ABSTRACT
Background: Amino acids are well known to be key effectors of gut protein turnover. We recently reported that enteral delivery of proteins markedly stimulated global duodenal protein synthesis in carbohydrate-fed healthy humans, but specifically affected proteins remain unknown.
Objective: We aimed to assess the influence of an enteral protein supply on the duodenal mucosal proteome in carbohydrate-fed humans.
Design: Six healthy volunteers received for 5 h, on 2 occasions and in random order, either an enteral infusion of maltodextrins alone (0.25 g · kg−1 · h−1) mimicking the fed state or maltodextrins with a protein powder (0.14 g proteins · kg−1 · h−1). Endoscopic duodenal biopsy specimens were then collected and frozen until analysis. A 2-dimensional polyacrylamide gel electrophoresis–based comparative proteomics analysis was then performed, and differentially expressed proteins (at least ±1.5-fold change; Student’s t test, P < 0.05) were identified by mass spectrometry. Protein expression changes were confirmed by Western blot analysis.
Results: Thirty-two protein spots were differentially expressed after protein delivery compared with maltodextrins alone: 28 and 4 spots were up- or downregulated, respectively. Among the 22 identified proteins, 11 upregulated proteins were involved either in the cytoskeleton (ezrin, moesin, plastin 1, lamin B1, vimentin, and β-actin) or in protein biosynthesis (glutamyl-prolyl–transfer RNA synthetase, glutaminyl–transfer RNA synthetase, elongation factor 2, elongation factor 1δ, and eukaryotic translation and initiation factor 3 subunit f).
Conclusions: Enteral delivery of proteins altered the duodenal mucosal proteome and mainly stimulated the expression of proteins involved in cytoskeleton and protein biosynthesis. These results suggest that protein supply may affect intestinal morphology by stimulating actin cytoskeleton remodeling. This trial was registered at clinicaltrials.gov as NCT01254110.
Keywords: cytoskeleton, duodenum, enteral nutrition, proteins, proteome
INTRODUCTION
In the human small intestinal mucosa, protein renewal approaches 50% per day (1), a value much higher than that observed in other tissues such as muscle or liver. Alteration of gut protein turnover may thus be involved in the occurrence of intestinal injury (i.e., enhanced intestinal permeability) (2). In contrast, gut protein metabolism can also be an interesting therapeutic or nutritional target to improve intestinal functions as during intestinal adaptation, for instance (3).
Interestingly, the administration of specific nutrients is able to affect gut protein metabolism (4–6). Amino acids, which are key effectors of gut protein turnover, have been tested extensively in vivo and in vitro, either alone or in combination with other nutrients (7). Glutamine, which is associated with less infectious complications in critically ill patients (8), stimulated protein synthesis in the duodenal mucosa of hypercatabolic dogs (9) or fasted humans (1) but not in the jejunal mucosa of malnourished rats (10). In carbohydrate-fed humans, glutamine affected neither protein synthesis nor proteasome activities in the duodenal mucosa (11) but modulated ubiquitination of several heat shock proteins (12). Arginine and leucine have been shown to stimulate protein synthesis in the small intestine of pigs (13–15) but not in the duodenal mucosa of healthy volunteers (16, 17). Surprisingly, the effects of different amino acid or protein mixtures have been poorly documented in the intestine compared with all the data obtained in muscle (18–20).
Adegoke et al. reported that luminal amino acids reduced mucosal protein synthesis in isolated jejunal segments of piglets (21) that was not observed after intravenous infusion (22). In contrast, hyperaminoacidemia markedly enhanced the protein fractional synthesis rate (FSR)11 in the ileal mucosa of postoperative patients (23). In healthy humans, intravenous infusion of amino acids did not appear to stimulate the synthesis of pancreatic and mucosal proteins (24), whereas an enteral mixture of nonessential amino acids increased duodenal protein FSR compared with saline (1). Interestingly, recent data suggested that dietary proteins can improve intestinal permeability in patients with Crohn disease (25). In carbohydrate-fed humans, we recently reported that enteral delivery of proteins enhanced protein FSR in human duodenal mucosa (11). However, in this latter study, we failed to observe an activation of the mammalian target of rapamycin (mTOR) pathway (11, 26) that usually mediates the effects of amino acids on protein translation in intestinal cells (5, 27, 28) and muscle (29).
Thus, we aimed to assess, in the fed state, the effects of an enteral protein supply on the expression of duodenal mucosal proteins and to elucidate the modulated metabolic pathways in healthy humans by a proteomic approach.
METHODS
Clinical protocol and ethical authorizations
The current study was performed in accordance with the guidelines of the Centre for Clinical Investigations, after approval by the local ethics committee (North-west I, France). Six healthy men gave their written informed consent to participate. The subjects were in good general health and had no hepatic, renal, or cardiac dysfunction or any medical or surgical digestive history. The volunteers had a mean ± SEM age of 22.3 ± 1.0 y and BMI (in kg/m2) of 22.5 ± 0.5.
During the 3 d before the experimental trial, all subjects consumed a controlled diet providing 30 kcal and 0.9 g protein · kg−1 · d−1. On the morning of the study after an overnight fasting, the subjects received over 5 h, on 2 occasions, and in a random order by a nasogastric feeding tube either maltodextrins alone (control condition: 0.25 g · kg−1 · h−1; Lactalis Nutrition Santé), which mimicked a carbohydrate fed state, or maltodextrins with a protein powder (protein powder condition: 22.4 mg N · kg−1 · h−1 or 0.14 g protein · kg−1 · h−1; Protifar Plus, Nutricia Advanced Medical Nutrition). The infusion rate was 3.5 mL · kg−1 · h−1. The dose of protein powder was chosen to supply an amount of nitrogen similar to that delivered in our previous studies (1, 30). An upper endoscopy (model XQ20; Olympus) was performed 30 min after the end of enteral feeding. Eight biopsy samples were collected from the duodenal mucosa, immediately snap-frozen in liquid nitrogen, and stored at −80°C for comparative proteomic and Western blot analysis. Two additional biopsy specimens were fixed in formalin for histologic assessment.
