Abstract
Background/purpose
Oral lichen planus (OLP) is a chronic inflammatory disease of oral mucosa. The present study investigated the expression of nucleotide-binding oligomerization domain (NOD), a pivotal sensor protein of the innate immune system, in OLP.
Materials and methods
Oral mucosal biopsies were collected from 20 patients with OLP and 6 individuals with normal oral mucosa (NOM). The expression of NOD1 and NOD2 was determined using RT-PCR and immunohistochemistry in OLP and NOM samples.
Results
The mRNA expression of NOD1 and NOD2 was significantly higher in the OLP group than in the NOM group. The protein expression of NOD1 was marginally upregulated in all mucosal layers in the OLP group compared with that of the NOM group; however, the differences were not significant. The expression of NOD2 was elevated in infiltrating lymphocytes of the submucosal layer in the OLP group compared with the NOM group, but was undetected in other inflammatory disease, inflammatory fibrous hyperplasia (IFH). This study revealed the upregulation of NOD2 mRNA and protein in the OLP group, but not in the NOM group.
Conclusion
These findings suggest that NOD2 may play an important role in the pathogenesis of OLP and represents a new diagnostic and treatment target.
Keywords: NOD1, NOD2, Oral lichen planus
Introduction
Oral lichen planus (OLP) is a chronic inflammatory disease of the oral mucosa.1 The most commonly affected sites include buccal mucosa, gingiva, tongue, and vermilion portion of the lips; however, any intraoral site can be affected.2 A wide spectrum of mucosal alterations is observed with reticular, atrophic, erosive, papular, and occasionally bullous lesions. The lesions may be chronic and rarely subside spontaneously.3 The clinical characteristics range from asymptomatic to debilitating pain triggered by salty, spicy, or hot food.1 Although the etiology of OLP remains unknown, the disease is considered an autoimmune disorder, resulting from the apoptosis of basal cells of the epithelium induced by autocytotoxic CD8+ T lymphocytes.4 Currently, no specific cure for OLP exists and the management of lesions can be difficult. Most therapeutic modalities, including systemic and topical corticosteroids, topical retinoids, cyclosporine, tacrolimus, and pimecrolimus, aim for symptomatic relief.1
The innate immune system is the first line of host defense against pathogens, and imbalances in this system can contribute to the development of severe infectious disease, chronic inflammatory disease, and autoimmune disease.5 The innate immune system comprises several classes of pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain-like receptors (NLRs).6 TLRs, the best characterized PRRs, bind numerous exogenous and endogenous antigens by recognizing conserved pathogen-associated molecular patterns (PAMPs).1 Nucleotide-binding oligomerization (NOD) is representative of NLRs and comprises a C-terminal leucine-rich repeat domain that is involved in the recognition of conserved microbial patterns or other ligands.5, 6, 7, 8
NOD1 and NOD2 are major immune response proteins that act primarily as intracellular PRRs involved in the detection of cytoplasmic PAMPs and endogenous products of tissue injury.9 NOD1 detects meso-diaminopimelic acid containing peptidoglycans of gram-negative bacteria,10,11 whereas NOD2 detects muramyl dipeptide, the largest peptidoglycan motif common to gram-negative and gram-positive bacteria.8, 9, 10, 11
Differential expression of TLRs in epithelial cells has been reported in several oral mucosal diseases, including OLP.7,12,13 NOD1 has been linked to atopy and asthma, whereas NOD2 has been implicated in Crohn's disease, Blau syndrome, and inflammatory bowel disease.5,14, 15, 16 However, the relationship between NOD and OLP is currently unknown. Therefore, the present study explored the relationship between NOD and OLP via examination of the expression patterns of NOD1 and NOD2 in patients with OLP.
