Abstract
In countries where malaria transmission has decreased substantially, thanks to the scale-up of control interventions, malaria elimination may be feasible. Nevertheless, this goal requires new strategies such as the active detection and treatment of infected individuals. As the detection threshold for the currently used diagnostic methods is 100 parasites/μL, most low-density, asymptomatic infections able to maintain transmission cannot be detected. Identifying them by molecular methods such as PCR is a possible option but the field deployment of these tests is problematic. Isothermal amplification of nucleic acids (at a constant temperature) offers the opportunity of addressing some of the challenges related to the field deployment of molecular diagnostic methods. One of the novel isothermal amplification methods for which a substantial amount of work has been done is the loop-mediated isothermal amplification (LAMP) assay. The present review describes LAMP and several other isothermal nucleic acid amplification methods, such as thermophilic helicase-dependent amplification, strand displacement amplification, recombinase polymerase amplification and nucleic acid sequence-based amplification, and explores their potential use as high-throughput, field-based molecular tests for malaria diagnosis.
Keywords: malaria elimination, isothermal amplification, PCR, tHDA, LAMP, NASBA, SDA, RPA
Introduction
Detecting Plasmodium parasites in peripheral blood, in both febrile patients and asymptomatic carriers, is essential for any malaria control programme aiming at decreasing local transmission and achieving the pre-elimination status.1 The WHO recently launched a global initiative known as T3 (‘Test. Treat. Track.’), aimed at supporting malaria-endemic countries in their efforts to achieve universal coverage with diagnostic testing, antimalarial treatment and surveillance.2 Though malaria has been eliminated from Europe and North America with no other diagnostic tool than microscopy,3 this seems more difficult in tropical countries, including sub-Saharan Africa, because of the substantial number of individuals harbouring a malaria infection at extremely low densities, undetectable by microscopy.4 The currently available tools for diagnosing malaria include microscopy, parasite antigen/enzyme detection kits [commonly referred to as rapid diagnostic tests (RDTs)] and molecular tools (discussed in Cordray and Richards-Kortum5), each of them with specific advantages and limitations and with the potential of being deployed at different levels of the health system (microscopy and RDTs more peripherally than molecular tools).5,6 Indeed, microscopy and RDTs remain the only feasible options at health facility or lower level, e.g. community case management of malaria.6 These two diagnostic tools have limitations. For example, the absence of the pfhrp-2 gene (encoding the HRP-2 protein, to which most antigen detection tests are directed) in Plasmodium falciparum populations from some endemic areas resulted in a substantial proportion of false negatives.7 Moreover, both microscopy and RDTs cannot detect parasite densities <100 parasites/μL, particularly in field conditions, while asymptomatic carriers have a much lower parasite density.8 The latter can be identified by nucleic acid amplification tests (NAATs), often by PCR, one of the most widely used molecular methods for the detection and identification of infectious diseases. The detection limit of PCR, which is ∼1 parasite/μL depending on the assay type,9,10 is lower than either RDTs or microscopy and PCR is therefore able to detect asymptomatic malaria carriers who may be targeted for treatment.11 In some settings, asymptomatic sexual and asexual stages of P. falciparum infections may persist and sustain transmission at very low parasite densities, below the threshold of detection by microscopy or RDTs and in reach only of molecular methods such as PCR.12–14 However, PCR-based assays are the least feasible to perform in field settings as they are prone to contamination, heavily influenced by the purity of the sample nucleic acid content thus relying on purified extracts and requiring cold storage facilities for reagents.5,15,16 Therefore, until recently, molecular diagnostic methods required specialized equipment and personnel and could not be carried out outside reference facilities. The possibility of amplifying DNA at isothermal temperatures without the need of a thermocycling apparatus has created the opportunity of performing molecular diagnostic tests at a more peripheral level17 and thus of improving the management of infectious diseases, especially in resource-limited settings.15,16,18 Recent guidelines published by WHO recommend that diagnostic devices for resource-limited settings should be ASSURED: Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free and Deliverable to end users.19,20 This paper reviews some molecular isothermal amplification techniques (with the ASSURED criteria in mind) with the aim of determining their possible deployment for high-throughput detection of asymptomatic P. falciparum carriers in the field, e.g. in mass screening and treatment campaigns.
Sample preparation for field-deployable molecular assays
Preparing the sample for molecular assays is a bottleneck for NAATs because it involves lengthy processes, often performed manually.18,21,22 DNA extraction for molecular assays is a critical step, with different methods resulting in different yield and quality of the nucleic acid.23,24 One of the major problems, particularly when processing large quantities of samples, is the inhibition of the PCR by the haem (from red blood cells) obtained from the crude DNA extraction.25 Ideally, the DNA extraction method should have the following characteristics: rapid preparation and high throughput; high reliability; production of good-quality DNA for long-term storage; avoidance of cross-contamination; and reasonable costs.23
Several isothermal technologies may fulfil these requirements as they appear less affected by the inhibitory effect of blood products and have similar sensitivity and specificity as standard PCR when DNA is extracted using crude methods such as heat treatment.18,26,27
PCR adaptations for detection of malaria parasites in the field
New approaches for simplifying PCR for field settings, including assays that are less prone to inhibition, have been explored.28–30 The palm-held PCR device by Ahram Biosystems31 features three stages of specially structured heat blocks that are maintained at temperatures suitable for each of the three PCR steps and the sample is circulated across the high- and low-temperature zones inside the sample tube. Field PCR units and portable real-time PCR systems with freeze-dried reagents have been developed by Biofire Diagnostics for mobile analytical laboratories and field hospitals for pathogen detection.32 Lab-on-chip, point-of-care diagnostic assays are also being designed for the diagnosis of disease pathogens.33 However, these assays are still relatively expensive for resource-limited settings and are not entirely infrastructure-free. Post-amplification detection is also being simplified with the development of PCR–nucleic acid lateral-flow immune assays.34
Isothermal amplification of nucleic acids
Based on new findings on DNA/RNA synthesis, non-PCR-based methods of nucleic acid amplification involving the use of accessory proteins and mimicking in vitro nucleic acid amplification have been developed.35 Isothermal approaches can facilitate rapid target amplification through single-temperature incubation, reducing the system complexity compared with PCR-based methods. Established isothermal amplification methods have different degrees of complexity (multiple enzymes or primers) and also varying sensitivity and specificity.36 A major advantage of isothermal amplification methods is the simplicity of endpoint determination, often by visual observation, which allows assays to be run in a closed system, thus reducing the risk of post-amplification contamination.20,37,38 The choice of which reaction to use is primarily driven by the target of interest to be amplified. Amplification time, reaction temperature, tolerance to substances in biological samples, length of the target, initial heat denaturation and complexity (i.e. the number of enzymes and primers required) are all factors to be considered when choosing an appropriate method.39 In the past two decades, several isothermal amplification methods for detecting pathogens, including malaria parasites, have been used (Table 1).
Table 1.