Protein extraction and 2-dimensional electrophoresis separation
Protein extraction and 2-dimensional electrophoresis (2DE) separation were performed as previously described (31). Briefly, endoscopic samples were homogenized in ice-cold lysis buffer containing 7 mol urea/L, 2 mol thiourea/L, 4% (wt:vol) 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate, 50 mmol dithiothreitol/L, 25 mmol spermine tetrahydrochloride/L, 0.5% (vol:vol) immobilized pH gradient buffer pH 3–10 nonlinear (GE Healthcare), and a protease inhibitor cocktail (P2714; Sigma-Aldrich). Total protein extract (40 μg for silver staining or 500 μg for Coomassie Brilliant Blue G-250 staining) was used to rehydrate Immobiline DryStrip gels (pH 3–10 nonlinear 18 cm; GE Healthcare). Proteins were resolved in the first dimension by isoelectric focusing for a total of 50,000 V · h by using the Ettan IPGphor 3 (GE Healthcare). After equilibration of the strips, the second dimension was performed on 8–16% polyacrylamide gradient gels (20 cm × 18 cm × 1 mm) and was run on an Ettan Daltsix vertical system (GE Healthcare). The 2-dimensional (2D) gels were silver-stained (PlusOne Silver Staining kit; GE Healthcare) or dyed after the Coomassie Brilliant Blue G-250 staining method (31).
2D image analysis
The 2D gel images were captured by scanning silver-stained gels with an ImageScanner II (GE Healthcare). Differential analysis was performed by using ImageMaster 2D Platinum v5.0 software (GE Healthcare) for spot detection, quantification, matching, and comparative analysis. Each tissue sample was subjected to 2DE 3 times to minimize run-to-run variation, and each set of 3 gels was compared by using ImageMaster to confirm the nonappearance of statistically differential spots within the set of gels. The most representative gel (gel migration, spot definition, and spot number) of each set was used to test the influence of protein powder compared with the control condition. The expression level was determined by the relative volume of each spot in the gel and expressed as %volume, calculated as spot volume/Σvolumes of all spots resolved in the gel. Variations in abundance were calculated as the ratio of mean values of %volume for a group of spots between the 2 conditions. Only spots with a %volume variation ratio >1.5 were considered relevant. The corresponding P values were determined by using the paired Student’s t test (significance level P < 0.05) after spot %volume log-transformation.
In-gel trypsin digestion and protein identification by liquid chromatography–electrospray ionization–tandem mass spectrometry
The protein spots of interest were excised from CBB-stained 2D gels by using the Ettan Spot Picker (GE Healthcare), and automated in-gel digestion of proteins was performed on the Ettan Digester (GE Healthcare) as previously described (32). SpeedVac-dried peptide extracts were resuspended in 10 μL of 5% (vol:vol) acetonitrile/0.1% (vol:vol) formic acid and then analyzed with a nano-LC1200 system coupled to a quadrupole time-of-flight (Q-TOF) 6520 mass spectrometer equipped with a nanospray source and an HPLC–chip cube interface (Agilent Technologies). Briefly, peptides were enriched and desalted on a 360-nL RP-C18 trap column and separated on a Polaris (3 μm particle size) C18 column (150 mm long × 75 μm inner diameter; Agilent Technologies). A 25-min linear gradient (3–80% acetonitrile in 0.1% formic acid) at a flow rate of 320 nL/min was used, and the eluent was analyzed with a Q-TOF mass spectrometer as previously described (31). For protein identification, tandem mass spectrometry peak lists were extracted and compared with the protein databases by using the MASCOT Daemon version 2.2.2 (Matrix Science) search engine. The searches were performed with the following specific parameters: enzyme specificity, trypsin; one missed cleavage permitted; no fixed modifications; variable modifications, methionine oxidation, cysteine carbamidomethylation, serine, tyrosine, and threonine phosphorylation; monoisotopic; peptide charge, 2+ and 3+ mass tolerance for precursor ions, 20 ppm; mass tolerance for fragment ions, 0.06 Da; nano-electrospray Q-TOF as instrument; taxonomy, human; database, UniProtKB/Swiss-Prot v55.6 (390,696 sequences; 140,503,634 residues). Protein hits were automatically validated if they satisfied one of the following criteria: identification with at least 2 top-ranking peptides (bold and red), each with a MASCOT score of >32 (P < 0.01), or at least 2 top-ranking peptides (bold and red), each with a MASCOT score of >24 (P < 0.05). To evaluate false-positive rates, all the initial database searches were performed by using the “decoy” option of MASCOT. Results were considered relevant if the false-positive rate never exceeded 1%.
Gene ontology–based analysis
The PANTHER classification system (version 8.1; http://www.pantherdb.org) was used to categorize the identified proteins according to their molecular function and biological process. To identify significantly enriched biological themes and functional groups in the list of identified proteins, we performed functional annotation clustering analysis by using the DAVID gene ontology (GO) tool (33). UniProt accession numbers of identified proteins were uploaded and mapped against the Homo sapiens reference data set. The following databases were used: PANTHER, Swiss-Pro/Protein Information Resource Keywords, Kyoto Encyclopedia of Genes and Genomes, and Reactome pathways. Only annotation clusters with enrichment scores (ESs) >1.3 (i.e., P < 0.05) and associated terms with P < 0.05 for the modified Fisher’s test and P < 0.15 for Benjamini-Hochberg correction, respectively, were considered significant (33).
Western blot analysis
Total protein extracts (20 μg) were resolved in 4–20% SDS-PAGE gels and electrotransferred onto nitrocellulose membrane (GE Healthcare) as previously described (31). After transfer, membranes were soaked in TBS-T solution [10 mmol Tris-HCl/L pH 7.4, 150 mmol NaCl/L, 0.2% (vol:vol) Tween 20] with 3% (wt:vol) bovine serum albumin for 2 h at room temperature. Blots were washed 3 times for 10 min with TBS-T and subsequently incubated overnight at 4°C in TBS-T and 3% (wt:vol) bovine serum albumin with various specific primary antibodies: mouse monoclonal antibodies anti–elongation factor 2 (EF-2; sc-166409), anti-ezrin (sc-32759), anti-moesin (sc-136268), anti–stress-70 protein (mitochondrial; sc-133137), anti–T-complex protein 1δ (sc-137092), anti–serine/threonine-protein phosphatase 5 (sc-136046), or anti–elongation factor 1δ (EF-1δ sc-130371); goat polyclonal antibodies anti–N-myc downstream-regulated gene 1 protein (sc-19464), anti–claudin 3 (sc-17660), or anti–eukaryotic translation initiation factor 3 subunit F (eIF3f; sc-30247) from Santa Cruz Biotechnology; mouse monoclonal antibodies anti–β-actin (A5441) from Sigma-Aldrich; rabbit monoclonal antibodies anti–heat shock protein 90α (8165, Cell Signaling Technology); and rabbit polyclonal antibodies anti–plastin 1 (AP13147b; Abgent), anti–zonula occludens protein-1 (ZO-1) (40-2200; Life Technologies), anti-occludin (71-1500; Life Technologies), or anti–claudin 1 (51-9000; Life Technologies). Membranes were washed 3 times for 10 min with TBS-T, incubated with horseradish peroxidase–conjugated goat anti–mouse (sc-2005, 1:5000; Santa Cruz Biotechnology), donkey anti–goat (sc-2020, 1:5000; Santa Cruz Biotechnology), or swine anti–rabbit (P0399, 1:5000; Dako) IgG for 1 h at room temperature. After 3 additional washes, immunocomplexes were revealed by using the enhanced chemiluminescence detection system. Protein bands were quantified by densitometry with ImageScanner III and ImageQuant TL software (GE Healthcare). The blots were reprobed with a goat anti-GAPDH polyclonal antibody (SAB2500451; Sigma-Aldrich) as a loading control.