Materials and methods
Participants and samples
The present study was approved by the institutional ethics committee of Wonkwang University Daejeon Dental Hospital (W-1301/001-001). Written, informed consent was obtained from all participants. Overall, 20 cases of OLP were retrieved from the records of patients who had visited the Department of Oral Medicine and Orofacial Pain at Wonkwang University Daejeon Dental Hospital. All patients were clinically and histologically diagnosed with OLP, using the diagnostic criteria of van der Meij and van der Waal.17 The patients included 9 males and 11 females, with a mean age of 52.3 years. None of them had other immune-related diseases. Patients with current or previous history of steroid therapy were excluded. Six biopsy specimens of normal oral mucosa (NOM) were collected from healthy volunteers without systemic or oral mucosal disease (3 males and 3 females), with an age distribution similar to that of the OLP group. The tissue samples were collected via a 6-mm punch biopsy from the buccal mucosa following the administration of local anesthesia in both the NOM and OLP groups. Half of the tissue was frozen in liquid nitrogen and stored at −80 °C until reverse transcription-polymerase chain reaction (RT-PCR) assay was performed. The other half was fixed in 10% buffered formalin and embedded in paraffin for histological analysis. In addition, six paraffin-embedded biopsy specimens (3 males and 3 females) were collected from patients with inflammatory fibrous hyperplasia (IFH) with an age distribution similar to that of the OLP group for comparative analysis of immune expression of NOD.
Histopathology
Tissue samples were fixed in 10% buffered formalin for more than 24 h, followed by dehydration of samples in an alcohol-xylene series and were then embedded in paraffin wax. From each block, 2-μm sections were prepared and hematoxylin (Vector, Burlingame, CA, USA) and eosin (Vector) (H&E) staining was performed for histological examination.
Reverse transcription-polymerase chain reaction (RT-PCR)
The expression of NOD1 and NOD2 was measured using RT-PCR. Human cementoblasts (HCEM) participate in the innate immune system by expressing NODs and were used as a positive control.18 Tissues obtained from the buccal mucosa were lysed using TRIzol reagent (Invitrogen, California, Carlsbad, USA). One μg of total RNA was reverse transcribed into complementary DNA (cDNA), and PCR was performed using the Superscript One-step RT-PCR with platinum Taq kit (Invitrogen). The following primer sets were used: human NOD1 (F: 5ʹ-ACATCCGCAATACTCAGTGTCTG-3ʹ, R: 5ʹ-ACGCTTTCTCTGAGTGAGCA-3ʹ), human NOD2 (F: 5ʹ-GAATGTGCTCTTCACTGCGAGCAA-3ʹ, R: 5ʹ-AGCATGACGTTC TTTGCCAGCA-3ʹ), and GAPDH (F: 5ʹ-CCAAGGTCATCCATGACAACTTTG-3ʹ, R: 5ʹ-GTCA TACCAGGAAATGAGCTTGACA-3ʹ). The reaction was conducted under the following conditions: amplification at 94 °C for 3 min followed by 35 cycles after an initial denaturation step at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s, and a final extension step at 72 °C for 10 min with all primers. Thereafter, PCR products were electrophoresed on a 1.5% acrylamide gel and visualized using a gel documentation system (Bio-rad, Hercules, CA, USA). Densitometric analysis of visual bands was performed using an automated digitizing software program (Multi Gauge V3.0; Fuji Film Co., Minato-ku, Tokyo, Japan), and the intensity of bands was normalized to the expression of an endogenous control GAPDH signal.
Immunohistochemistry
Paraffin-embedded tissues were used to identify NOD1 and NOD2 expression. The tissue sections were incubated in a solution with 3% H2O2/methanol for 30 min at room temperature (RT) to block peroxidase activity. Antigen retrieval was performed in a microwave-heated citrate buffer (pH 6.0) for 20 min, followed by 1% bovine serum albumin/phosphate buffered saline (RT, 30 min) to block non-specific immunoglobulin binding. Thereafter, these tissue sections were incubated with anti-NOD1 (1:100; NB100-56878, Novus Bio-logicals, Centennial, CO, USA) and anti-NOD2 (1:100; NB500-253, Novus Biologicals) antibodies overnight at 4 °C. The sections were incubated with anti-rabbit secondary antibody solution for 1 h at RT, followed by incubation with avidin-biotin peroxidase complex reagent (Vector) for 30 min. The organic compound 3,3′-diaminobenzidine tetrahydrochloride (DAB; Vector) was used as a chromogen for 10 min. Finally, the tissue sections were counterstained with hematoxylin. Isotype-negative controls were represented by normal rabbit IgG isotype control (Novus Biologicals) diluted at concentrations equivalent to the primary antibodies. NOD-positive stained cells were counted from five randomly selected areas under high-power field (400 × magnification) using a microscope coupled to a digital camera (DMI4000B; Leica Microsystems, Wetzlar, Germany). The percentage of positive cells among the total cells was calculated for each selected area. The proportion of positive staining cells was scored as follows: negative (0, −), weak (<10%, +), moderate (10–50%, ++), or strong (>50%, +++).
