway within the bone marrow microenvironment. Specifically, we found a relative overexpression of Wnt signaling inhibitors in the humerus compared to femur, with a resultant increase in WNT5A expression in femur marrow. Targeted inhibition of Wnt within CM from bone marrow stromal cells and knockdown of the Wnt receptors FZD4 or FZD8 significantly reduced prostate cancer cell invasion. Furthermore, small molecule inhibition of JNK, which is an important component of the noncanonical Wnt signaling pathway, also significantly inhibited tumor invasion.
While prior studies have sought to better understand the mechanisms that drive prostate cancer progression and resistance to treatment, only recently has there been an improved understanding of the role of Wnt signaling in prostate cancer metastasis. This has primarily been focused on the canonical pathway, but recent data suggest that the noncanonical Wnt pathway may be just as important, particularly in the promotion of prostate cancer metastasis [15], [17], [21], [38], [39]. We recently demonstrated that WNT5A expression in circulating tumor cells from men with mCRPC is strongly associated with mortality, even after adjusting for standard clinical parameters [17]. Additionally, Miyamoto and colleagues have demonstrated a strong link between WNT5A signaling and antiandrogen resistance in mCRPC [15]. While there are many regulators of Wnt activity, WIF1 has been shown to be critically important in regulating Wnt activity and reversing EMT, consistent with our data suggesting decreased WIF1 expression in the hindlimb marrow and decreased cell invasion in the presence of WIF1 overexpression [40]. These data support the potential importance of Wnt signaling as a driver of prostate cancer metastasis to bone.
Deregulation of JNK kinases has been implicated in the progression of human disease, including diabetes, neurological conditions, and cancer [41], [42], [43], [44]. Therefore, there has been a significant interest in the development of small molecule inhibitors to target the JNK family for pharmacological inhibition [44], [45]. Furthermore, activation of the Wnt signaling pathway and its colocalization with AR play a role in acquisition of androgen independence in CRPC, and Wnt signaling inhibition in this setting has been shown to resensitize prostate cancer cells to enzalutamide [46]. Thus, current high-throughput approaches are being utilized to facilitate screening of Wnt pathways for the development of targeted therapeutics, and numerous ongoing preclinical and early phase trials show promising results for eventual clinical drug delivery against Wnt components [47].
Our current study is limited by the lack of an attributable mechanism for augmented Wnt signaling within the hindlimb. While an abundance of other research has implicated numerous microenvironment factors that regulate tumor progression at distant sites, there are relatively little data specific to Wnt signaling in this context [48], [49], [50]. Further in vivo modulation of Wnt regulation of the bone microenvironment will be necessary to help confirm the causal relationship between Wnt signaling and prostate cancer bone tropism. Finally, validation in patient bone marrow aspirates will be key to confirming the importance of this pathway for driving prostate cancer bone metastasis in men with advanced prostate cancer. The ultimate impact of Wnt modulation in prostate cancer patients remains unclear, and there are also data suggesting a potential prodormancy effect of Wnt signaling in prostate cancer bone metastases [51].
These limitations notwithstanding, the data presented here indicate that Wnt regulation in the bone marrow is key driver of prostate cancer bone metastasis. While further investigation will be necessary, our study supports the potential importance of Wnt targeting as a therapeutic strategy in recurrent or advanced prostate cancer.
Acknowledgments
Acknowledgements
Funding
Supported by a Department of Defense Physician Research Training Award (W81XWH-14-1-0287) and the Prostate Cancer Foundation (T.M.M., R.S.T., Y.Q., F.C.). T. M. M. is also supported by the A. Alfred Taubman Medical Research Institute. F. C. receives support from a Career Enhancement Award from the NIH/NCI Prostate Cancer Specialized Program in Research Excellence (SPORE) at the University of Michigan, sub-award F036250. This work was also supported by the NIH: National Cancer Institute P01CA093900-10 and R01CA240991-01 and the University of Michigan Comprehensive Cancer Center Prostate SPORE (P50CA 069568).
Author contributions
Y. W., U. S., Y. Q., T. M. M., R. T., and F. C. conceived the study and analyses; Y. W., U. S., Y. Q., T. M. R., J. A. B., and H. Z. performed experiments, data processing, and data analysis with assistance from all authors; U. S., Y. W., Y. Q., T. M. M., and F. C. wrote the manuscript. All authors discussed results, commented, edited, and agreed on the submission of the manuscript. The authors also thank the University of Michigan Sequencing Core for their expertise and assistance.