Statistical analysis
The results are expressed as means ± SEMs and were compared by using GraphPad Prism 5.0 (GraphPad Software). To evaluate the effects of protein powder at 5 h (data from biopsy specimens), we assessed statistical analysis by using Student’s t test for paired data. For all, P < 0.05 was considered significant.
RESULTS
Histologic assessment
Histologic examination of duodenal biopsy specimens revealed no signs of mucosal lesions in any subjects.
Effects of protein supplementation on human duodenal mucosal proteome
Image analysis detected approximately 2150 protein spots per 2D gel, out of which 75% were successfully matched throughout all gels. Representative 2D gels from both conditions are shown in Figure 1. Thirty-two protein spots were differentially and significantly expressed in response to enteral protein delivery (Student’s t test, P < 0.05, Figure 1): 28 spots were upregulated (range: 1.50- to 4.04-fold), whereas 4 spots showed a reduced expression (with changes from 2.65- to 5.55-fold). These protein spots were analyzed by using liquid chromatography coupled with electrospray ionization–tandem mass spectrometry, and 27 spots (84%) were identified (Table 1). Overall, a total of 22 nonredundant proteins were obtained. Indeed, 2 proteins, glutamyl-prolyl–transfer RNA (tRNA) synthetase (spots 1–5, Figure 1) and vimentin (spots 23–24, Figure 1), were detected at multiple spot positions that could be due to protein isoforms (i.e., difference in maturation state, degradation, and/or posttranslational modifications). However, in that case, the expression variation for a given protein was similar between the spots (Table 1). Interindividual heterogeneity in the expression levels of altered proteins was low (Supplemental Table 1), and the order of the trials (control–protein powder or protein powder–control) did not affect the proteomics data (Supplemental Table 2).
FIGURE 1.
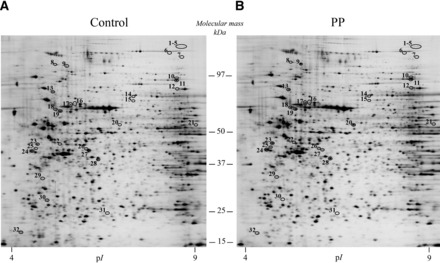
Silver-stained 2-dimensional gel images representing total proteins extracted from human duodenal mucosal biopsy samples after maltodextrin (A) and protein-supplemented maltodextrin (B) enteral perfusion. Differentially expressed proteins (i.e., at least ±1.5-fold modulated; circled spots with a number) were determined by statistical analysis (paired Student’s t test, P < 0.05) and correspond to the samples analyzed by liquid chromatography–tandem mass spectrometry. Protein identification results are depicted in Table 1. pI, isoelectric point; PP, protein powder.
TABLE 1.
Human duodenal mucosal proteome changes in response to protein infusion1
| Theoretical | |||||||||
| Spot number | Swiss-Prot accession number | Protein name | pI | Mr, kDa | Fold change2 | P value3 | Score on MASCOT | Sequence coverage, % | Peptide hit |
| 1, 2, 3, 4, 5 | P07814 | Glutamyl-prolyl-tRNA synthetase | 7.03 | 170.5 | 2.38, 1.75, 1.77, 1.75, 1.71 | 0.0108, 0.0395, 0.0296, 0.0478, 0.0391 | 78, 58, 32, 48, 38 | 2,1, 2, 2, 1 | 4, 2, 4, 2, 2 |
| 6 | Q92878 | DNA repair protein RAD50 | 6.48 | 153.8 | 1.84 | 0.0036 | 49 | 2 | 4 |
| 7, 8, 9 | — | Undetermined | — | — | 1.68, 2.04, 1.50 | 0.0273, 0.0155, 0.0058 | — | — | — |
| 10 | Q9H2F5 | Enhancer of polycomb homolog 1 | 8.77 | 93.4 | 1.50 | 0.0026 | 45 | 1 | 2 |
| 11 | P13639 | EF-2 | 6.41 | 95.3 | 1.65 | 0.0066 | 450 | 16 | 13 |
| 12 | P47897 | Glutaminyl-tRNA synthetase | 6.71 | 87.7 | 1.50 | 0.0415 | 163 | 9 | 7 |
| 13 | P07900 | HSP90α | 4.94 | 84.6 | 1.64 | 0.0019 | 487 | 16 | 12 |
| 14 | P15311 | Ezrin | 5.94 | 69.4 | 1.53 | 0.0044 | 254 | 11 | 8 |
| 15 | P26038 | Moesin | 6.08 | 67.8 | 2.26 | 0.0052 | 107 | 13 | 9 |
| 16 | P38646 | GRP-75 | 5.87 | 73.6 | 1.73 | 0.0060 | 627 | 30 | 19 |
| 17 | P38606 | V-type proton ATPase catalytic subunit A | 5.35 | 68.3 | 1.61 | 0.0105 | 480 | 22 | 14 |
| 18 | P20700 | Lamin B1 | 5.11 | 66.4 | 1.50 | 0.0410 | 1043 | 34 | 21 |
| 19 | Q14651 | Plastin 1 | 5.33 | 70.3 | 1.50 | 0.0004 | 263 | 3 | 3 |
| 20 | P53041 | PP5 | 5.88 | 56.8 | 1.82 | 0.0445 | 165 | 34 | 19 |
| 21 | P50991 | TCP-1δ | 7.96 | 57.9 | 1.91 | 0.0317 | 454 | 47 | 25 |
| 22 | Q6IB45 | eIF3f | 5.24 | 37.5 | 1.68 | 0.0009 | 197 | 9 | 3 |
| 23, 24 | P08670 | Vimentin | 5.06 | 53.6 | 1.73, 1.51 | 0.0008, 0.0003 | 117, 77 | 13, 10 | 6, 5 |
| 25 | — | Undetermined | — | — | 1.80 | 0.0464 | — | — | — |
| 26 | Q92597 | Protein NDRG1 | 5.49 | 42.8 | 1.50 | 0.0354 | 118 | 21 | 5 |
| 27 | Q969T7 | 7-Methylguanosine phosphate-specific 5′-nucleotidase | 6.21 | 33.6 | −5.55 | 0.0426 | 58 | 2 | 2 |
| 28 | P60709 | β-Actin | 5.29 | 41.7 | 4.04 | 0.0458 | 200 | 18 | 6 |
| 29 | P29692 | EF-1δ | 4.90 | 31.1 | 1.85 | 0.0041 | 65 | 12 | 2 |
| 30 | P62258 | 14-3-3 protein ɛ | 4.63 | 29.2 | −4.80 | 0.0028 | 286 | 28 | 7 |
| 31 | P09525 | Annexin A4 | 5.84 | 35.9 | −4.30 | 0.0018 | 221 | 22 | 7 |
| 32 | — | Undetermined | — | — | −2.65 | 0.0293 | — | — | — |
EF-1δ, elongation factor 1δ EF-2, elongation factor 2; eIF3f, eukaryotic translation initiation factor 3 subunit F; GRP-75, stress-70 protein (mitochondrial); HSP90α, heat shock protein 90α Mr, molar mass; NDRG1, N-myc downstream-regulated gene 1 protein; pI, isoelectric point; PP5, serine/threonine-protein phosphatase 5; TCP-1δ, T-complex protein 1δ tRNA, transfer RNA.