Statistical analysis
Statistical analysis was performed using a statistical software package (SPSS version 20; SPSS, Inc., Chicago, IL, USA). The significance of associations between the groups was assessed using a nonparametric Mann–Whitney U test, and a P value of <0.05 was considered statistically significant.
Results
Histopathology
The histopathological characteristics were analyzed using the H&E-stained buccal mucosa samples. In the OLP group, H&E-stained slides showed a hyperkeratotic and acanthotic epithelium, which was further characterized via destruction of basal cell layer, exocytosis of lymphocytes in the epithelium, and a band-like infiltration of inflammatory cells (predominantly lymphocytes) in the lamina propria, all of which were consistent with OLP (Fig. 1).
Figure 1.
Histopathology of oral mucosal tissues stained with hematoxylin and eosin (A, B) normal oral mucosa (NOM); (C, D) oral lichen planus (OLP). Photomicrographs were obtained at 100 × magnification. Scale bar = 100 μm.
mRNA expression of NOD1 and NOD2 in NOM and OLP
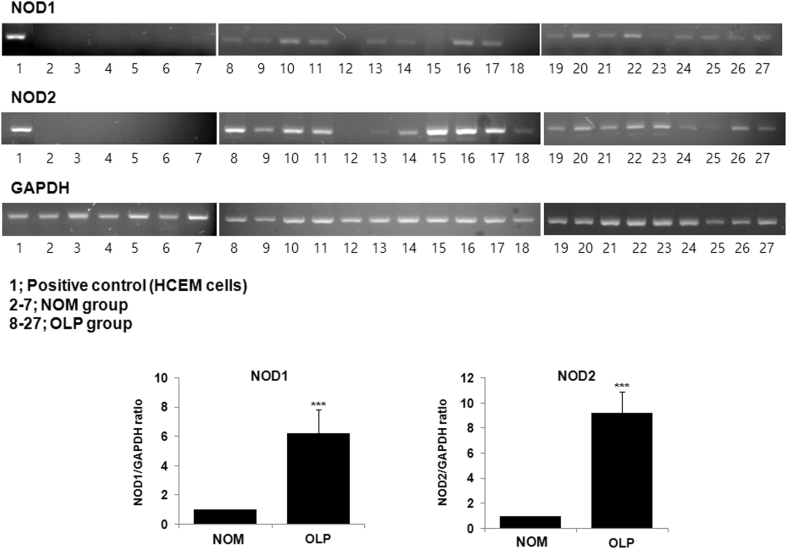
The expression of NOD1 and NOD2 genes was analyzed in the NOM and OLP groups using RT-PCR. Human cementoblast (HCEM) cells were used as a positive control. As shown in Fig. 2, NOD1 and NOD2 were significantly expressed in the OLP group, whereas neither gene was expressed significantly in the NOM group (P < 0.001). In particular, a strong expression of NOD2 was observed in the OLP sample. These findings demonstrated a significant relationship between NOD and OLP.
Figure 2.
Gene expression analysis of nucleotide-binding oligomerization domain (NOD) 1 and NOD2. Total RNAs were extracted from individual tissues. cDNA was synthesized using RT-PCR. HCEM cells were used as a positive control. 1, Positive control (HCEM cells); 2–7, normal oral mucosa (NOM) group; 8–27, oral lichen planus (OLP) group. The levels of gene expression are presented relative to GAPDH within each sample. Data are shown as median with interquartile range. ***P < 0.001 compared with the NOM group using Mann–Whitney U test.
