The genetic hallmark of Burkitt lymphoma (BL) is a translocation juxtaposing the MYC-oncogene to one of the three immunoglobulin (IG) loci.1 There is ongoing controversy whether IG-MYC negative “true” BL exist. Various studies have identified 2-22% of BL as “MYC-negative”.2–5 A small subset of these cases might nowadays be assigned to the recently described provisional entity of MYC-negative “Burkitt like lymphoma with 11q aberration” (abbreviated herein mnBLL,11q,).6 Nevertheless, the lack of detection of an IG-MYC translo-cation in a considerable number of cases might also be due to the genomic variability of the IG-MYC fusion and the limitations of the techniques applied for their detection. Indeed, classical molecular technologies like PCR and Southern blot analysis have been repeatedly shown to be hampered by the scattering of the breakpoints on both affected chromosomes. Thus, the existence of an IG-MYC translocation is nowadays usually examined applying interphase fluorescence in situ hybridization (FISH). As discussed elsewhere7,8 the scattering of breakpoints, the existence of insertions of 8q24 fragments into one of the IG loci or vice versa and the presence of non-IG-MYC translocations in lymphomas other than BL renders the application of only a MYC break-apart FISH assay insufficient for thorough diagnostics. Instead, the additional application of immunoglobulin heavy chain (IGH) IGH-MYC, immunoglobulin kappa (IGK) IGK-MYC and immunoglobulin lambda (IGL) IGL-MYC fusion probes is required to define a case as “IG-MYC negative”. However, even when applying the whole collection of these FISH probes, a subset of lymphomas with typical clinical presentation, morphologic features and even gene expression pattern of BL remains in which an IG-MYC fusion cannot be detected. By employing whole genome sequencing (WGS) to study a case of molecular cytogenetically IG-MYC negative lymphoma with typical features of BL, we here extend prior studies using FISH and fiber FISH to detect insertions of MYC into the (IGH) locus and vice versa,2 and show that they can carry cryptic insertions of MYC into IG loci which are not detectable by FISH but are functionally equivalent to the typical Burkitt translocations.
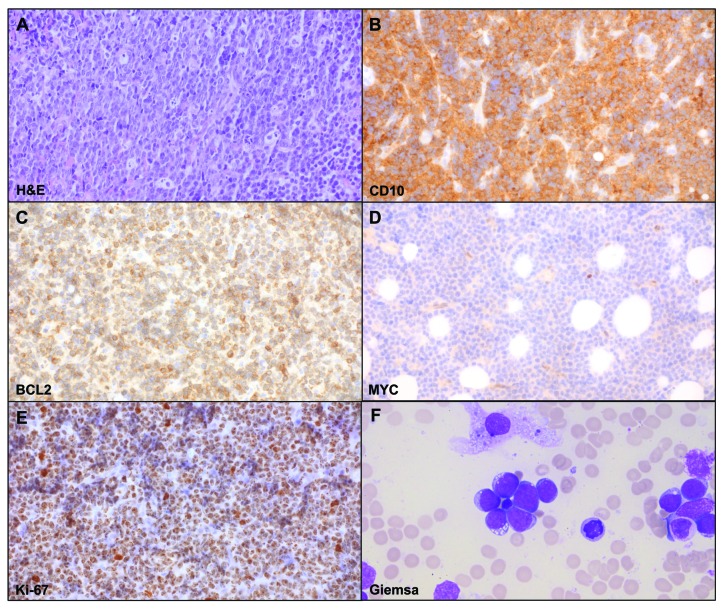
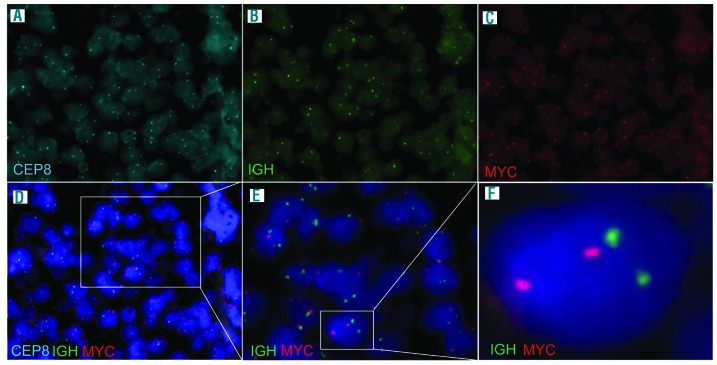
A 9-year-old Caucasian boy presented with an enlarged cervical lymph node (LN), exhaustion as well as weight loss. Routine staging prior to treatment, including chest X-ray, abdominal and LN ultrasound, cranial, cervical, abdominal and pelvic magnetic resonance imaging (MRI), revealed cervical and supraclavicular lymphadenopathy and possible bone marrow (BM) involvement. The biopsy of the cervical LN lacked typical LN structure. It showed a dense diffuse infiltrate with cohesive areas and starry-sky pattern. The tumor cells resembled small centroblasts and were CD20 and CD10 positive but terminal deoxynucleotidyl transferase (TdT) and Eppstein-Barr virus (EBV) negative. The proliferation marker Ki-67 was positive in around 100% of the cells (Figure 1). BCL2 expression was detectable in the majority of the tumor area (80%) but was variable in the extent of staining and the staining intensity. BCL2 positivity in BL has been described in up to 23% of cases with a total of 3% showing BCL2 expression in >75% of tumor cells.9 MYC protein expression determined by the anti c-MYC antibody [Y69] (Abcam) was only weak (Figure 1). BM aspirates showed 50% French-American-British (FAB) L3-blasts with atypical morphologic features (Figure 1F and Online Supplementary Figure S1). Based on these findings the diagnosis of BL was considered. To corroborate this diagnosis, we applied MYC break-apart as well as IGH-MYC (Figure 2), IGK-MYC and