Isothermal molecular-based tests for diagnosis of pathogens
| Isothermal amplification assay | Pathogen | Isothermal condition | Gold standard/ref test | Sensitivity or limit of detection | Specificity | Reference | |
|---|---|---|---|---|---|---|---|
| 1 | tHDA | Clostridium difficile | 65°C for 60 min | PCR | 100% | 100% | 79 |
| 2 | tHDA | herpes simplex virus | 64°C for 60 min | ELVIS shell vial assay | 100% | 96.30% | 78 |
| 3 | tHDA | H. pylori | 95°C for 2 min; 60°C for 1 h | culture; histology | 90%; 96.6% | 95.7%; 96.8% | 74 |
| 4 | tHDA | H. pylori | 95°C for 2 min; 60°C for 1 h | culture; histology | 92.5%; 100% | 95.4%; 98.8% | 75 |
| 5 | tHDA | Staphylococcus aureus; MRSA | 95°C for 5 min; 60°C for 1 h | biochemical and genotyping methods | 100%; 100% | 100%; 98% | 76 |
| 6 | tHDA | Ebola virus | 65°C for 120 min | ND | 0.2 pg of total human RNA for GAPDH detection; 3.2 copies of Ebola virus-armoured RNA | ND | 73 |
| 7 | tHDA | HIV | 65°C for 75 min | ND | 50 copies/assay | ND | 77 |
| 8 | tHDA | N. gonorrhoeae | 65°C for 60 min | Abbott CT/GC kit | 100% | 100% | 115 |
| 9 | tHDA | Plasmodium spp.; P. falciparum; P. vivax | 64°C for 90 min | microscopy/NASBAMT | 97% | 100% | 81 |
| 10 | LAMP | P. falciparum | 60°C for 120 min | PCR | 95% | 99% | 26 |
| 11 | LAMP | P. falciparum | 65°C for 120 min | PCR | 76%–79% | 58%–89% | 45 |
| 12 | LAMP | Plasmodium spp. | 60°C for 100 min; inactivation 80°C for 2 min | microscopy | 98.50% | 94.30% | 46 |
| 13 | LAMP | P. falciparum | 63°C for 90 min | microscopy | 97.80% | 85.70% | 44 |
| 14 | LAMP | Plasmodium spp. | 63°C for 90 min | microscopy; PCR | 96.7%; 98.9% | 91.7%; 100% | 6 |
| 15 | LAMP | Plasmodium spp. | 60°C for 100 min; inactivation 80°C for 2 min | microscopy | 98.30% | 100% | 116 |
| 16 | LAMP | Plasmodium spp.; P. falciparum | 65°C for 40 min; inactivation 80°C for 5 min | PCR | 93.9%; 93.3% | 100%; 100% | 25 |
| 17 | LAMP | P. falciparum; P. vivax | 60°C for 60 min | composite ref (LAMP, Mx, PCR) | 100%; 100% | 100%; 100% | 58 |
| 18 | LAMP | Plasmodium spp.; P. falciparum | 65°C for 40 min; inactivation 80°C for 5 min | nPCR | 98.4%; 97% | 98.1%; 99.2% | 59 |
| 19 | LAMP | P. falciparum | 65°C for 90 min; inactivation 80°C for 5 min | RT–PCR | 100%; 100% | 98.1%; 100% | 47 |
| 20 | NASBA | Trypanosoma brucei | 65°C for 2 min; 41°C for 92 min | microscopy | 10 parasites/mL | 100% | 117 |
| 21 | NASBA | Leishmania | 65°C for 2 min; 41°C for 92 min | microscopy | 93.3%; 98.6% | 100%; 100% | 118 |
| 22 | NASBA | Plasmodium spp. | 65°C for 4 min; 41°C for 2 h | microscopy | 97.40% | 80.90% | 67 |
| 23 | NASBA | Plasmodium spp. | 65°C for 4 min; 41°C for 2 h | microscopy | 100% | 94% | 68 |
| 24 | NASBA | astrovirus | 65°C for 5 min; 41°C for 95 min | RT–PCR | 66 | ||
| 25 | NASBA | P. falciparum | 65°C for 4 min; 41°C for 2 h | microscopy | 119 | ||
| 26 | NASBA | P. falciparum | 65°C for 4 min; 41°C for 2 h | real-time QT-PCR | 69 | ||
| 27 | NASBA | M. tuberculosis | 42°C for 1 h | culture | 85.70% | 95.50% | 120 |
| 28 | NASBA | hepatitis A virus | 65°C for 5 min; 40 ± 1°C for 150 min | ND | 1 pfu | ND | 121 |
| 29 | RPA | MRSA | 37°C for 60 min | ND | 2 copies/rxn | ND | 87 |
| 30 | RPA | MRSA | 39°C for 1 h | ND | ND | 88 | |
| 31 | RPA | RVFV | 42°C for 20 min | ND | 102 | ||
| 32 | SDA | M. tuberculosis | 95°C for 2 min; 40°C for 2 h | culture and smear at positive threshold of 2.4 and 15.5 M. tuberculosis organisms per rxn | 100%; 95% | 84%; 96% | 122 |
| 33 | SDA | M. tuberculosis | BD ProbeTec™ ET System | culture | 96.10% | 100% | 111 |
| 34 | SDA | E. coli | 95°C for 2 min; 53°C for 35 min; 95°C for 5 min | PCR | 4.3 cfu/5 μL | 100% | 110 |
| 35 | SDA | N. gonorrhoeae; C. trachomatis | BD ProbeTec™ ET System | composite ref (PCR and LCR) | 90%; 95% | 100%; 100% | 123 |
| 36 | SDA | N. gonorrhoeae; C. trachomatis | 52.5°C for 1 h | culture or composite ref of any two molecular methods (PCR, SDA, LCR) | 100%; 92% | 100%; 99% | 124 |
| 37 | SDA | N. gonorrhoeae; C. trachomatis | BD ProbeTec™ ET System | composite ref of two or three (PCR, SDA, TMA) | 97.1% or 100%; 92.2% or 100% | 98.8% or 96%; 96.47% or 89.6% | 125 |
| 38 | SDA | N. gonorrhoeae; C. trachomatis | BD ProbeTec™ ET System | 100%; 95.3% | 99.7%; 99.3% | 126 | |
| 39 | SDA | M. tuberculosis | BD ProbeTec™ ET System | culture | 77.80% | 97.70% | 109 |
| 40 | SDA | N. gonorrhoeae | BD ProbeTec™ ET System | microscopy | 98.10% | 100% | 127 |
| 41 | SDA | N. gonorrhoeae; C. trachomatis | BD ProbeTec™ ET System | culture | 96.2%; 92% | 98.4%; 94.9% | 128 |
GADPH, glyceraldehyde-3-phosphate dehydrogenase; LCR, ligase chain reaction; Mx, microscopy; NASBAMT, NASBA malaria test; ND, not determined; rxn, reaction.
Isothermal amplification methods currently used in malaria diagnosis
Loop-mediated isothermal amplification (LAMP)
LAMP, first described in 2000,40 can be performed with simplified and inexpensive specimen processing, under isothermal conditions in a simple heating block or water bath. Furthermore, it can be formatted for visual detection without the need for instrumentation, which is a major advantage for its field deployment.41,42 LAMP is a one-step amplification reaction that employs self-recurring strand-displacement synthesis primed by a specially designed set of primers identifying six distinct sequences on the target DNA.40 Detailed reviews (principles, primer set-up and design) are available elsewhere.37,38,41–43 At costs ranging from USD <1.00/test (using ready-made reaction mixture prepared with individual reagents) to USD 5.31/test (using commercially available reaction mixture),38 LAMP assays proffer to be cheaper than PCR assays. However, one major shortfall of the LAMP assay is the complexity in primer and assay design and optimization (Table 2).
Table 2.