+, upregulated; −, downregulated.
Paired Student’s t test.
Biological relevance of differentially expressed proteins
PANTHER GO analysis allowed us to elucidate the different functions and processes in which the 22 proteins identified are putatively involved. According to the molecular function analysis (Figure 2), most of the proteins were related with binding (26.9%), catalytic activity (26.9%), and structural molecule activity (23.1%). Concerning biological processes, the identified proteins were mainly involved in metabolic (23.9%), cellular (17.4%), and developmental (13%) processes and in cellular component organization (13%) (Figure 2).
FIGURE 2.
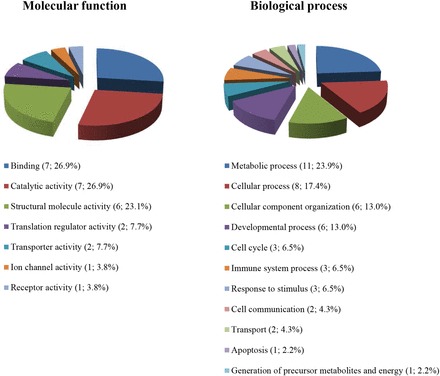
Pie chart depicting the functional classification of proteins differentially expressed in human duodenal mucosa after 5 h of protein infusion. Using the online resource PANTHER classification system, proteins of interest were categorized into the broad GO classes of biological process and molecular function. For each category, the number of identified proteins and their percentage of the total number of proteins in the pie chart are indicated for each GO term. GO, gene ontology.
To expand the GO analysis, we next analyzed the proteomic data with the DAVID functional annotation tool. DAVID uses GO and other data sources to cluster proteins based on the shared annotations to similarity clusters. DAVID analysis identified 3 significant annotation clusters (Table 2), of which the highest ranked cluster had an ES of 2.09 and was represented by 2 terms relating to molecular chaperones. The molecular chaperone cluster accounted for 18% of the identified proteins and included T-complex protein 1δ, 14-3-3 protein ɛ, and 2 members of the heat shock protein family [heat shock protein 90α and stress-70 protein (mitochondrial)]. These proteins play an important role in protein folding, transport, and degradation. We also discovered that cytoskeletal proteins were significantly enriched among identified proteins and showed higher expression after protein supplementation. The cytoskeleton protein cluster (ES = 1.90, Table 2) was made up from 4 actin cytoskeleton proteins (ezrin, moesin, plastin 1, and β-actin) and 2 intermediate filament proteins (lamin B1 and vimentin). The last DAVID annotation cluster had an ES of 1.87 and was represented by 3 terms relating to protein metabolism (Table 2). This was supported from analysis of individual functional categories by using DAVID Swiss-Prot Protein Information Resource keywords, which revealed that 5 proteins involved in protein biosynthesis were highly enriched in our protein set (23.3-fold enrichment, P = 8.6E-04). Interestingly, the expression of these proteins was markedly increased in duodenal mucosa after protein supplementation. Three of these are elongation or initiation factors (EF-1δ, EF-2, and eIF3f), whereas 2 others are enzymes that catalyze the ligation of amino acids to their cognate tRNAs (glutamyl-prolyl-tRNA synthetase and glutaminyl-tRNA synthetase).
TABLE 2.
DAVID functional annotation clusters generated from the list of proteins differentially expressed in human duodenal mucosa after 5 h of protein infusion
| Annotation category | Term | Enrichment score1 | Protein count | P value2 | Benjamini3 | Related genes4 |
| Annotation cluster 1 | ||||||
| PANTHER_MF_ALL | MF00077: chaperone | 2.09 | 4 | 0.0018 | 0.066 | CCT4, HSPA9, HSP90AA1, YWHAE |
| SP_PIR_KEYWORDS | Chaperone | 3 | 0.013 | 0.14 | CCT4, HSPA9, HSP90AA1 | |
| Annotation cluster 2 | ||||||
| SP_PIR_KEYWORDS | Actin binding | 1.90 | 3 | 0.00086 | 0.011 | EZN, MSN, PLS1 |
| PANTHER_BP_ALL | BP00286: cell structure | 6 | 0.0023 | 0.096 | ACTB, EZN, LMNB1, MSN, PLS1, VIM | |
| PANTHER_MF_ALL | MF00091: cytoskeletal protein | 6 | 0.0052 | 0.097 | ACTB, EZN, LMNB1, MSN, PLS1, VIM | |
| PANTHER_MF_ALL | MF00261: actin binding cytoskeletal protein | 4 | 0.015 | 0.14 | ACTB, EZN, MSN, PLS1 | |
| Annotation cluster 3 | ||||||
| SP_PIR_KEYWORDS | Protein biosynthesis | 1.87 | 5 | 0.000046 | 0.00086 | EEF1D, EEF2, EIF3F, EPRS, QARS |
| REACTOME_PATHWAY | REACT_17015: metabolism of proteins | 5 | 0.0037 | 0.033 | ACTB, CCT4, EEF1D, EEF2, EIF3F | |
| PANTHER_MF_ALL | MF00071: translation factor | 3 | 0.0093 | 0.11 | EEF1D, EEF2, EIF3F |
Fold enrichment of the cluster.