IGL-MYC fusion FISH probes. Remarkably, with none of these assays could any IG-MYC fusion or any other kind of MYC rearrangement be detected. Breaks within the BCL2 and BCL6 loci were also not identified. The results of these analyses prompted us to investigate the differential diagnosis of a mnBLL,11q, but interphase FISH analyses excluded the typical 11q aberration. Hence, although the morphology, the immunophenotype and the clinical presentation were all in line with the diagnosis of BL, the lack of a detectable IG-MYC fusion along with the weak MYC protein expression questioned this diagnosis. Therefore, we took diffuse-large B-cell lymphoma (DLBCL) as differential diagnosis into account, although BM infiltration with L3-blasts is rarely observed in this entity.10 At this time point, the patient was treated according to the non-Hodgkin lymphoma Berlin-Frankfurt-Münster (NHL-BFM) registry 2012 guidelines analog to NHL-BFM treatment published earlier in the risk group R4.11
Figure 1.
Morphologic features of cervical lymph node biopsy showing a mature germinal center B cell lymphoma immunophenotype. (A) Hematoxylin and eosin (H&E) preparation showing a diffuse infiltrate with starry-sky and cohesive growth pattern (magnification 400x). Immunohistochemistry shows (B) expression of CD10, (C) heterogeneous expression of BCL2 with strong positivity of single cells, (D) weak MYC expression (magnification 400x) and (E) strong Ki-67 positivity in about 100% of cells. (F) displays the photomicrograph of the Giemsa staining of the bone marrow aspirate (magnification 200x).
Figure 2.
Fluorescence in situ hybridization on paraffin embedded tumor sections using the LSI IGH/MYC/CEP 8 Tri-Color Dual Fusion Probe (Abbott/Vysis). (A) Signal pattern for the centromere 8 (CEP8) part of the probe. (B) Signal pattern for the immunoglobulin heavy chain (IGH) part of the probe. (C) Signal pattern for the MYC part of the probe and (D-F) picture merging signal patterns of all parts of the probe in different magnifications. Results show no significant amount of colocalized signals indicative for an IGH-MYC fusion. Note that due to sectioning artefacts, nuclei can either lack signals, or in case of overlaying nuclei, can give the impression of extra signals per nuclei.
For further genetic characterization of this unusual B-cell lymphoma, we performed immunoglobulin heavy chain gene (IGHV) sequence analysis. This revealed a monoclonal, productive IG-rearrangement with 4.4% of the IGHV sequence being mutated, supporting the diagnosis of a clonal B-cell lymphoma. Moreover, we analyzed the chromosomal imbalances performing OncoScan SNP-array (Affymetrix, Santa Clara, CA, USA). We could not identify, besides a sub-clonal loss of chromosome Y, any chromosomal imbalances. In particular, in agreement with the FISH findings, we could not detect any alterations on chromosome 11q (Online Supplementary Figure S2). The lack of any autosomal imbalances is unusual for DLBCL, which usually show a rather complex karyotype whereas in BL only few secondary chromosomal alterations are normally detected, if at all.12,13
Recently, we and others have described alterations in the ID3 gene to be present in up to 70-80% of BL,13–16 whereas these mutations are rare in other mature B-cell lymphomas.17 Hence, using the LN biopsy, we screened the ID3 gene for mutations applying Sanger sequencing. Indeed, we detected sub-clonal stopgain and missense ID3 mutations located on independent alleles. The occurrence of sub-clonal ID3 mutations in BL has been already described.13 Both mutations, not reported as polymorphisms, were located within the functional basic helix loop helix domain13,14 (Online Supplementary Table S1 and Online Supplementary Figure S3).