Comparison between established PCR and novel isothermal amplification assays
| PCR | LAMP | NASBA | tHDA | RPA | SDA | ||
|---|---|---|---|---|---|---|---|
| 1 | sample processing prior to amplification | nucleic acid extraction required | amplification from crude samples possible | nucleic acid extraction required | amplification from crude samples possible | nucleic acid extraction required | amplification from crude samples possible |
| 2 | cost | USD 7–8 per sample129 | USD <1–5.338 | equivalent to PCR | relatively cheaper than PCR | relatively cheaper than PCR | relatively cheaper than PCR |
| 3 | ease of use/simplicity of operation | complex | relatively easier to set up than PCR | relatively easier to set up than PCR | relatively easier to set up than PCR | relatively easier to set up than PCR | relatively easier to set up than PCR |
| 4 | skill/training required | high | moderate | high | moderate | moderate | moderate |
| 5 | stability of reagents | cold chain required for enzymes | cold chain required for enzymes | cold chain required for enzymes | cold chain required for enzymes | reagents available as dry pellets | cold chain required for enzymes |
| 6 | amplification time | ∼2 h | 30–60 min | ∼2 h | ∼60 min | ∼60 min | 30–60 min |
| 7 | simplicity of design | complex primer design and assay optimization | complex primer design and assay optimization | complex primer design and assay optimization | complex primer design and assay optimization | complex primer design and assay optimization | complex primer design and assay optimization |
| 8 | principle | high temperature and thermostable polymerase | thermophilic strand displacement polymerase | reverse transcription and strand displacement polymerase | helicase and thermophilic strand displacement polymerase | recombinase–polymerase complex | thermophilic strand displacement polymerase |
| 9 | test temperature | varying | 65°C | 41°C | 60–65°C | 37°C | 40°C |
| 10 | risk of contamination | potential risk minimized by proper set up | potential risk minimized by proper set up | potential risk minimized by proper set up | potential risk minimized by proper set up | potential risk minimized by proper set up | potential risk minimized by proper set up |
| 11 | quality assurance control | possible | possible | possible | possible | possible | possible |
| 12 | post-amplification detection | electrophoresis; fluorescence detection | naked eye; turbidity measurement; electrophoresis | fluorescence detection | lateral-flow strip; electrophoresis; fluorescence detection | fluorescence detection; electrophoresis | fluorescence detection; electrophoresis |
| 13 | sensitivity | high | high | high | high | high | high |
| 14 | specificity | high | high | high | high | high | high |
| 15 | limit of detection (Plasmodium) | 1–5 parasites/μL | 1–5 parasites/μL | <1 parasite/μL | unknown | unknown | unknown |
| 16 | Plasmodium species identification | yes | yes | yes | yes | possible | possible |
| 17 | identification of sexual and asexual forms | yes | yes | yes | possible | possible | possible |
| 18 | high throughput | yes | yes | yes | yes | yes | yes |
| 19 | instrumentation requirement | thermocycler | heating block or water bath | real-time cycler | heating block or water bath | real-time cycler; heating block or water bath | real-time cycler; heating block or water bath |
| 20 | infrastructure requirement | electricity | electricity and exothermal chemical devices | electricity | electricity | electricity | electricity |
| 21 | field tested | no | yes | yes | yes | no | no |
| 22 | product developer | various | Eiken Group, Japan | Cangene Corporation, Canada | Biohelix Corporation, USA | TwistDx, USA | Becton Dickinson and Co., USA |
The use of LAMP for the diagnosis of various diseases, including human malaria, has been investigated extensively.6,25,26,44–57 A lower limit of detection in the range of 5–10 parasites/μL has been reported,25,46 while sensitivity and specificity, when compared with an 18S rRNA gene PCR assay using crude DNA extracts, were 95% and 99%, respectively (Table 1).26 However, a subsequent study using the same PCR and LAMP protocols reported much lower estimates of sensitivity and specificity, i.e. 76% and 90%, respectively.45 Taking microscopy as the gold standard and with different primer sets targeting the same gene, the sensitivity and specificity were 96% and 94%, respectively.46 In 130 field samples collected in Thailand and using a composite reference diagnosis for each sample, i.e. two out of three tests [microscopy, nested PCR (nPCR) and LAMP] giving the same result, LAMP had 100% sensitivity and specificity for P. falciparum infection.58 With primers targeting the mitochondrial genome and nPCR assay as the reference test, the sensitivity and specificity were 93% and 100%, respectively.25 In a recent study in Uganda, the sensitivity and specificity of LAMP P. falciparum primers were 98% and 98%, respectively, and 97% and 99% for the genus primers.59 In a recent comparison of an optimized LAMP protocol against a highly sensitive three-well nPCR reference assay (in which amplification in any well of three replicates per sample was counted as positive), LAMP showed a sensitivity of 90% compared with 51% for microscopy.60 LAMP has also been reported for the detection of other Plasmodium species including Plasmodium vivax,46,61Plasmodium malariae, Plasmodium ovale,46 human Plasmodium knowlesi infection62,63 and gametocytes by reverse transcription (RT–LAMP),47 showing the ease of adaptability of this method.
Nucleic acid sequence-based amplification (NASBA)
NASBA, first described by Kievits et al.64 in 1991, is a homogeneous, isothermal nucleic acid amplification method that is particularly suited to RNA targets in a double-stranded DNA background. A cocktail of three enzymes (reverse transcriptase, T7 RNA polymerase and RNase H) acting in concert allows the rapid amplification of target sequences by >108-fold without the use of expensive thermal-cycling equipment, the end product being a single-stranded RNA antisense to the original RNA template.65 Since there is no DNA denaturation step in NASBA, contaminating genomic or proviral DNA (the precursor or latent form of a virus integrated into the genetic material of a host cell) is not amplified. However, the extent of the reaction cannot be controlled by adjusting the number of cycles and the likelihood of non-specific interactions is increased because the amplification temperature cannot exceed 41°C without the risk of enzymatic denaturation.66
The reliability of the NASBA process has been tested by sequencing the RNA product directly from a NASBA reaction, with 90% of the sequences readable, an excellent result considering the AT richness of the P. falciparum genome.65 For detection and semi-quantification of malaria parasites and species identification, NASBA, compared with microscopy, had a sensitivity of 97% and specificity of 81%.67 Another study detecting products of NASBA amplification by electrochemiluminescence reported a sensitivity of 100% and specificity of 94%.68 Schneider et al.69 reported a significant correlation between parasite quantification results by real-time quantitative NASBA (QT-NASBA) and real-time quantitative PCR (QT-PCR). QT-NASBA has also been used to determine gametocyte carriage (prevalence and density) with the ability of detecting gametocyte densities as low as 0.02–0.1/μL.69,70 The cost of NASBA assays has not been determined. However, considering that NASBA requires similar consumables and infrastructure as PCR, it is expected that assay costs would be similar (Table 2).
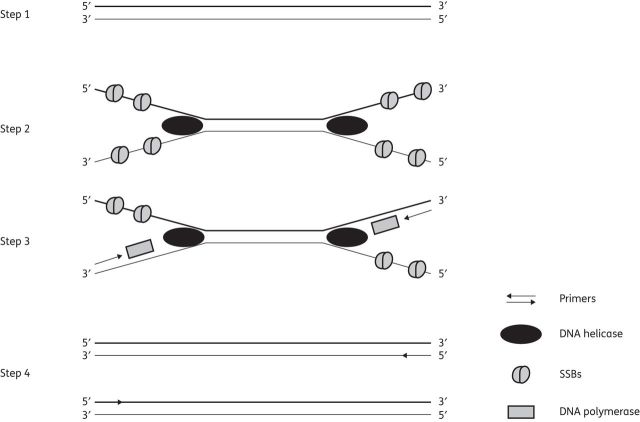
Thermophilic helicase-dependent amplification (tHDA)
In this system, which was first described in 2004,71 strands of duplex DNA are separated by a DNA helicase and coated by single-stranded DNA-binding proteins (SSBs). Sequence-specific primers hybridize to each border of the target DNA and DNA polymerases extend the primers annealed to the templates to produce a double-stranded DNA (Figure 1). The two newly synthesized double-stranded DNA products are subsequently used as substrates by DNA helicases, entering the next round of the reaction. Thus, a simultaneous chain reaction proceeds, resulting in the exponential amplification of the selected target sequence.71 tHDA amplifies nucleic acid targets efficiently at 65°C and requires fewer protein components than the mesophilic HDA platform, which is performed at 37°C.72 However, because both methods are isothermal and do not require thermocycling, they present a relatively cheaper platform than PCR.
Figure 1.