Footnotes
One Sentence Summary: Noncanonical Wnt signaling plays an important role in determining prostate cancer metastatic tropism.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100747.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Bubendorf L, Schöpfer A, Wagner U. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum. Pathol. 2000;31(5):578–583. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- 2.Vahid Reza Dabbagh Kakhki AUKAAURSAUA-SMAUMT-K. Pattern and distribution of bone metastases in common malignant tumors. 2013;16(2):66–69–66-69. [DOI] [PubMed]
- 3.Taichman RS, Loberg RD, Mehra R. The evolving biology and treatment of prostate cancer. J. Clin. Invest. 2007;117(9):2351–2361. doi: 10.1172/JCI31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe LR, Brown AMC. Wnt signaling and breast cancer. Cancer Biology & Therapy. 2004;3(1):36–41. doi: 10.4161/cbt.3.1.561. [DOI] [PubMed] [Google Scholar]
- 5.Rheinbay E, Suvà Mario L, Gillespie Shawn M. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3(5):1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki H, Watkins DN, Jair K-W. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 2004;36:417. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 7.Uematsu K, He B, You L. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene. 2003;22:7218. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 8.Reifenberger J, Knobbe CB, Wolter M. Molecular genetic analysis of malignant melanomas for aberrations of the WNT signaling pathway genes CTNNB1, APC. ICAT and BTRC. Int J Cancer. 2002;100(5):549–556. doi: 10.1002/ijc.10512. [DOI] [PubMed] [Google Scholar]
- 9.Su H-Y, Lai H-C, Lin Y-W. Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int. J. Cancer. 2010;127(3):555–567. doi: 10.1002/ijc.25083. [DOI] [PubMed] [Google Scholar]
- 10.Rampias T, Boutati E, Pectasides E. Activation of Wnt signaling pathway by human papillomavirus E6 and E7 oncogenes in HPV16-positive oropharyngeal squamous carcinoma cells. Mol. Cancer Res. 2010 doi: 10.1158/1541-7786.MCR-09-0345. http://mcr.aacrjournals.org/content/early/2010/03/07/1541-7786.MCR-09-0345.abstract [DOI] [PubMed] [Google Scholar]
- 11.Polakis P. Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 2012;4(5). [DOI] [PMC free article] [PubMed]
- 12.Yokoyama NN, Shao S, Hoang BH. Wnt signaling in castration-resistant prostate cancer: implications for therapy. American journal of clinical and experimental urology. 2014;2(1):27–44. [PMC free article] [PubMed] [Google Scholar]
- 13.Kypta RM, Waxman J. Wnt/β-catenin signalling in prostate cancer. Nature Reviews Urology. 2012;9:418. doi: 10.1038/nrurol.2012.116. [DOI] [PubMed] [Google Scholar]
- 14.Robinson D, Van Allen Eliezer M, Wu Y-M. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto DT, Zheng Y, Wittner BS. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Dehm SM, Hillman DW. A prospective genome-wide study of prostate cancer metastases reveals association of wnt pathway activation and increased cell cycle proliferation with primary resistance to abiraterone acetate–prednisone. Ann. Oncol. 2018;29(2):352–360. doi: 10.1093/annonc/mdx689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singhal U, Wang Y, Henderson J. Multigene profiling of CTCs in mCRPC identifies a clinically relevant prognostic signature. Mol. Cancer Res. 2018 doi: 10.1158/1541-7786.MCR-17-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Truica CI, Byers S, Gelmann EP. β-Catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 2000;60(17):4709. [PubMed] [Google Scholar]
- 19.Yang F, Li X, Sharma M. Linking β-catenin to androgen-signaling pathway. J. Biol. Chem. 2002;277(13):11336–11344. doi: 10.1074/jbc.M111962200. [DOI] [PubMed] [Google Scholar]
- 20.Bisson I, Prowse DM. WNT signaling regulates self-renewal and differentiation of prostate cancer cells with stem cell characteristics. Cell Res. 2009;19:683. doi: 10.1038/cr.2009.43. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi S, Watanabe T, Okada M. Noncanonical Wnt signaling mediates androgen-dependent tumor growth in a mouse model of prostate cancer. Proc. Natl. Acad. Sci. 2011;108(12):4938. doi: 10.1073/pnas.1014850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajan P, Sudbery IM, Villasevil MEM. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur. Urol. 2014;66(1):32–39. doi: 10.1016/j.eururo.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komiya Y, Habas R. Wnt signal transduction pathways. Organ. 2008;4(2):68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goessling W, North TE, Loewer S. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136(6):1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamanaka H, Moriguchi T, Masuyama N. JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 2002;3(1):69. doi: 10.1093/embo-reports/kvf008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guise TA, Mohammad KS, Clines G, et al. Basic mechanisms responsible for osteolytic and osteoblastic bone metastases. Clinical Cancer Research 2006;12(20):6213s. [DOI] [PubMed]
