ABSTRACT
Intestinal infections with F4 enterotoxigenic Escherichia coli (ETEC) are worldwide an important cause of diarrhea in neonatal and recently weaned pigs. Adherence of F4 ETEC to the small intestine by binding to specific receptors is mediated by F4 fimbriae. Porcine aminopeptidase N (ANPEP) was recently identified as a new F4 receptor. In this study, 7 coding mutations and 1 mutation in the 3′ untranslated region (3' UTR)were identified in ANPEP by reverse transcriptase (RT–) PCR and sequencing using 3 F4 receptor-positive (F4R+) and 2 F4 receptor-negative (F4R–) pigs, which were F4 phenotyped based on the MUC4 TaqMan, oral immunization, and the in vitro villous adhesion assay. Three potential differential mutations (g.2615C > T, g.8214A > G, and g.16875C > G) identified by comparative analysis between the 3 F4R+ and 2 F4R– pigs were genotyped in 41 additional F4 phenotyped pigs. However, none of these 3 mutations could be associated with F4 ETEC susceptibility. In addition, the RT-PCR experiments did not reveal any differential expression or alternative splicing in the small intestine of F4R+ and F4R– pigs. In conclusion, we hypothesize that the difference in F4 binding to ANPEP is due to modifications in its carbohydrate moieties.
Keywords: aminopeptidase N, F4 enterotoxigenic Escherichia coli, F4 phenotyping, F4 receptor, mutation, pig
INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) expressing F4 fimbriae are worldwide a major cause of diarrhea in neonatal and recently weaned pigs (Do et al., 2006; Zhang et al., 2007; Amezcua et al., 2008; Madoroba et al., 2009; de la Fe Rodriguez et al., 2011). The bacteria colonize the small intestine of the pig by binding with their F4 fimbriae to specific receptors and produce enterotoxins, inducing diarrhea (Jones and Rutter, 1972; Alexander, 1994). The F4 fimbriae, composed of major and minor subunit structures, exist in 3 serological variants, namely F4ab, F4ac, and F4ad (Orskov et al., 1964; Guinee and Jansen, 1979; Mooi and de Graaf, 1985). These variants differ in the amino acid composition of the major fimbrial subunit FaeG, which has adhesive properties and recognizes glycoconjugates on the surface of enterocytes (Kearns and Gibbons, 1979; Sellwood, 1980; Bakker et al., 1992; Van den Broeck et al., 1999a). Although different F4 receptor profiles were observed due to different F4 ETEC adhesion phenotypes, no causal mutation in previous proposed candidate genes has yet been identified (Ren et al., 2012; reviewed in Schroyen et al., 2012).
Recently, aminopeptidase N (ANPEP) has been found to act as an endocytotic F4 receptor by comparative proteomic analysis of the brush border proteins in F4ac receptor-positive and F4ac receptor-negative pigs (Melkebeek et al., 2012). Aminopeptidase N, belonging to the M1 family of zinc metallopeptidase, is a 936 amino acid membrane glycoprotein and is widely expressed on the surface of various cell types, including porcine enterocytes (Delmas et al., 1992; Rawlings and Barrett., 1993; Olsen et al., 1997). The gene encoding ANPEP (GenBank: NC_010449.4; Supplementary Fig. 1) is located on chromosome 7 and is composed of 20 exons (Poulsen et al., 1991).
The purpose of this study was to investigate whether a mutation in ANPEP could be associated with F4 ETEC susceptibility in pig.
MATERIAL AND METHODS
Animals, Sample Collection, and F4 Phenotyping
Forty-six mixed-breed pigs from 7 different litters were used in this study (Table 1). Blood samples were collected before euthanasia at 6 to 18 wk of age in EDTA blood tubes and stored at –20°C. After euthanasia, mid-jejunum samples for RNA analysis were collected and washed 3 times with Krebs–Henseleit buffer (0.12 M NaCl, 0.014 M KCl, 0.001 M KH2PO4, and 0.025 M NaHCO3, pH 7.4). Next, they were frozen in liquid nitrogen and stored at –80°C. Mid-jejunum samples for the in vitro villous adhesion assay were washed twice with ice cold Krebs-Henseleit buffer followed by 1 washing step with Krebs–Henseleit buffer containing 1% (vol/vol) formaldehyde. Villi were then scraped from the mucosa and stored as mentioned in Van den Broeck et al. (1999b).
Table 1.