Schematic representation of tHDA. Steps 1 and 2: DNA helicase binds to double-stranded DNA and begins unwinding while SSBs attach to stabilize the single strands. Step 3: sequence-specific primers bind and DNA polymerase synthesizes new DNA strands in the 5′→3′ direction. Step 4: duplex DNA strands formed serve as a template for another round of amplification.
The tHDA platform has been successfully used for the detection of several pathogens.73 When compared with culture methods for detection of Helicobacter pylori, tHDA-ELISA had a sensitivity and specificity of 90% and 96%, respectively, and a higher sensitivity (97%) and specificity (97%) when compared with histology.74 However, when using colorimetric detection with gold nanoparticle probes, the sensitivity was higher (93% compared with culture methods and 100% with histology) and specificity similar or higher (95% compared with culture methods and 99% with histology detection).75 The sensitivity and specificity for the detection of different pathogens was higher when amplified DNA was applied onto a vertical-flow strip embedded in a disposable cassette (Table 1).76–80 Recently, clinical and analytical performance of tHDA for Plasmodium detection and species-level identification in blood samples was reported, with overall sensitivity of 97% (95% CI, 87%–99%) and specificity of 100% (95% CI, 85%–100%).81
Adaption of the tHDA on a microarray platform can be carried out directly on the surface of a glass slide by immobilizing one primer on the glass substrate and leaving the corresponding primer in solution. With the helicase unwinding the DNA template, the resulting DNA single strand is able to anneal at the immobilized primer and will be subsequently elongated by the DNA polymerase. Labelling the corresponding primer with a reporter allows successful detection of amplified targets from the microarray platform by laser scanning or total internal reflection fluorescence technologies.82
Other isothermal amplification assays with potential for use in malaria diagnosis
In recent years, a vast array of isothermal amplification methods targeting DNA, RNA or both have been developed or used for the diagnosis of pathogens. These are T7 promoter-driven amplifications: transcription-mediated amplification (TMA),83 single primer isothermal amplification,84 strand displacement methods such as strand displacement amplification (SDA) and smart amplification (SmartAmp),85,86 recombinase polymerase amplification (RPA),87,88 isothermal and chimeric primer-initiated amplification of nucleic acids,89 self-sustained sequence replication reaction,90 exponential amplification reaction,91 cross-priming amplification,92,93 rolling circle amplification94 and the genome exponential amplification reaction technique.95 Table 2 summarizes the comparison of some important features between PCR and the isothermal amplification methods described in this review. Most of these assays require multiple enzymes (two or more), rigorous optimization, a heat source and post-amplification analysis.
Based on the WHO ASSURED criteria for diagnostic assays for resource-limited settings, two of the isothermal amplification methods, which are deemed more advanced towards field implementation, namely RPA and SDA, were selected for further discussion. Their suitability for deployment as field-based molecular tests for malaria parasite detection was based on assay chemistry, simplicity of design and operation, cost-efficiency and robustness.
RPA
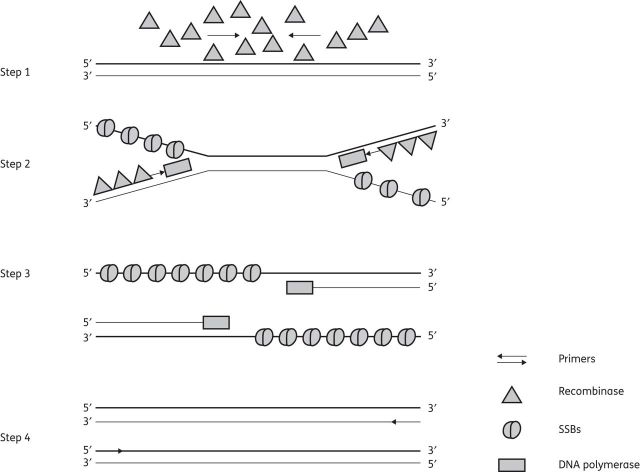
In RPA, the isothermal amplification of specific DNA fragments is achieved by the binding of opposing oligonucleotide primers to template DNA and their extension by a DNA polymerase. Global melting of the template is not required for the primers to be directed to their complementary target sequences. Instead, RPA employs recombinase–primer complexes to scan double-stranded DNA and facilitate strand exchange at cognate sites.96–98 The resulting structures are stabilized by SSBs interacting with the displaced template strand, thus preventing the ejection of the primer by branch migration (Figure 2).99 Recombinase disassembly leaves the 3′-end of the oligonucleotide accessible to a strand-displacing DNA polymerase, in this case the large fragment of Bacillus subtilis PolI (Bsu),100 and primer extension ensues. Exponential amplification is accomplished by the cyclic repetition of this process.
Figure 2.
Schematic representation of RPA. Step 1: primers and recombinases form a complex that targets a homologous DNA sequence. Step 2: DNA polymerase synthesizes a new strand by displacing the complementary strand (strand exchange). SSBs help stabilize the displaced single strands. Step 3: the parent DNA strand separates and synthesis continues to form two new duplex DNA strands. Step 4: duplex DNA strands formed serve as a template for another round of amplification.
Key to RPA is the establishment of a dynamic reaction environment that balances the formation and disassembly of recombinase–primer filaments.87 The reaction system is provided in a stabilized dried format, which may permit transportation and limited storage without refrigeration even though long-term storage under refrigeration is still recommended.101 RPA has also been recently reported in a SlipChip microfluidic platform that enables multistep manipulation in parallel with large numbers of small volumes, consisting of two plates containing wells and ducts that can be brought into contact and moved relatively to one another to manipulate fluids by creating and breaking fluidic paths.88 Recently, a highly sensitive isothermal RPA assay for the detection of Rift Valley fever virus (RVFV) RNA on a mobile device was published, though the assay is yet to be validated on clinical samples.102
SDA
SDA is based on the ability of a restriction enzyme to nick or cut the unmodified strand of a hemiphosphorothioate form of its recognition site as well as the ability of a DNA polymerase to initiate replication at the ‘cut site’ and displace the downstream non-template strand. Primers containing recognition sites for the nicking restriction enzyme bind to opposite strands of target DNA at positions flanking the sequence to be amplified. The target fragment is exponentially amplified by coupling sense and antisense reactions in which strands displaced from the sense reaction serve as a target for the antisense reaction and vice versa.103 The method consists of two parts (Figure 3): (i) a target generation process that makes copies of the target sequence flanked by enzyme restriction sites; and (ii) the exponential amplification of these modified target sequences by repeated nicking, strand displacement and priming of displaced strands.104
Figure 3.
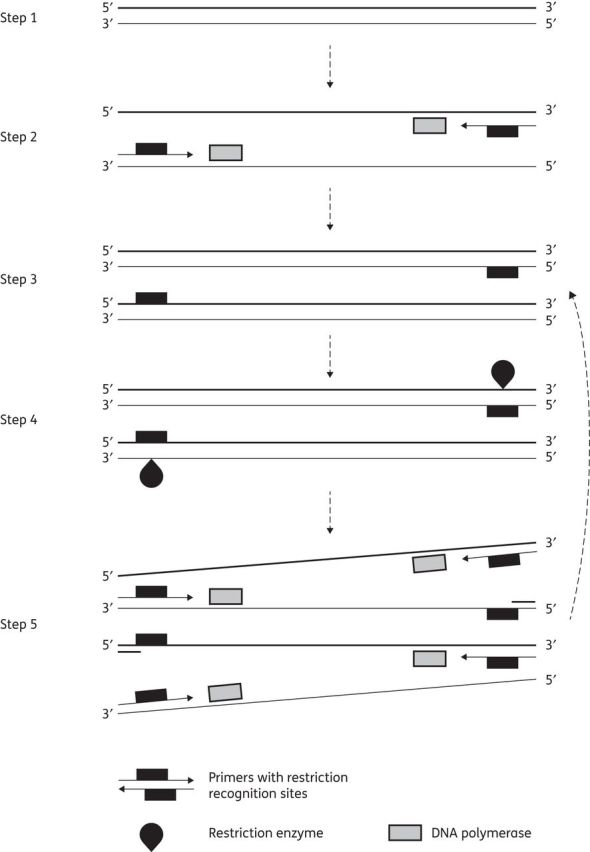
Schematic representation of SDA. Step 1: denaturation of double-stranded DNA. Steps 2 and 3: primers bind on each strand while DNA polymerase extends to produce double-stranded DNA with a modified (hemiphosphorodioate) recognition site. Step 4: a restriction enzyme cleaves the unmodified strand of the newly synthesized double-stranded DNA, displacing it. Step 5: DNA polymerase synthesizes a new strand from the cleaved DNA strand for another round of restriction digest and polymerization.