The lack of chromosomal imbalances and the presence of ID3 mutations further supported the diagnosis of BL on the genetic level. Thus, we considered the possibility that the lymphoma harbors a cryptic IG-MYC alteration not detectable by FISH. Therefore, we subjected DNA from the diagnostic BM sample to WGS. Most intriguingly, we detected in addition to an intact MYC allele a copy number gain of 8,643 bp in size of chr8:128,750,275-128,758,918 bp (hg19) encompassing the exons 2 and 3 (and, thus, the whole coding region) of the MYC gene (Online Supplementary Figure S4-5) which was inserted into the IGH locus (insertion break-junctions: chr14:106,177,155 bp and chr14:106,177,167 bp, hg19). Sanger sequencing (Figure 3A) verified the two break-point-junctions of the insertion into the IGH locus in the BM sample subjected to WGS and also in the LN biopsy. The MYC gene was inserted within the IGHA1 switch region (Figure 3B) likely due to an aberrant class switch recombination. Remarkably, the IGHA switch regions are more frequently affected by chromosomal breakpoints of IGH-MYC translocations in BL than by IGH translocation in other germinal center derived B-cell lymphomas.18,19 By the insertion, the MYC gene is juxtaposed to one of the IGH enhancers in a similar way as in a typical Burkitt translocation t(8;14) (Figure 3B). Thus, the insertion should result in deregulated MYC expression similar to typical IGH-MYC fusions. However, as outlined above, immunohistochemistry showed only weak MYC protein expression (Figure 1D). A recent study has shown that mutations within the epitope binding site of the Y69 anti-MYC antibody, especially at amino acid position 11 of the MYC protein (based on ENSP00000259523 this is an asparagine [N]), changes the epitope and hence can result in MYC negativity by immunohistochemistry.20 In line, mining the WGS data, we detected mutations in the MYC gene leading to amino acid changes at positions 4 (chr8:128,750,519A>G, p.N4S) and 12 (chr8:128,750, 543A>C, p.Y12S) of the MYC protein (Online Supplementary Table S1). These changes, are located immediately adjacent to the N11 polymorphism and within the Y69 target epitope as defined by the vendor (first 100 amino acids of MYC). Hence, it is likely that these mutations interfere with Y69 binding and, thus, could explain the weak MYC protein staining. Finally, we mined the WGS data for mutations in genes frequently altered in BL.13–15 Besides changes of MYC, we identified mutations in SMARCA4 and BCL6. Strikingly, by WGS of the BM sample we failed to identify the ID3 mutations detected by targeted sequencing in the LN biopsy. In line, some of the MYC mutations identified in the BM biopsy were not detected in the LN biopsy and the other way around by Sanger sequencing (Online Supplementary Table S1 and Online Supplementary Figure S6) but not those at position N4 and Y12 as described above. This indicates that, although both lymphoma manifestations share the same MYC insertion into the IGH locus and, thus, stem from the same ancestor clone, they accumulated different mutations during tumor progression (Online Supplementary Figure S7). In this regard, it is worth noting that both MYC and ID3 mutations in BL based on their sequence properties have been shown to stem from an AID-driven aberrant somatic hypermutation mechanism.13
Figure 3.
Overview of the MYC gene insertion into the immunoglobulin heavy chain locus. (A) Verification of the breakpoint junctions by Sanger sequencing of the MYC gene locus insertion within the immunoglobulin heavy chain (IGH) locus in the lymph node (LN) as well as bone marrow (BM) tumor biopsy. The sequences confirmed the insertion of an 8,643 bp long sequence from chr8:128,750,275-128,758,918 bp, which was inserted between chr14:106,177,155-106,177,167 bp deleting 11 bp between the breakpoints. Moreover, at the second breakpoint five nucleotides were introduced. (B) Schematic outline of the MYC insertion into the IGH locus. Based on whole genome sequencing (WGS) data we could show that part of the MYC gene, which carries the coding exons 2 and 3, is gained and inserted IGHA1 the IGH locus within the switch region of IGHA1 (IGHA1 switch region chr14:106,715,033-106,178,629bp, hg19, Hübschmann et al., unpublished).
In summary, considering the morphological features, the immunophenotype, the genomic landscape as well as the finding of the MYC-insertion within the IGH locus, we conclude that the patient indeed can be diagnosed with BL carrying a cryptic MYC insertion within the IGH locus. Given that IG-MYC fusions are also recurrent in other B-cell neoplasms, it is intriguing to speculate, that molecular cytogenetically cryptic insertions do also occur in other B-cell lymphoma subtypes. Besides this, we can learn two additional lessons from this rare and unusual case: First, the case teaches us that in case molecular cytogenetic techniques do not indicate the occurrence of an IG-MYC translocation in an otherwise typical BL, the analysis of the ID3 mutation status as well as the chromosomal imbalance pattern can be helpful for establishing the diagnosis. In the Online Supplementary Figure S8 we propose how this could be integrated into the diagnostic work up for such cases. Secondly, the case teaches us that the frequency of MYC-negative BL (besides mnBLL,11q) might be overestimated due to the fact that cryptic MYC-insertions within the IG locus (or vice versa) are not searched for or not detected with the current routinely applied molecular cytogenetic approaches. WGS or targeted sequencing of potential structural variant break-point regions might be required to resolve the MYC status in such cases.