Detailed list of the pigs used in this study
| Information pig | ANPEP mutations1 | F4 binding profiling | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F4 adhesion4 | |||||||||||
| Pig | Litter | Sow | Boar | M1 | M2 | M3 | MUC4 2 | IR3 | ab | ac | ad |
| 1 | 1 | Sow A | Boar A | C/C | A/G | C/C | SS | + | +++ | +++ | + |
| 2 | 1 | Sow A | Boar A | C/C | A/G | C/C | SS | + | +++ | +++ | + |
| 3 | 1 | Sow A | Boar A | C/T | A/G | C/G | SS | + | +++ | +++ | +++ |
| 4 | 1 | Sow A | Boar A | C/C | A/G | C/C | SR | + | +++ | +++ | + |
| 5 | 1 | Sow A | Boar A | C/C | A/A | C/C | SS | + | +++ | +++ | + |
| 6 | 2 | Sow B | Boar B | C/C | A/A | C/C | SS | + | +++ | +++ | + |
| 7 | 2 | Sow B | Boar B | C/C | A/G | C/C | SR | + | +++ | +++ | +++ |
| 8 | 2 | Sow B | Boar B | C/C | G/G | C/C | SS | + | +++ | +++ | +++ |
| 9 | 3 | Sow C | Boar A | C/C | A/A | C/C | SS | + | + | +++ | +++ |
| 10 | 3 | Sow C | Boar A | C/C | A/G | C/C | SR | + | +++ | +++ | +++ |
| 11 | 5 | Sow E | Boar D | C/T | G/G | C/G | SR | + | +++ | +++ | +++ |
| 12 | 5 | Sow E | Boar D | C/C | G/G | C/C | SR | + | + | +++ | +++ |
| 13 | 5 | Sow E | Boar D | C/T | G/G | C/G | SR | + | + | +++ | +++ |
| 14 | 5 | Sow E | Boar D | C/C | G/G | C/C | SR | + | + | +++ | +++ |
| 15 | 5 | Sow E | Boar D | C/C | G/G | C/C | SR | + | +++ | +++ | +++ |
| 16 | 6 | Sow F | Boar E | C/C | A/G | C/C | SR | + | + | +++ | + |
| 17 | 6 | Sow F | Boar E | C/C | A/G | C/C | SR | + | +++ | +++ | + |
| 18 | 6 | Sow F | Boar E | C/T | A/G | C/C | SR | + | + | +++ | + |
| 19 | 7 | Sow G | Boar F | C/C | A/A | C/C | SR | + | +++ | +++ | +++ |
| 20 | 7 | Sow G | Boar F | C/C | A/G | C/C | SR | + | + | +++ | + |
| 21 | 7 | Sow G | Boar F | C/C | A/G | C/C | SR | + | + | +++ | + |
| 22 | 4 | Sow D | Boar C | C/C | A/G | C/C | RR | – | – | – | – |
| 23 | 4 | Sow D | Boar C | C/C | G/G | C/C | RR | – | – | – | – |
| 24 | 4 | Sow D | Boar C | C/C | A/G | C/C | RR | – | – | – | – |
| 25 | 4 | Sow D | Boar C | C/C | G/G | C/C | RR | – | – | – | – |
| 26 | 5 | Sow E | Boar D | C/T | G/G | C/G | RR | – | – | – | – |
| 27 | 5 | Sow E | Boar D | C/C | A/G | C/C | RR | – | – | – | – |
| 28 | 1 | Sow A | Boar A | C/C | A/G | C/C | RR | + | +++ | +++ | +++ |
| 29 | 2 | Sow B | Boar B | C/C | A/G | C/C | RR | + | +++ | + | +++ |
| 30 | 2 | Sow B | Boar B | C/C | G/G | C/C | RR | + | +++ | +++ | +++ |
| 31 | 2 | Sow B | Boar B | C/C | A/G | C/C | RR | + | + | + | +++ |
| 32 | 4 | Sow D | Boar C | C/C | G/G | C/C | RR | + | + | +++ | + |
| 33 | 4 | Sow D | Boar C | C/C | A/A | C/C | RR | + | + | +++ | +++ |
| 34 | 4 | Sow D | Boar C | C/T | A/G | C/G | RR | + | + | +++ | + |
| 35 | 4 | Sow D | Boar C | C/C | G/G | C/C | RR | + | +++ | + | +++ |
| 36 | 5 | Sow E | Boar D | C/C | G/G | C/C | RR | + | + | + | + |
| 37 | 7 | Sow G | Boar F | C/C | A/G | C/C | RR | + | + | + | + |
| 38 | 4 | Sow D | Boar C | C/C | G/G | C/C | RR | + | – | – | – |
| 39 | 4 | Sow D | Boar C | C/C | A/G | C/C | RR | + | – | – | – |
| 40 | 6 | Sow F | Boar E | C/C | A/G | C/C | RR | + | – | – | + |
| 41 | 6 | Sow F | Boar E | C/C | A/G | C/C | RR | + | – | – | – |
| 42 | 7 | Sow G | Boar F | C/C | A/G | C/C | RR | + | – | – | + |
| 43 | 4 | Sow D | Boar C | C/C | G/G | C/C | RR | – | + | +++ | + |
| 44 | 4 | Sow D | Boar C | C/T | G/G | C/G | RR | – | + | +++ | +++ |
| 45 | 5 | Sow E | Boar D | C/C | G/G | C/C | RR | – | + | +++ | +++ |
| 46 | 3 | Sow C | Boar A | C/C | A/G | C/C | SR | + | – | – | + |
1 ANPEP mutations are defined as follows: M1= g.2615C > T; M2 = g.8214A > G; M3 = g.16875C > G.
2 MUC4 genotypes are defined as follows: SS denotes homozygous susceptible, SR denotes heterozygous susceptible, and RR denotes homozygous resistant.
3Immune response (IR) is defined as follows: + denotes positive immune response on oral immunization with F4 fimbriae and – denotes negative immune response.
4F4 enterotoxigenic Escherichia coli adhesion is defined as follows: +++ denotes strong adhesion, + denotes weak adhesion, and – denotes no adhesion.
The MUC4 TaqMan assay was performed as described by Nguyen et al. (2013) and is based on the g.8227G > C mutation of MUC4 (Genbank: DQ848681) associated with F4ab/ac ETEC susceptibility (Jorgensen et al., 2004).