Despite the seemingly complicated sequence of events, SDA operates under a very simple protocol. Target DNA is heat denatured in the presence of all reagents except the restriction enzyme and polymerase; amplification then proceeds at 40°C after cooling and addition of the enzymes.85 Amplified products may then be detected by a variety of methods.105 The original SDA process was not very efficient and it has been improved by incorporating a thermostable polymerase and a different exonuclease to increase the yield and rate of amplification. These new conditions allow a 1010-fold amplification of target after 15 min at 60°C.106,107 SDA can also be used to detect RNA by incorporating a reverse transcription step.108 Varying sensitivities and specificities have been reported for diagnosis of Mycobacterium tuberculosis, Neisseria gonorrhoeae, Chlamydia trachomatis and enterohaemorrhagic Escherichia coli (Table 1).109–111
Discussion and conclusions
Isothermal amplification techniques have potential for the field diagnosis of malaria infection. They eliminate the need for a costly and power-intensive thermocycler, produce results in a short time (from 30 min to 1 h), can be used to process large numbers of samples required for active surveillance and are capable of detecting infections of <1 parasite/μL of blood, of both sexual and asexual stages.25,46,69,70,112 The sensitivity and specificity of these techniques are comparable to those of PCR-based diagnostics.5,37 However, it should be noted that isothermal amplification techniques, though relatively cheaper than PCR, are not totally equipment- or infrastructure-free but rather have a better potential for field deployment due to their simplified amplification conditions; thus, alternative heat sources are being explored.20
One major limitation for the field deployment of these isothermal amplification assays is the endpoint detection of amplified products. Real-time fluorescence or turbidity measurement is the most reliable method of post-amplification detection.18 With LAMP assays, positive samples can be visualized and identified with the naked eye as a result of the white precipitate of magnesium pyrophosphate formed during the reaction.41 Adaptation of the vertical-flow strip embedded in a disposable cassette or lab-on-chip portable devices for endpoint detection of amplified product with the isothermal amplification assays would be more user-friendly and field deployable.77,81,87
The Foundation for Innovative New Diagnostics has been working with the Hospital for Tropical Diseases in London and Eiken Chemical Company (Japan) in the development of a simplified LAMP assay for the diagnosis of malaria. Prototypes of this test have been compared with PCR using samples from febrile patients in two clinical trials, one in London (travellers) and the other in an endemic setting in Uganda.59,60,113 This places LAMP at the forefront of all the isothermal amplification assays with the potential to replace PCR in the nearest future. Commercial LAMP reaction kits have also been developed recently for numerous viral, bacterial and protozoan pathogens.37 The combination of LAMP into a ‘lab on a chip’ with diagnosis performed on a single-use device would offer a sensitive alternative to microscopy and RDTs.25,114
As the global malaria map continues to shrink and elimination is seriously being considered in certain territories, active detection of asymptomatic carriers may be scaled up in these regions. Therefore, a robust field-deployable, molecular-based assay that is able to give results comparable to laboratory-based assays, using crude sample sources and simple end product detection, would be the best approach to handle the large number of samples that would be generated. The development of isothermal amplification techniques has paved the way for this.
Transparency declarations
None to declare.
Acknowledgements
We would like to thank Professor Peter Verbruggen for adapting the figures in Illustrator.
References
- 1.malERA Consultative Group on Diagnoses and Diagnostics. A research agenda for malaria eradication: diagnoses and diagnostics. PLoS Med. 2011;8:e1000396. doi: 10.1371/journal.pmed.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2012. T3: Test. Treat. Track. Scaling up Diagnostic Testing, Treatment and Surveillance for Malariahttp://www.who.int/malaria/publications/atoz/test_treat_track_brochure.pdf .
- 3.Wongsrichanalai C, Barcus MJ, Muth S, et al. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT) Am J Trop Med Hyg. 2007;77:119–27. [PubMed] [Google Scholar]
- 4.Lindblade KA, Steinhardt L, Samuels A, et al. The silent threat: asymptomatic parasitemia and malaria transmission. Expert Rev Anti Infect Ther. 2013;11:623–39. doi: 10.1586/eri.13.45. [DOI] [PubMed] [Google Scholar]
- 5.Cordray MS, Richards-Kortum RR. Emerging nucleic acid-based tests for point-of-care detection of malaria. Am J Trop Med Hyg. 2012;87:223–30. doi: 10.4269/ajtmh.2012.11-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucchi NW, Demas A, Narayanan J, et al. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS One. 2010;5:e13733. doi: 10.1371/journal.pone.0013733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koita OA, Doumbo OK, Ouattara A, et al. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg. 2012;86:194–8. doi: 10.4269/ajtmh.2012.10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15:66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snounou G, Viriyakosol S, Zhu XP, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–20. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 10.Rougemont M, Van Saanen M, Sahli R, et al. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–43. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.malERA Consultative Group on Monitoring, Evaluation and Surveillance. A research agenda for malaria eradication: monitoring, evaluation, and surveillance. PLoS Med. 2011;8:e1000400. doi: 10.1371/journal.pmed.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nwakanma D, Kheir A, Sowa M, et al. High gametocyte complexity and mosquito infectivity of Plasmodium falciparum in the Gambia. Int J Parasitol. 2008;38:219–27. doi: 10.1016/j.ijpara.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Karl S, Gurarie D, Zimmerman PA, et al. A sub-microscopic gametocyte reservoir can sustain malaria transmission. PLoS One. 2011;6:e20805. doi: 10.1371/journal.pone.0020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO. 2012. Disease Surveillance for Malaria Controlhttp://apps.who.int/iris/bitstream/10665/44851/1/9789241503341_eng.pdf?ua=1 .