- 28.Jung YH, Shiozawa Y, Wang JC. Prevalence of prostate cancer metastases after intravenous inoculation provides clues into the molecular basis of dormancy in the bone marrow microenvironment. Neoplasia. 2012;14(5):429–439. doi: 10.1596/neo.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Hobbs B, Collin F. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 30.Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995;85(4):997–1005. [PubMed] [Google Scholar]
- 31.Niknafs YS, Pandian B, Gajjar T, et al. MiPanda: a resource for analyzing and visualizing next-generation sequencing transcriptomics data. Neoplasia (New York, N.Y.) 2018;20(11):1144–1149. [DOI] [PMC free article] [PubMed]
- 32.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allinen M, Beroukhim R, Cai L. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6(1):17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Morrissey C, Vessella RL. The role of tumor microenvironment in prostate cancer bone metastasis. J. Cell. Biochem. 2007;101(4):873–886. doi: 10.1002/jcb.21214. [DOI] [PubMed] [Google Scholar]
- 35.Taichman RS, Cooper C, Keller ET. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62(6):1832–1837. [PubMed] [Google Scholar]
- 36.Sun Y-X, Schneider A, Jung Y. Skeletal localization and neutralization of the SDF-1(CXCL12)/CXCR4 axis blocks prostate cancer metastasis and growth in osseous sites in vivo. J. Bone Miner. Res. 2005;20(2):318–329. doi: 10.1359/JBMR.041109. [DOI] [PubMed] [Google Scholar]
- 37.Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat. Rev. Cancer. 2011;11:411. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandsmark E, Hansen AF, Selnæs KM. A novel non-canonical Wnt signature for prostate cancer aggressiveness. Oncotarget. 2016;8(6):9572–9586. doi: 10.18632/oncotarget.14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto H, Oue N, Sato A. Wnt5a signaling is involved in the aggressiveness of prostate cancer and expression of metalloproteinase. Oncogene. 2010;29(14):2036–2046. doi: 10.1038/onc.2009.496. [DOI] [PubMed] [Google Scholar]
- 40.Yee DS, Tang Y, Li X. The Wnt inhibitory factor 1 restoration in prostate cancer cells was associated with reduced tumor growth, decreased capacity of cell migration and invasion and a reversal of epithelial to mesenchymal transition. Mol. Cancer. 2010;9:162. doi: 10.1186/1476-4598-9-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirosumi J, Tuncman G, Chang L. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 42.Yarza R, Vela S, Solas M. c-Jun N-terminal kinase (JNK) signaling as a therapeutic target for Alzheimer's disease. Front. Pharmacol. 2016;6:321. doi: 10.3389/fphar.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunot S, Vila M, Teismann P, et al. JNK-mediated induction of cyclooxygenase 2 is required for neurodegeneration in a mouse model of Parkinson's disease. Proc. Natl. Acad. Sci. U. S. A. 2004;101(2):665. [DOI] [PMC free article] [PubMed]
- 44.Bubici C, Papa S. JNK signalling in cancer: in need of new, smarter therapeutic targets. Br. J. Pharmacol. 2014;171(1):24–37. doi: 10.1111/bph.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang T, Inesta-Vaquera F, Niepel M. Discovery of potent and selective covalent inhibitors of JNK. Chem. Biol. 2012;19(1):140–154. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, Cheng L, Li J. Inhibition of the Wnt/β-catenin pathway overcomes resistance to enzalutamide in castration-resistant prostate cancer. Cancer Res. 2018;78(12):3147. doi: 10.1158/0008-5472.CAN-17-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang K, Wang X, Zhang H. The evolving roles of canonical WNT signaling in stem cells and tumorigenesis: implications in targeted cancer therapies. Lab. Investig. 2015;96:116. doi: 10.1038/labinvest.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cackowski FC, Eber MR, Rhee J. Mer tyrosine kinase regulates disseminated prostate cancer cellular dormancy. J. Cell. Biochem. 2017;118(4):891–902. doi: 10.1002/jcb.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sosa MS, Parikh F, Maia AG. NR2F1 controls tumour cell dormancy via SOX9- and RARβ-driven quiescence programmes. Nat. Commun. 2015;6:6170. doi: 10.1038/ncomms7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linde N, Casanova-Acebes M, Sosa MS. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat. Commun. 2018;9(1):21. doi: 10.1038/s41467-017-02481-5. http://europepmc.org/abstract/MED/29295986 http://europepmc.org/articles/PMC5750231?pdf=render http://europepmc.org/articles/PMC5750231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren D, Dai Y, Yang Q, Zhang X. Wnt5a induces and maintains prostate cancer cells domancy in bone. J. Exp. Med. 2019;216:428. doi: 10.1084/jem.20180661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures