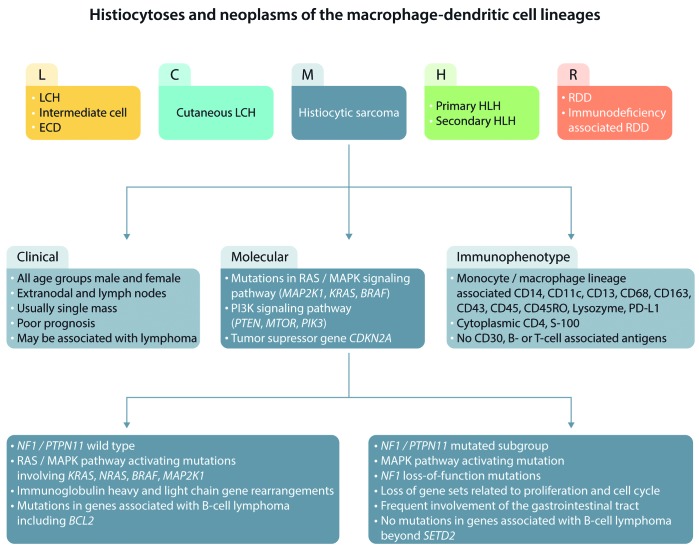
A revised classification of histiocytoses and their neoplasms was long overdue when members of the Histiocyte Society suggested dividing them into five groups, designated L (Langerhans cell), C (Cutaneous), M (Malignant), R (Rosai-Dorfman), and H (Hemophagocytic) (Figure 1).1 As an example, the L group, which includes Langerhans cell histiocytosis and Erdheim Chester disease, is characterized by mutations in the MAPK pathway and BRAF V600E. In contrast histiocytic sarcomas in the M group remain an elusive category. These rare and highly malignant neoplasms occur at all ages, and frequently involve extranodal sites including skin, soft tissues, and the gastrointestinal tract. There are few objective criteria for diagnosis other than expression of histiocyte markers (CD68, CD163, CD4, lysozyme, CD21, CD35, S100) and exclusion of other tumors by a panel of antibodies including, but not limited to, S100, keratins, EMA, Melan-A, HMB45, and B- and T-lymphoid markers.2 Further complications arise in the apparent plasticity between histiocytic sarcomas and other malignancies, such as follicular lymphomas, demonstrated by translocations, immunoglobulin gene rearrangements, or mutational analysis.3 Because of these pitfalls, histiocytic sarcoma has been vastly over diagnosed, with mimics including T-cell and other lymphomas with histiocyte-rich backgrounds, and, in particular, CD30 positive anaplastic large cell lymphomas. Clearly, any progress in diagnosing and treating these aggressive neoplasms will depend on identifying specific molecular and other markers for their accurate diagnosis.
Figure 1.
Clinical, immunophenotypic, and molecular changes in histiocytic sarcoma and relation to other histiocytoses. LCH: Langerhans cell histiocytosis; HLH: hemophagocytic lymphohistiocytosis; RDD: Rosai Dorfman Disease; ECD: Erdheim-Chester disease.
Until recently, molecular analysis of histiocytic sarcomas has given confusing results, and there have been no consistent cytogenetic abnormalities. Mutations involving the RAS-MAPK signaling pathway, BRAF V600E mutations, as activation of PI3K and the tumor suppressor gene CDKN2A4 have been most frequently reported, and there have been no reports of ALK translocations. Some cases, particularly in patients with associated B-cell lymphomas, have demonstrated immunoglobulin heavy or light chain gene rearrangements.
In this issue of Haematologica, Egan et al. performed genomic profiling of 21 primary histiocytic sarcomas, and in addition to confirming frequent alterations in the RAS/MAPK pathway, identified a novel intra-chromosomal transcript between exon 12 of TTYH3 and exon 8 of BRAF on chromosome 7. Moreover, differential expression analysis identified two distinct molecular subgroups with distinct molecular profiles and associated clinical characteristics based on the presence or absence of NF1/PTP11 mutations.5 Cases that had NF1/PTPN11 mutations had a predilection for gastrointestinal tract involvement. This group was characterized by loss of gene sets related to cellular proliferation and the cell cycle. Further data suggest that a subset of these cases involve co-occurring NF1 and PTPN11 mutations, suggesting that activating mutations of PTPN11 may synergize with NF1 mutations to potentiate oncogenesis. None of these cases had abnormalities in genes associated with B-cell lymphomas, besides SETD2, or had B-cell-related gene rearrangements.
In contrast, cases with wild-type NF1/PTP11 had RAS/MAPK pathway activating mutations, and were unrelated to site of presentation, and had activating mutations involving KRAS, NRAS, BRAF, and MAP2K1. This group alone demonstrated an association with immunoglobulin gene rearrangements and genes associated with B-cell lymphomas, including BCL2 rearrangement. This molecular profile offers an intriguing insight into the relationship between histiocytic sarcoma and B-cell lymphoma. The ability to identify subsets of histiocytic sarcoma according to their molecular profiles offers a potential for improvement in the diagnosis and targeted therapy of these neoplasms that has so far foiled conventional pathological examination.
References
- 1.Emile JF, Abla O, Fraitag S, et al. Revised classification of histiocytoses and neoplasms of the macrophage-dendritic cell lineages. Blood. 2016;127(22):2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pileri SA, Grogan TM, Harris NL, et al. Tumours of histiocytes and accessory dendritic cells: an immunohistochemical approach to classification from the International Lymphoma Study Group based on 61 cases. Histopathology. 2002;41(1):1–29. [DOI] [PubMed] [Google Scholar]
- 3.Feldman AL, Arber DA, Pittaluga S, et al. Clonally related follicular lymphomas and histiocytic/dendritic cell sarcomas: evidence for transdifferentiation of the follicular lymphoma clone. Blood. 2008;111(12):5433–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shanmugam V, Griffin GK, Jacobsen ED, Fletcher CDM, Sholl LM, Hornick JL. Identification of diverse activating mutations of the RAS-MAPK pathway in histiocytic sarcoma. Mod Pathol. 2019;32(6):830–843. [DOI] [PubMed] [Google Scholar]
- 5.Egan C, Nicolae A, Lack J, et al. Genomic profiling of primary histiocytic sarcoma reveals two molecular subgroups. Haematologica. 2020;105(4):951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]