Immunohistochemical analysis of NOD1 and NOD2 in NOM and OLP
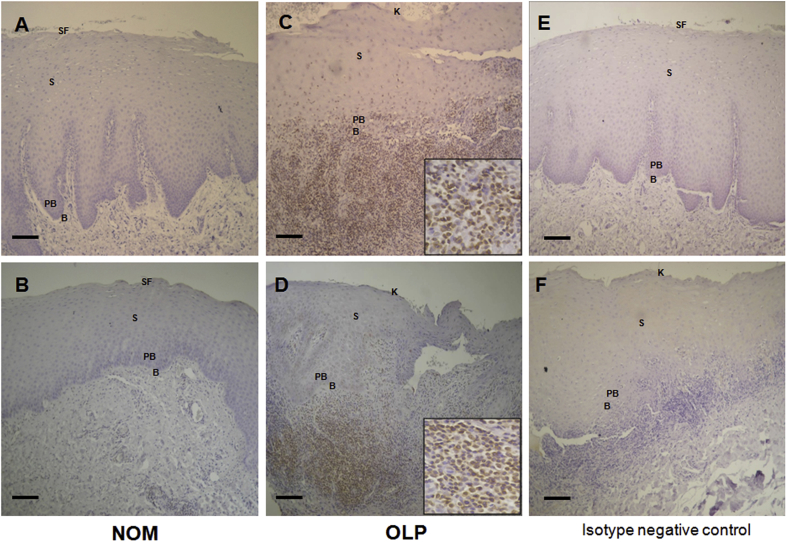
To measure the levels of NOD1 and NOD2 proteins, immunohistochemistry was performed in the NOM and OLP groups. As shown in Fig. 3, moderate and high expression of NOD1 was observed in the NOM and the OLP groups, respectively. Moreover, the expression of NOD1 was observed in the basal and parabasal layers in both the NOM (mild) and the OLP (moderate) groups. The expression of NOD1 in the OLP group was marginally higher than in the NOM group; however, the differences were not significant. Moreover, no expression of NOD1 in the lymphocytes was observed in the OLP group. The expression of NOD2 was markedly increased in the OLP group; however, almost no expression was found in the NOM group (Fig. 4). Compared with the NOM group, a mild expression of NOD2 in the basal and parabasal layers (P < 0.05) and a strong expression of NOD2 in the infiltrating lymphocytes of the submucosal layer (P < 0.001) were observed in the OLP group. The differences in the expression of NOD1 and NOD2 are summarized in Table 1.
Figure 3.
Immunohistochemical analysis of nucleotide-binding oligomerization domain (NOD) 1 expression (A, B) normal oral mucosa (NOM); (C, D) oral lichen planus (OLP); (E) isotype negative control of NOM; (F) isotype negative control of OLP. No signal is detected in the negative control sections using normal rabbit IgG. Photomicrographs were obtained at 100 × magnification. B: Basal layer; PB: Parabasal layer; S: Spinous layer; SF: Superficial layer; K: Keratinized layer. Scale bar = 100 μm.
Figure 4.
Immnohistochemical analysis of nucleotide-binding oligomerization domain (NOD) 2 expression (A, B) normal oral mucosa (NOM); (C, D) oral lichen planus (OLP); (E) isotype negative control of NOM; (F) isotype negative control of OLP. No signal was detected in the negative controls using normal rabbit IgG. Photomicrographs were obtained at 100 × magnification (Insert x 400). B: Basal layer; PB: Parabasal layer; S: Spinous layer; SF: Superficial layer; K: Keratinized layer. Scale bar = 100 μm.
Table 1.
Expression of nucleotide-binding oligomerization domain (NOD) 1 and NOD2 in normal oral mucosa (NOM) and oral lichen planus (OLP).
| NOM group | % of positive cells | OLP group | % of positive cells | |||
|---|---|---|---|---|---|---|
| NOD1 | Spinous and superfical layer | 44.0 ± 16.0 | ++ | Spinous and keratinized layer | 62.4 ± 12.8 | +++ |
| Basal and parabasal layer | 8.2 ± 4.7 | + | Basal and parabasal layer | 12 ± 5.7 | ++ | |
| Lymphocytes | 0.0 ± 0.0 | – | Lymphocytes | 0.0 ± 0.0 | – | |
| NOD2 | Spinous and superfical layer | 0.0 ± 0.0 | – | Spinous and keratinized layer | 0.0 ± 0.0 | – |
| Basal and parabasal layer | 0.0 ± 0.0 | – | Basal and parabasal layer | 7.9 ± 3.1* | + | |
| Lymphocytes | 0.0 ± 0.0 | – | Lymphocytes | 84.2 ± 6.6*** | +++ | |
−; negative (0), +; mild (<10%), ++; moderate (10%–50%), +++; strong (>50%).