For the oral immunization, pigs were orally given 1 mg of F4ac fimbriae (strain Gis26) in 10 mL PBS on 3 consecutive days and once again at 15 d after primary immunization. Blood was collected before immunization and at 15 and 21 d after immunization from the jugular vein to determinate seropositive in F4-specific ELISA.
The presence of the F4 receptor was determined by performing the in vitro villous adhesion assay for the 3 F4 variants (F4ab/ac/ad; Van den Broeck et al., 1999b; see Table 1).
Experimental and animal management procedures were approved by the animal care and ethics committee of the Faculty of Veterinary Medicine, University of Ghent (EC2010/042), Heidestraat, Belgium.
Deoxyribonucleic Acid Isolation, RNA Isolation, and cDNA Synthesis
Deoxyribonucleic acid isolation of porcine blood for MUC4 mutation detection and ANPEP mutation screening was performed as described by Van Poucke et al. (2005).
Total RNA isolation of mid-jejunum samples of 5 pigs (pig 5, 8, 15, 23, and 26) was performed by using the Aurum Total RNA Fatty and Fibrous Tissue kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the instructions of the manufacturer. Residual DNA was removed with an on-column deoxyribonuclease (DNase) treatment. The concentration of total RNA (between 400 and 1,600 ng/μL) and the optical density (OD)260/280 ratio for purity (between 2.09 and 2.14) were measured by the Nanodrop ND-1000 Spectrophotometer (Isogen Life Science, De Meern, The Netherlands). Ribonucleic acid quality was assessed by comparison of the 28S and the 18S ribosomal bands from 1 μg of total RNA on a 0.8% agarose gel stained with ethidium bromide. All RNA samples were confirmed to be DNA free by a minus reverse transcriptase (RT–) PCR with Sus scrofa (Sscr) ANPEP ± 5 primers on 20 ng RNA (Supplementary Table 1). A 10-μL PCR mix containing 0.5 U FastStart Taq DNA polymerase (Roche Diagnostics, Brussels, Belgium), 10x FastStart Buffer with 20 mM MgCl2 (Roche Diagnostics, Brussels, Belgium), 200 μM deoxynucleotide triphosphates (Bioline, London, UK), 0.5 μM primers, and the RNA template was used. Positive and negative controls were included. Polymerase chain reaction conditions were 3.5 min at 95°C, 30 cycles with denaturation at 95°C for 30 s, annealing at 60°C for 30 s and elongation at 72°C for 1 min, and 1 cycle at 72°C for 4 min. The PCR product was examined by gel electrophoresis.
Then, cDNA was synthesized from 1 μg high quality RNA with the Improm-II Reverse Transcriptase kit (Promega, Merelbeke, Belgium) using Random and Oligo dT primers (each 0.5 μg per reaction), verified by PCR (similar to the minus RT-PCR but with 10x diluted cDNA as a template and 40 PCR cycles), and examined by gel electrophoresis.
Structural Mutation Detection of ANPEP by reverse transcriptase-Polymerase Chain Reaction and Sequencing
Complementary DNA was used for amplifying the whole coding sequence of ANPEP in 4 overlapping amplicons with SscrANPEP ± 1, 2, 3, and 4 primers designed with Primer3Plus (Untergasser et al., 2007; Supplementary Table 1) in 3 F4 receptor-positive (F4R+) piglets (pig 5, 8, and 15) and 2 F4 receptor-negative (F4R–) piglets (pig 23 and 26). The composition of the PCR mix and the PCR conditions with 40 PCR cycles were similar as described above (see Supplementary Table 1 for annealing temperatures). Polymerase chain reaction products were examined by gel electrophoresis for differential gene expression and alternative splicing. After purification of the PCR products with Exonuclease I (4 U) and Antarctic Phosphatase (2 U; New England Biolabs, Ipswich, UK) at 37°C for 30 min and 80°C for 15 min, 1 μL PCR product was directly sequenced with 2 pmol of the corresponding ANPEP primers and an additional sequence primer (See Supplementary Table 1) using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Nieuwerkerk a/d Ijssel, The Netherlands). The sequence reactions were analyzed on a 16-capillary 3130xl DNA Analyzer (Applied Biosystems) according to the manufacturer's protocol. Sequences were processed using BioEdit software (Hall, 1999).
Mutation Screening by Polymerase Chain Reaction-Restriction Fragment Length Polymorphism
Potential differential mutations (g.2615C > T, g.8214A > G, and g.16875C > G) found in the structural mutation detection were genotyped in 41 additional F4-phenotyped pigs. Polymerase chain reaction-RFLP assays were developed for screening the mutations on genomic DNA. The primers, restriction enzymes (New England Biolabs), and cutting positions are listed in Supplementary Table 2. Digested PCR products were analyzed on 3% agarose gels.