- 15.Peeling RW, Mabey D. Point-of-care tests for diagnosing infections in the developing world. Clin Microbiol Infect. 2010;16:1062–9. doi: 10.1111/j.1469-0691.2010.03279.x. [DOI] [PubMed] [Google Scholar]
- 16.Pai NP, Vadnais C, Denkinger C, et al. Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries. PLoS Med. 2012;9:e1001306. doi: 10.1371/journal.pmed.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karami A. A review of the current isothermal amplification techniques: applications, advantages and disadvantages. J Glob Infect Dis. 2011;3:293–302. [Google Scholar]
- 18.Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–50. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mabey D, Peeling RW, Ustianowski A, et al. Diagnostics for the developing world. Nat Rev Microbiol. 2004;2:231–40. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 20.LaBarre P, Hawkins KR, Gerlach J, et al. A simple, inexpensive device for nucleic acid amplification without electricity—toward instrument-free molecular diagnostics in low-resource settings. PLoS One. 2011;6:e19738. doi: 10.1371/journal.pone.0019738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dineva MA, MahiLum-Tapay L, Lee H. Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. Analyst. 2007;132:1193–9. doi: 10.1039/b705672a. [DOI] [PubMed] [Google Scholar]
- 22.Al-Soud WA, Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–93. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henning L, Felger I, Beck HP. Rapid DNA extraction for molecular epidemiological studies of malaria. Acta Trop. 1999;72:149–55. doi: 10.1016/s0001-706x(98)00090-4. [DOI] [PubMed] [Google Scholar]
- 24.Sultan DM, Khalil MM, Abdouh AS, et al. Imported malaria in United Arab Emirates: evaluation of a new DNA extraction technique using nested PCR. Korean J Parasitol. 2009;47:227–33. doi: 10.3347/kjp.2009.47.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polley SD, Mori Y, Watson J, et al. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol. 2010;48:2866–71. doi: 10.1128/JCM.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poon LL, Wong BW, Ma EH, et al. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem. 2006;52:303–6. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 27.Chen JH, Lu F, Lim CS, et al. Detection of Plasmodium vivax infection in the Republic of Korea by loop-mediated isothermal amplification (LAMP) Acta Trop. 2010;113:61–5. doi: 10.1016/j.actatropica.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Kermekchiev MB, Barnes WM. Direct DNA amplification from crude clinical samples using a PCR enhancer cocktail and novel mutants of Taq. J Mol Diagn. 2010;12:152–61. doi: 10.2353/jmoldx.2010.090070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuehrer HP, Fally MA, Habler VE, et al. Novel nested direct PCR technique for malaria diagnosis using filter paper samples. J Clin Microbiol. 2011;49:1628–30. doi: 10.1128/JCM.01792-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor BJ, Martin KA, Arango E, et al. Real-time PCR detection of Plasmodium directly from whole blood and filter paper samples. Malar J. 2011;10:244. doi: 10.1186/1475-2875-10-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahram Biosystems. Palm PCR. http://www.ahrambio.com/products_palmpcr_technology.html .
- 32.BioFire. R.A.P.I.D.® BioDetection System. http://biofiredefense.com/rapid/
- 33.bigtec Labs. Trueprep-MAG and Truelab Uno. http://www.bigteclabs.com/product.html#fragment-10 .
- 34.Mens PF, Moers AP, de Bes LM, et al. Development, validation and evaluation of a rapid PCR-nucleic acid lateral flow immuno-assay for the detection of Plasmodium and the differentiation between Plasmodium falciparum and Plasmodium vivax. Malar J. 2012;11:279. doi: 10.1186/1475-2875-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleos Nucleot Nucl. 2008;27:224–43. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- 36.Chang CC, Chen CC, Wei SC, et al. Diagnostic devices for isothermal nucleic acid amplification. Sensors. 2012;12:8319–37. doi: 10.3390/s120608319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdul-Ghani R, Al-Mekhlafi AM, Karanis P. Loop-mediated isothermal amplification (LAMP) for malarial parasites of humans: would it come to clinical reality as a point-of-care test? Acta Trop. 2012;122:233–40. doi: 10.1016/j.actatropica.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Han ET. Loop-mediated isothermal amplification test for the molecular diagnosis of malaria. Expert Rev Mol Diagn. 2013;13:205–18. doi: 10.1586/erm.12.144. [DOI] [PubMed] [Google Scholar]
- 39.Asiello PJ, Baeumner AJ. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip. 2011;11:1420–30. doi: 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- 40.Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parida MM. Rapid and real-time detection technologies for emerging viruses of biomedical importance. J Biosci. 2008;33:617–28. doi: 10.1007/s12038-008-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parida M, Sannarangaiah S, Dash PK, et al. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18:407–21. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eiken Chemical Co. Ltd. The Principle of LAMP Method, ver.7.2. http://loopamp.eiken.co.jp/e/lamp/anim.html .
- 44.Yamamura M, Makimura K, Ota Y. Evaluation of a new rapid molecular diagnostic system for Plasmodium falciparum combined with DNA filter paper, loop-mediated isothermal amplification, and melting curve analysis. Jpn J Infect Dis. 2009;62:20–5. [PubMed] [Google Scholar]
- 45.Paris DH, Imwong M, Faiz AM, et al. Loop-mediated isothermal PCR (LAMP) for the diagnosis of falciparum malaria. Am J Trop Med Hyg. 2007;77:972–6. [PubMed] [Google Scholar]
- 46.Han ET, Watanabe R, Sattabongkot J, et al. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol. 2007;45:2521–8. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buates S, Bantuchai S, Sattabongkot J, et al. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) for clinical detection of Plasmodium falciparum gametocytes. Parasitol Int. 2010;59:414–20. doi: 10.1016/j.parint.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Xia JF, Yan XF, Yu H, et al. Simple and rapid detection of human enterovirus 71 by reverse-transcription and loop-mediated isothermal amplification: cryopreservation affected the detection ability. Diagn Microbiol Infect Dis. 2011;71:244–51. doi: 10.1016/j.diagmicrobio.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Chen L, Yin X, et al. Application of DNA-based diagnostics in detection of schistosomal DNA in early infection and after drug treatment. Parasit Vectors. 2011;4:164. doi: 10.1186/1756-3305-4-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeda H, Kokeguchi S, Fujimoto C, et al. Detection of periodontal pathogen Porphyromonas gingivalis by loop-mediated isothermal amplification method. FEMS Immunol Med Microbiol. 2005;43:233–9. doi: 10.1016/j.femsim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Misawa Y, Yoshida A, Saito R, et al. Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J Infect Chemother. 2007;13:134–40. doi: 10.1007/s10156-007-0508-9. [DOI] [PubMed] [Google Scholar]
- 52.Yamazaki W, Ishibashi M, Kawahara R, et al. Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of Vibrio parahaemolyticus. BMC Microbiol. 2008;8:163. doi: 10.1186/1471-2180-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatibana BT, Sano A, Uno J, et al. Detection of Paracoccidioides brasiliensis gp43 gene in sputa by loop-mediated isothermal amplification method. J Clin Lab Anal. 2009;23:139–43. doi: 10.1002/jcla.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thekisoe OM, Rodriguez CV, Rivas F, et al. Detection of Trypanosoma cruzi and T. rangeli infections from Rhodnius pallescens bugs by loop-mediated isothermal amplification (LAMP) Am J Trop Med Hyg. 2010;82:855–60. doi: 10.4269/ajtmh.2010.09-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang T, Liu J, Deng YQ, et al. Development and evaluation of a reverse transcription-loop-mediated isothermal amplification assay for rapid detection of enterovirus 71. J Clin Microbiol. 2011;49:870–4. doi: 10.1128/JCM.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bearinger JP, Dugan LC, Baker BR, et al. Development and initial results of a low cost, disposable, point-of-care testing device for pathogen detection. IEEE Trans Biomed Eng. 2011;58:805–8. doi: 10.1109/TBME.2010.2089054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pyrc K, Milewska A, Potempa J. Development of loop-mediated isothermal amplification assay for detection of human coronavirus-NL63. J Virol Methods. 