Significant comparison of NOM and OLP (*, P < 0.05; ***, P < 0.001, Mann–Whitney U test).
Immunohistochemical analysis of NOD1 and NOD2 in IFH
Using immunohistochemistry, the expression of NOD1 and NOD2 was measured in IFH tissues. As shown in Fig. 5, the faint expression of NOD1 in the mucosal layer was partly observed in the IFH group. However, no expression of NOD2 was detected in the mucosal layer or the infiltrating lymphocytes in IFH tissues.
Figure 5.
Immnohistochemical analysis of nucleotide-binding oligomerization domain (NOD) 1 and NOD2 in inflammatory fibrous hyperplasia (IFH) tissues (A, B) expression of NOD1; (C, D) expression of NOD2; (E, F) isotype negative control. No signal was detected in the negative controls using normal rabbit IgG. Photomicrographs were obtained at 100 × magnification. B: Basal layer; PB: Parabasal layer; S: Spinous layer; K: Keratinized layer. Scale bar = 100 μm.
Discussion
OLP is a relatively common condition, with severe forms causing persistent mucosal erosions and ulceration. Although its pathogenesis remains unclear, an immune-mediated response is typically considered. NOD function has been described variety of diseases, including chronic inflammatory and autoimmune diseases as well as cancer.19,20 Recent studies investigating NOD expression and function in the oral epithelium have reported that NOD plays an important role in the maintenance of homeostasis and progression of inflammatory oral diseases (e.g., pulpitis and periodontitis) via innate immune system.21, 22, 23 In the present study, the expression of NOD1 and NOD2 in OLP was investigated. First, the mRNA expression of NOD1 and NOD2 was examined using RT-PCR. The expression of NOD1 and NOD2 was highly upregulated in the OLP group but not in the NOM group, which was consistent with previous studies investigating this expression in other inflammatory oral diseases, suggesting that the expression of NOD may be related to OLP pathogenesis.
Immunohistochemistry was used to elucidate the levels of protein expression and the location of NOD1 and NOD2 in the NOM and OLP groups. Previous studies have reported the widespread expression of NOD1 in many cell types and tissues in vivo.5 In the present study, the protein expression of NOD1 was observed in both NOM and OLP tissues, unlike the mRNA expression of NOD1, and the level of NOD1 was marginally increased in OLP compared with NOM. The expression of NOD1 in NOM is consistent with a previous study that reported a robust expression of NOD1 in normal oral epithelial tissues.21 Bacterial infection is one proposed etiological factor of OLP.24,25 NOD1 mediates the recognition of peptidoglycan derived primarily from gram-negative bacteria. A previous study has shown that oral epithelial cells interact with bacteria in the normal flora of oral mucosal tissues. Another study found that although the expression of NOD1 was upregulated in the oral epithelial cells, the production of pro-inflammatory cytokines was not increased.26 Therefore, it appears that the expression of NOD1 is functionally activated in NOM. Although the total amount of bacteria detected within the epithelia of OLP tissues did not differ from the level in control tissues, the levels of bacteria detected within the basal layer of the epithelia was significantly increased in OLP tissues.25 In the present study, a marginally higher expression of NOD1 was observed in OLP compared with NOM in the mucosal layer; however, no significant differences were found between the NOM and OLP groups. Therefore, the induction of NOD1 may be related to increased bacterial invasion of oral mucosa in OLP; however, it might not be functionally involved in OLP. Further studies are needed to investigate the role of NOD1 in OLP.