RESULTS
F4 Phenotyping in Pigs
To obtain a reliable F4 phenotype, 3 tests were performed in 46 pigs, namely the MUC4 TaqMan assay, the oral immunization with F4 fimbriae and the in vitro villous adhesion assay (Table 1). In this study, all homozygous susceptible (SS) pigs (n = 7) for the MUC4 TaqMan assay were F4 seropositive and showed adhesion towards F4ab/ac/ad ETEC. Also, all heterozygous susceptible (SR) pigs (n = 15) were F4 seropositive: 14 pigs showed F4ab/ac/ad ETEC adhesion and 1 pig showed only F4ad ETEC adhesion. Twenty-four pigs were genotyped homozygous resistant (RR): 6 seronegative and 3 seropositive pigs showed no F4 ETEC adhesion, 10 seropositive and 3 seronegative pigs showed F4ab/ac/ad ETEC adhesion, and 2 seropositive pigs showed only F4ad ETEC adhesion. Only pigs (n = 21) with 3 positive tests (F4R+) and pigs (n = 6) with 3 negative tests (F4R–) were used in the structural mutation detection and screening.
Structural Mutation Detection of ANPEP
A total of 8 mutations were found in 3 F4R+ and 2 F4R– pigs (Table 2): 2 silent mutations (exon 1), 5 missense mutations (exon 1, 4, and 12), and 1 mutation in the beginning of the partially sequenced 3′ untranslated region (UTR; exon 20). The 5 missense mutations resulting in amino acid substitutions had no effect on the glycosylation sites predicted by NetNglyc 1.0 and NetOGlyc 4.0 (R. Gupta, E. Jung, S. Brunak, 2004, personal commmunication; Steentoft et al., 2013).
Table 2.
Result of ANPEP mutation detection in 3 F4 receptor-positive and 2 F4 receptor-negative pigs1
| Item2 | EX1 g.2380 C > T (Y33Y) | EX1 g.2419 G > A (Q46Q) | EX1 g.2602 C > A (F107L) | EX1 g.2603 A > C (I108L) | EX1 g.2615 C > T (P112S) | EX4 g.4328 C > T (P330S) | EX 12 g.8214 A > G (I621V) | EX 20 g.16875 C > G (–) |
|---|---|---|---|---|---|---|---|---|
| PIG 5 (R+) | TAC/TAC (Y33Y) | CAG/CAG (Q46Q) | TTA/TTA (L107L) | CTT/CTT (L108L) | CCC/CCC (P112P) | CCC/TCC (P330S) | ATC/ATC (I621I) | C/C |
| PIG 8 (R+) | TAC/TAC (Y33Y) | CAG/CAG (Q46Q) | TTA/TTA (L107L) | CTT/CTT (L108L) | CCC/CCC (P112P) | CCC/CCC (P330P) | GTC/GTC (V621V) | C/C |
| PIG 15 (R+) | TAC/TAT (Y33Y) | CAG/CAA (Q46Q) | TTA/TTA (L107L) | CTT/CTT (L108L) | CCC/CCC (P112P) | CCC/CCC (P330P) | GTC/GTC (V621V) | C/C |
| PIG 23 (R–) | TAC/TAC (Y33Y) | CAG/CAG (Q46Q) | TTA/TTA (L107L) | CTT/CTT (L108L) | CCC/CCC (P112P) | CCC/CCC (P330P) | GTC/GTC (V621V) | C/C |
| PIG 26 (R–) | TAC/TAT (Y33Y) | CAG/CAA (Q46Q) | TTA/TTA (L107L) | CTT/CTT (L108L) | CCC/TCC (P112S) | TCC/TCC (S330S) | GTC/GTC (V621V) | C/G |
1Mutation in codon and substituted amino acid are underlined.
2R+ denotes receptor-positive for F4 ETEC; R- denotes receptor-negative for F4 ETEC.
The mutation in the 3′ UTR was found in a region where no microRNA binding sites were detected by miRbase (Griffiths-Jones, 2004). Comparative analysis between F4R+ and F4R– identified 3 possible differential mutations (Table 2).
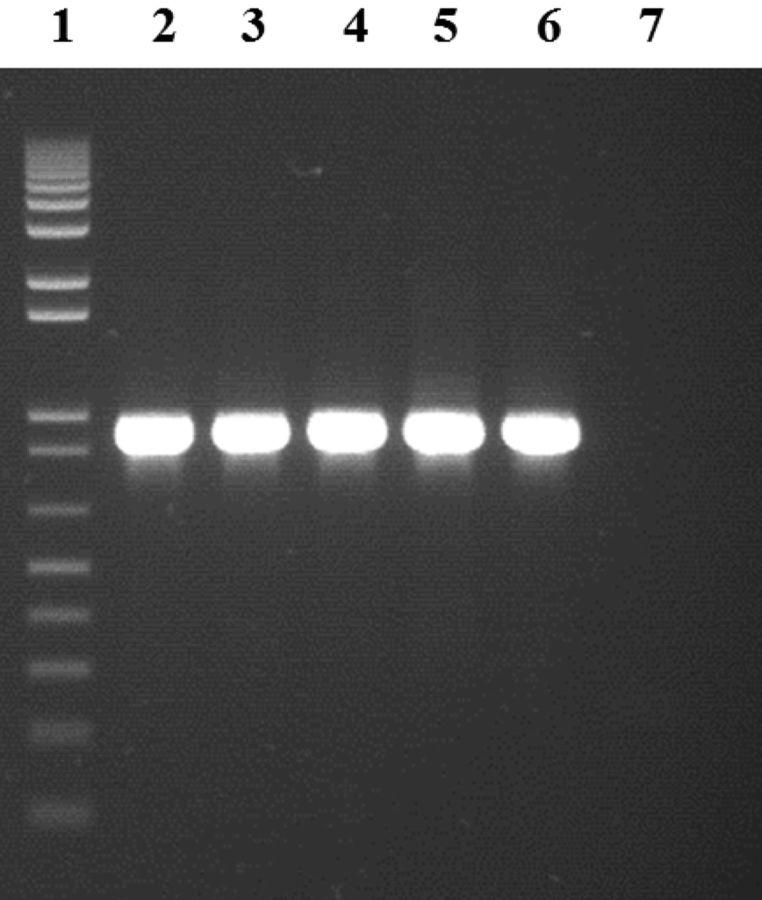
During the structural mutation detection, the expression of ANPEP in the small intestine was semiquantitative analyzed via gel electrophoresis of the RT-PCR products. No obvious expression difference or alternative splice variants were observed between F4R+ and F4R– pigs as illustrated in Fig. 1.