2011;175:133–6. doi: 10.1016/j.jviromet.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poschl B, Waneesorn J, Thekisoe O, et al. Comparative diagnosis of malaria infections by microscopy, nested PCR, and LAMP in northern Thailand. Am J Trop Med Hyg. 2010;83:56–60. doi: 10.4269/ajtmh.2010.09-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polley SD, Gonzalez IJ, Mohamed D, et al. Clinical evaluation of a loop-mediated amplification kit for diagnosis of imported malaria. J Infect Dis. 2013;208:637–44. doi: 10.1093/infdis/jit183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hopkins H, Gonzalez IJ, Polley SD, et al. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J Infect Dis. 2013;208:645–52. doi: 10.1093/infdis/jit184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel JC, Oberstaller J, Xayavong M, et al. Real-time loop-mediated isothermal amplification (RealAmp) for the species-specific identification of Plasmodium vivax. PLoS One. 2013;8:e54986. doi: 10.1371/journal.pone.0054986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lau YL, Fong MY, Mahmud R, et al. Specific, sensitive and rapid detection of human Plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J. 2011;10:197. doi: 10.1186/1475-2875-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iseki H, Kawai S, Takahashi N, et al. Evaluation of a loop-mediated isothermal amplification method as a tool for diagnosis of infection by the zoonotic simian malaria parasite Plasmodium knowlesi. J Clin Microbiol. 2010;48:2509–14. doi: 10.1128/JCM.00331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kievits T, van Gemen B, van Strijp D, et al. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J Virol Methods. 1991;35:273–86. doi: 10.1016/0166-0934(91)90069-c. [DOI] [PubMed] [Google Scholar]
- 65.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–2. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 66.Tai JH, Ewert MS, Belliot G, et al. Development of a rapid method using nucleic acid sequence-based amplification for the detection of astrovirus. J Virol Methods. 2003;110:119–27. doi: 10.1016/s0166-0934(03)00108-3. [DOI] [PubMed] [Google Scholar]
- 67.Smits HL, Gussenhoven GC, Terpstra W, et al. Detection, identification and semi-quantification of malaria parasites by NASBA amplification of small subunit ribosomal RNA sequences. J Microbiol Methods. 1997;28:65–75. [Google Scholar]
- 68.Schallig HD, Schoone GJ, Lommerse EJ, et al. Usefulness of quantitative nucleic acid sequence-based amplification for diagnosis of malaria in an academic hospital setting. Eur J Clin Microbiol Infect Dis. 2003;22:555–7. doi: 10.1007/s10096-003-0985-4. [DOI] [PubMed] [Google Scholar]
- 69.Schneider P, Wolters L, Schoone G, et al. Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. J Clin Microbiol. 2005;43:402–5. doi: 10.1128/JCM.43.1.402-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bousema T, Okell L, Shekalaghe S, et al. Revisiting the circulation time of Plasmodium falciparum gametocytes: molecular detection methods to estimate the duration of gametocyte carriage and the effect of gametocytocidal drugs. Malar J. 2010;9:136. doi: 10.1186/1475-2875-9-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.An L, Tang W, Ranalli TA, et al. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J Biol Chem. 2005;280:28952–8. doi: 10.1074/jbc.M503096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goldmeyer J, Kong H, Tang W. Development of a novel one-tube isothermal reverse transcription thermophilic helicase-dependent amplification platform for rapid RNA detection. J Mol Diagn. 2007;9:639–44. doi: 10.2353/jmoldx.2007.070012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gill P, Amini M, Ghaemi A, et al. Detection of Helicobacter pylori by enzyme-linked immunosorbent assay of thermophilic helicase-dependent isothermal DNA amplification. Diagn Microbiol Infect Dis. 2007;59:243–9. doi: 10.1016/j.diagmicrobio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 75.Gill P, Alvandi HA, Abdul-Tehrani H, et al. Colorimetric detection of Helicobacter pylori DNA using isothermal helicase-dependent amplification and gold nanoparticle probes. Diagn Microbiol Infect Dis. 2008;62:119–24. doi: 10.1016/j.diagmicrobio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 76.Goldmeyer J, Li H, McCormac M, et al. Identification of Staphylococcus aureus and determination of methicillin resistance directly from positive blood cultures by isothermal amplification and a disposable detection device. J Clin Microbiol. 2008;46:1534–6. doi: 10.1128/JCM.02234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang W, Chow WH, Li Y, et al. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low-resource settings. J Infect Dis. 2010;201(Suppl 1):S46–51. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim HJ, Tong Y, Tang W, et al. A rapid and simple isothermal nucleic acid amplification test for detection of herpes simplex virus types 1 and 2. J Clin Virol. 2011;50:26–30. doi: 10.1016/j.jcv.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chow WH, McCloskey C, Tong Y, et al. Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic Clostridium difficile. J Mol Diagn. 2008;10:452–8. doi: 10.2353/jmoldx.2008.080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jordan JA, Ibe CO, Moore MS, et al. Evaluation of a manual DNA extraction protocol and an isothermal amplification assay for detecting HIV-1 DNA from dried blood spots for use in resource-limited settings. J Clin Virol. 2012;54:11–4. doi: 10.1016/j.jcv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 81.Li Y, Kumar N, Gopalakrishnan A, et al. Detection and species identification of malaria parasites by isothermal tHDA amplification directly from human blood without sample preparation. J Mol Diagn. 2013;15:634–41. doi: 10.1016/j.jmoldx.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andresen D, von Nickisch-Rosenegk M, Bier FF. Helicase-dependent amplification: use in OnChip amplification and potential for point-of-care diagnostics. Expert Rev Mol Diagn. 2009;9:645–50. doi: 10.1586/erm.09.46. [DOI] [PubMed] [Google Scholar]
- 83.Pasternack R, Vuorinen P, Miettinen A. Evaluation of the Gen-Probe Chlamydia trachomatis transcription-mediated amplification assay with urine specimens from women. J Clin Microbiol. 1997;35:676–8. doi: 10.1128/jcm.35.3.676-678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kurn N, Chen P, Heath JD, et al. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin Chem. 2005;51:1973–81. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 85.Walker GT, Fraiser MS, Schram JL, et al. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–6. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mitani Y, Lezhava A, Kawai Y, et al. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat Methods. 2007;4:257–62. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- 87.Piepenburg O, Williams CH, Stemple DL, et al. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen F, Davydova EK, Du W, et al. Digital isothermal quantification of nucleic acids via simultaneous chemical initiation of recombinase polymerase amplification reactions on SlipChip. Anal Chem. 2011;83:3533–40. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uemori T, Mukai H, Takeda O, et al. Investigation of the molecular mechanism of ICAN, a novel gene amplification method. J Biochem. 2007;142:283–92. doi: 10.1093/jb/mvm137. [DOI] [PubMed] [Google Scholar]
- 90.Guatelli JC, Whitfield KM, Kwoh DY, et al. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Natl Acad Sci USA. 1990;87:1874–8. doi: 10.1073/pnas.87.5.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Ness J, Van Ness LK, Galas DJ. Isothermal reactions for the amplification of oligonucleotides. Proc Natl Acad Sci USA. 2003;100:4504–9. doi: 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fang R, Li X, Hu L, et al. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol. 2009;47:845–7. doi: 10.1128/JCM.01528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yulong Z, Xia Z, Hongwei Z, et al. Rapid and sensitive detection of Enterobacter sakazakii by cross-priming amplification combined with immuno-blotting analysis. Mol Cell Probes. 2010;24:396–400. doi: 10.1016/j.mcp.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 94.Lizardi PM, Huang X, Zhu Z, et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–32. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 95.Prithiviraj J, Hill V, Jothikumar N. Rapid detection of microbial DNA by a novel isothermal genome exponential amplification reaction (GEAR) assay. Biochem Biophys Res Commun. 2012;420:738–42. doi: 10.1016/j.bbrc.2012.03.055. [DOI] [PubMed] [Google Scholar]