P value from the modified Fisher’s exact test (Expression Analysis Systematic Explorer score).
P value adjusted for multiple testing according to the Benjamini-Hochberg model.
Genes encoding the differentially expressed proteins in human: ACTB, actin, β CCT4, chaperonin containing TCP1, subunit 4 (δ); EEF1D, eukaryotic translation elongation factor 1δ EEF2, eukaryotic translation elongation factor 2; EIF3F, eukaryotic translation initiation factor 3, subunit F; EPRS, glutamyl-prolyl–transfer RNA synthetase; EZN, ezrin; HSPA9, heat shock 70-kDa protein 9; HSP90AA1, heat shock protein 90-kDa α; LMNB1, lamin B1; MSN, moesin; PLS1, plastin 1; QARS, glutaminyl–transfer RNA synthetase; VIM, vimentin; YWHAE, 14-3-3 protein ɛ.
Western blotting analysis of differentially expressed proteins
To validate the data from proteomic analysis, we performed Western blot analysis on duodenal mucosal extracts. Protein concentrations of EF-1δ, EF-2, eIF3f, ezrin, stress-70 protein (mitochondrial), heat shock protein 90α, moesin, N-myc downstream-regulated gene 1 protein, plastin 1, serine/threonine-protein phosphatase 5, T-complex protein 1δ, and β-actin were markedly higher in the duodenal mucosa after protein supplementation (Figure 3). These protein expression changes were consistent with the results given by 2DE analysis and thus confirmed the proteomic findings.
FIGURE 3.
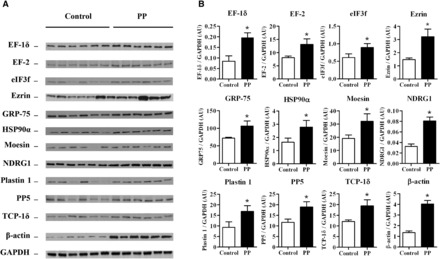
Effects of protein supplementation on EF-1δ, EF-2, eIF3f, ezrin, GRP-75, HSP90α, moesin, NDRG1, plastin 1, PP5, TCP-1δ, and β-actin expression in human duodenal mucosa. Representative immunoblots (A) and densitometric analysis of EF-1δ, EF-2, eIF3f, ezrin, GRP 75, HSP90α, moesin, NDRG1, plastin 1, PP5, TCP-1δ, and β-actin expression (B) in the duodenal mucosa of healthy volunteers after 5 h of enteral maltodextrins alone (control) or with protein supplementation. Values are means ± SEMs (n = 6). *Significantly different from the control group, P < 0.05 (paired Student’s t test). AU, arbitrary units; EF-1δ, elongation factor 1δ EF-2, elongation factor 2; eIF3f, eukaryotic translation initiation factor 3 subunit F; GRP-75, stress-70 protein (mitochondrial); HSP90α, heat shock protein 90α NDRG1, N-myc downstream-regulated gene 1 protein; PP, protein powder; PP5, serine/threonine-protein phosphatase 5; TCP-1δ, T-complex protein 1δ.
DISCUSSION
In the present study, we showed in carbohydrate-fed humans that an enteral delivery of proteins modified the duodenal mucosal proteome, particularly by enhancing the expression of several proteins involved in the cytoskeleton and protein synthesis.
Protein supplementation significantly enhanced ezrin, moesin, plastin 1, β-actin, lamin B1, and vimentin expression in human duodenal mucosa, suggesting that enteral protein supply may affect intestinal morphology, particularly the actin cytoskeleton dynamics (Figure 4). To our knowledge, no previous data have shown an effect of enteral proteins on the intestinal cellular cytoskeleton. However, Montoya et al. (34) have reported an alteration of duodenal villus-crypt architecture in rats consuming a protein-free diet. In patients with Crohn disease, oral supplementation with whey proteins improved intestinal permeability and duodenal mucosal morphology that was also observed with glutamine supplementation (25). Interestingly, amino acids, which are constituents of proteins and signaling molecules, have been shown to affect the expression of cytoskeleton-related proteins. Indeed, under apoptotic conditions, a high dose of glutamine altered the human ileocecal adenocarcinoma cell proteome by modulating the expression of proteins involved in cytoskeleton organization (35). In the jejunum of adult rats, glutamine preserved the actin cytoskeleton and protected the mucosal barrier against ischemia-reperfusion injury (36), whereas arginine promoted an increased permeability and a disruption of the actin cytoskeleton (37). Although the actin cytoskeleton is a key element in cell and tissue organization (38, 39), it also plays a critical role in cell-cell adhesion and maintenance of the intestinal epithelial barrier (40). The tight junction protein ZO-1 establishes notably a link between actin cytoskeleton and other tight junction proteins such as occludin (41). In our study, protein supplementation did not affect ZO-1, occludin, claudin 1, and claudin 3 expression in human duodenal mucosa (Supplemental Figure 1), suggesting that small intestinal permeability remained unchanged.
FIGURE 4.
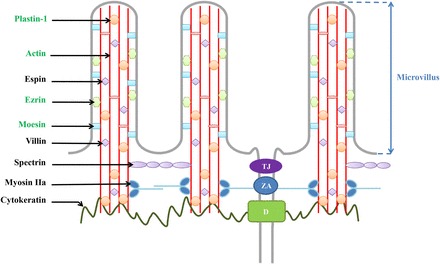
Intervention levels of ezrin, moesin, plastin 1, and β-actin in the apical structure of human duodenal epithelial cells. Proteins in green are upregulated after protein supplementation. These modifications suggest an actin cytoskeleton remodeling in human duodenal mucosa. D, desmosome; TJ, tight junction; ZA, zonula adherens.