NOD2 has been found in the oral cavity, macrophages, and keratinocytes and is highly expressed in normal oral epithelia.5,21 Compared with previous studies, the present study showed no expression of NOD2 in NOM tissues. In addition to the role of NOD in the innate immune response to bacterial infections, mounting evidence suggests that the expression of NOD1 and NOD2 influences adaptive immune response.27 Histopathologically, OLP is characterized by band-like dense lymphocytic infiltration at the interface between the mucosa and submucosa.24,25 The infiltrated lymphocytes in OLP include mainly CD4+ and CD8+ T cells.28 Recent studies suggest that NOD1 and NOD2 influence T-cell differentiation by increasing cytokine synthesis by dendritic cells.29, 30, 31 NOD2 is functionally active in the T cells of humans, and the expression of NOD2 is higher in activated/memory CD4+ T cells as well as is inducible following T-cell receptor ligation.32 In the present study, intense expression of NOD2, such as mRNA expression, and site-specific induction were observed in OLP tissues. In the OLP group, a significant expression of NOD2 was detected in lymphocytes and a weak expression in the basal and parabasal layers. However, no expression was observed in the mucosal layer (spinous and keratinized layer). Therefore, the results suggest that NOD2 is upregulated in infiltrating lymphocytes and can contribute to the triggering system for the inflammatory response of T cells in OLP. To confirm whether OLP is characterized by an intense expression of NOD2 in lymphocyte infiltration, the expression of NOD2 was analyzed in IFH via immunohistochemistry. OLP and IFH are oral mucosal lesions with similar histopathological characteristics, such as intense subepithelial inflammatory infiltration.33 The result demonstrated that no NOD2 was expressed within the infiltrating lymphocytes in IFH, suggesting that the induction of NOD2 in infiltrating lymphocytes was a characteristic feature of OLP.
In conclusion, the results of the present study suggest that the strong expression of NOD2 in infiltrating lymphocytes plays an important etiological role in the pathogenesis of OLP. Therefore, NOD2 may represents a new diagnostic and therapeutic target in OLP. Further studies are needed to investigate the function of NOD in OLP as well as the treatment modalities available to regulate NOD expression and function.
Declaration of Competing Interest
The authors have no conflict of interest to declare.
Acknowledgements
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2012R1A1A2041317, NRF-2016R1D1A3B03931034, NRF-2016R1D1A1B01006388).
References
- 1.Au J., Patel D., Campbell J.H. Oral lichen planus. Oral Maxillofac Surg Clin N Am. 2013;25:93–100. doi: 10.1016/j.coms.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Sciubba J.J. Autoimmune oral mucosal diseases: clinical, etiologic, diagnostic, and treatment considerations. Dent Clin N Am. 2011;55:89–103. doi: 10.1016/j.cden.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Crincoli V., Di Bisceglie M.B., Scivetti M. Oral lichen planus: update on etiopathogenesis, diagnosis and treatment. Immunopharmacol Immunotoxicol. 2011;33:11–20. doi: 10.3109/08923973.2010.498014. [DOI] [PubMed] [Google Scholar]
- 4.Scully C., Carrozzo M. Oral mucosal disease: lichen planus. Br J Oral Maxillofac Surg. 2008;46:15–21. doi: 10.1016/j.bjoms.2007.07.199. [DOI] [PubMed] [Google Scholar]
- 5.Correa R.G., Milutinovic S., Reed J.C. Roles of NOD1 (NLRC1) and NOD2 (NLRC2) in innate immunity and inflammatory diseases. Biosci Rep. 2012;32:597–608. doi: 10.1042/BSR20120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson M.R., Kaminski J.J., Kurt-Jones E.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janardhanam S.B., Prakasam S., Swaminathan V.T. Differential expression of TLR-2 and TLR-4 in the epithelial cells in oral lichen planus. Arch Oral Biol. 2012;57:495–502. doi: 10.1016/j.archoralbio.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Inohara N., Chamaillard M., McDonald C. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 9.Philpott D.J., Girardin S.E. Nod-like receptors: sentinels at host membranes. Curr Opin Immunol. 2010;22:428–434. doi: 10.1016/j.coi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Chamaillard M., Hashimoto M., Horie Y. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 11.Girardin S.E., Boneca I.G., Carneiro L.A. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 12.Ohno S., Tateishi Y., Tatemoto Y. Enhanced expression of Toll-like receptor 2 in lesional tissues and peripheral blood monocytes of patients with oral lichen planus. J Dermatol. 2012;38:335–344. doi: 10.1111/j.1346-8138.2010.00956.x. [DOI] [PubMed] [Google Scholar]