Figure 1.
Agarose gel of Sus scrofa aminopeptidase N +/-3 (primer) ± 3 PCR products using cDNA as a template representing the absence of differential gene expression and alternative splice variants in the small intestine of F4 receptor-positive (F4R+) and F4 receptor-negative (F4R–) pigs. Lane 1: 1Kb Plus DNA ladder (Invitrogen, Carlsbad, CA, USA); lane 2: F4R+ pig 5; lane 3: F4R+ pig 8; lane 4: F4R+ pig 15; lane 5: F4R– pig 23; lane 6: F4R– pig 26; lane 7: water.
Mutation Screening in 41 Additional F4-Phenotyped Piglets
Three mutations (g.2615C > T, g.8214A > G, and g.16875C > G) were screened by PCR-RFLP in 41 additional F4 phenotyped pigs (Table 1). No TT homozygotes (g.2615C > T) and no GG homozygotes (g.16875C > G) were observed in this study. When comparing F4R+ (n = 21) and F4R– pigs (n = 6), AA homozygotes of g.8214A > G mutation were absent in the F4R– pigs (Table 3). The AA genotype was also absent in the 3 seropositive pigs with RR genotype and with no F4 ETEC adhesion (Table 1).
Table 3.
Genotype frequency of g.2615C > T (M1), g.8214A > G (M2), and g.16875C > G (M3) mutations in the different F4 enterotoxigenic Escherichia coli (ETEC) adhesion groups of 21 F4 receptor-positive and 6 F4 receptor-negative pigs
| F4 ETEC adhesion1 | F4ab +++ | F4ab + | F4ab – | F4ac +++ | F4ac + | F4ac – | F4ad +++ | F4ad + | F4ad – | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | CC | 22 | 11/22 (50%) | 6/22 (27%) | 5/22 (23%) | 17/22 (77%) | / | 5/22 (23%) | 8/22 (36%) | 9/22 (41%) | 5/22 (23%) |
| CT | 5 | 2/5 (40%) | 2/5 (40%) | 1/5 (20%) | 4/5 (80%) | / | 1/5 (20%) | 3/5 (60%) | 1/5 (20%) | 1/5 (20%) | |
| TT | 0 | / | / | / | / | / | / | / | / | / | |
| M2 | AA | 4 | 3/4 (75%) | 1/4 (25%) | 0/4 (0%) | 4/4 (100%) | / | 0/4 (0%) | 2/4 (50%) | 2/4 (50%) | 0/4 (0%) |
| AG | 14 | 7/14 (50%) | 4/14 (29%) | 3/14 (21%) | 11/14 (79%) | / | 3/14 (21%) | 3/14 (21%) | 8/14 (57%) | 3/14 (21%) | |
| GG | 9 | 3/9 (33%) | 3/9 (33%) | 3/9 (33%) | 6/9 (66%) | / | 3/9 (33%) | 6/9 (66%) | 0/9 (0%) | 3/9 (33%) | |
| M3 | CC | 23 | 11/23 (48%) | 7/23 (30%) | 5/23 (22%) | 18/23 (78%) | / | 5/23 (22%) | 8/23 (35%) | 10/23 (43%) | 5/23 (22%) |
| CG | 4 | 2/4 (50%) | 1/4 (25%) | 1/4 (25%) | 3/4 (75%) | / | 1/4 (25%) | 3/4 (75%) | 0/4 (0%) | 1/4 (25%) | |
| GG | 0 | / | / | / | / | / | / | / | / | / | |
1F4 ETEC adhesion is defined as follows: +++ denotes strong adhesion, + denotes weak adhesion, and – denotes no adhesion.
The other genotypes (g.2615C > T: CC and CT; g.8214A > G: AG and GG; and g.16875C > G: CC and CG) were present in both F4R+ and F4R– pigs.
DISCUSSION
The F4 binding profile of 46 pigs was determined based on the MUC4 TaqMan assay, the oral immunization with purified F4ac fimbriae and the in vitro villous adhesion assay for the 3 F4 variants. The results of the MUC4 TaqMan assay confirm that the g.8227G > C mutation of MUC4 is associated with F4ab/ac ETEC susceptibility (Jorgensen et al., 2004). Nevertheless, the genotypes, especially the RR genotype, were not completely consistent with the results of the oral immunization and the in vitro villous adhesion assay. Ten of the RR pigs (41.67%) became seropositive and showed adhesion towards F4ab/ac ETEC. This result confirms previous findings that the g.8227G > C mutation is a marker but not the actual causative mutation (Rasschaert et al., 2007; Li et al., 2008). Recently, a new strongly associated marker (g.28784 T > C) has been identified in MUC13 and has shown a very high accuracy for distinguishing the F4ac susceptible and resistant pigs (Ren et al., 2012).