Among cytoskeletal proteins upregulated after protein supplementation, ezrin and plastin 1 have been reported to play a crucial role in microvilli formation and nutrient absorption (42, 43) (Figure 4). Grimm-Günter et al. (44) showed an increased fragility of the intestinal epithelium and a decreased transepithelial resistance in plastin 1 knockout mice, suggesting that plastin 1 was an important regulator of brush-border microvilli morphology. Interestingly, during intestinal adaptation, hyperplasia has been observed and is associated with better outcome by increasing exchange and absorptive surface (45). We can thus speculate that protein infusion may stimulate actin cytoskeleton remodeling in the duodenal mucosa without affecting intestinal permeability, leading to increase the brush-border exchange surface. Additional experiments should evaluate the effects of enteral proteins in patients with short bowel syndrome because ingestion of whey proteins improved the intestinotrophic action of glucagon-like peptide 2 in parenterally fed rats (46).
In the present study, protein supplementation also enhanced the expression of EF-1δ, EF-2, eIF3f, glutamyl-prolyl-tRNA synthetase, and glutaminyl-tRNA synthetase in human duodenal mucosa. These data are in agreement with and extend our previous report showing that enteral supply of proteins enhanced duodenal protein FSR in carbohydrate-fed humans (11). Although eIF3f is a protein involved in apoptotic signaling (47) and translation initiation (48, 49), its role in intestinal cells has been poorly documented until now. Only one study has shown that eIF3f expression was significantly decreased in small intestine tumors (50). Interestingly, eIF3f has been shown to regulate protein synthesis and skeletal muscle mass. Indeed, mTOR/raptor complex bound to the scaffold protein eIF3f during muscle hypertrophy, which phosphorylated S6K1 and regulated downstream effectors of mTOR (51). Conversely, in muscles undergoing atrophy, an accumulation of the inactive form of S6K1 after the degradation of eIF3f by the E3 ubiquitin ligase, named muscle atrophy F-box (MAFbx), has been observed (51). In our previous study, neither mTOR nor S6K1 was affected by the protein supply in the duodenal mucosa (11), suggesting that the effects of intestinal eIF3f on protein synthesis were not linked to a regulation of the mTOR pathway. eIF3f has also been reported to be a positive regulator of Notch signaling (52), which is involved in intestinal development and cell turnover (53). The putative role of eIF3f in intestinal cells should be thus evaluated in further experiments.
In conclusion, protein supplementation altered the expression of several duodenal mucosal proteins, which are mainly involved in the cytoskeleton and protein synthesis. Protein supplementation may stimulate actin cytoskeleton remodeling in the duodenal mucosa without affecting intestinal permeability. Further investigations are needed to evaluate whether oral protein supplementation may contribute to promote intestinal adaptation.
Supplementary Material
Acknowledgments
We thank the staffs of the Clinical Investigation Centre (CIC 1404) and the Endoscopy Unit for their assistance with volunteer inclusion and infusion procedures and with the endoscopic procedure, respectively.
The authors’ responsibilities were as follows—AG, JB, PC, and DV: analyzed the data; A-FC: recruited healthy volunteers; SL: performed endoscopy and sampling; AC: prepared enteral solutions; and AG, PD, and MC: designed the experiment, analyzed the results, and wrote the manuscript. None of the authors declared a conflict of interest. The funders of this study had no involvement in the design, implementation, analysis, or interpretation of the data.
ABBREVIATIONS
- EF-1δ
elongation factor 1δ
- EF-2
elongation factor 2
- eIF3f
eukaryotic translation initiation factor 3 subunit F
- ES
enrichment score
- FSR
fractional synthesis rate
- GO
gene ontology
- mTOR
mammalian target of rapamycin
- Q-TOF
quadrupole time-of-flight
- tRNA
transfer RNA
- ZO-1
zonula occludens protein-1
- 2D
2-dimensional
- 2DE
2-dimensional electrophoresis
FOOTNOTES
Supported by the French Agency for Research (Agence Nationale pour la Recherche, ANR-07-PNRA-020) and the Nutricia Research Foundation.
REFERENCES
- 1. Coëffier M, Claeyssens S, Hecketsweiler B, Lavoinne A, Ducrotté P, Déchelotte P.. Enteral glutamine stimulates protein synthesis and decreases ubiquitin mRNA level in human gut mucosa. Am J Physiol Gastrointest Liver Physiol 2003;285:G266–73. [DOI] [PubMed] [Google Scholar]
- 2. Coëffier M, Gloro R, Boukhettala N, Aziz M, Lecleire S, Vandaele N, Antonietti M, Savoye G, Bôle-Feysot C, Déchelotte P, et al. Increased proteasome-mediated degradation of occludin in irritable bowel syndrome. Am J Gastroenterol 2010;105:1181–8. [DOI] [PubMed] [Google Scholar]
- 3. Walther A, Coots A, Nathan J, Kocoshis S, Tiao G.. Physiology of the small intestine after resection and transplant. Curr Opin Gastroenterol 2013;29:153–8. [DOI] [PubMed] [Google Scholar]
- 4. Le Bacquer O, Laboisse C, Darmaun D.. Glutamine preserves protein synthesis and paracellular permeability in Caco-2 cells submitted to “luminal fasting.” Am J Physiol Gastrointest Liver Physiol 2003;285:G128–36. [DOI] [PubMed] [Google Scholar]
- 5. Boukhettala N, Claeyssens S, Bensifi M, Maurer B, Abed J, Lavoinne A, Déchelotte P, Coëffier M.. Effects of essential amino acids or glutamine deprivation on intestinal permeability and protein synthesis in HCT-8 cells: involvement of GCN2 and mTOR pathways. Amino Acids 2012;42:375–83. [DOI] [PubMed] [Google Scholar]
- 6. Bauchart-Thevret C, Cui L, Wu G, Burrin DG.. Arginine-induced stimulation of protein synthesis and survival in IPEC-J2 cells is mediated by mTOR but not nitric oxide. Am J Physiol Endocrinol Metab 2010;299:E899–909. [DOI] [PubMed] [Google Scholar]