- 96.Yonesaki T, Ryo Y, Minagawa T, et al. Purification and some of the functions of the products of bacteriophage T4 recombination genes, uvsX and uvsY. Eur J Biochem. 1985;148:127–34. doi: 10.1111/j.1432-1033.1985.tb08816.x. [DOI] [PubMed] [Google Scholar]
- 97.Shibata T, Cunningham RP, DasGupta C, et al. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc Natl Acad Sci USA. 1979;76:5100–4. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Formosa T, Alberts BM. Purification and characterization of the T4 bacteriophage uvsX protein. J Biol Chem. 1986;261:6107–18. [PubMed] [Google Scholar]
- 99.Harris LD, Griffith JD. Formation of D loops by the UvsX protein of T4 bacteriophage: a comparison of the reaction catalyzed in the presence or absence of gene 32 protein. Biochemistry. 1988;27:6954–9. doi: 10.1021/bi00418a042. [DOI] [PubMed] [Google Scholar]
- 100.Okazaki T, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XV. Purification and properties of a polymerase from Bacillus subtilis. J Biol Chem. 1964;239:259–68. [PubMed] [Google Scholar]
- 101.TwistDX. Recombinase Polymerase Amplification: A Breakthrough Alternative to PCR. http://www.twistdx.co.uk/our_technology/
- 102.Euler M, Wang Y, Nentwich O, et al. Recombinase polymerase amplification assay for rapid detection of Rift Valley fever virus. J Clin Virology. 2012;54:308–12. doi: 10.1016/j.jcv.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 103.Walker GT. Empirical aspects of strand displacement amplification. PCR Methods Appl. 1993;3:1–6. doi: 10.1101/gr.3.1.1. [DOI] [PubMed] [Google Scholar]
- 104.Walker GT, Nadeau JG, Spears PA, et al. Multiplex strand displacement amplification (SDA) and detection of DNA sequences from Mycobacterium tuberculosis and other mycobacteria. Nucleic Acids Res. 1994;22:2670–7. doi: 10.1093/nar/22.13.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Spargo CA, Haaland PD, Jurgensen SR, et al. Chemiluminescent detection of strand displacement amplified DNA from species comprising the Mycobacterium tuberculosis complex. Mol Cell Probes. 1993;7:395–404. doi: 10.1006/mcpr.1993.1058. [DOI] [PubMed] [Google Scholar]
- 106.Spargo CA, Fraiser MS, Van Cleve M, et al. Detection of M. tuberculosis DNA using thermophilic strand displacement amplification. Mol Cell Probes. 1996;10:247–56. doi: 10.1006/mcpr.1996.0034. [DOI] [PubMed] [Google Scholar]
- 107.Monis PT, Giglio S. Nucleic acid amplification-based techniques for pathogen detection and identification. Infect Genet Evol. 2006;6:2–12. doi: 10.1016/j.meegid.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nycz CM, Dean CH, Haaland PD, et al. Quantitative reverse transcription strand displacement amplification: quantitation of nucleic acids using an isothermal amplification technique. Anal Biochem. 1998;259:226–34. doi: 10.1006/abio.1998.2641. [DOI] [PubMed] [Google Scholar]
- 109.Mazzarelli G, Rindi L, Piccoli P, et al. Evaluation of the BDProbeTec ET system for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary samples: a multicenter study. J Clin Microbiol. 2003;41:1779–82. doi: 10.1128/JCM.41.4.1779-1782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ge B, Larkin C, Ahn S, et al. Identification of Escherichia coli O157:H7 and other enterohemorrhagic serotypes by EHEC-hlyA targeting, strand displacement amplification, and fluorescence polarization. Mol Cell Probes. 2002;16:85–92. doi: 10.1006/mcpr.2001.0389. [DOI] [PubMed] [Google Scholar]
- 111.Visca P, De Mori P, Festa A, et al. Evaluation of the BDProbeTec strand displacement amplification assay in comparison with the AMTD II direct test for rapid diagnosis of tuberculosis. Clin Microbiol Infect. 2004;10:332–4. doi: 10.1111/j.1198-743X.2004.00818.x. [DOI] [PubMed] [Google Scholar]
- 112.Mens PF, Schoone GJ, Kager PA, et al. Detection and identification of human Plasmodium species with real-time quantitative nucleic acid sequence-based amplification. Malar J. 2006;5:80. doi: 10.1186/1475-2875-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.González IJ, Polley S, Hopkins H, et al. Molecular diagnosis for screening and elimination of malaria: performance of the first commercially available malaria LAMP test. Malar J. 2012;11(Suppl 1):O30. [Google Scholar]
- 114.Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62–9. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tong Y, Lemieux B, Kong H. Multiple strategies to improve sensitivity, speed and robustness of isothermal nucleic acid amplification for rapid pathogen detection. BMC Biotechnol. 2011;11:50. doi: 10.1186/1472-6750-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sirichaisinthop J, Buates S, Watanabe R, et al. Evaluation of loop-mediated isothermal amplification (LAMP) for malaria diagnosis in a field setting. Am J Trop Med Hyg. 2011;85:594–6. doi: 10.4269/ajtmh.2011.10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mugasa CM, Laurent T, Schoone GJ, et al. Nucleic acid sequence-based amplification with oligochromatography for detection of Trypanosoma brucei in clinical samples. J Clin Microbiol. 2009;47:630–5. doi: 10.1128/JCM.01430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mugasa CM, Laurent T, Schoone GJ, et al. Simplified molecular detection of Leishmania parasites in various clinical samples from patients with leishmaniasis. Parasit Vectors. 2010;3:13. doi: 10.1186/1756-3305-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schoone GJ, Oskam L, Kroon NC, et al. Detection and quantification of Plasmodium falciparum in blood samples using quantitative nucleic acid sequence-based amplification. J Clin Microbiol. 2000;38:4072–5. doi: 10.1128/jcm.38.11.4072-4075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gill P, Ramezani R, Amiri MV, et al. Enzyme-linked immunosorbent assay of nucleic acid sequence-based amplification for molecular detection of M. tuberculosis. Biochem Biophys Res Commun. 2006;347:1151–7. doi: 10.1016/j.bbrc.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 121.Abd el-Galil KH, el-Sokkary MA, Kheira SM, et al. Real-time nucleic acid sequence-based amplification assay for detection of hepatitis A virus. Appl Environ Microbiol. 2005;71:7113–6. doi: 10.1128/AEM.71.11.7113-7116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Down JA, O'Connell MA, Dey MS, et al. Detection of Mycobacterium tuberculosis in respiratory specimens by strand displacement amplification of DNA. J Clin Microbiol. 1996;34:860–5. doi: 10.1128/jcm.34.4.860-865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Van Dyck E, Ieven M, Pattyn S, et al. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by enzyme immunoassay, culture, and three nucleic acid amplification tests. J Clin Microbiol. 2001;39:1751–6. doi: 10.1128/JCM.39.5.1751-1756.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cosentino LA, Landers DV, Hillier SL. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by strand displacement amplification and relevance of the amplification control for use with vaginal swab specimens. J Clin Microbiol. 2003;41:3592–6. doi: 10.1128/JCM.41.8.3592-3596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bachmann LH, Johnson RE, Cheng H, et al. Nucleic acid amplification tests for diagnosis of Neisseria gonorrhoeae and Chlamydia trachomatis rectal infections. J Clin Microbiol. 2010;48:1827–32. doi: 10.1128/JCM.02398-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chan EL, Brandt K, Olienus K, et al. Performance characteristics of the Becton Dickinson ProbeTec System for direct detection of Chlamydia trachomatis and Neisseria gonorrhoeae in male and female urine specimens in comparison with the Roche Cobas System. Arch Pathol Lab Med. 2000;124:1649–52. doi: 10.5858/2000-124-1649-PCOTBD. [DOI] [PubMed] [Google Scholar]
- 127.Ryan C, Kudesia G, McIntyre S, et al. BD ProbeTec ET assay for the diagnosis of gonorrhoea in a high-risk population: a protocol for replacing traditional microscopy and culture techniques. Sex Transm Infect. 2007;83:175–9. doi: 10.1136/sti.2006.021816. discussion 9–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Van Der Pol B, Ferrero DV, Buck-Barrington L, et al. Multicenter evaluation of the BDProbeTec ET System for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J Clin Microbiol. 2001;39:1008–16. doi: 10.1128/JCM.39.3.1008-1016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Boonma P, Christensen PR, Suwanarusk R, et al. Comparison of three molecular methods for the detection and speciation of Plasmodium vivax and Plasmodium falciparum. Malar J. 2007;6:124. doi: 10.1186/1475-2875-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]