Three RR pigs (12.5%) showed in vitro no F4 ETEC adhesion and became nevertheless seropositive. Blotting of brush border membrane proteins of these pigs with F4 fimbriae displayed similar F4 binding patterns as the F4R– group described in Nguyen et al. (2013). It has been described that oral administration of F4ac antigens to F4R– pigs (based on the in vitro villous adhesion test) can prime the immune system resulting in a secondary antibody response following a subsequent intramuscular immunization (Van den Broeck et al., 2002). Looking at the F4-specific serum IgA response of these piglets 7 d after boosting a weaker response occurred (from a titer of 7 to 30), in comparison with F4R+ piglets (titer increased until 43). Consequently, we hypothesize that, at least during the second oral immunization, F4ac antigens boosted the immune system via small wounds in the mouth or mucosa of these pigs.
The positive immune response with F4ac fimbriae in F4adR+ pigs (2 RR pigs and 1 SR pig) can either be explained as for 3 seropositive F4 adhesion-negative RR pigs (described above) or might be due the high homology in the protein sequences of the F4 fimbrial subunit protein in the 3 F4 variants (Gaastra et al., 1981, 1983; Josephsen et al., 1984).
One seropositive SR pig (6.67%) and 2 seronegative RR pigs (8.3%) showed no F4ab/ac ETEC adhesion. Three seronegative RR pigs (12.5%) were F4ab/ac/ad ETEC adhesion positive. Although the in vitro villous adhesion test has been proven to be reliable (Van den Broeck et al., 1999b), it has been described that the correlation between in vitro F4 ETEC adhesion to isolated brush border vesicles and F4 ETEC susceptibility is not absolute (Francis et al., 1998).
Having well-characterized case-control groups is necessary to find a strong genotype–phenotype correlation in genetic association studies. We propose to use the MUC4 TaqMan assay, the oral immunization, and the in vitro villous adhesion assay for F4 phenotyping the case-control groups in linkage studies for F4 ETEC.
Because ANPEP was recently identified as an endocytotic receptor for F4ac ETEC (Melkebeek et al., 2012), the cDNA sequence of ANPEP was investigated. Although binding of F4ab/ad ETEC to ANPEP is still unknown, pigs susceptible or resistant towards the 3 F4 ETEC variants were used in this study.
First, a structural mutation detection and screening were performed for finding a mutation that could alter the binding site or change the protein structure and therefore sterically blocking the binding site. Previously, it has been shown that ANPEP is a functional cell surface receptor for group I coronaviruses and that this interaction is dependent on species-specific amino acid differences in the receptor protein (Delmas et al., 1992, 1994; Yeager et al., 1992; Tresnan et al., 1996; Benbacer et al., 1997; Li et al., 2007; Tusell et al., 2007). For instance, a T742V or a T742R substitution in feline ANPEP, a common receptor for group I coronaviruses, abrogated the receptor activity for feline enteric coronavirus, canine coronavirus, and porcine transmissible gastroenteritis coronavirus (Tresnan and Holmes, 1998; Tusell et al., 2007).
Comparative analysis of F4R+ (n = 21) and F4R– pigs (n = 6) in the ANPEP mutation screening showed that no structural mutation in ANPEP is associated with F4 ETEC susceptibility.
Second, alternative splicing could play a role in F4 ETEC susceptibility by removing or adding specific domains of the F4 fimbriae binding site or by altering the steric structure of ANPEP. Although human ANPEP is subjected to alternative splicing (Dybkaer et al., 2001), we observed no porcine ANPEP alternative splice variants in the small intestine during structural mutation detection.
Third, a regulatory mutation could explain susceptibility or resistance to F4 ETEC by altering the ANPEP expression in the small intestine. According to Nam and Lee (2010), the density of ANPEP could play an important role in contributing to an efficient porcine epidemic diarrhea virus infection. Because an obvious difference in expression between F4R+ and F4R– pigs was expected, the expression of ANPEP was only semiquantitatively analyzed during structural mutation detection. We could conclude that no apparent expression difference was observed, hence the F4ac susceptibility is not governed by a regulatory mutation.
Previously, other candidate receptors for the 3 F4 variants were suggested, all glycoconjugates (reviewed in Van den Broeck et al., 2000). The carbohydrate moiety of these glycoconjugates appeared to be necessary for establishing adhesion with the F4 adhesin (Erickson et al., 1992; Grange and Mouricout, 1995; Grange et al., 1999; Coddens et al., 2011).
For binding to ANPEP, it was shown that F4ac ETEC, like porcine transmissible gastroenteritis CV, is binding on the carbohydrate moieties of ANPEP in a sialic acid-dependent manner (Schultze et al., 1996; Melkebeek et al., 2012). Given the results described above, we hypothesize that the susceptibility towards F4 ETEC mediated by aminopeptidase N is not due to modifications in the protein itself but due to modifications in the carbohydrate structures attached to it.