- 7. Bertrand J, Goichon A, Déchelotte P, Coëffier M.. Regulation of intestinal protein metabolism by amino acids. Amino Acids 2013;45:443–50. [DOI] [PubMed] [Google Scholar]
- 8. Déchelotte P, Hasselmann M, Cynober L, Allaouchiche B, Coëffier M, Hecketsweiler B, Merle V, Mazerolles M, Samba D, Guillou YM, et al. L-alanyl-L-glutamine dipeptide–supplemented total parenteral nutrition reduces infectious complications and glucose intolerance in critically ill patients: the French controlled, randomized, double-blind, multicenter study. Crit Care Med 2006;34:598–604. [DOI] [PubMed] [Google Scholar]
- 9. Humbert B, Nguyen P, Dumon H, Deschamps J-Y, Darmaun D.. Does enteral glutamine modulate whole-body leucine kinetics in hypercatabolic dogs in a fed state? Metabolism 2002;51:628–35. [DOI] [PubMed] [Google Scholar]
- 10. Tannus AFS, Darmaun D, Ribas DF, Oliveira JED, Marchini JS.. Glutamine supplementation does not improve protein synthesis rate by the jejunal mucosa of the malnourished rat. Nutr Res 2009;29:596–601. [DOI] [PubMed] [Google Scholar]
- 11. Coëffier M, Claeyssens S, Bôle-Feysot C, Guérin C, Maurer B, Lecleire S, Lavoinne A, Donnadieu N, Cailleux A-F, Déchelotte P.. Enteral delivery of proteins stimulates protein synthesis in human duodenal mucosa in the fed state through a mammalian target of rapamycin-independent pathway. Am J Clin Nutr 2013;97:286–94. [DOI] [PubMed] [Google Scholar]
- 12. Bertrand J, Goichon A, Chan P, Azhar S, Lecleire S, Donnadieu N, Vaudry D, Cailleux A-F, Déchelotte P, Coëffier M.. Enteral glutamine infusion modulates ubiquitination of heat shock proteins, Grp-75 and Apg-2, in the human duodenal mucosa. Amino Acids 2014;46:1059–67. [DOI] [PubMed] [Google Scholar]
- 13. Corl BA, Odle J, Niu X, Moeser AJ, Gatlin LA, Phillips OT, Blikslager AT, Rhoads JM.. Arginine activates intestinal p70(S6k) and protein synthesis in piglet rotavirus enteritis. J Nutr 2008;138:24–9. [DOI] [PubMed] [Google Scholar]
- 14. Murgas Torrazza R, Suryawan A, Gazzaneo MC, Orellana RA, Frank JW, Nguyen HV, Fiorotto ML, El-Kadi S, Davis TA.. Leucine supplementation of a low-protein meal increases skeletal muscle and visceral tissue protein synthesis in neonatal pigs by stimulating mTOR-dependent translation initiation. J Nutr 2010;140:2145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin Y, Yao K, Liu Z, Gong M, Ruan Z, Deng D, Tan B, Liu Z, Wu G.. Supplementing L-leucine to a low-protein diet increases tissue protein synthesis in weanling pigs. Amino Acids 2010;39:1477–86. [DOI] [PubMed] [Google Scholar]
- 16. Coëffier M, Claeyssens S, Bensifi M, Lecleire S, Boukhettala N, Maurer B, Donnadieu N, Lavoinne A, Cailleux A-F, Déchelotte P.. Influence of leucine on protein metabolism, phosphokinase expression, and cell proliferation in human duodenum1,3. Am J Clin Nutr 2011;93:1255–62. [DOI] [PubMed] [Google Scholar]
- 17. Claeyssens S, Lecleire S, Leblond J, Marion R, Hecketsweiler B, Lavoinne A, Ducrotté P, Déchelotte P, Coëffier M.. Lack of effect of acute enteral arginine infusion on whole-body and intestinal protein metabolism in humans. Dig Dis Sci 2007;52:1826–32. [DOI] [PubMed] [Google Scholar]
- 18. Volpi E, Kobayashi H, Sheffield-Moore M, Mittendorfer B, Wolfe RR.. Essential amino acids are primarily responsible for the amino acid stimulation of muscle protein anabolism in healthy elderly adults. Am J Clin Nutr 2003;78:250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koopman R, Walrand S, Beelen M, Gijsen AP, Kies AK, Boirie Y, Saris WHM, van Loon LJC.. Dietary protein digestion and absorption rates and the subsequent postprandial muscle protein synthetic response do not differ between young and elderly men. J Nutr 2009;139:1707–13. [DOI] [PubMed] [Google Scholar]
- 20. Pennings B, Boirie Y, Senden JMG, Gijsen AP, Kuipers H, van Loon LJC.. Whey protein stimulates postprandial muscle protein accretion more effectively than do casein and casein hydrolysate in older men. Am J Clin Nutr 2011;93:997–1005. [DOI] [PubMed] [Google Scholar]
- 21. Adegoke OA, McBurney MI, Samuels SE, Baracos VE.. Luminal amino acids acutely decrease intestinal mucosal protein synthesis and protease mRNA in piglets. J Nutr 1999;129:1871–8. [DOI] [PubMed] [Google Scholar]
- 22. Adegoke OAJ, McBurney MI, Samuels SE, Baracos VE.. Modulation of intestinal protein synthesis and protease mRNA by luminal and systemic nutrients. Am J Physiol Gastrointest Liver Physiol 2003;284:G1017–26. [DOI] [PubMed] [Google Scholar]
- 23. Rittler P, Kuppinger D, Krick M, Demmelmair H, Koletzko B, Jauch K-W, Hartl WH.. Differential regulation of protein synthesis in hepatic and intestinal tissues by amino acids: studies in patients recovering from major abdominal operations. Surgery 2009;146:113–21. [DOI] [PubMed] [Google Scholar]
- 24. O’Keefe SJ, Lemmer ER, Ogden JM, Winter T.. The influence of intravenous infusions of glucose and amino acids of pancreatic enzyme and mucosal protein synthesis in human subjects. JPEN J Parenter Enteral Nutr 1998;22:253–8. [DOI] [PubMed] [Google Scholar]
- 25. Benjamin J, Makharia G, Ahuja V, Anand Rajan KD, Kalaivani M, Gupta SD, Joshi YK.. Glutamine and whey protein improve intestinal permeability and morphology in patients with Crohn’s disease: a randomized controlled trial. Dig Dis Sci 2012;57:1000–12. [DOI] [PubMed] [Google Scholar]
- 26. Kimball SR.. Does enteral protein administration stimulate duodenal mucosa protein synthesis through an mTORC1-independent signaling pathway? Am J Clin Nutr 2013;97:235–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xi P, Jiang Z, Dai Z, Li X, Yao K, Zheng C, Lin Y, Wang J, Wu G.. Regulation of protein turnover by L-glutamine in porcine intestinal epithelial cells. J Nutr Biochem 2012;23:1012–7. [DOI] [PubMed] [Google Scholar]