Acknowledgments
We thank the KinderKrebsInitiative Buchholz Holm-Seppensen for infrastructural support. We thank the High-Throughput Sequencing Unit of the Genome and Proteome Core Facility of the German Cancer Research Center (DKFZ, Heidelberg) for excellent technical support.
Footnotes
Funding: this study has been supported by the German Ministry of Science and Education (BMBF) in the framework of the MMML- MYC-SYS project (036166B), the ICGC MMML-Seq and ICGC DE Mining projects (01KU1002A-J and 01KU1505G, respectively). The NHL-BFM-Registry 2012 is supported by a grant from the Deutsche Kinderkrebsstiftung (DKS 2014.11 A/B).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20(40):5595–5610. [DOI] [PubMed] [Google Scholar]
- 2.Haralambieva E, Schuuring E, Rosati S, et al. Interphase fluorescence in situ hybridization for detection of 8q24/MYC breakpoints on routine histologic sections: validation in Burkitt lymphomas from three geographic regions. Genes Chromosomes Cancer. 2004;40(1):10–18. [DOI] [PubMed] [Google Scholar]
- 3.Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354(23):2431–2442. [DOI] [PubMed] [Google Scholar]
- 4.Leucci E, Cocco M, Onnis A, et al. MYC translocation-negative classical Burkitt lymphoma cases: an alternative pathogenetic mechanism involving miRNA deregulation. J Pathol. 2008;216(4):440–450. [DOI] [PubMed] [Google Scholar]
- 5.Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354(23):2419–2430. [DOI] [PubMed] [Google Scholar]
- 6.Salaverria I, Martin-Guerrero I, Wagener R, et al. A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood. 2014; 123(8):1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerma EG, Siebert R, Kluin PM, Baudis M. Translocations involving 8q24 in Burkitt lymphoma and other malignant lymphomas: a historical review of cytogenetics in the light of todays knowledge. Leukemia. 2009;23(2):225–234. [DOI] [PubMed] [Google Scholar]
- 8.Aukema SM, Siebert R, Schuuring E, et al. Double-hit B-cell lymphomas. Blood. 2011;117(8):2319–2331. [DOI] [PubMed] [Google Scholar]
- 9.Masqué-Soler N, Szczepanowski M, Kohler CW, et al. Clinical and pathological features of Burkitt lymphoma showing expression of BCL2–an analysis including gene expression in formalin-fixed paraffin-embedded tissue. Br J Haematol. 2015;171(4):501–508. [DOI] [PubMed] [Google Scholar]
- 10.Swerdlow S, et al. in WHO Classification of Haematopoietic and Lymphoid Tissues (ed Swerdlow S, et al.), International Agency for Research on Cancer, Lyon, France, 2017. [Google Scholar]
- 11.Woessmann W, Seidemann K, Mann G, et al. The impact of the methotrexate administration schedule and dose in the treatment of children and adolescents with B-cell neoplasms: a report of the BFM Group Study NHL-BFM95. Blood. 2005;105(3):948–958. [DOI] [PubMed] [Google Scholar]
- 12.Scholtysik R, Kreuz M, Klapper W, et al. Detection of genomic aberrations in molecularly defined Burkitt’s lymphoma by array-based, high resolution, single nucleotide polymorphism analysis. Haematologica. 2010;95(12):2047–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter J, Schlesner M, Hoffmann S, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. [DOI] [PubMed] [Google Scholar]
- 14.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohde M, Bonn BR, Zimmermann M, et al. Relevance of ID3-TCF3-CCND3 pathway mutations in pediatric aggressive B-cell lymphoma treated according to the non-Hodgkin Lymphoma Berlin-Frankfurt-Münster protocols. Haematologica. 2017;102(6):1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy A, Zhang J, Davis NS, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 2017;171(2):481–494.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taub R, Kirsch I, Morton C, et al. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA. 1982;79(24):7837–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López C, Kleinheinz K, Aukema SM, et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat Commun. 2019;10(1):1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collinge B, Chong L, Ben-Neriah S, et al. Deciphering discordance between MYC mRNA and MYC IHC in DLBCL: the role of MYC exon 2 mutations and N11S polymorphism. Blood. 2017; 130(1):3994. [Google Scholar]