LITERATURE CITED
- Alexander T. J. L. 1994. Neonatal diarrhoea in pigs. In: Gyles C. L. editor, Escherichia coli in domestic animals and humans. Cab International, Wallingford, UK: p. 151–169. [Google Scholar]
- Amezcua R., Friendship R. M., Dewey C. E. 2008. An investigation of the presence of Escherichia coli O149:K91:F4 on pig farms in southern Ontario and the use of antimicrobials and risk factors associated with the presence of this serogroup. Can. Vet. J. 49:39–45. [PMC free article] [PubMed] [Google Scholar]
- Bakker D., Willemsen P. T., Simons L. H., van Zijderveld F. G., de Graaf F. K. 1992. Characterization of the antigenic and adhesive properties of FaeG, the major subunit of K88 fimbriae. Mol. Microbiol. 6:247–255. [DOI] [PubMed] [Google Scholar]
- Benbacer L., Kut E., Besnardeau L., Laude H., Delmas B. 1997. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 71:734–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coddens A., Valis E., Benktander J., Ångström J., Breimer M. E., Cox E., Teneberg S. 2011. Erythrocyte and porcine intestinal glycosphingolipids recognized by F4 fimbriae of enterotoxigenic Escherichia coli. PLoS ONE 6:e23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fe Rodriguez P. Y., Coddens A., Del Fava E., Abrahantes J. C., Shkedy Z., Martin L. O. M., Muñoz E. C., Duchateau L., Cox E., Goddeeris B. M. 2011. High prevalence of F4+ and F18+ Escherichia coli in Cuban piggeries as determined by serological survey. Trop. Anim. Health Prod. 43:937–946. [DOI] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L'Haridon R., Vogel L. K., Sjöström H., Noren O., Laude H. 1992. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature 357:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Sjöström H., Noren O., Laude H. 1994. Further characterization of aminopeptidase N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342:293–298. [DOI] [PubMed] [Google Scholar]
- Do T. N., Cu P. H., Nguyen H. X., Au T. X., Vu Q. N., Driesen S. J., Townsend K. M., Chin J. J., Trott D. J. 2006. Pathotypes and serogroups of enterotoxigenic Escherichia coli isolated from pre-weaning pigs in north Vietnam. J. Med. Microbiol. 55:93–99. [DOI] [PubMed] [Google Scholar]
- Dybkaer K., Kristensen J. S., Pedersen F. S. 2001. Single site polymorphisms and alternative splicing of the human CD13 gene – Different splicing frequencies among patients with acute myeloid leukaemia and healthy individuals. Br. J. Haematol. 112:691–696. [DOI] [PubMed] [Google Scholar]
- Erickson A. K., Willgohs J. A., McFarland S. Y., Benfield D. A., Francis D. H. 1992. Identification of two porcine brush border glycoproteins that bind the K88ac adhesin of Escherichia coli and correlation of these glycoproteins with the adhesive phenotype. Infect. Immun. 60:983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D. H., Grange P. A., Zeman D. H., Baker D. R., Sun R., Erickson A. K. 1998. Expression of mucin-type glycoprotein K88 receptors strongly correlates with piglet susceptibility to K88(+) enterotoxigenic Escherichia coli, but adhesion of this bacterium to brush borders does not. Infect. Immun. 9:4050–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaastra W., Klemm P., de Graaf F. K. 1983. The nucleotide sequence of the K88ad protein subunit of porcine enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 18:177–183. [Google Scholar]
- Gaastra W., Mooi F. R., Stuitje A. R., de Graaf F. K. 1981. The nucleotide sequence of the gene encoding the K88ab protein subunit of porcine enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 12:41–46. [Google Scholar]
- Grange P. A., Erickson A. K., Levery S. B., Francis D. H. 1999. Identification of an intestinal neutral glycosphingolipid as a phenotypic-specific receptor for K88ad fimbrial adhesin of Escherichia coli. Infect. Immun. 37:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange P. A., Mouricout M. A. 1995. Transferrin associated with the porcine intestinal mucosa is a receptor specific for K88ab fimbriae of Escherichia coli. Infect. Immun. 64:606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. 2004. The microRNA registry. Nucleic Acids Res. 32:D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinee P. A. M., Jansen W. H. 1979. Behavior of Escherichia coli K antigens K88ab, K88ac, and K88ad in immunoelectrophoresis, double diffusion, and hemagglutination. Infect. Immun. 23:700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41:95–98. [Google Scholar]
- Jones G. W., Rutter J. M. 1972. Role of the K88 antigen in the pathogenesis of neonatal diarrhea caused by Escherichia coli in piglets. Infect. Immun. 6:918–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen C. B., Cirera S., Archibald A. L., Anderson L., Fredholm M., Edfors-Lilja I. 2004. Porcine polymorphisms and methods for detecting them. International application published under the patent cooperation treaty (PCT). PCT/DK2003/000807 or WO2004/048606A2. [Google Scholar]
- Josephsen J., Hansen F., de Graaf F. K., Gaastra W. 1984. The nucleotide sequence of the protein subunit of the K88ac fimbriae of porcine enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 25:301–306. [Google Scholar]
- Kearns M., Gibbons R. A. 1979. The possible nature of pig intestinal receptor for the K88 antigen of Escherichia coli. FEMS Microbiol. Lett. 6:165–168. [Google Scholar]