- 28. Nakajo T, Yamatsuji T, Ban H, Shigemitsu K, Haisa M, Motoki T, Noma K, Nobuhisa T, Matsuoka J, Gunduz M, et al. Glutamine is a key regulator for amino acid–controlled cell growth through the mTOR signaling pathway in rat intestinal epithelial cells. Biochem Biophys Res Commun 2005;326:174–80. [DOI] [PubMed] [Google Scholar]
- 29. Peyrollier K, Hajduch E, Blair AS, Hyde R, Hundal HS.. L-leucine availability regulates phosphatidylinositol 3-kinase, p70 S6 kinase and glycogen synthase kinase-3 activity in L6 muscle cells: evidence for the involvement of the mammalian target of rapamycin (mTOR) pathway in the L-leucine–induced up-regulation of system A amino acid transport. Biochem J 2000;350:361–8. [PMC free article] [PubMed] [Google Scholar]
- 30. Coëffier M, Claeyssens S, Lecleire S, Leblond J, Coquard A, Bôle-Feysot C, Lavoinne A, Ducrotté P, Déchelotte P.. Combined enteral infusion of glutamine, carbohydrates, and antioxidants modulates gut protein metabolism in humans. Am J Clin Nutr 2008;88:1284–90. [DOI] [PubMed] [Google Scholar]
- 31. Goichon A, Coëffier M, Claeyssens S, Lecleire S, Cailleux A-F, Bôle-Feysot C, Chan P, Donnadieu N, Lerebours E, Lavoinne A, et al. Effects of an enteral glucose supply on protein synthesis, proteolytic pathways, and proteome in human duodenal mucosa. Am J Clin Nutr 2011;94:784–94. [DOI] [PubMed] [Google Scholar]
- 32. Goichon A, Chan P, Lecleire S, Coquard A, Cailleux A-F, Walrand S, Lerebours E, Vaudry D, Déchelotte P, Coëffier M.. An enteral leucine supply modulates human duodenal mucosal proteome and decreases the expression of enzymes involved in fatty acid beta-oxidation. J Proteomics 2013;78:535–44. [DOI] [PubMed] [Google Scholar]
- 33. Huang W, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 34. Montoya CA, Leterme P, Lalles J-P.. A protein-free diet alters small intestinal architecture and digestive enzyme activities in rats. Reprod Nutr Dev 2006;46:49–56. [DOI] [PubMed] [Google Scholar]
- 35. Deniel N, Marion-Letellier R, Charlionet R, Tron F, Leprince J, Vaudry H, Ducrotté P, Déchelotte P, Thébault S.. Glutamine regulates the human epithelial intestinal HCT-8 cell proteome under apoptotic conditions. Mol Cell Proteomics 2007;6:1671–9. [DOI] [PubMed] [Google Scholar]
- 36. Kozar RA, Schultz SG, Bick RJ, Poindexter BJ, DeSoignie R, Moore FA.. Enteral glutamine but not alanine maintains small bowel barrier function after ischemia/reperfusion injury in rats. Shock 2004;21:433–7. [DOI] [PubMed] [Google Scholar]
- 37. Kozar RA, Verner-Cole E, Schultz SG, Sato N, Bick RJ, Desoignie R, Poindexter BJ, Moore FA.. The immune-enhancing enteral agents arginine and glutamine differentially modulate gut barrier function following mesenteric ischemia/reperfusion. J Trauma 2004;57:1150–6. [DOI] [PubMed] [Google Scholar]
- 38. Miyoshi J, Takai Y.. Structural and functional associations of apical junctions with cytoskeleton. Biochim Biophys Acta. 2008;1778:670–91. [DOI] [PubMed] [Google Scholar]
- 39. Rottner K, Stradal TEB.. Actin dynamics and turnover in cell motility. Curr Opin Cell Biol 2011;23:569–78. [DOI] [PubMed] [Google Scholar]
- 40. Louvard D, Kedinger M, Hauri HP.. The differentiating intestinal epithelial cell: establishment and maintenance of functions through interactions between cellular structures. Annu Rev Cell Biol 1992;8:157–95. [DOI] [PubMed] [Google Scholar]
- 41. Fanning AS, Jameson BJ, Jesaitis LA, Anderson JM.. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem 1998;273:29745–53. [DOI] [PubMed] [Google Scholar]
- 42. Casaletto JB, Saotome I, Curto M, McClatchey AI.. Ezrin-mediated apical integrity is required for intestinal homeostasis. Proc Natl Acad Sci USA 2011;108:11924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fehon RG, McClatchey AI, Bretscher A.. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol 2010;11:276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grimm-Günter E-MS, Revenu C, Ramos S, Hurbain I, Smyth N, Ferrary E, Louvard D, Robine S, Rivero F.. Plastin 1 binds to keratin and is required for terminal web assembly in the intestinal epithelium. Mol Biol Cell 2009;20:2549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi P, Guo J, Erwin CR, Warner BW.. IGF-2 mediates intestinal mucosal hyperplasia in retinoblastoma protein (Rb)–deficient mice. J Pediatr Surg 2013;48:1340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu X, Murali SG, Holst JJ, Ney DM.. Whey protein potentiates the intestinotrophic action of glucagon-like peptide-2 in parenterally fed rats. Am J Physiol Regul Integr Comp Physiol 2009;297:R1554–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi J, Hershey JWB, Nelson MA.. Phosphorylation of the eukaryotic initiation factor 3f by cyclin-dependent kinase 11 during apoptosis. FEBS Lett 2009;583:971–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiao H, Xu LH, Yamada Y, Liu DX.. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS ONE 2008;3:e1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Masutani M, Sonenberg N, Yokoyama S, Imataka H.. Reconstitution reveals the functional core of mammalian eIF3. EMBO J 2007;26:3373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shi J, Kahle A, Hershey JWB, Honchak BM, Warneke JA, Leong SPL, Nelson MA.. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene 2006;25:4923–36. [DOI] [PubMed] [Google Scholar]
- 51. Csibi A, Cornille K, Leibovitch M-P, Poupon A, Tintignac LA, Sanchez AMJ, Leibovitch SA.. The translation regulatory subunit eIF3f controls the kinase-dependent mTOR signaling required for muscle differentiation and hypertrophy in mouse. PLoS ONE 2010;5:e8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moretti J, Chastagner P, Gastaldello S, Heuss SF, Dirac AM, Bernards R, Masucci MG, Israël A, Brou C.. The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol 2010;8:e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vooijs M, Liu Z, Kopan R.. Notch: architect, landscaper, and guardian of the intestine. Gastroenterology 2011;141:448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