- 13.Siponen M., Kauppila J.H., Soini Y. TLR4 and TLR9 are induced in oral lichen planus. J Oral Pathol Med. 2012;41:741–747. doi: 10.1111/j.1600-0714.2012.01169.x. [DOI] [PubMed] [Google Scholar]
- 14.Franchi L., Park J.H., Shaw M.H. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 15.Clarke T.B., Davis K.M., Lysenko E.S. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niess J.H., Klaus J., Stephani J. NOD2 polymorphism predicts response to treatment in Crohn's disease - first steps to a personalized therapy. Dig Dis Sci. 2012;57:879–886. doi: 10.1007/s10620-011-1977-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Meij E.H., van der Waal I. Lack of clinicopathologic correlation in the diagnosis of oral lichen planus based on the presently available diagnostic criteria and suggestions for modifications. J Oral Pathol Med. 2003;32:507–512. doi: 10.1034/j.1600-0714.2003.00125.x. [DOI] [PubMed] [Google Scholar]
- 18.Ahn M.Y., Yoon H.E., Park J.H. Characterization of NODs and TLRs in innate immune response of human cementoblast cells. Oral Dis. 2013;19:374–380. doi: 10.1111/odi.12012. [DOI] [PubMed] [Google Scholar]
- 19.Lange C., Hemmrich G., Klostermeier U.C. Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol. 2011;28:1687–1702. doi: 10.1093/molbev/msq349. [DOI] [PubMed] [Google Scholar]
- 20.Antosz H., Osiak M. NOD1 and NOD2 receptors: integral members of the innate and adaptive immunity system. Acta Biochim Pol. 2013;60:351–360. [PubMed] [Google Scholar]
- 21.Sugawara Y., Uehara A., Fujimoto Y. Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. J Dent Res. 2006;85:524–529. doi: 10.1177/154405910608500609. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y.Y., Chan C.H., Hung S.L. Up-regulation of nucleotide-binding oligomerization domain 1 in inflamed human dental pulp. J Endod. 2011;37:1370–1375. doi: 10.1016/j.joen.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Jeon D.I., Park S.R., Ahn M.Y. NOD1 and NOD2 stimulation triggers innate immune responses of human periodontal ligament cells. Int J Mol Med. 2012;29:699–703. doi: 10.3892/ijmm.2012.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roopashree M.R., Gondhalekar R.V., Shashikanth M.C. Pathogenesis of oral lichen planus - a review. J Oral Pathol Med. 2010;39:729–734. doi: 10.1111/j.1600-0714.2010.00946.x. [DOI] [PubMed] [Google Scholar]
- 25.Choi Y.S., Kim Y., Yoon H.J. The presence of bacteria within tissue provides insights into the pathogenesis of oral lichen planus. Sci Rep. 2016;6:29186. doi: 10.1038/srep29186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uehara A., Fujimoto Y., Fukase K. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol. 2007;44:3100–3111. doi: 10.1016/j.molimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Caruso R., Warner N., Inohara N. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan K., Hegarty A.M., Hodgson T. Aetiology, diagnosis and treatment of oral lichen planus. Br J Hosp Med. 2014;75:492–496. doi: 10.12968/hmed.2014.75.9.492. [DOI] [PubMed] [Google Scholar]
- 29.Fritz J.H., LeBourhis L., Sellge G. Nod1- mediated innate immune recognition of peptidoglycan contributes to the onset of adaptive immunity. Immunity. 2007;26:445–459. doi: 10.1016/j.immuni.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Liu D., Rhebergen A.M., Eisenbarth S.C. Licensing adaptive immunity by NOD-like receptors. Front Immunol. 2013;4:486. doi: 10.3389/fimmu.2013.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz H., Posselt G., Wurm P. TLR8 and NOD signaling synergistically induce the production of IL-1beta and IL-23 in monocyte-derived DCs and enhance the expression of the feedback inhibitor SOCS2. Immunobiology. 2013;218:533–542. doi: 10.1016/j.imbio.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Zanello G., Goethel A., Forster K. Nod2 activates NF-kB in CD4+ T cells but its expression is dispensable for T cell-induced colitis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monteiro B.V., Pereira Jdos S., Nonaka C.F. Immunoexpression of Th17-related cytokines in oral lichen planus. Appl Immunohistochem Mol Morphol. 2015;23:409–415. doi: 10.1097/PAI.0000000000000096. [DOI] [PubMed] [Google Scholar]