- Li B. X., Ge J. W., Li Y. L. 2007. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 365:166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Li Y., Qiu X., Niu X., Liu Y., Zhang Q. 2008. Identification and screening of gene(s) related to susceptibility to enterotoxigenic Escherichia coli F4ab/ac in piglets. Asian-Aust. J. Anim. Sci. 21:489–493. [Google Scholar]
- Madoroba E., Van Driessche E., De Greve H., Mast J., Ncube I., Read J., Beeckmans S. 2009. Prevalence of enterotoxigenic Escherichia coli virulence genes from scouring piglets in Zimbabwe. Trop. Anim. Health Prod. 41:1539–1547. [DOI] [PubMed] [Google Scholar]
- Melkebeek V., Rasschaert K., Bellot P., Tilleman K., Favoreel H., Deforce D., De Geest B. G., Goddeeris B. M., Cox E. 2012. Targeting aminopeptidase N, a newly identified receptor for F4ac fimbriae, enhances the intestinal mucosal immune response. Mucosal Immunol. 5:635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F. R., de Graaf F. K. 1985. Molecular biology of fimbriae of enterotoxigenic Escherichia coli. Curr. Top. Microbiol. Immunol. 188:119–138. [DOI] [PubMed] [Google Scholar]
- Nam E., Lee C. 2010. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Vet. Microbiol. 144:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. U., Goetstouwers T., Coddens A., Van Poucke M., Peelman L., Deforce D., Melkebeek V., Cox E. 2013. Differentiation of F4 receptor profiles in pigs based on their mucin 4 polymorphism, responsiveness to oral F4 immunization and in vitro binding of F4 to villi. Vet. Immunol. Immunopathol. 152:93–100. [DOI] [PubMed] [Google Scholar]
- Olsen J., Kokholm K., Noren O., Sjöström H. 1997. Structure and expression of aminopeptidase N. Adv. Exp. Med. Biol. 421:47–57. [DOI] [PubMed] [Google Scholar]
- Orskov I., Orskov F., Sojka W. J., Wittig W. 1964. K antigens K88ab(L) and K88ac(L) in E. coli. Acta Pathol. Microbiol. Scand. 62:439–447. [DOI] [PubMed] [Google Scholar]
- Poulsen P. H., Thomsen P. D., Olsen J. 1991. Assignment of the porcine aminopeptidase N (PEPN) gene to chromosome 7cen—-q21. Cytogenet. Cell Genet. 57:44–46. [DOI] [PubMed] [Google Scholar]
- Rasschaert K., Verdonck F., Goddeeris B. M., Duchateau L., Cox E. 2007. Screening of pigs resistant to F4 enterotoxigenic Escherichia coli (ETEC) infection. Vet. Microbiol. 123:249–253. [DOI] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J. 1993. Evolutionary families of peptidases. Biochem. J. 290:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Yan X., Ai H., Zhang Z., Huang X., Ouyang J., Yang M., Yang H., Han P., Zeng W., Chen Y., Guo Y., Xiao S., Ding N., Huang L. 2012. Susceptibility towards enterotoxigenic Escherichia coli F4ac diarrhea is governed by the MUC13 gene in pigs. PLoS ONE 7:e44573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroyen M., Stinckens A., Verhelst R., Niewold T., Buys N. 2012. The search for the gene mutations underlying enterotoxigenic Escherichia coli F4ab/ac susceptibility in pigs: A review. Vet. Res. 43:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Krempl C., Ballesteros M. L., Shaw L., Schauer R., Enjuanes L., Herrler G. 1996. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 70:5634–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellwood R. 1980. The interaction of the K88 antigen with porcine intestinal epithelial cell brush borders. Biochim. Biophys. Acta 632:326–335. [DOI] [PubMed] [Google Scholar]
- Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., Levery S. B., Clausen H. 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresnan D. B., Holmes K. V. 1998. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv. Exp. Med. Biol. 440:69–75. [DOI] [PubMed] [Google Scholar]
- Tresnan D. B., Levis R., Holmes K. V. 1996. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 70:8669–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusell S. M., Schittone S. A., Holmes K. V. 2007. Mutational analysis of aminopeptidase N, a receptor for several group I coronaviruses, identifies key determinants of viral host range. J. Virol. 81:1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. A. M. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broeck W., Bouchaut H., Cox E., Goddeeris B. M. 2002. Receptor-independent priming of the systemic immune system of pigs by low oral doses of F4 fimbriae. Vet. Immunol. Immunopathol. 85:171–178. [DOI] [PubMed] [Google Scholar]
- Van den Broeck W., Cox E., Goddeeris B. M. 1999a. Receptor-specific binding of purified F4 to isolated villi. Vet. Microbiol. 68:255–263. [DOI] [PubMed] [Google Scholar]
- Van den Broeck W., Cox E., Oudega B., Goddeeris B. M. 1999b. Receptor-dependent immune responses in pigs after oral immunization with F4 fimbriae. Infect. Immun. 67:520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broeck W., Cox E., Oudega B., Goddeeris B. M. 2000. The F4 fimbrial antigen of Escherichia coli and its receptors. Vet. Microbiol. 71:223–244. [DOI] [PubMed] [Google Scholar]
- Van Poucke M., Vandesompele J., Mattheeuws M., Van Zeveren A., Peelman L. J. 2005. A dual fluorescent multiprobe assay for prion protein genotyping in sheep. BMC Infect. Dis. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C. L., Ashmun R. A., Williams R. K., Cardellichio C. B., Shapiro L. H., Look A. T., Holmes K. V. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhao M., Ruesch L., Omot A., Francis D. 2007. Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet. Microbiol. 123:145–152. [DOI] [PubMed] [Google Scholar]